Abstract
The general theories of molecular evolution depend on relatively arbitrary assumptions about the relative distribution and rate of advantageous, deleterious, neutral, and nearly neutral mutations. The Fisher geometrical model (FGM) has been used to make distributions of mutations biologically interpretable. We explored an FGM-based molecular model to represent molecular evolutionary processes typically studied by nearly neutral and selection models, but in which distributions and relative rates of mutations with different selection coefficients are a consequence of biologically interpretable parameters, such as the average size of the phenotypic effect of mutations and the number of traits (complexity) of organisms. A variant of the FGM-based model that we called the static regime (SR) represents evolution as a nearly neutral process in which substitution rates are determined by a dynamic substitution process in which the population’s phenotype remains around a suboptimum equilibrium fitness produced by a balance between slightly deleterious and slightly advantageous compensatory substitutions. As in previous nearly neutral models, the SR predicts a negative relationship between molecular evolutionary rate and population size; however, SR does not have the unrealistic properties of previous nearly neutral models such as the narrow window of selection strengths in which they work. In addition, the SR suggests that compensatory mutations cannot explain the high rate of fixations driven by positive selection currently found in DNA sequences, contrary to what has been previously suggested. We also developed a generalization of SR in which the optimum phenotype can change stochastically due to environmental or physiological shifts, which we called the variable regime (VR). VR models evolution as an interplay between adaptive processes and nearly neutral steady-state processes. When strong environmental fluctuations are incorporated, the process becomes a selection model in which evolutionary rate does not depend on population size, but is critically dependent on the complexity of organisms and mutation size. For SR as well as VR we found that key parameters of molecular evolution are linked by biological factors, and we showed that they cannot be fixed independently by arbitrary criteria, as has usually been assumed in previous molecular evolutionary models.
Keywords: molecular evolution, balanced mutation theory, fluctuating environment, population size, complexity, Fisher’s geometrical model
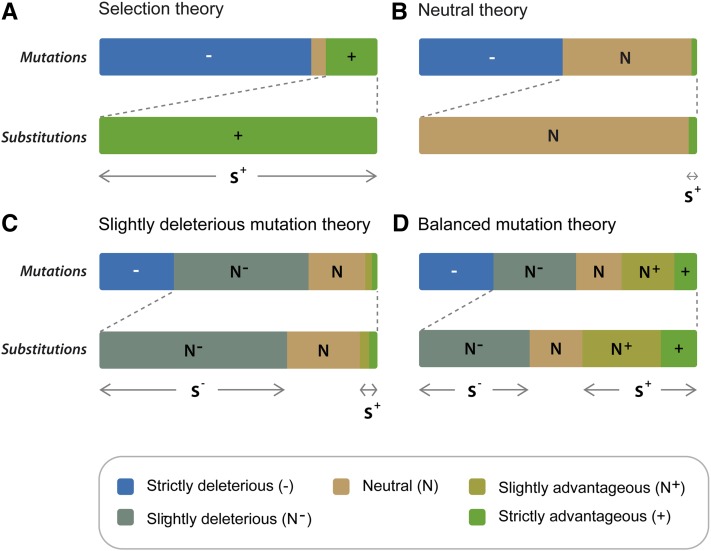
THE nearly neutral theory of molecular evolution (Ohta and Kimura 1971; Ohta 1972, 1973, 1977, 1992, 1996), as is generally understood, affirms that the vast majority of amino acid substitutions are slightly deleterious; hence, it has been called the slightly deleterious mutation theory (Figure 1C) (Ohta and Kimura 1971; Ohta 1972, 1973, 1977, 1987, 1996; Kimura 1983; Gillespie 1995, 2004; Kreitman 1996). In the original exponential “shift” model of Ohta (1977) selection coefficients are chosen at random from an exponential probability distribution, and the population mean fitness shifts back when a mutation is fixed (see also Ohta and Gillespie 1996). This model was modified by Kimura (1979), who proposed the gamma shift model, to overcome Ohta’s previous assumptions that imply a rate of substitution that is too low when population size is increased above moderate values (see also Nielsen and Yang 2003). Later, Ohta and Tachida (1990) and Tachida (1991) developed another kind of nearly neutral model to relax some criticized assumptions (see below), but those models produced a very different prediction than that of slightly deleterious mutation models. In fact, Gillespie (1994, 1995) uncovered that in these later models, known as the house-of-cards or “fixed” models, only half of the substitutions are deleterious and the other half are advantageous (see also Tachida 1996, 2000). Thus, these kinds of nearly neutral models may be subsumed in a different category that we will call the balanced mutation theory (Figure 1D). Nevertheless, these later models were strongly criticized because the rate of substitution becomes 0 starting from a small effective population size (specifically, substitution stops when 2Nσs > 4, where σs is the standard deviation of the selection coefficients, and N is the effective population size); that is, except for a narrow window of 2Nσs these models predict even lower rates of substitution than those generated by the original exponential shift model (Gillespie 1994, 1995, 1999; Ohta and Gillespie 1996). Thus the nearly neutral theory continued to emphasize the substitution of slightly deleterious mutations (Ohta 1992, 1996, 2007; Gillespie 1995, 2004; Eyre-Walker et al. 2002; Sella and Hirsh 2005; Gu 2007a,b; Harris 2010). On the other hand, selection models (Figure 1A), in which natural selection rather than genetic drift is the main force causing substitutions, commonly depend on fluctuating environments that are required to continue evolution and may explain some of the empirical phenomena found in molecular data (Ohta and Gillespie 1996). Nevertheless, it has been recognized that there is a lack of a general model of molecular evolution that can account for all major molecular phenomena (Ohta and Gillespie 1996; Kreitman 1996; Nielsen and Yang 2003). Here we propose that a reinterpretation of recent developments of molecular evolutionary models based on Fisher’s (1930) geometrical framework (Gu 2007a,b; Su et al. 2010; Razeto-Barry et al. 2011; Razeto-Barry and Maldonado 2011) may offer an alternative modeling framework that gives a better account of molecular evolutionary phenomena than previous models.
Figure 1 .
(A) Darwin’s (and neo-Darwinian selection) theory postulated the existence of deleterious (−) and advantageous (+) changes, but Darwin recognized the existence of neutral changes (Bernardi 2007). Deleterious mutations are immediately rejected by negative (or purifying) selection and neutral mutations are neglected. All the substitutions have a positive selection coefficient s > 0. (B) The neutral theory (Kimura 1968, 1983) postulated the existence of an important fraction of neutral mutations (N) and a very small fraction of advantageous mutations. Neutral mutations are fixed by random drift and constitute the majority of substitutions. A very small minority of substitutions have s > 0. (C) The slightly deleterious mutation theory (Ohta 1972, 1992; Kimura 1979) is a nearly neutral theory that included mutations between neutral and advantageous (N+), as well as between neutral and deleterious (N−). These nearly neutral mutations are fixed by random drift too, and constitute, with the neutral, the majority of substitutions. The majority of substitutions have s < 0. (D) The balanced mutation theory also incorporates slightly deleterious mutations (N−), but also postulates an important fraction of advantageous (compensatory) mutations fixed after the fixations of slightly deleterious mutations. Compensatory mutations constitute an important fraction of substitutions. 50% of no neutral substitutions have s > 0 and 50% have s < 0 (adapted from Ohta 1992; Bromham and Penny 2003; Bernardi 2007).
