Abstract
Alterations in the pancreatic fluid proteome of individuals with chronic pancreatitis may offer insights into the development and progression of the disease. The endoscopic pancreas function test (ePFT) can safely collect large volumes of pancreatic fluid that are potentially amenable to proteomic analyses using difference gel electrophoresis (DiGE) coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS). Pancreatic fluid was collected endoscopically using the ePFT method following secretin stimulation from three individuals with severe chronic pancreatitis and three chronic abdominal pain controls. The fluid was processed to minimize protein degradation and the protein profiles of each cohort, as determined by DiGE and LC-MS/MS, were compared. This DiGE-LC-MS/MS analysis reveals proteins that are differentially expressed in chronic pancreatitis compared to chronic abdominal pain controls. Proteins with higher abundance in pancreatic fluid from chronic pancreatitis individuals include: actin, desmoplankin, alpha-1-antitrypsin, SNC73, and serotransferrin. Those of relatively lower abundance include carboxypeptidase B, lipase, alpha-1-antichymotrypsin, alpha-2-macroglobulin, Arp2/3 subunit 4, glyceraldehyde-3-phosphate dehydrogenase, and protein disulfide isomerase. Endoscopic collection (ePFT) in tandem with DiGE-LC-MS/MS is a suitable approach for pancreatic fluid proteome analysis, however, further optimization of our protocol, as outlined herein, may improve proteome coverage in future analyses.
Keywords: pancreas, pancreas juice, pancreatic function test, biomarkers
Introduction
Chronic pancreatitis is characterized by chronic inflammation and progressive scarring leading to irreversible functional damage to the pancreas, resulting in pain, malabsorption of fat and protein, loss of endocrine function, and in some cases, pancreatic carcinoma [1; 2]. Currently diagnosis of this progressively debilitating disease is limited to moderate and advanced disease stages. A better understanding of chronic pancreatitis pathogenesis is necessary to elucidate mechanisms of early disease.
The application of proteomics to the study of pancreatic disease may accelerate the discovery of physiologically- and clinically-relevant biomarkers. Moreover, the analysis of a proximal body fluid, such as pancreatic fluid, increases the likelihood of biomarker discovery in the context of a particular diseased organ (i.e., the pancreas). Pancreatic fluid is an excellent clinical specimen for such analyses, as its protein composition is of lower complexity compared to serum and because the proteins in pancreatic fluid predominantly originate from the exocrine pancreas [3; 4].
We have developed and described an endoscopic pancreatic function test (ePFT) that collects pancreatic fluid without cannulation of the pancreatic duct and without the risk of procedure-related injury [5; 6; 7]. The ePFT collection method replaces the Dreiling tube with an upper endoscope [5]. Dreiling tubes allow for the collection of duodenal and gastric fluid during pancreatic function testing (PFT) [8], but the placement of Dreiling tubes, can be time consuming, cumbersome, and requires fluoroscopy. In addition, traditional endoscopic retrograde cholangiopancreatography (ERCP) has also been utilized to collect pancreatic fluid directly from the pancreatic duct [9]. Similarly, ERCP is highly invasive and is also associated with significant risks (5–10%) for the development of acute pancreatitis in patients [10; 11].
In contrast to the traditional pancreatic fluid collection methods, ePFT leads to significantly less morbidity for the patients, lowers cost and allows for the collection of larger volumes of fluid. Furthermore, the ePFT is now considered an acceptable alternative for the assessment of pancreas secretory physiology [12; 13]. It is possible that some of the proteins collected are from gastric or duodenal origin. However, duodenal protein secretion is minimal, and the efflux of gastric fluid is limited by placing the patient in the left lateral decubitus position during the procedure. Moreover, both fluids, (collectively known as gastroduodenal fluid [14]) are evacuated prior to ePFT and any remnants are subsequently diluted by the protein-rich secretin-stimulated pancreatic secretions. As the ePFT collection method is a valuable tool for acquiring pancreatic fluid, even from individuals without pancreas-related disease, we aim to evaluate this technique, coupled with DiGE, as a suitable approach to investigate differential protein secretion in the pancreatic fluid of individuals with chronic pancreatitis and chronic abdominal pain.
Difference gel electrophoresis (DiGE), followed by tandem mass spectrometry (MS/MS), may be a valuable strategy to study and understand better the abnormalities in the proteomic profiles of individuals with chronic pancreatitis. In DiGE, two samples can be compared by labeling each with a specific fluorescent dye which has unique spectral properties. Following two-dimensional gel separation, gels are imaged at each dye-specific wavelength. The merged images reveal overlapping or uniquely labeled gel spots, which can be excised and in-gel digested. The resulting peptides can be analyzed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) for protein identification. We apply this mass spectrometry-based proteomics strategy to investigate protein differences in pancreatic fluid from individuals with severe chronic pancreatitis and chronic abdominal pain controls. Although our long-term goals involve the elucidation of biomarkers of early chronic pancreatitis, we have chosen to investigate two extremes of the disease so as to gain insight into what proteins to target, as no early state diagnosis is possible currently.
The primary objectives of our exploratory investigation are as follows: 1) collect pancreatic fluid with the ePFT method after secretin stimulation, 2) analyze pancreatic fluid using DiGE, 3) determine the identity of proteins that are differentially secreted in the pancreatic fluid of chronic pancreatitis and chronic abdominal pain controls using LC-MS/MS techniques, and 4) assess the utility of DiGE-LC-MS/MS analysis for the differential proteome analysis of pancreatic fluid.
The methodology established herein enables further comparative analysis of the proteins in the ePFT-collected pancreatic fluid secretome of healthy individuals and those with pancreatic disease, thus broadening our knowledge of pancreatic disease pathogenesis.
Materials and Methods
Study Design and Setting
Proteomic analysis of endoscopically collected pancreatic fluid in an academic center.
Study Population
This protocol was approved by the Institutional Review Board at Brigham and Women’s Hospital (IRB # 2007-P-002480/1). The study population (Table 1) included adult patients seen in the Center for Pancreatic Diseases at Brigham and Women’s Hospital for abdominal pain and dyspepsia. Subjects were referred to the Center for Pancreatic Disease to eliminate pancreas etiologies for their gastrointestinal symptoms. All subjects underwent the following: 1) comprehensive history and physical examination, 2) review of radiologic and endoscopic data, and 3) upper endoscopy with ePFT followed by mucosal biopsy. The diagnosis of chronic pancreatitis was deemed definitive according to the M-ANNHEIM (Multiple risk factors, Alcohol, Nicotine, Nutrition, Hereditary factors, Efferent duct factors, Immunological factors, and - Miscellaneous and metabolic factors) classification [15]. The M-ANNHEIM classification is a standardized system designed to classify chronic pancreatitis according to etiology, clinical staging, and severity of the disease [15]. This system considered clinical imaging data resulting from a wide array of laboratory test results, including ultrasound (US), endoscopic ultrasound (EUS), magnetic resonance imaging (MRI), computed tomography (CT) as well as other risk factors [15; 16].
Table 1.
Demographic and clinical data for study cohort.
| ID | Reason for referral |
Age (yr) |
Gender | CT Scan | MRI | Pancreatic elastase – 1 (units/mg) |
EUS Score (0–9) |
ePFT Peak Bicarbonate (meq/L) |
Pancreatic fluid protein conc. (mg/mL) |
Endoscopic Biopsy (gastric, duodenum) |
|---|---|---|---|---|---|---|---|---|---|---|
| CAP1 | abdominal pain | 28 | male | normal | normal | n/a | 3 | 84 | 1.2 | normal |
| CAP2 | abdominal pain | 53 | female | normal | normal | 148 (diarrhea) | 1 | 92 | 0.8 | gastropathy |
| CAP3 | abdominal pain | 54 | male | normal | normal | 202 | n/a | 114 | 0.6 | normal |
| CP1 | chronic pancreatitis | 64 | female | atrophy, calcifications, strictured and dilated duct | atrophy, dilated and strictured duct, filling effects, low T1 signal, dilated side branches | 221 | n/a | 37 | 1.1 | duodenitis, gastropathy |
| CP2 | chronic pancreatitis | 60 | female | atrophy, calcifications, strictured and dilated duct | n/a | n/a | 8 | 54 | 0.6 | normal |
| CP3 | chronic pancreatitis | 52 | female | n/a | atrophy, dilated and strictured duct, low T1 signal, dilated side branches | <15 | 5 | 38 | 0.7 | normal |
CT, indicates computed tomography; MRI, magnetic resonance imaging; EUS, endoscopic ultrasound; ePFT, endoscopic pancreas function test; CP, chronic pancreatitis; conc., concentration; n/a, not available.
Materials
ChiRhoStim® synthetic human secretin was from ChiRhoClin (Burtonsville MD). Other reagents and solvents were from Sigma-Aldrich (St. Louis, MO) and Burdick & Jackson (Morristown, NJ), respectively.