Theoretical studies of molecular evolution assume predefined distributions of selection coefficients of mutants (e.g., Ohta 1973, 1977; Kimura 1979; Ohta and Tachida 1990; Gillespie 1993, 1994). Thus the rate and proportion of different types of mutations are dependent only upon the population size and some parameters of the distribution of the selection coefficients (typically and σs, the mean and the standard deviation of selection coefficients, respectively). However, in real processes the distribution of mutant selection coefficients is determined by the operation of the evolutionary dynamics and therefore it should not be assumed a priori (Sella and Hirsh 2005). Moreover, choosing a specified distribution is somewhat ad hoc because it lacks a clear-cut biological interpretation (Gu 2007b).
The Fisher geometrical model (FGM) has been used to make distributions of mutations biologically interpretable (Martin and Lenormand 2006a,b, 2008; Gu 2007a,b). Given a distribution of the size of phenotypic effects of mutations (r), in the FGM the distribution of mutant selection coefficients is determined by geometrical relations between the number of traits of organisms and the distance to an optimum trait combination. In turn, this distance depends on the phenotypic traits of the population, the environmental changes (Orr 1998, 1999, 2000), and the fixed drift load of the population (Hartl and Taubes 1998; Poon and Otto 2000; Sella and Hirsh 2005; Tenaillon et al. 2007; Sella 2009), which allows exploration of relationships between parameters that may be linked to biologically interpretable factors and thus should not be independently specified by arbitrary criteria. For example, Gu (2007a,b) used the FGM to model the slightly deleterious mutation theory under the shift model framework, finding a natural explanation for Kimura’s gamma function of selection coefficients, which would be related to the number n of phenotypic dimensions influenced by the mutations of a gene (gene pleiotropy). Thus Gu found that the gamma distribution case used by Kimura corresponds to n = 1 phenotypic dimensions and the exponential distribution of Ohta corresponds to n = 2 (Gu 2007a).
Gu’s work also allows the study of the relationship between molecular evolution and population size in the FGM. However, the assumptions of Gu’s model in the FGM inherit the problems of the original shift models, which were strongly criticized because of their biologically unreasonable assumptions (Tachida 1991; Ohta 1992; Gillespie 1995; Ohta and Gillespie 1996; Razeto-Barry et al. 2011). For instance, shift models require that all mutations be deleterious. Thus, when a deleterious mutation gets fixed, all subsequent mutations must be less fit than the fixed mutation. Shift models allow simplifying assumptions that were used in Gu’s work to make the problem analytically tractable. However, more realistic assumptions can be approached with computer simulations (as in the house-of-cards model). Here we relaxed the assumptions of Gu’s model in the FGM and developed a model that supports a balanced mutation theory of molecular evolution, which we call the static regime (SR). The steady state in the FGM has been understood as a nearly neutral evolutionary process (Hartl and Taubes 1996; Sella and Hirsh 2005; Sella 2009) and it is in some aspects similar to the house-of-cards nearly neutral model (Ohta and Tachida 1990; Tachida 1991, 1996). We explored the possible interpretations and evolutionary consequences of this model through simulations in the Fisher geometrical framework both in a balanced steady state (the SR) and in an interplay between adaptive processes and balanced steady states in a randomly fluctuating environment, henceforth the variable regime (VR). We found that when molecular evolution, both in a fluctuating environment (VR) and in the steady state (SR), is biologically interpreted in the FGM it does not need the arbitrary independent specification of some evolutionarily relevant parameters because they are locked in relationships that depend upon biologically interpretable factors. We found other differences with previous nearly neutral models that overcome unrealistic properties of these previous models, supporting the SR and VR as potentially good models for representing general processes of molecular evolution.
The Model
Model assumptions
The FGM represents a population as a point in an n-dimensional space of states, in which each axis represents a different organismal phenotypic trait and the origin represents the optimum state of a population given a specific environmental condition (Orr 1998, 2000; Welch and Waxman 2003). Mutations are represented as random vectors isotropically distributed in this hyperspace (see below). Vectors that approach the origin are advantageous and those pointing away from the optimum are deleterious; the selection coefficients of the vectors are determined according to a Gaussian fitness function centered on the optimum (which, without loss of generality, takes a fitness value of 1). Environmental fluctuations are represented as optimum shifts (Barton 2001; Gu 2007b; Razeto-Barry et al. 2011). In contrast to the nearly neutral shift models (see also Gu 2007a,b), in our model when a mutation is fixed the phenotype of individuals in the population is modified, acquiring a new fitness value; thus subsequent mutations start from the new phenotypic state (Razeto-Barry et al. 2011).
Given that a previously fixed mutation in a population changes the phenotypic starting point of each new mutation, the probability distributions of new mutations are shifting permanently, wandering in the Cartesian n-dimensional space. Because of their difficult analytical tractability, models in which population fitness fluctuates as a result of mutant fixations are analyzed by computer simulations (Ohta and Tachida 1990; Tachida 1991, 1996, 2000; Gillespie 1995; Razeto-Barry et al. 2011). We simulated asexual populations under the assumption of weak mutation (Nu < 1, where u is the genomic mutation rate); thus the evolutionary process is depicted as a succession of fixations and neglects the effects of polymorphisms. We followed the methods of Razeto-Barry et al. (2011) to model molecular evolutionary processes in the framework of the FGM but with the difference that, in contrast to previous models (Gu 2007a,b; Su et al. 2010; Razeto-Barry et al. 2011; Razeto-Barry and Maldonado 2011), the number of dimensions n is not interpreted as the number of orthogonal traits affected by mutations in a gene, but rather as the number of orthogonal traits affected by mutations in the entire organismal genome.
The usual FGM-based modeling of evolutionary processes is the analysis of a bout of adaptation after a sudden, recent environmental shift of the optimum (Orr 1998, 1999, 2000; Welch and Waxman 2003), but the fate of an adaptive bout is to reach a fitness plateau (Silander et al. 2007), which is characterized by a balanced steady state in which molecular evolution does not stop (Hartl and Taubes 1996). This steady state has been studied while maintaining a fixed optimum (Hartl and Taubes 1998; Poon and Otto 2000; Sella and Hirsh 2005; Tenaillon et al. 2007; Sella 2009). In the VR (distinguished from SR) we modeled temporally fluctuating random optimum shifts; thus the evolutionary process was an alternation between adaptive bouts and balanced steady states that are determined by the variability of the environmental changes. Thus obviously the VR converges to the SR when environmental variability and amplitude tend to zero.
We studied the rate of molecular evolution as the ratio between the substitution and mutation rate (k/u), which is usually measured by the ratio between synonymous and nonsynonymous substitution rates (dN/dS), under the assumption that synonymous substitutions are almost neutral. In contrast to Gu (2007a,b) we utilized a top-down approach to isotropic random vector generation (Poon and Otto 2000); that is, we directly specified only the distribution of total mutation length and did not specify the marginal distributions along each axis. Thus a change in the number of dimensions does not affect the total length of the mutation’s effects, which allows us to distinguishing between the effect of dimensionality and the effect of mutation size on the molecular evolutionary rate (see Razeto-Barry et al. 2011). We followed Gu’s bottom-up approach for the random shifts of optimum values, but corrected the amplitudes of shifts by requiring that amplitudes be equal for different numbers of dimensions (see Razeto-Barry et al. 2011). Following Kimura (1983) and Orr (1998), the distribution used for mutation magnitudes was uniform.
Simulations
Simulations were performed with Monte Carlo methods by which vectors are generated randomly with a uniform distribution of vector magnitudes (from 0 to r). The total effect of a gene mutation (mutation size ) was measured as the Euclidean distance , where is the value of coordinate i of the mutant, is the value of the present phenotype (wild type), and i = 1, …, n. Assuming for simplicity that effective and census population sizes are equal, these changes are fixed with probability , where N is the effective population size and s is the selection coefficient of the mutation (Crow and Kimura 1970).
Fitness values follow the Gaussian function , where z is the distance to the optimum point. Selection coefficients are defined as , where wmut is the fitness of the mutant and w+ is the fitness of the wild type. We obtained the ratio between substitution rate and mutation rate (k/u) for different conditions, varying population size (N), complexity (number of dimensions, n), average size of phenotypic effects of mutations (hereafter mutation size, ), and (defined below) amplitude (σa) and variability (τ) of optimum shifts.
Random optimum shifts were simulated such that , where v and λ are the number and the expected number of changes in a time interval, respectively. Time intervals between consecutive changes (t) followed an exponential distribution , where τ = 1/λ is the expected time between optimum changes. Assuming a constant mutation rate per gene per individual per generation (u), the expected time between environmental changes is (Razeto-Barry et al. 2011). To compare processes with different effective population size in environments with equivalent times between changes, we then set τ ∝ N. The amplitudes of environmental changes were obtained by , where zi are the coordinates of the new optimum which were independently chosen from a Gaussian distribution centered at the origin of the coordinates, i.e., , where σa represents the standard deviation of the amplitudes of environmental changes. We set to guarantee that any relationship between evolutionary rate and dimensionality will be due to the effect of dimensionality itself and not because of a correlation between dimensionality and amplitude of environmental changes (see Razeto-Barry et al. 2011).
Strictly advantageous substitutions are defined as substitutions that comply with s > 1/N, i.e., advantageous substitutions fixed by positive selection; effectively neutral substitutions are defined as , i.e., substitutions fixed mainly by random drift; and strictly deleterious substitutions are defined as s < −1/N, i.e., deleterious substitutions fixed by drift in spite of the strong negative selection against them. When we denote mutations or substitutions with s > 0 and s < 0 we simply speak of “advantageous” or “deleterious,” respectively.
Results
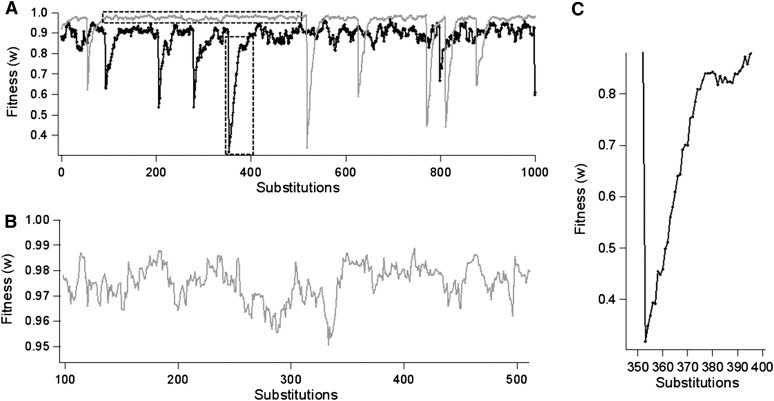
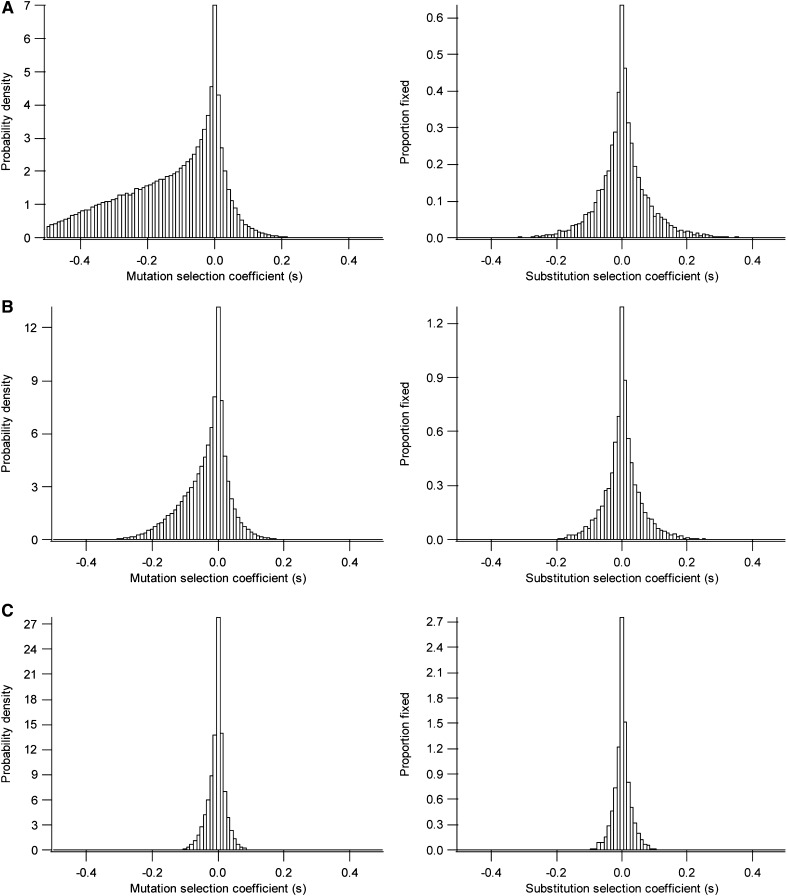
In Figure 2 we show a trial of the substitution process used for simulations with random environmental variability (the VR). After a shift of the optimum, the population suffers a burst of adaptive substitutions (Figure 2C) until it achieves a balanced steady state (Figure 2B). In the balanced steady state, the population remains dynamically around a suboptimum equilibrium fitness that is lower for smaller population sizes (Hartl and Taubes 1998; Poon and Otto 2000; Tenaillon et al. 2007) and in which a fluctuating substitution process occurs (Figure 2B). In the SR the ratio between advantageous (s > 0) and deleterious (s < 0) mutations depends on the size of mutations (Figure 3, left) with a leptokurtic distribution skewed toward more negative values for higher mutation sizes. The proportion of advantageous mutations tends to a maximum value of 0.5 for smaller mutation sizes (Figure 3C, left). The distribution of selection coefficients of substitutions (Figure 3, right) follows a leptokurtic but symmetric distribution independently of the mutation size. That is, the expected proportion of advantageous substitutions is always 0.5. The mean and standard deviation (σs) of the selection coefficients of mutations were calculated according to different mutation sizes, both for the steady state (SR) and for fluctuating environmental evolutionary processes (VR) (Figure 4). The mean selection coefficient decreases while the standard deviation increases with the increase of mutation size under all conditions (Figure 4A). The curves are clearly differentiated for smaller population sizes (N = 10 and N = 100), but they are very similar for larger population sizes (N = 100, 1000 and 10,000) (Figure 4A), and the absolute magnitude of the coefficient of variation of the selection coefficients approaches a value of one (Figure 4B). Curves are only slightly sensitive to other parameters such as to the number of dimensions and environmental variability (see Figure 4).