Experimental Workflow
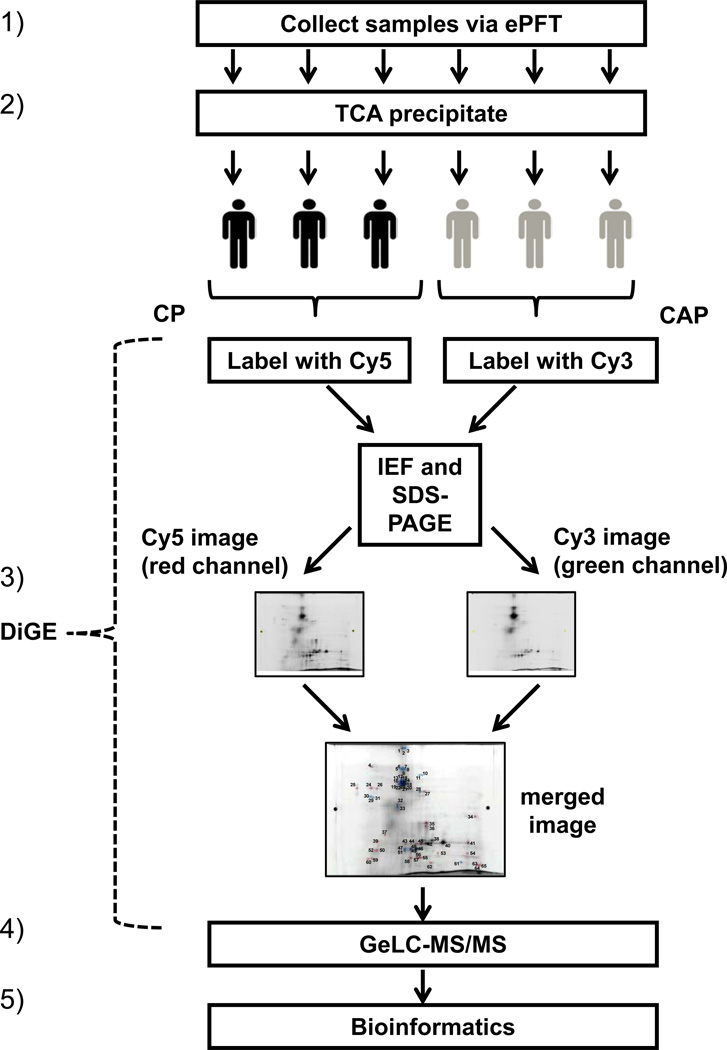
The overall analysis is shown in Figure 1: 1) ePFT sample collection, 2) trichloroacetic acid precipitation, 3) DiGE analysis, 4) in-gel tryptic digestion followed by liquid chromatography-coupled tandem mass spectrometry (GeLC-MS/MS), and 5) bioinformatic analysis.
Figure 1. Experimental workflow.
The procedure was as follows: (1) pancreatic fluid was collected from chronic pancreatitis subjects (n=3) and chronic abdominal pain controls (n=3) with the ePFT method, (2) proteins were extracted with TCA precipitation, (3) DiGE was performed with differentially-labeled samples from the two cohorts, (4) gel spots indicating differentially abundant proteins were excised and processed using GeLC-MS/MS techniques, and (5) bioinformatics analysis of the data was performed.
Pancreatic Fluid Collection (ePFT method)
The ePFT procedure was performed in three major stages, as follows: 1) pre-procedural assessment, 2) endoscopic procedure and 3) post-procedural assessment/recovery.
A. Pre-procedural assessment
Prior to upper endoscopy, all study subjects underwent a history and physical examination including list of allergies, medications, substance use/abuse, vital signs, and physical examination. Pre-procedural sedation review included airway assessment based on Mallampati airway scale and American Society of Anesthesiologists Physical Status Classification (ASA Class). All study subjects in this protocol had a Mallampati score of B, Class 2 and ASA Class II or better.
B. Procedure
Endoscopic collection was performed at Brigham and Women’s Hospital endoscopy unit as follows: 1) the patient was placed in the left lateral decubitus position with slight head elevation. 2) The posterior pharynx was sprayed with topical cetacaine spray. 3) A sedation and analgesia bolus was administered. 4) Further sedation doses were given if necessary for patient comfort. 5) After the sedation bolus, a bite-block was placed. 6) Esophagogastroduodenoscopy (EGD) was performed using a standard (10 mm) or thin (6 mm) gastroscope for visualization of the esophagus, stomach, and duodenum (2 to 5 minutes). 7) Gastroduodenal fluid [14] was aspirated (approximately 1 minute) as completely as possible through the gastroscope. 8) A test dose of synthetic human secretin (ChiRhoStim®) was administered and patients were monitored for anaphylaxis or adverse reaction, followed by a standard weight-based intravenous bolus (0.2 µg/kg) given over 1 minute. 9) Pancreatic fluid was aspirated from the descending duodenum at specific timed intervals (0 to 60 minutes) following hormonal stimulation and stored on ice. Only the 30-minute time point for each patient was used for the ensuing analysis. Samples were divided and sent to the Brigham and Women’s Hospital Biochemistry Laboratory for measurement of electrolyte profiles and the Proteomics Center at Children’s Hospital Boston for proteomic analysis. Approximately 5–10 mL of pancreatic fluid was collected from each patient at the 30 minute time point. Biopsies of the stomach and duodenum were obtained to eliminate microscopic gastrointestinal disease, such as Helicobacter pylori or celiac sprue, as a cause of dyspepsia/abdominal pain.
C. Post-procedural Assessment / Recovery
Study participants recovered and were discharged from the endoscopy unit based on hospital procedural sedation guidelines assessing level of consciousness, vital signs, oxygen saturation, alertness, gag reflex, degree of nausea, and ability to ambulate.
Pancreatic Fluid Biochemical Analysis
Pancreatic fluid samples were frozen at −80°C and stored until analysis; all measurements were conducted within two weeks of sample collection in the CLIA-certified Brigham and Women's Hospital Clinical Chemistry Laboratory with an AU640 (Olympus America, Center Valley, PA) automated chemistry analyzer. Total bicarbonate was measured by the two-step phosphoenolpyruvate carboxylase-malate dehydrogenase enzymatic-photometric method [17]. The mean peak bicarbonate concentration from previously published studies in secretin-stimulated pancreatic fluid is 103 ± 11 meq/l [18]. The cut-off point of 80 meq/L was two standard deviations below the mean and considered abnormal [19].
Pancreatic Fluid Sample Preparation for DiGE and Mass Spectrometry
Pancreatic fluid was processed as described previously [20]. In brief, pancreatic fluid samples were collected on ice, centrifuged at 4°C at 14,000 rpm to remove cellular debris, and aliquoted (500 µL) prior to storage at −80°C. Protein concentration was determined using the BioRAD protein assay (BioRAD, Hercules, CA) according to the manufacturer’s instructions. The protein concentration of pancreatic fluid typically ranges from 0.6–1.2 mg/mL. The proteins were extracted from an aliquot of pancreatic fluid via precipitation with the addition of 12.5% trichloroacetic acid (TCA) [21]. This process limits protein degradation by instantaneously deactivating enzymes and removing salts that will interfere with the subsequent electrophoresis. Approximately 50 µg of pancreatic fluid protein from three different subjects were pooled, so that a total of 150 µg of protein could be labeled with the appropriate Cy dye.
Difference Gel Electrophoresis (DiGE) Analysis
The lyophilized samples were transported on dry ice to the W. M. Keck facility at Yale University (New Haven, CT). The samples were dissolved in buffer (7M urea, 2M thiourea, 4% CHAPS (w/v), 25 mM Tris, pH 8.6 at 4°C). The pancreatic fluid from the chronic abdominal pain (CAP) controls was labeled with Cy-3 dye and the fluid from the chronic pancreatitis (CP) subjects was labeled with Cy-5 dye.
For the first dimension isoelectric focusing gel, the labeled samples were pooled and mixed with 400 µL rehydration buffer containing 7 M urea, 2M thiourea, 4% CHAPS (w/v), 1% DTT (w/v), 2% (v/v) Pharmalytes pH 3–10 and a trace amount of bromophenyl blue, and loaded onto 24 cm pH 3–10 linear IPG strips (G.E. Healthcare) by reswelling for 3 hours at 20°C. Isoelectric focusing was performed on an Ettan IPGphor 3 (GE Healthcare) for approximately 60 kVh at 20°C, 50 µA/strip using the following voltage gradient: (i) 1 h at 30 V, (ii) 1 h at 500V, (iii) 1 h at 1000V, then approximately an 8 h linear gradient to 8000V continuing until reaching 60kVh. After focusing, IPG strips were incubated with an equilibration buffer containing 6M urea, 10mM Tris (pH 6.8), 30% glycerol (w/v), 1% SDS (w/v), and 2% DTT for 15 minutes at room temperature. This solution was replaced with equilibration buffer containing 5% iodoacetamide for another 10 minutes. For the second dimension gel, IPG strips were applied to 22 × 24 cm SDS-PAGE gels (12% T, 2.6% C) (Jule, Inc., Milford, CT), which were run overnight at 125V (constant) and 15 °C in an Ettan DALT twelve electrophoresis chamber (GE Healthcare, Fairfield, CT).
For image acquisition, gels were scanned using a Typhoon 9410 Imager (GE Healthcare, Fairfield, CT). Cy3 images were scanned using a 532 nm laser and an emission filter of 580 nm. Cy5 images were scanned using a 633 nm laser and a 670 nm Band Pass 30 Hz emission filter. Photomultiplier voltage was adjusted for each channel to minimize any signal saturation. All gels were scanned at 100µm resolution, and images were further processed using ImageQuant V5.0 (GE Healthcare, Fairfield, CT) prior to analyses on DeCyder (GE Healthcare, Fairfield, CT) software. Gel image analysis was performed using DeCyder v6.5 (GE Healthcare, Fairfield, CT). Spot detection was conducted on image pairs consisting of each sample from the same gel. These two images overlay and allow direct measurement (if applicable) of volume ratios of spots between the standard and the sample. Selected spots were excised using an Ettan Spot Picker instrument (GE Healthcare, Fairfield, CT).
Mass spectrometry
Mass spectrometry analysis of excised gel plugs was performed at the Proteomics Center at Children’s Hospital Boston. Proteins in each gel plug were digested in-gel with trypsin [22; 23]. The extracted peptides from each gel plug were subjected to peptide fractionation using reversed-phase high performance liquid chromatography (HPLC; Thermo Scientific) and the gradient-eluted peptides were analyzed by an in-line LTQ FT mass spectrometer (Thermo Scientific). The liquid chromatography columns (15 cm × 100 µm ID) were packed in-house (Magic C18, 5 µm, 100 Å, Michrom BioResources, Auburn, CA, into PicoTips, New Objective, Woburn, MA). Samples were analyzed with a 30 minute linear gradient (0–35% acetonitrile with 0.2% formic acid) with a constant flow rate of 400 nL/min. Data were acquired in a data-dependent manner, in which MS/MS fragmentation was performed on the six most intense peaks of every full MS scan.
Bioinformatics and Data Analysis
All data generated from the gel spots were searched against the IPI-human database (v3.36) using the Mascot search engine (v.2.204; Matrix Science). One miscleavage per peptide was allowed and mass tolerances of ± 10 ppm (monoisotopic) for precursor and of ± 0.8 Da for fragment ions were used, as was default for LTQ FT-ICR data analysis. Variable amino acid modifications: deamidation (Asn/Gln), pyro-glutamate (N-terminal Glu/Gln), CyDye-Cy5 (Cys), CyDye-Cy3 (Cys) and oxidation (Met). Mascot search results were combined using in-house-developed software. In strict compliance with a set of recommendations [24; 25; 26] proposed by the major proteomic journals, we present the following protein identification validation method that minimizes false positives and reports only high confidence identifications. Our false discovery rate (FDR) was 1% at the peptide level as determined by searching the same dataset against the target database and a decoy database; the latter featured the concatenated forward and reversed amino acid sequences of all the entries in the IPI-human database (v3.36) [27; 28]. We applied the following stringent identification criteria for protein identifications to ensure a false positive rate of ≤ 0.1% at the protein level: 1) a minimum of 2 unique peptides was required for protein identification, 2) each peptide had a score equal or greater than the 1% FDR cut-off (see above), and 3) each matched peptide corresponds to the highest scoring peptide for that MS/MS spectra. Gene ontology (GO) classification of subcellular localization and biological function were extracted from the UniProt database [29].
Results
Demographics of study population
Pancreatic fluid was safely collected via secretin-stimulated ePFT from the duodenum of all six (6) subjects (Table 1). Of these individuals, three were diagnosed with definite chronic pancreatitis (CP) and the remaining three served as chronic abdominal pain (CAP) controls, without evidence of pancreatic dysfunction and structural abnormalities. The three individuals with chronic pancreatitis were classified as such according to the M-ANNHEIM classification [15]. Conversely, the M-ANNHEIM criteria for chronic pancreatitis were not met for any of the three chronic abdominal pain controls. The mean peak bicarbonate levels of the individuals with chronic pancreatitis and chronic abdominal pain controls were 43 and 97 meq/L, respectively (p-value=0.0070).
Difference gel electrophoresis (DiGE) analysis revealed differentially-imaged gel spots
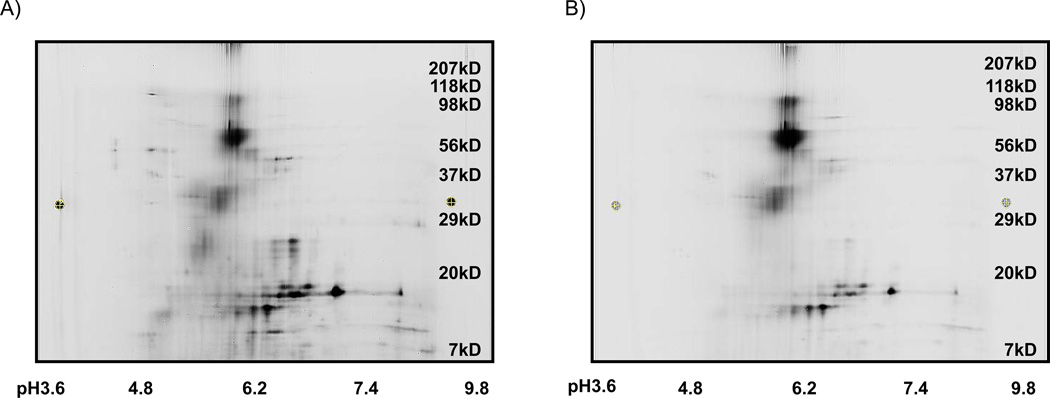
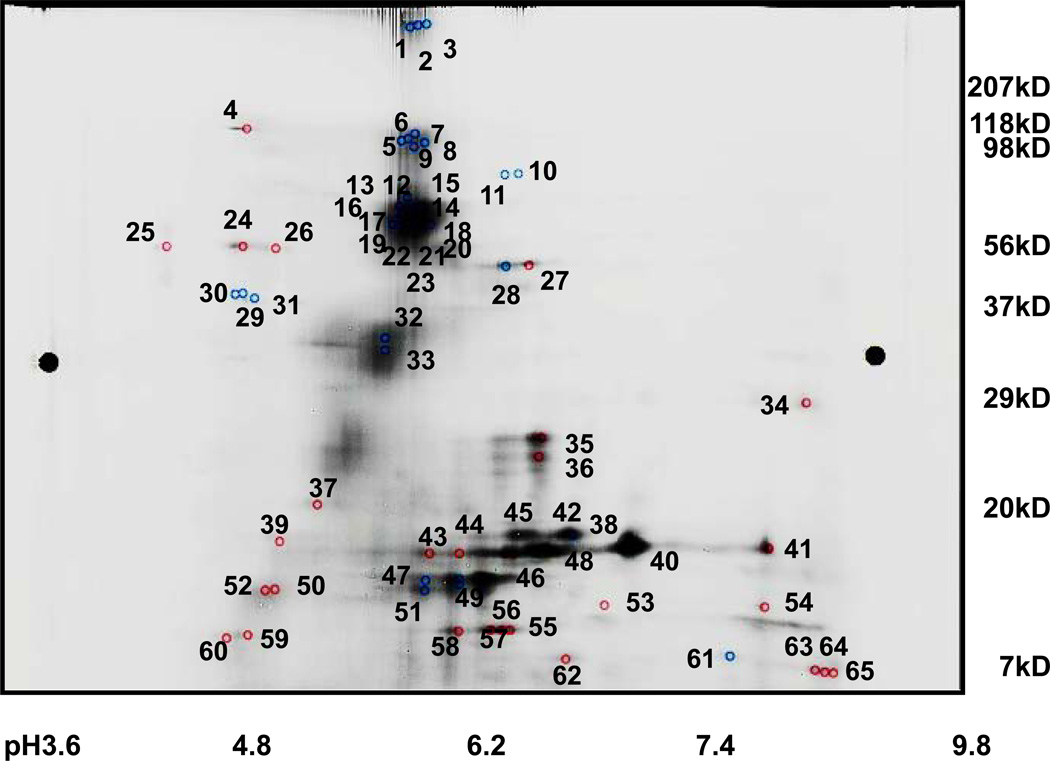
The image of the Cy3-conjugated chronic abdominal pain sample (Figure 2A) differed substantially from that of the Cy5-conjugated chronic pancreatitis sample (Figure 2B). When the two images were superimposed, 65 spots were identified as being differently abundant in the two samples (Figure 3). Of these 65 protein spots, 30 were more intense in the samples originating from the chronic abdominal pain controls, while 35 were more intense for chronic pancreatitis fluid samples.
Figure 2. DiGE images for chronic abdominal pain and chronic pancreatitis pancreatic fluid samples.
A) Image of Cy-3-labeled chronic abdominal pain pancreatic fluid sample. B) Image of Cy5-labeled chronic pancreatitis pancreatic fluid sample. The molecular weight (kDa) of the proteins may be approximated using the scale on the right of the gel and the values corresponding to the pI on the IPG strips are listed below the gel image.
Figure 3. Overlay of DiGE images.
The numerical values indicate gel spots that were excised and processed via GeLC-MS/MS. These numbers correspond to those in the leftmost column of Table 2.
Using the intensity measurement at the appropriate wavelength, the DeCyder software (GE Healthcare, Fairfield, CT) calculated the ratios (Cy5/Cy3 and −Cy3/Cy5) of the proteins in each spot with respect to the sample of origin. Of the 35 spots that were of higher intensity in the chronic pancreatitis subjects, 23 had greater than 1.5-fold increase over the controls, of which 9 had greater than a 3-fold difference. Similarly, when examining the 30 spots that showed relatively lower abundance in chronic pancreatitis subject samples, 29 had greater than a 1.5-fold decrease over the controls, of which 9 had greater than a 3-fold decrease (Table 2). Mass spectrometry-based proteomics techniques were used to identify these differentially expressed proteins.
Table 2.