Figure 2 .
(A) Fitness of the evolutionary process for a sequence of substitutions with a randomly shifting optimum (variable geometric model, VR). The thick line corresponds to population size of N = 50 (τ = 5 × 105), and the thin line corresponds to N = 200 (τ = 105). (B) Zoom of the balanced steady state for N = 200 indicated by a dashed horizontal rectangle in A. (C) Zoom of the adaptive bout for N = 50 indicated by a dashed vertical rectangle in A. Simulation parameters were n = 20 dimensions, mutation size , shift amplitude σa = 0.85.
Figure 3 .
Probability density of selection coefficients of mutations (left) and the proportion fixed (right) in the balanced steady state (SR) model for different average mutation sizes: (A) , (B) , (C) . General parameters used were N = 10, n = 5 . Each distribution was constructed with 104 substitutions.
Figure 4 .
(A) Mean selection coefficient (solid line and right scale) and standard deviation of selection coefficients (left scale) for different mutation sizes (). Dotted lines correspond to effective population size N = 10 and dashed lines to N = 100; there are also triangles (N = 1000) and circles (N = 10,000), which are indistinguishable. Lines are superposed for different dimensions (n = 3, n = 30) and different environmental variability and amplitude (fixed and shifting optimum with τ = 104, σa = 0.425). (B) Coefficient of variation of selection coefficients for different mean step sizes. N = 1000 (dashed line); N = 10 (dotted line).
With no environmental variability (the SR), we found a negative relationship between total and effectively neutral, evolutionary rate and population size (Figure 5). The decrease is greater for larger mutation sizes (Figure 5). Strictly advantageous and strictly deleterious substitution rates are similar and decrease with increasing population sizes for larger mutation sizes (Figure 5, A and B), have a maximum for intermediate sizes (Figure 5C), and increased for small mutation sizes (Figure 5D). With environmental variability (the VR), the expected rate of advantageous substitutions (s > 0) is greater than that of deleterious substitutions (s < 0) and the relationship between evolutionary rate and population size is more complex (Figure 6). For small population sizes, the total evolutionary rate decreases with increase of population size except when mutation size is very small and the number of dimensions is large (Figure 6D). The decrease in total substitution rate reaches a plateau with smaller population sizes when mutation size is greater (Figure 6, A and B). The value of this plateau, in which the total substitution rate remains equal for different population sizes, is larger for lower mutation sizes and higher number of dimensions. The strictly advantageous substitution rate increases with population size, and the increase is stronger for smaller mutation size. However, the critical population size where the strictly advantageous rate exceeds the effectively neutral rate is larger for smaller mutation sizes (Figure 6, A and B).
Figure 5 .
Ratio between substitution rate and mutation rate in relation to population size for different sizes of mutation effects in the SR. (A) Average size (,σs ≈ 0.13). (B) (,σs ≈ 0.055), (C) (,σs ≈ 0.13), (D) (,σs ≈ 0). Dashed lines correspond to the total substitution rate. The upper continuous lines represent to effectively neutral substitutions. The lower continuous lines correspond to strictly advantageous and strictly deleterious substitution rates, which are indistinguishable. All plots were obtained for dimensions n = 2, n = 10, n = 30, but all are perfectly superposed and indistinguishable. Each simulation corresponds to 2 × 104 substitutions in the steady state.
Figure 6 .
Ratio between substitution rate and mutation rate vs. population size for different sizes of mutation effects in the VR. (A) Average size (,σs ≈ 0.13). (B) (,σs ≈ 0.055), (C) (,σs ≈ 0.13), (D) (,σs ≈ 0). Dashed lines correspond to the total substitution rate. The upper continuous lines correspond to neutral substitutions. The lower continuous lines correspond to a strictly advantageous (s > 1/N) substitution rate; dotted lines correspond to a strictly deleterious (s < −1/N) substitution rate. All plots were obtained for dimensions n = 2 (red), n = 10 (black), n = 30 (blue). Each simulation corresponds to 2 × 104 substitutions. The environmental variability parameters were σa = 0.85, τ = 500.N, which on the average result in a mean fitness w = 0.7 and a mean load L = 0.3. Each curve corresponds to 140,000 substitutions.
Discussion
The SR showed similarities to the house-of-cards or “fixed” model of molecular evolution (see Ohta and Tachida 1990; Tachida 1991), in which evolution is an alternating process with half of the substitutions being advantageous and the other half deleterious (Figure 3) (Tachida 1991; Gillespie 1994, 1995; Sella and Hirsh 2005). Most of these advantageous mutations would be compensatory, i.e., intragenic or intergenic mutations that restore the fitness loss due to previous deleterious mutations (Poon and Otto 2000). However, the SR model overcomes some problems of the house-of-cards model. It has been claimed that the house-of-cards model is not a plausible model of molecular evolution because the substitution rate is a rapidly decreasing (typically concave) function of the strength of selection (2Nσs), which stops when 2Nσs > 4 (Tachida 1991; Gillespie 1994, 1995; Ohta and Gillespie 1996). That is, it cannot contain enough mutations that behave effectively as neutral when the population size is large (Kimura 1979, 1983). By contrast, in the SR the relationship between substitution rate and population size (and thus the strength of selection) is convex (as in the shifting models; see Gillespie 1994, 1995) and evolution does not stop even for 2Nσs ≈ 260 (e.g., in Figure 5A σs ≈ 0.13, and when N = 1000, k/u ≈ 0.05). Tachida (1996) showed that in the house-of-cards model substitutions continue to occur even when 2Nσs = 20 if the distribution of the selection coefficients of mutations is uniform, making this model more plausible. However, the assumption of a uniform distribution is not realistic according to the current data that show leptokurtic selection coefficient distributions (e.g., Keightley 1994; Lynch et al. 1999; Orr 2010), as in our SR. Therefore, the SR may be considered to be a plausible nearly neutral model of molecular evolution without the apparent deficiencies of previous models, giving new support to the balanced nearly neutral models.