Proteins identified with the highest Mascot scores following LC-MS/MS analysis from each excised DiGE gel spot. The “dye ratio” represents Cy5/Cy3 and −Cy3/C5 intensity ratios for proteins of higher and lower abundance, respectively, in chronic pancreatitis (Cy5) with respect to chronic abdominal pain controls (Cy3).
| Spot# | Mascot result | Calculated | Estimate from gel | dye ratio |
|||||
|---|---|---|---|---|---|---|---|---|---|
| protein name | IPI number | score | peptides | MW (kDa) | pI | MW (kDa) | pI | ||
| 1 | Desmoplakin | IPI00013933 | 426 | 57 | 332 | 6.4 | 250 | 5.7 | 1.4277 |
| 2 | Serum albumin | IPI00745872 | 541 | 28 | 69 | 5.9 | 250 | 5.7 | 2.7033 |
| 3 | Serum albumin | IPI00745872 | 176 | 16 | 69 | 5.9 | 250 | 5.7 | 2.3701 |
| 4 | Endoplasmin | IPI00027230 | 262 | 28 | 92 | 4.8 | 98 | 4.8 | −2.7721 |
| 5 | Serum albumin | IPI00745872 | 441 | 25 | 69 | 5.9 | 98 | 5.7 | 3.0376 |
| 6 | Serum albumin | IPI00745872 | 328 | 23 | 69 | 5.9 | 98 | 5.7 | 2.9618 |
| 7 | Serum albumin | IPI00745872 | 443 | 28 | 69 | 5.9 | 98 | 5.7 | 3.5638 |
| 8 | Serum albumin | IPI00745872 | 446 | 30 | 69 | 5.9 | 98 | 5.7 | 2.6246 |
| 9 | Serum albumin | IPI00745872 | 441 | 26 | 69 | 5.9 | 98 | 5.7 | 3.6763 |
| 10 | TF Serotransferrin | IPI00022463 | 174 | 15 | 77 | 6.8 | 85 | 6.2 | 4.7389 |
| 11 | TF Serotransferrin | IPI00022463 | 71 | 4 | 77 | 6.8 | 85 | 6.2 | 6.2057 |
| 12 | Serum albumin | IPI00745872 | 309 | 19 | 69 | 5.9 | 56 | 5.7 | 2.7793 |
| 13 | Serum albumin | IPI00745872 | 477 | 34 | 69 | 5.9 | 56 | 5.7 | 3.4765 |
| 14 | Serum albumin | IPI00745872 | 567 | 34 | 69 | 5.9 | 56 | 5.7 | 2.858 |
| 15 | Serum albumin | IPI00745872 | 550 | 40 | 69 | 5.9 | 56 | 5.7 | 3.2367 |
| 16 | SNC73 protein | IPI00386879 | 63 | 5 | 53 | 6.5 | 56 | 5.7 | 2.7797 |
| 17 | SNC73 protein | IPI00386879 | 82 | 5 | 53 | 6.5 | 56 | 5.7 | 2.1518 |
| 18 | TF Serotransferrin | IPI00022463 | 620 | 42 | 77 | 6.8 | 56 | 5.7 | 2.476 |
| 19 | Serum albumin | IPI00745872 | 433 | 34 | 69 | 5.9 | 56 | 5.7 | 3.7413 |
| 20 | Serum albumin | IPI00745872 | 613 | 39 | 69 | 5.9 | 56 | 5.7 | 2.7902 |
| 21 | Actin | IPI00021439 | 108 | 6 | 42 | 5.3 | 56 | 5.7 | 1.8866 |
| 22 | Serum albumin | IPI00745872 | 466 | 31 | 69 | 5.9 | 56 | 5.7 | 4.0114 |
| 23 | Serum albumin | IPI00745872 | 346 | 30 | 69 | 5.9 | 56 | 5.7 | 2.8934 |
| 24 | Serum albumin | IPI00745872 | 346 | 30 | 69 | 5.9 | 56 | 4.8 | −4.208 |
| 25 | Protein disulfide-isomerase | IPI00010796 | 184 | 16 | 57 | 4.8 | 56 | 4.5 | −4.401 |
| 26 | Alpha-1-antichymotrypsin | IPI00550991 | 72 | 3 | 51 | 5.3 | 56 | 4.8 | −3.9172 |
| 27 | Pancreatic alpha-amylase | IPI00025476 | 342 | 22 | 58 | 6.6 | 50 | 6.2 | −1.4011 |
| 28 | Pancreatic alpha-amylase | IPI00025476 | 436 | 20 | 58 | 6.6 | 50 | 6.4 | 1.0452 |
| 29 | Alpha-amylase 2B | IPI00021447 | 111 | 4 | 58 | 6.6 | 37 | 4.8 | 1.5795 |
| 30 | Alpha-1-antitrypsin | IPI00553177 | 229 | 10 | 47 | 5.4 | 37 | 4.8 | 1.7298 |
| 31 | Alpha-1-antitrypsin | IPI00553177 | 115 | 10 | 47 | 5.4 | 37 | 4.8 | 1.4306 |
| 32 | Serum albumin | IPI00745872 | 99 | 12 | 69 | 5.9 | 34 | 5.5 | 1.7838 |
| 33 | Serum albumin | IPI00745872 | 255 | 19 | 69 | 5.9 | 34 | 5.5 | 1.792 |
| 34 | Glyceraldehyde-3-phosphate dehydrogenase | IPI00219018 | 81 | 12 | 36 | 8.6 | 29 | 8.0 | −2.1575 |
| 35 | Pancreatic alpha-amylase | IPI00025476 | 132 | 9 | 58 | 6.6 | 25 | 6.4 | −2.4613 |
| 36 | Alpha-amylase 2B | IPI00021447 | 146 | 6 | 58 | 6.6 | 25 | 6.4 | −3.0721 |
| 37 | Serum albumin | IPI00745872 | 251 | 20 | 69 | 5.9 | 18 | 5.0 | −2.5469 |
| 38 | Pancreatic alpha-amylase | IPI00025476 | 158 | 7 | 58 | 6.6 | 20 | 6.4 | 1.163 |
| 39 | Pancreatic alpha-amylase | IPI00025476 | 127 | 5 | 58 | 6.6 | 17 | 5.8 | −1.1942 |
| 40 | Trypsin-1 | IPI00011694 | 287 | 28 | 27 | 6.1 | 16 | 7.2 | −1.5565 |
| 41 | Serum albumin | IPI00745872 | 130 | 7 | 69 | 5.9 | 16 | 7.7 | −1.1998 |
| 42 | Trypsin-1 | IPI00011694 | 283 | 29 | 27 | 6.1 | 16 | 6.5 | −1.5206 |
| 43 | Serum albumin | IPI00745872 | 180 | 12 | 69 | 5.9 | 16 | 6.2 | −1.2733 |
| 44 | Pancreatic alpha-amylase | IPI00025476 | 132 | 6 | 58 | 6.6 | 16 | 6.4 | −1.3937 |
| 45 | Trypsin-1 | IPI00011694 | 67 | 6 | 27 | 6.1 | 16 | 6.6 | −1.1982 |
| 46 | Pancreatic alpha-amylase | IPI00025476 | 173 | 7 | 58 | 6.6 | 12 | 6.2 | 1.4043 |
| 47 | Serum albumin | IPI00745872 | 175 | 15 | 69 | 5.9 | 12 | 5.8 | 2.5705 |
| 48 | Alpha-amylase 2B | IPI00021447 | 108 | 4 | 58 | 6.6 | 12 | 5.8 | 1.9996 |
| 49 | Alpha-amylase 2B | IPI00021447 | 148 | 5 | 58 | 6.6 | 12 | 5.8 | 1.9578 |
| 50 | Serum albumin | IPI00745872 | 181 | 16 | 69 | 5.9 | 10 | 4.8 | −1.7044 |
| 51 | Serum albumin | IPI00745872 | 211 | 20 | 69 | 5.9 | 12 | 5.8 | 1.876 |
| 52 | Serum albumin | IPI00745872 | 191 | 16 | 69 | 5.9 | 10 | 4.8 | −1.7595 |
| 53 | Pancreatic triacylglycerol lipase | IPI00027720 | 58 | 4 | 51 | 6.3 | 10 | 7.0 | −2.1418 |
| 54 | Carboxypeptidase B | IPI00009826 | 106 | 4 | 47 | 6.2 | 10 | 7.8 | −1.8589 |
| 55 | Pancreatic alpha-amylase | IPI00025476 | 82 | 3 | 58 | 6.6 | 15 | 6.0 | −2.7219 |
| 56 | Alpha-amylase 1 precursor | IPI00025476 | 70 | 4 | 58 | 6.6 | 15 | 6.0 | −2.2757 |
| 57 | Serum albumin | IPI00745872 | 155 | 12 | 69 | 5.9 | 15 | 6.0 | −3.0823 |
| 58 | Pancreatic alpha-amylase | IPI00025476 | 141 | 7 | 58 | 6.6 | 15 | 6.0 | −1.992 |
| 59 | Alpha-2-macroglobulin | IPI00478003 | 71 | 6 | 163 | 6.0 | 7 | 4.4 | −4.1582 |
| 60 | Serum albumin | IPI00745872 | 137 | 13 | 69 | 5.9 | 7 | 4.4 | −3.8268 |
| 61 | Trypsin-1 | IPI00011694 | 84 | 6 | 27 | 6.1 | 8 | 7.6 | 1.1487 |
| 62 | Trypsin-1 | IPI00011694 | 88 | 6 | 27 | 6.1 | 8 | 6.6 | −1.45 |
| 63 | Serum albumin | IPI00745872 | 85 | 10 | 69 | 5.9 | 7 | 8.7 | −2.6158 |
| 64 | Arp2/3 complex 20 kDa subunit | IPI00554811 | 92 | 6 | 20 | 5.3 | 7 | 8.7 | −3.2895 |
| 65 | Serum albumin | IPI00745872 | 211 | 15 | 69 | 5.9 | 7 | 8.7 | −3.2561 |
Abbreviations: Spot#, gel spot as numbered in Figure 3; score, Mascot score; peptides, number of peptides identified per protein; MW, molecular weight; pI, isoelectric point.
Mass spectrometry analysis identified different proteins in chronic pancreatitis and control pancreatic fluid
All 65 spots determined to be differentially-imaged were excised and subjected to LC-MS/MS for protein identification. In Table 2, we listed the proteins with the most significant Mascot score corresponding to each spot (as numbered in Figure 3). In addition, we included the international protein index (IPI) number, the Mascot score, and the number of peptides for each protein identified using Mascot in Table 2. Likewise, we noted the theoretical isoelectric point (pI) and molecular weight (MW) of each protein, as well as the corresponding values that were estimated from the two-dimensional gel. Finally, the “dye ratio” was listed, which represents either the Cy5/Cy3 or −Cy3/C5 ratios for proteins of higher or lower abundance, respectively, in the pancreatic fluid of chronic pancreatitis individuals compared to chronic abdominal pain controls.
The proteins of higher abundance in pancreatic fluid from chronic pancreatitis individuals with respect to chronic abdominal pain controls included cytoskeletal proteins: actin and desmoplankin, and extracellular proteins: alpha-1-antitrypsin, SNC73, and serotransferrin. Those proteins that were of relatively lower abundance included known secreted pancreatic enzymes: carboxypeptidase B and lipase; and secreted protease inhibitors: alpha-1-antichymotrypsin and alpha-2-macroglobulin. In addition, the cytoplasmic proteins actin-related protein (Arp) 2/3 complex subunit 4 and glyceraldehyde-3-phosphate dehydrogenase, as well as endoplasmin and protein disulfide isomerase, which were localized to the endoplasmic reticulum, were also found to be of lower abundance in the pancreatic fluid of chronic pancreatitis individuals with respect to chronic abdominal pain controls.
Proteolytic fragments of amylase and serum albumin were identified in both samples
Several proteins listed in Table 2, such as amylase and serum albumin, were determined to be of both relatively higher and lower abundance in the pancreatic fluid of chronic pancreatitis patients as a result of the specific gel spot from which the protein was identified. Although apparently contradictory, these proteins were identified in multiple spots on the gel typically below their nominal molecular weight, and were likely to have undergone differential proteolysis. Our current aim, however, was to identify proteins which were differentially abundant in the pancreatic fluid of chronic pancreatitis individuals compared with chronic abdominal pain controls, however future studies may investigate this differential proteolysis or optimize further the 2DGE protocol to prevent or lessen the degree of proteolysis. Table 3 listed those proteins which were consistently either relatively higher or lower in abundance in fluid specimens from the chronic pancreatitis subjects. In addition, we included in this table the subcellular localization and biological function of these proteins according to their gene ontology classifications in the UniProt database [29].
Table 3.
Proteins consistently of either higher or lower abundance in pancreatic fluid from chronic pancreatitis individuals with respect to chronic abdominal pain controls.
| dye ratio (mean) |
abundance in CP |
protein name | IPI number | subcellular localization |
biological function |
spot #(s) |
|---|---|---|---|---|---|---|
| 1.8866 | higher | Actin | IPI00021439 | cytoskeleton | structural | 21 |
| 1.5802 | higher | Alpha-1-antitrypsin | IPI00553177 | extracellular | protease inhibitor | 30, 31 |
| 1.4277 | higher | Desmoplakin | IPI00013933 | cytoskeleton | structural | 1 |
| 2.4658 | higher | SNC73 protein | IPI00386879 | extracellular | n/a | 16, 17 |
| 4.4735 | higher | TF Serotransferrin | IPI00022463 | extracellular | cell proliferation | 10, 12, 18 |
| −3.2895 | lower | Arp2/3 complex 20 kDa subunit | IPI00554811 | cytoskeleton | structural | 64 |
| −3.9172 | lower | Alpha-1-antichymotrypsin | IPI00550991 | extracellular | protease inhibitor | 26 |
| −4.1582 | lower | Alpha-2-macroglobulin | IPI00478003 | extracellular | protease inhibitor | 59 |
| −2.2757 | lower | Alpha-amylase 1 precursor | IPI00025476 | extracellular | enzyme | 56 |
| −1.8589 | lower | Carboxypeptidase B | IPI00009826 | extracellular | enzyme | 54 |
| −2.7721 | lower | Endoplasmin | IPI00027230 | endoplasmic reticulum | chaperone | 4 |
| −2.1575 | lower | Glyceraldehyde-3-phosphate dehydrogenase | IPI00219018 | cytoplasm | metabolic enzyme | 34 |
| −2.1418 | lower | Pancreatic triacylglycerol lipase | IPI00027720 | extracellular | enzyme | 53 |
| −4.4010 | lower | Protein disulfide-isomerase | IPI00010796 | endoplasmic reticulum | enzyme | 25 |
Discussion
We successfully use the ePFT collection method and DiGE coupled with LC-MS/MS to identify proteins from pancreatic fluid. DiGE analysis reveals differentially imaged gel spots when pancreatic fluid from chronic pancreatitis and chronic abdominal pain individuals are compared. Differential imaging analysis reveals 65 spots to be more intense in either the Cy5 (CP) or Cy3 (CAP) image. Of these protein spots, 30 are more intense in the samples originating from the chronic abdominal pain controls and 35 are more intense from the chronic pancreatitis subject samples. Using tandem mass spectrometry analysis, we successfully identified differentially-abundant proteins in pancreatic fluid of individuals with chronic pancreatitis and chronic abdominal pain controls.
Proteins that were consistently identified with higher abundance in the pancreatic fluid of chronic pancreatitis subjects, with respect to the control group, are listed at the top of Table 3. Using gene ontology analysis, we determine these proteins to be classified as either secreted or have roles in cellular structure and movement. Among the secreted proteins is serotransferrin, which is a protein responsible for the transport of iron from sites of absorption and heme degradation to sites of storage and utilization [30]. A proteomic analysis of serotransferrin in blood has shown association with pancreatic cancer [31]. Also of higher abundance is SNC73, a protein that shares homology with immunoglobulins. Although SN73 has not been previously associated with pancreatitis, it has been found to be a potential biomarker of colorectal cancer and other human epithelial carcinomas [32]. Another intracellularly secreted protein, alpha-1-antitrypsin (serpin A1), is a serine protease inhibitor that can bind covalently and inactivate trypsin, and is often associated with pancreatic dysfunction [33]. In addition to these secreted proteins, we also identify desmoplakin, a major cytoskeletal component of desmosomes that are part of intercellular junctions that tightly link adjacent cells [34]. Finally, actin, which is a ubiquitous protein involved in cell motility and structure [35], is also identified via LC-MS/MS analysis. Actin and cell-to-cell interaction proteins (e.g., desmoplakin) have been shown to have a role in the development of chronic pancreatitis via stellate cell activation [36; 37; 38].
Similarly, proteins that were consistently identified with lower abundance in the pancreatic fluid of chronic pancreatitis subjects, with respect to our control group, are listed in the lower portion of Table 3. These proteins include both those localized intracellularly and those which are secreted. Many of these proteins have been shown to have implications in pancreatic function. Intracellular proteins include metabolic enzymes protein disulfide isomerase and glyceraldehyde-3-phosphate dehydrogenase, as well as an Arp 2/3 complex subunit and endoplasmin. Protein disulfide-isomerase (PDI) generally is localized to the endoplasmic reticulum and catalyzes the formation, breakage, and rearrangement of disulfide bonds between cysteine residues [39; 40]. Along with its function in the transport and processing of secreted proteins, endoplasmin has been identified as a molecular chaperone, and is also a heat shock protein [41]. Both PDI and endoplasmin have been previously identified as being exported from the endoplasmic reticulum in the rat exocrine pancreas [42]. Similarly, glyceraldehyde-3-phosphate dehydrogenase partakes in membrane trafficking in the early secretory pathway [43]. It has been shown that differentially expressed alternative splice isoforms of glyceraldehyde-3-phosphate dehydrogenase are detectable in plasma and is a potential candidate biomarker of pancreatic cancer [44]. Likewise, the Arp 2/3 complex subunit 4, plays a role in cellular movement. This protein functions in the binding of the Arp2/3 complex to actin and is involved in regulation of actin polymerization [45]. Future studies may aim to further investigate the roles of these proteins in the development of chronic pancreatitis in in vitro and animal model systems.
In addition to these intracellular proteins, certain secreted protease inhibitor inhibitors are also of relatively lower abundance in the pancreatic fluid of chronic pancreatitis subjects. Protease inhibitors are of relevance to chronic pancreatitis, as dysfunction in pancreatic enzymes is a hallmark of the disease. Alpha-1-antichymotrypsin is a serine protease inhibitor, which inactivates neutrophil cathepsin G and mast cell chymase [46]. Similarly, alpha-2-macroglobulin, which is a large protein, can inactivate a variety of proteases, including serine-, cysteine-, aspartic- and metalloproteases [47; 48]. Both alpha-1-antichymotrypsin and alpha-2-macroglobulin may have significant implications in the inactivation of certain proteases that are secreted from the pancreata of chronic pancreatitis subjects. As expected, various pancreatic enzymes are also of lower abundance in the pancreatic fluid of chronic pancreatitis subjects. As enzyme deficiency is a characteristic sign of chronic pancreatitis, the pancreatic enzymes, trypsin-1, carboxypeptidase B, alpha-amylase 1, and triacylglycerol lipase, are generally of lower abundance in the pancreatic fluid from chronic pancreatitis subjects. A correlation may exist between these relatively lower abundance enzymes and inhibitors; however, analyses of these binary or multiplexed interactions must be performed before conclusions regarding these data can be drawn.