This difference between the SR and house-of-cards models is explained because, in contrast to the house-of-cards model, evolution does not stop because of low strength of selection in the SR, since in the FGM it is possible to overshoot the optimum phenotypic value; i.e., mutations directed to the phenotype with highest fitness in the FGM can decrease the fitness (because they can overshoot the optimum value) and thus more mutations can behave as effectively neutral. Contrarily, in the house-of-cards model all mutations directed toward higher fitness confer higher fitness if they are fixed, because advantageous mutations can take unlimited positive selection coefficients. The consequence is that molecular evolution in the house-of-cards models tends to stop because the pressure toward higher fitness decreases the number of possible further advantageous mutations.
The evolutionary role of compensatory mutations is not completely understood and there are few theoretical models in evolutionary biology in which compensatory mutations are explicitly incorporated (Poon et al. 2005). It has been suggested that the current evidence for a high rate of advantageous mutations fixed by positive selection, ∼50% or more (Fay et al. 2002; Bierne and Eyre-Walker 2004; Eyre-Walker 2006; Bachtrog 2008), could be explained as the effect of compensatory mutations (Kondrashov et al. 2002; DePristo et al. 2005; Pál et al. 2006; Camps et al. 2007). This idea was proposed earlier by Hartl and Taubes (1996) using the FGM framework, stating that in the steady state there is “selection without adaptation,” i.e., positive selection but upholding only the status quo in a balance between deleterious mutations and later advantageous compensatory mutations. In contrast with this assumption, we found that the proportion of advantageous (compensatory) mutations fixed by positive selection (i.e., strictly advantageous mutations) is very low in the steady state (much lower than 50%; see Figure 5). The explanation for this result is that compensatory mutations are abundant and thus come after one or a very small number of deleterious mutations previously fixed by drift, and thus both are of the same order of magnitude; i.e., both are mainly effectively neutral (see Figure 5). In other words, given that in the SR not more than 50% of substitutions are advantageous and the distribution of selection coefficients of substitutions is leptokurtic (Figure 3), necessarily only a small proportion of advantageous substitutions (s > 0) could be strictly advantageous (s > 1/N). Thus, given the small selection coefficients of advantageous mutations, it is difficult to explain the high rate of mutations fixed by positive selection (Fay et al. 2002; Bierne and Eyre-Walker 2004; Eyre-Walker 2006; Bachtrog 2008). It is possible that if compensatory mutations were very rare, on the average several deleterious substitutions could be fixed before an advantageous mutation compensated the previous effect of the deleterious ones. In this case, a higher proportion of compensatory substitutions could be of greater size and strictly advantageous. By contrast, our model assumes a high rate of compensatory mutations, which is in agreement with current studies (Poon and Otto 2000; Whitlock et al. 2003; Poon et al. 2005). For example, Poon et al.’s (2005) study in viruses, prokaryotes, and eukaryotes revealed that on average there are 11.8 compensatory mutations per deleterious mutation. Our conclusions seem robust in this scenario.
Both the slightly deleterious mutation and the balanced mutation theories (Figure 1C,D) have been considered within the nearly neutral theory. In turn, the distinction between nearly neutral models and selection models seems to be somewhat unclear (see Ohta 1996; Kreitman 1996; Ohta and Gillespie 1996). The main difference between the nearly neutral and selection theories is that the former predicts a negative relationship between evolution and population size whereas the latter predicts the contrary (Ohta 1996). The role of population size in population genetic models of molecular evolution was examined by Gillespie (1999), who defined three domains according to the relationship between substitution rate and population size. In Ohta’s domain, the rate of substitution decreases with increasing population size, while in Kimura’s domain, the rate of substitution remains close to the mutation rate, while in Darwin’s domain, the rate of substitution increases with increasing population size. Our SR verified an inverse relationship between the substitution rate and population size (Figure 5) (see also Gu 2007a); thus the SR falls under Ohta’s domain. Because this inverse relationship is essential to explaining the protein molecular clock under the nearly neutral theory (see Ohta 1992), the SR may also be understood as a nearly neutral model. However, it is important to distinguish the SR from the slightly deleterious mutation theory (Figure 1C), which is the model most commonly associated with the nearly neutral theory (Gillespie 1995, 2004; Ohta 1996). The differences between the slightly deleterious mutation models and the SR are important in their predictions about both mutations and substitutions. There are two major differences: (i) the mutation assumptions of the SR involve a higher fraction of advantageous (mainly compensatory) mutations than the slightly deleterious mutation models and (ii) the predictions of the SR imply a much greater fraction of advantageous substitutions than the slightly deleterious mutation models; thus the SR predicts 50% advantageous substitutions (Figure 1D and Figure 3, right; see also Gillespie 1995; Sella and Hirsh 2005).
Furthermore, the SR can relate the evolutionary rate to the average size of phenotypic effects of mutations (mutation size). In the SR the total evolutionary rate is determined mainly by the effectively neutral mutations and is greater for smaller mutation sizes (Figure 5), which is consistent with the decrease of the mean selection coefficients of larger mutations (Figure 4A). However, when the rate of strictly advantageous and strictly deleterious substitutions are described separately, we found a complex relationship with population size for different ranges of mutation sizes: a decrease with population size for larger mutation sizes (Figure 5A,B), a maximum for intermediate sizes (Figure 5C), and an increase with population size for smaller mutation sizes (Figure 5D). This trend to increase the rate of strictly advantageous substitutions (with the corresponding balance by deleterious substitutions) with population size when mutation size is low may be explained as follows. As mentioned before, the decrease of the evolutionary rate with population size is the commonly expected behavior under the nearly neutral framework, because larger population sizes imply strong selection against deleterious mutations, decreasing the substitution rate of deleterious mutations and thus also decreasing the rate of advantageous compensatory mutations. Although we found this pattern for the total molecular rate, we found that for lower mutation sizes the rate of strictly advantageous substitutions increases with population size (Figure 5D, lower curve), which is not typically predicted by nearly neutral models. This occurs because for small mutation sizes, the SR implies that a high proportion of nearly neutral mutations (50%) are advantageous (Figure 3C). In this situation, the distribution of selection coefficients of mutations is symmetrical (Figure 3C) and thus the increase of population size has the effect of increasing the strength of selection equally for advantageous and deleterious mutations. Therefore, given that the probability of fixation is higher for advantageous than deleterious mutations, the effect of the increment in the strength of selection when the distribution of mutations is symmetrical is the increase of the rate of strictly advantageous mutations (with the respective balance of strictly deleterious ones) (Figure 5D).