As illustrated in Table 2, some proteins, mainly amylase and serum albumin, were determined to be both of relatively higher and lower abundance in chronic pancreatitis compared to chronic abdominal pain controls depending on the specific spot which was analyzed by DiGE-LC-MS/MS. At first, these data appear contradictory; however, plausible explanations exist for these apparent discrepancies. For instance, due to the degree of activity of endogenous proteolytic enzymes in a particular sample of pancreatic fluid, different breakdown products may have been produced. More specifically, the fragmented proteins may be indicative of cohort-specific enzymatic activity of pancreatic fluid. For example, although the theoretical molecular weight of amylase is approximately 58 kDa, it is rarely identified above 50 kDa by our LC-MS/MS analysis. There were several gel spots below 20 kDa which were identified as amylase, and are likely to be cleavage products. Likewise, serum albumin is abundant in pancreatic fluid and was identified to be the highest-scoring protein in several gel spots. Cleavage products of serum albumin, are also prevalent in the lower molecular weight region (<20 kDa) of the gel. Further peptidomic-based investigation [49; 50; 51] may be warranted to compare differences in the endogenous activity of specific pancreas-secreted enzymes in pancreatic disease and non-diseased cohorts, as impairment of these enzymes may be indicative of underlying pancreatic dysfunction..
Unlike one-dimensional SDS-PAGE, 2DGE-based methods (e.g. DiGE) require a substantial amount of time in which the protein solution must equilibrate with the IEF gel strip. Although fractionation occurs under denaturing conditions, it still may be possible for proteolysis to occur during sample processing [52; 53], as is particularly true for the protease-rich pancreatic fluid. To improve the robustness of our DiGE analysis, methods of reducing degradation products using protease inhibitors early in sample preparation for example, may be integrated into our working protocol to decrease the degree of endogenous proteolysis. In regard to pancreatic fluid studies, there is evidence both for and against the addition of protease inhibitors early in the study [12, 27, 32, 33].
In addition, there are several other technical aspects of our DiGE approach that can be further optimized to effectively remove highly abundant proteins, such as albumin. Although we identify some differentially-expressed proteins in the pancreatic fluid of chronic pancreatitis compared to chronic abdominal pain controls, we do realize that future analyses would benefit from an increased depth of differentially-expressed proteins. Samples were pooled as to control for patient-to-patient variability, as we aimed to investigate the global difference between cohorts, rather than between individuals [54; 55]. However, the high abundance of albumin in our pancreatic fluid samples may be exacerbated by the pooling of samples. To control for this drawback, albumin depletion may be performed using antibodies or dye-based columns, as is often performed with blood samples [56; 57]. A major caveat of albumin removal, is that it functions endogenously as a carrier of less abundant, but more significant proteins and these potential markers of disease may be lost during the depletion process [58]. As such, the denaturation of the protein sample may be a prerequisite to disrupt protein binding to albumin prior to depletion. Nevertheless, by targeted depletion, one adds another step to the sample handling process, one which may increase protein loss and sample variability. However, such optimizations may be necessary if low abundant proteins are to be studied.
There are several advantages to using DiGE for the proteomic analysis of pancreatic fluid, such as i) inherent two-dimensional sample fractionation, ii) better reproducibility, and iii) increased sensitivity and dynamic range. In addition, DiGE resolves some of the drawbacks traditionally associated with two-dimensional gel electrophoresis (2DGE), including gel-to-gel variation, as two or three samples can be conjugated to a specific fluorescent dye (Cy2, Cy3, or Cy5) and analyzed on a single gel [59]. However, along with the aforementioned benefits, the use of DiGE in pancreatic fluid proteomics is also subject to several limitations. Such caveats include: (i) spots containing multiple proteins, (ii) poor spot resolution at high pI values, (iii) very acidic and very basic proteins not being well represented, (iv) very small or very large proteins not being well-resolved, (v) irreproducibility of gels among experiments, and (vi) potential proteolysis resulting from sample handling [60].
Using our methodology, we discovered large changes between the two samples, however for future experiments, higher sensitivity could be attained with improved focusing in the first dimension. Although attempts were made with TCA precipitation to eliminate any non-proteinacious matter, the first dimension focusing showed sub-optimal, diffuse protein pattern. Potential reasons for the diffuse protein patterns include the possibility that non-protein contaminant (bound or unbound to protein) such as nucleic acid may be present, that residual salt present in the sample may have limited the focusing time and not allow steady state to be achieved, or that the samples were not completely solubilized. Improved focusing would enable us to detect more subtle changes in abundance, as opposed to only the major changes that we see in the present study. However, given that we observed dramatic differences between the samples even with the sub-optimal resolution, we chose to identify those proteins which showed these differences, as it would be beneficial to identify potential biomarkers that are abundant and readily detected.
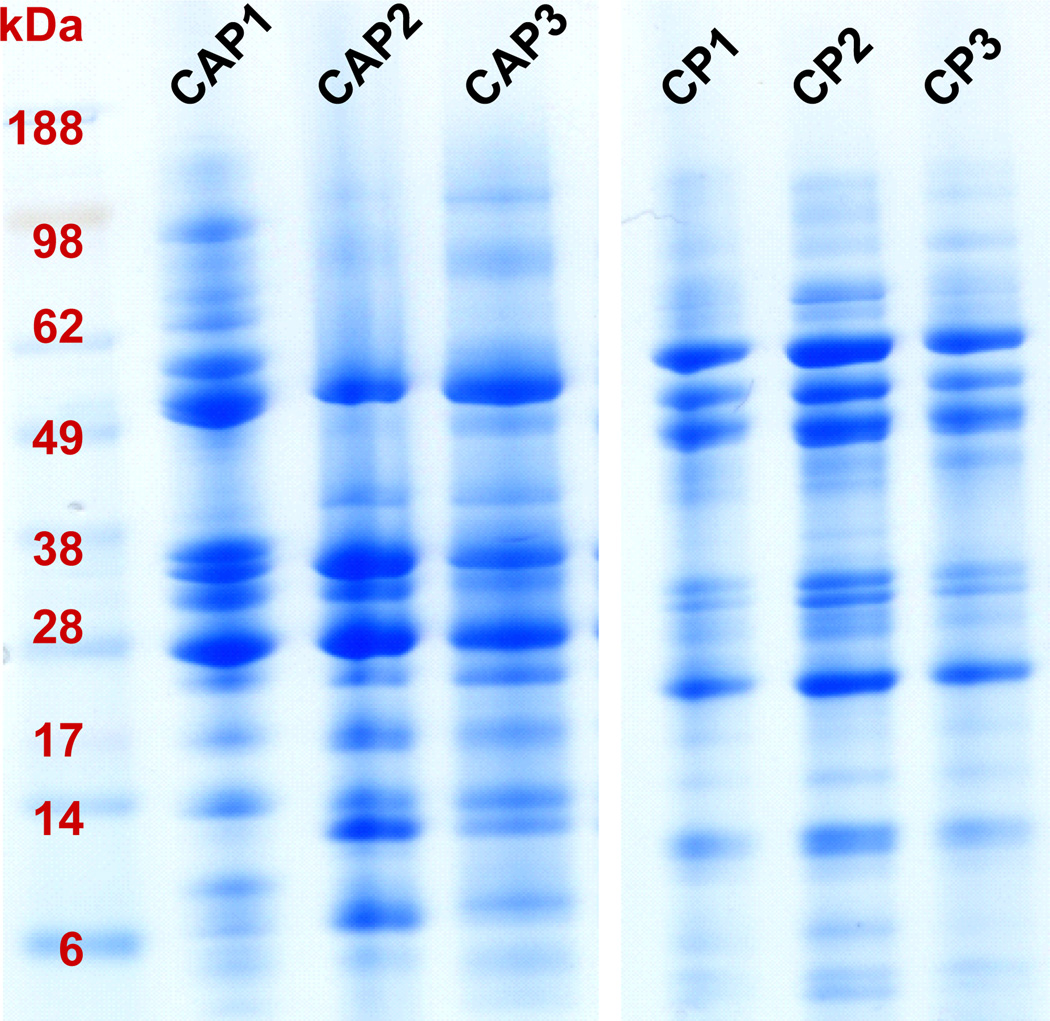
It was our intention to use 2DGE to fractionate, isolate, relatively quantify, and identify proteins that differ in the pancreatic fluid between cohorts of chronic pancreatitis and chronic abdominal pain. However, as pancreatic fluid appears to have a proteome of only several hundred proteins, separation by electrophoretic mobility may be sufficient for fractionation to identify differentially abundant proteins. It follows that the use of one-dimensional gel electrophoresis (1DGE) may provide a technically simpler and more robust method of analyzing pancreatic fluid for downstream mass spectrometry analysis. The proteome of pancreatic fluid is relatively small in comparison to many cellular proteomes, thus the analysis thereof is less likely to benefit from the added dimension of fractionation by isoelectric point, unless certain isoforms or co-migrating proteins require further fractionation. Also, many of the caveats for 2DGE, as discussed above, do not apply to 1DGE. Following our previously–established protocol [21], we analyzed the 6 individual samples via 1DGE, as is depicted in Figure 4. These 1DGE images illustrate sharp, distinct protein banding patterns. We have since pursued the use of 1DGE fractionation of pancreatic fluid proteins in a large scale comparative study investigating chronic pancreatitis (manuscript in preparation). Moreover, the strategy of coupling 1DGE with mass spectrometry is currently the most widely used method of mass spectrometry-based protein identification for pancreatic fluid [20; 61; 62; 63; 64; 65; 66].
Figure 4. 1DGE protein fractionation.
Each gel lane represents ePFT-collected pancreatic fluid that has been TCA precipitated from a particular patient (Table 1). CAP, chronic abdominal pain; CP, chronic pancreatitis.
In summary, using DiGE-LC-MS/MS, we have demonstrated the feasibility to identify differentially-abundant proteins from ePFT-collected pancreatic fluid of chronic pancreatitis subjects and chronic abdominal pain controls. As a result of the enzyme insufficiency associated with chronic pancreatitis, many of the proteins of relatively lower abundance in the pancreatic fluid from chronic pancreatitis individuals with respect to that of chronic abdominal pain controls are proteolytic enzymes, as is expected. Although evidence supports that 1DGE may be superior to DiGE, we conclude that DiGE does have a role in the proteomic analysis of pancreatic fluid. Further optimization of the methodology that we have described herein enables the development of future comparative analyses of proteins from ePFT-collected fluid, and thereby can further broaden our knowledge of pancreatic disease pathogenesis.
ACKNOWLEDGMENTS
Funds were provided by the Harvard Digestive Diseases Center (NIH 5 P30 DK034854-24) and the NIH/NIDDK NRSA Fellowship (NIH NIDDK 1 F32 DK085835-01A1). In addition, we would like to thank the Burrill family for their generous support through the Burrill Research Grant. We would also like to thank members of the Steen Lab at Children’s Hospital Boston, in particular John FK Sauld and Dominic Winter, and Kate Repas from the Center for Pancreatic Disease at Brigham and Women’s Hospital for their technical assistance and critical reading of the manuscript.
LIST OF ABBREVIATIONS
- 2DGE
two-dimensional gel electrophoresis
- CP
chronic pancreatitis
- CAP
chronic abdominal pain
- DiGE
difference imaging gel electrophoresis
- ePFT
endoscopic pancreatic function test
- GO
gene ontology
- IEF
isoelectric focusing
- IPG
immobilized pH gradient
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
- MW
molecular weight
- pI
isoelectric point
- TCA
trichloroacetic acid
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
JP carried out the experiments and drafted the original manuscript. JP and DC conceived of the study. JP, DC, PB and HS participated in its design and coordination. DC and LL collected the specimens and assisted in experimental design. All authors helped to draft the manuscript and approved the final manuscript.
Contributor Information
Joao A. Paulo, Department of Pathology, Children’s Hospital Boston and Harvard Medical School, Boston, MA; Proteomics Center at Children’s Hospital Boston, Boston, MA; Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Linda S. Lee, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Peter A. Banks, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
Hanno Steen, Department of Pathology, Children’s Hospital Boston and Harvard Medical School, Boston Proteomics Center at Children’s Hospital Boston, Boston, MA.
Darwin L. Conwell, Center for Pancreatic Disease, Division of Gastroenterology, Hepatology and Endoscopy, Brigham and Women’s Hospital and Department of Medicine, Harvard Medical School, Boston, MA.
REFERENCES
- 1.Witt H, Apte MV, Keim V, Wilson JS. Chronic pancreatitis: challenges and advances in pathogenesis, genetics, diagnosis, and therapy. Gastroenterology. 2007;132:1557–1573. doi: 10.1053/j.gastro.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed SA, Wray C, Rilo HL, Choe KA, Gelrud A, Howington JA, Lowy AM, Matthews JB. Chronic pancreatitis: recent advances and ongoing challenges. Curr Probl Surg. 2006;43:127–238. doi: 10.1067/j.cpsurg.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Paulo J, Vaezzadeh A, Conwell D, Lee R, Steen H. Sample Handling of Body Fluids for Proteomics. In: Ivanov A, Lazarev A, editors. Sample Preparation in Biological Mass Spectrometry. New York, NY: Springer; 2011. [Google Scholar]
- 4.Paulo JA, Lee LS, Wu B, Banks PA, Steen H, Conwell DL. Mass spectrometry-based proteomics of endoscopically collected pancreatic fluid in chronic pancreatitis research. Proteomics Clin Appl. 2011 doi: 10.1002/prca.201000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conwell DL, Zuccaro G, Jr, Vargo JJ, Morrow JB, Obuchowski N, Dumot JA, Trolli PA, Burton A, O'Laughlin C, Van Lente F. An endoscopic pancreatic function test with cholecystokinin-octapeptide for the diagnosis of chronic pancreatitis. Clin Gastroenterol Hepatol. 2003;1:189–194. doi: 10.1053/cgh.2003.50028. [DOI] [PubMed] [Google Scholar]
- 6.Conwell DL, Zuccaro G, Jr, Vargo JJ, Trolli PA, Vanlente F, Obuchowski N, Dumot JA, O'Laughlin C. An endoscopic pancreatic function test with synthetic porcine secretin for the evaluation of chronic abdominal pain and suspected chronic pancreatitis. Gastrointest Endosc. 2003;57:37–40. doi: 10.1067/mge.2003.14. [DOI] [PubMed] [Google Scholar]
- 7.Wu B, Conwell DL. The endoscopic pancreatic function test. Am J Gastroenterol. 2009;104:2381–2383. doi: 10.1038/ajg.2008.181. [DOI] [PubMed] [Google Scholar]
- 8.Stevens T, Conwell DL, Zuccaro G, Jr, Van Lente F, Lopez R, Purich E, Fein S. A prospective crossover study comparing secretin-stimulated endoscopic and Dreiling tube pancreatic function testing in patients evaluated for chronic pancreatitis. Gastrointest Endosc. 2008;67:458–466. doi: 10.1016/j.gie.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 9.Andriulli A, Loperfido S, Napolitano G, Niro G, Valvano MR, Spirito F, Pilotto A, Forlano R. Incidence rates of post-ERCP complications: a systematic survey of prospective studies. Am J Gastroenterol. 2007;102:1781–1788. doi: 10.1111/j.1572-0241.2007.01279.x. [DOI] [PubMed] [Google Scholar]
- 10.Wang P, Li ZS, Liu F, Ren X, Lu NH, Fan ZN, Huang Q, Zhang X, He LP, Sun WS, Zhao Q, Shi RH, Tian ZB, Li YQ, Li W, Zhi FC. Risk factors for ERCP-related complications: a prospective multicenter study. Am J Gastroenterol. 2009;104:31–40. doi: 10.1038/ajg.2008.5. [DOI] [PubMed] [Google Scholar]
- 11.Cheng CL, Sherman S, Watkins JL, Barnett J, Freeman M, Geenen J, Ryan M, Parker H, Frakes JT, Fogel EL, Silverman WB, Dua KS, Aliperti G, Yakshe P, Uzer M, Jones W, Goff J, Lazzell-Pannell L, Rashdan A, Temkit M, Lehman GA. Risk factors for post-ERCP pancreatitis: a prospective multicenter study. Am J Gastroenterol. 2006;101:139–147. doi: 10.1111/j.1572-0241.2006.00380.x. [DOI] [PubMed] [Google Scholar]
- 12.Pollack BJ, Grendell JH. Where have all the dreiling tubes gone? Am J Gastroenterol. 2006;101:356–359. doi: 10.1111/j.1572-0241.2006.00398.x. [DOI] [PubMed] [Google Scholar]
- 13.Forsmark CE. The early diagnosis of chronic pancreatitis. Clin Gastroenterol Hepatol. 2008;6:1291–1293. doi: 10.1016/j.cgh.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Paulo JA, Lee LS, Wu B, Repas K, Banks PA, Conwell DL, Steen H. Proteomic analysis of endoscopically (endoscopic pancreatic function test) collected gastroduodenal fluid using in-gel tryptic digestion followed by LC-MS/MS. Proteomics Clin Appl. 2010;4:715–725. doi: 10.1002/prca.201000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider A, Lohr JM, Singer MV. The M-ANNHEIM classification of chronic pancreatitis: introduction of a unifying classification system based on a review of previous classifications of the disease. J Gastroenterol. 2007;42:101–119. doi: 10.1007/s00535-006-1945-4. [DOI] [PubMed] [Google Scholar]
- 16.Diaconu BL, Ciobanu L, Mocan T, Pfutzer RH, Scafaru MP, Acalovschi M, Singer MV, Schneider A. Investigation of the SPINK1 N34S mutation in Romanian patients with alcoholic chronic pancreatitis. A clinical analysis based on the criteria of the M-ANNHEIM classification. J Gastrointestin Liver Dis. 2009;18:143–150. [PubMed] [Google Scholar]
- 17.Forrester RL, Wataji LJ, Silverman DA, Pierre KJ. Enzymatic method for determination of CO2 in serum. Clin Chem. 1976;22:243–245. [PubMed] [Google Scholar]
- 18.Stevens T, Conwell D, Zuccaro G, Van Lente F, Khandwala F, Hanaway P, Vargo JJ, Dumot JA. Analysis of pancreatic elastase-1 concentrations in duodenal aspirates from healthy subjects and patients with chronic pancreatitis. Dig Dis Sci. 2004;49:1405–1411. doi: 10.1023/b:ddas.0000042238.80040.cc. [DOI] [PubMed] [Google Scholar]
- 19.Stevens T, Conwell DL, Zuccaro G, Van Lente F, Khandwala F, Purich E, Vargo JJ, Fein S, Dumot JA, Trolli P, O'Laughlin C. Electrolyte composition of endoscopically collected duodenal drainage fluid after synthetic porcine secretin stimulation in healthy subjects. Gastrointest Endosc. 2004;60:351–355. doi: 10.1016/s0016-5107(04)01809-7. [DOI] [PubMed] [Google Scholar]