As mentioned before, contrary to previous studies of the FGM that have been focused separately on the adaptive process (Orr 1998, 1999, 2006; Welch and Waxman 2003; Griswold and Whitlock 2003) or on the steady state (Hartl and Taubes 1998; Poon and Otto 2000; Sella and Hirsh 2005; Tenaillon et al. 2007; Sella 2009), the VR is a model in which there is an interplay between adaptive and balanced steady-state processes (Figure 2; see also Razeto-Barry et al. 2011). Generally, selection models of molecular evolution assume environmental changes (Gillespie 1993; Ohta and Gillespie 1996), as for example, in the mutational landscape model (Gillespie 1984, 1991), NK model (Kauffman 1993), TIM model (Takahata et al. 1975), and SAS-CFF model (Gillespie 1991). The reason is that populations tend to evolve to a point where most mutations are deleterious through the substitution of advantageous mutations. The idea that permanent advantageous mutation fixation by positive selection could occur without optimum shifts probably comes from early findings in the vertebrate major histocompatibility complex and coevolutionary processes of pathogens that erroneously were taken as a model for the general evolution of proteins (Hughes 2007). Thus, a more general molecular evolutionary model would predict that when all mutationally accessible advantageous alleles are exhausted, the majority of newly arising mutations will be deleterious (Gillespie 1994) or nearly neutral (Hartl and Taubes 1996). Accordingly, in the VR the evolutionary rate increases due to temporal environmental fluctuations (Figure 6) compared to the rate without environmental fluctuations but with the same mutation sizes (Figure 5) (see also Razeto-Barry et al. 2011, Figure 2). Given that in the VR populations reach dynamic (i.e., nonstatic) steady state after adaptive bouts, evolution does not stop without environmental fluctuations (contrary to other models; see Ohta 1996). Nevertheless, given that compensatory mutations cannot explain the repeated substitutions by positive selection, and because positive selection tends to stop after some steps (Hughes 2007), the optimum shifts assumption is the only remaining reasonable hypothesis for maintaining a selection model at the molecular level.
The higher evolutionary rate in the VR compared to the SR is evidently due to an increase of strictly advantageous substitution rate (Figure 6, lower solid lines). Interestingly, this increase is greater for smaller mutation sizes, attaining values k/u > 1 (Figure 6D). Paradoxically, this does not imply that for smaller mutation sizes the evolutionary process becomes more influenced by natural selection. In fact, the rate of strictly advantageous substitutions exceeded the effectively neutral rate with larger mutation sizes even for low population size (Figure 6, A and B), but this did not occur with smaller mutation sizes (Figure 6D). The increase of the strictly advantageous substitution rate for smaller mutation sizes occurs because the adaptive process needs a larger number of advantageous substitutions to reach the steady state (see Razeto-Barry and Maldonado 2011). Interestingly, the evolutionary rate attains a plateau that does not depend on population size, but rather on the complexity (number of dimensions) of organisms (Figure 6, A and B). For greater organism complexity, the evolutionary rate increases, which may be understood as a consequence of Orr’s (2000) “cost of complexity”; that is, more complex organisms spend more time in adaptive processes than less complex ones, accumulating on the average a larger number of mutations with smaller adaptive contribution (see Razeto-Barry et al. 2011; Razeto-Barry and Maldonado 2011). The independence of substitution rate and population size in the plateau is due to the fact that the increase of strictly advantageous substitutions is accurately balanced by the decrease of effectively neutral substitutions (Figure 6). As the effectively neutral substitution rate decreases monotonically, the strictly advantageous rate increases to a plateau. This is consistent with recent evidence on the rate of strictly advantageous substitution that appears to be independent of population size (Bachtrog 2008). Bachtrog (2008) found only a slightly higher rate of strictly advantageous substitutions in Drosophila melanogaster than in D. miranda, in spite of the latter having an effective population size five times smaller than the former. Furthermore, more generally this may explain why the estimated proportion of amino acid variants driven to fixation by positive selection seems to differ among species with small population sizes but not much for species with very large effective population sizes, even though these differ by several orders of magnitude (Harris 2010, Tables 2 and 3).
As expected, the increase of environmental amplitude has an effect on the evolutionary rate equivalent to the increase of environmental variability in increasing the rate of advantageous substitution (Razeto-Barry et al. 2011), but biologically these phenomena are not completely equivalent. Indeed, an important assumption of the VR simulations is that populations do not go to extinction due to environmental fluctuations. In fact, populations cannot support a too-high level of load (i.e., suboptimum fitness value due to an environmental shift of great amplitude) (Haldane 1957). For example, in our simulations the mean load due to environmental fluctuations is L = 0.3 (i.e., a fitness value of w = 0.7, for σa = 0.85). That is on the order of the cost assumed by Ewens (2004); however, Haldane (1957) assumed a load of 0.1 on the basis of human data. This value is probably conservative and depends on population density (Nunney 2003). Overall, populations with high reproductive excess could bear the fitness decrease due to environmental changes (Nunney 2003; Ewens 2004). Another assumption of our model where the strength of the environmental change is important is that the fitness surface is Gaussian. This is an assumption usually justified because when a population is close to the optimum, a Gaussian fitness function is a good local approximation for many arbitrary fitness functions (Lande 1980); however, it could be less accurate under strong environmental change where the population is not so near the optimum (Martin and Lenormand 2006a). Finally, models relaxing the assumption of Nu < 1 should be developed in the future.
Contrary to the suggestion of Orr (1998) the uniform distribution we assumed for mutation sizes (see also Kimura 1983) is consistent with the majority of current empirical and theoretical evidence on distributions of the fitness effects of mutations. According to the majority of studies, the distributions of deleterious mutations (Keightley 1994; Lynch et al. 1999) and beneficial mutations (Sanjuán et al. 2004; Kassen and Bataillon 2006; Orr 2006, 2010) are leptokurtic. Note that all these studies (empirical and theoretical) dealt with the size of the fitness effects of mutations (selection coefficients), not with the size of the phenotypic effects of mutations. As is clear in Figure 3 (left), taking a uniform distribution of mutational (phenotypic) effects (r), the distribution of fitness effects (selection coefficients, s) is leptokurtic. In fact, when the average mutational size () is large enough (Figure 3A), the distribution of s among deleterious mutations is L-shaped rather than exponential, which coincides with the literature (Lynch et al. 1999), while the distribution of s among beneficial mutations is exponential-like, which also coincides with the literature (Sanjuán et al. 2004; Kassen and Bataillon 2006; Orr 2006, 2010). Note also that the exponential distribution of s arises from different mutational distributions of r, including the uniform (Orr 2006).
In addition to the mentioned relationships among substitution rate, mutation size, population size, and organismal complexity, some important parameters were found linked in the SR and the VR. For example, for both the SR and the VR, the absolute magnitude of the coefficient of variation of selection coefficients () approaches one as the mean step size increases when N ≥ 100 (e.g., Figure 4B, upper curve); that is, if the mean of the absolute value of the selection coefficients decreases, the standard deviation increases nearly in the same proportion. Recently, it has been realized that a benefit of the FGM is that it makes some of the distributions used in molecular evolution biologically interpretable (Sella and Hirsh 2005; Martin and Lenormand 2006a,b, 2008; Gu 2007a,b). A priori assumptions of a particular distribution of mutant selection coefficients have been considered inappropriate because the distribution determined by evolutionary dynamics will differ in important ways from distributions assumed a priori (Sella and Hirsh 2005). But another problem is that some parameters assumed in the general models (particularly and σs) are crucial by themselves for other biologically relevant issues and should be determined instead of just being assumed. For example, the proportion of deleterious to advantageous mutations (determined by and σs) is crucial to estimating the probability of extinction of populations and it is frequently fixed arbitrarily in the models due to the lack of other theoretical or empirical criteria to determine it (Whitlock 2000; Whitlock et al. 2003).
Other parameters are also linked in the SR. The ratio between advantageous and deleterious mutations is locked; i.e., it cannot take arbitrarily independent values. It yields a maximum value of 1 when mutation size tends to zero (Figure 3C, according to Fisher 1930). When mutation size is small, the limiting factor for the selection coefficient of deleterious and advantageous mutations is the size of mutations (Figure 3C), and a large proportion (50%) of nearly neutral mutations are advantageous. Interestingly, this fact was suggested verbally by Gillespie (1995) as a criticism to Ohta’s (1977, 1992) assumptions, based on Fisher’s (1930) classical result.
The results of simulations indicate that at least three of the conclusions of this study are well verified in the model. First, compensatory substitutions cannot take arbitrary values and necessarily a low proportion of compensatory substitutions are strictly advantageous. Second, the proportion of advantageous substitutions has a minimum of 0.5 (for very low environmental variability), and the proportion of advantageous mutations has a maximum of 0.5 (for very small mutation size). Third, the absolute magnitude of the coefficient of variation of selection coefficients approaches one. Again, these conclusions support the importance of obtaining values for these parameters by modeling the evolutionary process and not by a priori decisions (Sella and Hirsh 2005), which in turn, may give independent plausibility to some molecular evolutionary models rather than to others (Figure 1) when there are not enough data to judge.
Acknowledgments
We thank Associate Editor Lindi Wahl and two anonymous reviewers for their insightful suggestions. We also thank the Institute of Ecology and Biodiversity ICM-P05-002 and PFB-23-CONICYT, and FONDECYT 1090794 (R.A.V.), and the Institute of Philosophy and Complexity Science (IFICC). P.R.-B. acknowledges support from Doctoral Scholarship 21050901-CONICYT and Supporting Doctoral Thesis Scholarship 23070216-CONICYT.
Footnotes
Communicating editor: L. M. Wahl
Literature Cited
- Bachtrog D., 2008. Similar rates of protein adaptation in Drosophila miranda and D. melanogaster, two species with different current effective population sizes. BMC Evol. Biol. 8: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 2001. The role of hybridization in evolution. Mol. Ecol. 10: 551–568 [DOI] [PubMed] [Google Scholar]
- Bernardi G., 2007. The neoselectionist theory of genome evolution. Proc. Natl. Acad. Sci. USA 104(20): 8385–8390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N., Eyre-Walker A., 2004. The genomic rate of adaptive amino acid substitution in Drosophila. Mol. Biol. Evol. 21: 1350–1360 [DOI] [PubMed] [Google Scholar]
- Bromham L., Penny D., 2003. The modern molecular clock. Nat. Rev. Genet. 4: 216–224 [DOI] [PubMed] [Google Scholar]
- Camps M., Asael H., Lawrence A. L., 2007. Genetic constraints on protein evolution. Crit. Rev. Biochem. Mol. Biol. 42: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory. Harper & Row, New York [Google Scholar]
- DePristo M. A., Weinreich D. M., Hartl D. L., 2005. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat. Rev. Genet. 6: 678–687 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., 2006. The genomic rate of adaptive evolution. Trends Ecol. Evol. 21(10): 569–575 [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., Smith N. G. C., Gaffney D., 2002. Quantifying the slightly deleterious mutation model of molecular evolution. Mol. Biol. Evol. 19(12): 2142–2149 [DOI] [PubMed] [Google Scholar]
- Ewens W. J., 2004. Mathematical Population Genetics. Springer, Philadelphia, PA [Google Scholar]
- Fay J. C., Wyckoff G. J., Wu C. I., 2002. Testing the neutral theory of molecular evolution with genomic data from Drosophila. Nature 415: 1024–1026 [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1930. Genetical Theory of Natural Selection. Clarendon Press, Oxford. [Google Scholar]
- Gillespie J. H., 1984. Molecular evolution over the mutational landscape. Evolution 38: 1116–1129 [DOI] [PubMed] [Google Scholar]
- Gillespie J. H., 1991. The Causes of Molecular Evolution. Oxford University Press, Oxford [Google Scholar]
- Gillespie J. H., 1993. Substitution processes in molecular evolution. I. Uniform and clustered substitutions in a haploid model. Genetics 134: 971–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1994. Substitution processes in molecular evolution. III. Deleterious alleles. Genetics 138: 943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie J. H., 1995. On Ohta’s hypothesis: most amino acid substitutions are deleterious. J. Mol. Evol. 40: 64–69 [Google Scholar]
- Gillespie J. H., 1999. The role of population size in molecular evolution. Theor. Popul. Biol. 55: 145–156 [DOI] [PubMed] [Google Scholar]
- Gillespie J. H., 2004. Population Genetics, Ed. 2 Johns Hopkins Univ. Press, London [Google Scholar]
- Griswold C. K., Whitlock M. C., 2003. The genetics of adaptation: the roles of pleiotropy, stabilizing selection and drift in shaping the distribution of bidirectional fixed mutational effects. Genetics 165: 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., 2007a Stabilizing selection of protein function and distribution of selection coefficient among sites. Genetica 130: 93–97 [DOI] [PubMed] [Google Scholar]
- Gu X., 2007b Evolutionary framework for protein sequence evolution and gene pleiotropy. Genetics 175: 1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1957. The cost of natural selection. J. Genet. 55: 511–524 [Google Scholar]
- Harris E. E., 2010. Nonadaptive processes in primate and human evolution. Yearb. Phys. Anthropol. 53: 13–45 [DOI] [PubMed] [Google Scholar]
- Hartl D., Taubes C. H., 1996. Compensatory nearly neutral mutations: selection without adaptation. J. Theor. Biol. 182: 303–309 [DOI] [PubMed] [Google Scholar]
- Hartl D., Taubes C. H., 1998. Towards a theory of evolutionary adaptation. Genetica 103: 525–533 [PubMed] [Google Scholar]
- Hughes A. L., 2007. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity 99: 364–373 [DOI] [PubMed] [Google Scholar]
- Kassen R., Bataillon T., 2006. Distribution of fitness effects among beneficial mutations before selection in experimental populations of bacteria. Nat. Genet. 38: 484–488 [DOI] [PubMed] [Google Scholar]
- Kauffman S. A., 1993. The Origins of Order. Oxford University Press, Oxford [Google Scholar]
- Keightley P. D., 1994. The distribution of mutation effects on viability in Drosophila melanogaster. Genetics 138: 1315–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1968. Evolutionary rate at the molecular level. Nature 217: 624–626 [DOI] [PubMed] [Google Scholar]
- Kimura M., 1979. Model of effectively neutral mutations in which selective constraint is incorporated. Proc. Natl. Acad. Sci. USA 76: 3440–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., 1983. The Neutral Allele Theory of Molecular Evolution. Cambridge University Press, Cambridge [Google Scholar]
- Kondrashov A. S., Sunyaev S., Kondrashov F. A., 2002. Dobzhansky–Muller incompatibilities in protein evolution. Proc. Natl. Acad. Sci. USA 99: 14878–14883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M., 1996. The neutral theory is dead: long live the neutral theory. Bioessays 18(8): 678–683 [DOI] [PubMed] [Google Scholar]
- Lande R., 1980. The genetic covariance between characters maintained by pleiotropic mutations. Genetics 94: 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Blanchard J., Houle D., Kibota T., Schultz S., et al. , 1999. Perspective: spontaneous deleterious mutation. Evolution 53: 645–663 [DOI] [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2006a A general multivariate extension of Fisher’s geometrical model and the distribution of mutation fitness effects across species. Evolution 60(5): 893–907 [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2006b The fitness effect of mutations across environments: a survey in light of fitness landscape models. Evolution 60: 2413–2427 [PubMed] [Google Scholar]
- Martin G., Lenormand T., 2008. The distribution of beneficial and fixed mutation fitness effects close to an optimum. Genetics 179: 907–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Yang Z., 2003. Estimating the distribution of selection coefficients from phylogenetic data with applications to mitochondrial and viral DNA. Mol. Biol. Evol. 20(8): 1231–1239 [DOI] [PubMed] [Google Scholar]
- Nunney L., 2003. The cost of natural selection revisited. Ann. Zool. Fenn. 40: 185–194 [Google Scholar]
- Ohta T., 1972. Evolutionary rate of cistrons and DNA divergence. J. Mol. Evol. 1: 150–157 [DOI] [PubMed] [Google Scholar]
- Ohta T., 1973. Slightly deleterious mutant substitutions in evolution. Nature 246: 96–98 [DOI] [PubMed] [Google Scholar]
- Ohta T., 1977. Extension of the neutral mutation drift hypothesis, pp. 148–167 in Molecular Evolution and Polymorphism, edited by M. Kimura. National Institute of Genetics, Mishima, Japan [Google Scholar]
- Ohta T., 1987. Very slightly deleterious mutations and the molecular clock. J. Mol. Evol. 26: 1–6 [DOI] [PubMed] [Google Scholar]
- Ohta T., 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23: 263–286 [Google Scholar]
- Ohta T., 1996. The current significance and standing of neutral and nearly neutral theoies. BioEssays 18(8): 673–694 [DOI] [PubMed] [Google Scholar]
- Ohta T., 2007. Drift and selection in evolving interacting systems, pp. 285–298 in Structural Approaches to Sequence Evolution, chap. 13, edited by Bastolla U., Porto M., Roman H. E., Vendruscolo M. Springer, Berlin/Heidelberg [Google Scholar]
- Ohta T., Gillespie J. H., 1996. Development of neutral and nearly neutral theories. Theor. Popul. Biol. 49: 128–142 [DOI] [PubMed] [Google Scholar]
- Ohta T., Kimura M., 1971. On the constancy of the evolutionary rate of cistrons. J. Mol. Evol. 1: 18–25 [DOI] [PubMed] [Google Scholar]
- Ohta T., Tachida H., 1990. Theoretical study of near neutrality. I. Heterozygosity and rate of mutant substitution. Genetics 126: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr A. H., 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52(4): 935–949 [DOI] [PubMed] [Google Scholar]
- Orr A. H., 1999. The evolutionary genetics of adaptation: a simulation study. Genet. Res. Camb. 74: 207–214 [DOI] [PubMed] [Google Scholar]
- Orr A. H., 2000. Adaptation and the cost of complexity. Evolution 54: 13–20 [DOI] [PubMed] [Google Scholar]
- Orr A. H., 2006. The distribution of beneficial fitness effects among beneficial mutations in Fisher’s geometric model of adaptation. J. Theor. Biol. 238: 279–285 [DOI] [PubMed] [Google Scholar]
- Orr A. H., 2010. The population genetics of beneficial Mutations. Phil. Trans. R. Soc. B 365: 1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C., Papp B., Lercher M. J., 2006. An integrated view of protein evolution. Nat. Rev. Genet. 7: 337–348 [DOI] [PubMed] [Google Scholar]
- Poon A., Otto S. P., 2000. Compensating for our load of mutations: freezing the meltdown of small populations. Evolution 54: 1467–1479 [DOI] [PubMed] [Google Scholar]
- Poon A., Davis B. H., Chao L., 2005. The coupon collector and the suppressor mutation: estimating the number of compensatory mutations by maximum likelihood. Genetics 170: 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razeto-Barry P., Maldonado K., 2011. Adaptive cis-regulatory changes may involve few mutations. Evolution 65(11): 3332–3335 [DOI] [PubMed] [Google Scholar]
- Razeto-Barry P., Díaz J., Cotoras D., Vásquez R. A., 2011. Molecular evolution, mutation size and gene pleiotropy: a geometric reexamination. Genetics 187(3): 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R., Moya A., Elena S. F., 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl. Acad. Sci. USA 101: 8396–8401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G., 2009. An exact steady-state solution of Fisher’s geometric model and other models. Theor. Popul. Biol. 75: 30–34 [DOI] [PubMed] [Google Scholar]
- Sella G., Hirsh A. E., 2005. The application of statistical physics to evolutionary biology. Proc. Natl. Acad. Sci. USA 102(27): 9541–9546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silander O. K., Tenaillon O., Chao L., 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5(4): e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Zeng Y., Gu X. 2010. A preliminary analysis of gene pleiotropy estimated from protein sequences. J. Exp. Zool. 314B: 115–122 [Google Scholar]
- Tachida H., 1991. A study on a nearly neutral mutation model in finite populations. Genetics 128: 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachida H., 1996. Effects of the shape of distribution of mutant effect in nearly neutral mutation models. J. Genet. 75(1): 33–48 [Google Scholar]
- Tachida H., 2000. Molecular evolution in a multisite nearly neutral mutation model. J. Mol. Evol. 50: 69–81 [DOI] [PubMed] [Google Scholar]
- Takahata N., Ishii K., Matsuda H., 1975. Effect of temporal fluctuation of selection coefficient on gene frequency in a population. Proc. Natl. Acad. Sci. USA 72: 4541–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Silander O. K., Uzan J. P., Chao L., 2007. Quantifying organismal complexity using a population genetic approach. PLoS ONE 2: e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J. J., Waxman D., 2003. Modularity and the cost of complexity. Evolution 57(8): 1723–1734 [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., 2000. Fixation of new alleles and the extinction of small populations: drift load, beneficial alleles, and sexual selection. Evolution 54: 1855–1861 [DOI] [PubMed] [Google Scholar]
- Whitlock M. C., Grisworld C. K., Peters A. D., 2003. Compensating for the meltdown: the critical effective size of a population with deleterious and compensatory mutations. Ann. Zool. Fenn. 40: 169–183 [Google Scholar]