- 20.Paulo JA, Lee LS, Wu B, Repas K, Mortele KJ, Banks PA, Steen H, Conwell DL. Identification of pancreas-specific proteins in endoscopically (endoscopic pancreatic function test) collected pancreatic fluid with liquid chromatography--tandem mass spectrometry. Pancreas. 2010;39:889–896. doi: 10.1097/MPA.0b013e3181cf16f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulo JA, Lee LS, Wu B, Repas K, Banks PA, Conwell DL, Steen H. Optimized sample preparation of endoscopic collected pancreatic fluid for SDS-PAGE analysis. Electrophoresis. 2010;31:2377–2387. doi: 10.1002/elps.200900762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neubauer G, Mann M. Mapping of phosphorylation sites of gel-isolated proteins by nanoelectrospray tandem mass spectrometry: potentials and limitations. Anal Chem. 1999;71:235–242. doi: 10.1021/ac9804902. [DOI] [PubMed] [Google Scholar]
- 23.Steen H, Kuster B, Fernandez M, Pandey A, Mann M. Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Anal Chem. 2001;73:1440–1448. doi: 10.1021/ac001318c. [DOI] [PubMed] [Google Scholar]
- 24.Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A. The need for guidelines in publication of peptide and protein identification data: Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol Cell Proteomics. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Taylor GK, Goodlett DR. Rules governing protein identification by mass spectrometry. Rapid Commun Mass Spectrom. 2005;19:3420. doi: 10.1002/rcm.2225. [DOI] [PubMed] [Google Scholar]
- 26.Wilkins MR, Appel RD, Van Eyk JE, Chung MC, Gorg A, Hecker M, Huber LA, Langen H, Link AJ, Paik YK, Patterson SD, Pennington SR, Rabilloud T, Simpson RJ, Weiss W, Dunn MJ. Guidelines for the next 10 years of proteomics. Proteomics. 2006;6:4–8. doi: 10.1002/pmic.200500856. [DOI] [PubMed] [Google Scholar]
- 27.Elias JE, Gibbons FD, King OD, Roth FP, Gygi SP. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat Biotechnol. 2004;22:214–219. doi: 10.1038/nbt930. [DOI] [PubMed] [Google Scholar]
- 28.Moore RE, Young MK, Lee TD. Method for screening peptide fragment ion mass spectra prior to database searching. J Am Soc Mass Spectrom. 2000;11:422–426. doi: 10.1016/S1044-0305(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 29.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh L-SL. The Universal Protein Resource (UniProt) Nucl. Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F, Lum JB, McGill JR, Moore CM, Naylor SL, van Bragt PH, Baldwin WD, Bowman BH. Human transferrin: cDNA characterization and chromosomal localization. Proc Natl Acad Sci U S A. 1984;81:2752–2756. doi: 10.1073/pnas.81.9.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun ZL, Zhu Y, Wang FQ, Chen R, Peng T, Fan ZN, Xu ZK, Miao Y. Serum proteomic-based analysis of pancreatic carcinoma for the identification of potential cancer biomarkers. Biochim Biophys Acta. 2007;1774:764–771. doi: 10.1016/j.bbapap.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Geng LY, Shi ZZ, Dong Q, Cai XH, Zhang YM, Cao W, Peng JP, Fang YM, Zheng L, Zheng S. Expression of SNC73, a transcript of the immunoglobulin alpha-1 gene, in human epithelial carcinomas. World J Gastroenterol. 2007;13:2305–2311. doi: 10.3748/wjg.v13.i16.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 34.Bornslaeger EA, Corcoran CM, Stappenbeck TS, Green KJ. Breaking the connection: displacement of the desmosomal plaque protein desmoplakin from cell-cell interfaces disrupts anchorage of intermediate filament bundles and alters intercellular junction assembly. J. Cell Biol. 1996;134:985–1001. doi: 10.1083/jcb.134.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys. 2008;37:65–95. doi: 10.1146/annurev.biophys.37.032807.125912. [DOI] [PubMed] [Google Scholar]
- 36.Mathison A, Liebl A, Bharucha J, Mukhopadhyay D, Lomberk G, Shah V, Urrutia R. Pancreatic stellate cell models for transcriptional studies of desmoplasia-associated genes. Pancreatology. 2010;10:505–516. doi: 10.1159/000320540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masamune A, Watanabe T, Kikuta K, Shimosegawa T. Roles of pancreatic stellate cells in pancreatic inflammation and fibrosis. Clin Gastroenterol Hepatol. 2009;7:S48–S54. doi: 10.1016/j.cgh.2009.07.038. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu K. Pancreatic stellate cells: molecular mechanism of pancreatic fibrosis. J Gastroenterol Hepatol. 2008;23(Suppl 1):S119–S121. doi: 10.1111/j.1440-1746.2007.05296.x. [DOI] [PubMed] [Google Scholar]
- 39.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 40.Gleiter S, Bardwell JC. Disulfide bond isomerization in prokaryotes. Biochim Biophys Acta. 2008;1783:530–534. doi: 10.1016/j.bbamcr.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunati AM, Contri A, Muenchbach M, James P, Marin O, Pinna LA. GRP94 (endoplasmin) co-purifies with and is phosphorylated by Golgi apparatus casein kinase. FEBS Lett. 2000;471:151–155. doi: 10.1016/s0014-5793(00)01378-8. [DOI] [PubMed] [Google Scholar]
- 42.Takemoto H, Yoshimori T, Yamamoto A, Miyata Y, Yahara I, Inoue K, Tashiro Y. Heavy chain binding protein (BiP/GRP78) and endoplasmin are exported from the endoplasmic reticulum in rat exocrine pancreatic cells, similar to protein disulfide-isomerase. Arch Biochem Biophys. 1992;296:129–136. doi: 10.1016/0003-9861(92)90554-a. [DOI] [PubMed] [Google Scholar]
- 43.Tisdale EJ. Glyceraldehyde-3-phosphate dehydrogenase is phosphorylated by protein kinase Ciota/lambda and plays a role in microtubule dynamics in the early secretory pathway. J Biol Chem. 2002;277:3334–3341. doi: 10.1074/jbc.M109744200. [DOI] [PubMed] [Google Scholar]
- 44.Menon R, Zhang Q, Zhang Y, Fermin D, Bardeesy N, DePinho RA, Lu C, Hanash SM, Omenn GS, States DJ. Identification of novel alternative splice isoforms of circulating proteins in a mouse model of human pancreatic cancer. Cancer Res. 2009;69:300–309. doi: 10.1158/0008-5472.CAN-08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Egile C, Rouiller I, Xu XP, Volkmann N, Li R, Hanein D. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 2005;3:e383. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalsheker NA. Alpha 1-antichymotrypsin. Int J Biochem Cell Biol. 1996;28:961–964. doi: 10.1016/1357-2725(96)00032-5. [DOI] [PubMed] [Google Scholar]
- 47.Lin YC, Vaseeharan B, Ko CF, Chiou TT, Chen JC. Molecular cloning and characterisation of a proteinase inhibitor, alpha 2-macroglobulin (alpha2-M) from the haemocytes of tiger shrimp Penaeus monodon. Mol Immunol. 2007;44:1065–1074. doi: 10.1016/j.molimm.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 48.He H, McCartney DJ, Wei Q, Esadeg S, Zhang J, Foster RA, Hayes MA, Tayade C, Van Leuven F, Croy BA. Characterization of a murine alpha 2 macroglobulin gene expressed in reproductive and cardiovascular tissue. Biol Reprod. 2005;72:266–275. doi: 10.1095/biolreprod.104.029835. [DOI] [PubMed] [Google Scholar]
- 49.Sasaki K, Satomi Y, Takao T, Minamino N. Snapshot peptidomics of the regulated secretory pathway. Mol Cell Proteomics. 2009;8:1638–1647. doi: 10.1074/mcp.M900044-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quintana LF, Campistol JM, Alcolea MP, Banon-Maneus E, Sol-Gonzalez A, Cutillas PR. Application of label-free quantitative peptidomics for the identification of urinary biomarkers of kidney chronic allograft dysfunction. Mol Cell Proteomics. 2009;8:1658–1673. doi: 10.1074/mcp.M900059-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zougman A, Pilch B, Podtelejnikov A, Kiehntopf M, Schnabel C, Kumar C, Mann M. Integrated analysis of the cerebrospinal fluid peptidome and proteome. J Proteome Res. 2008;7:386–399. doi: 10.1021/pr070501k. [DOI] [PubMed] [Google Scholar]
- 52.Havanapan PO, Thongboonkerd V. Are protease inhibitors required for gel-based proteomics of kidney and urine? J Proteome Res. 2009;8:3109–3117. doi: 10.1021/pr900015q. [DOI] [PubMed] [Google Scholar]
- 53.Olivieri E, Herbert B, Righetti PG. The effect of protease inhibitors on the two-dimensional electrophoresis pattern of red blood cell membranes. Electrophoresis. 2001;22:560–565. doi: 10.1002/1522-2683(200102)22:3<560::AID-ELPS560>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 54.Diz AP, Truebano M, Skibinski DO. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis. 2009;30:2967–2975. doi: 10.1002/elps.200900210. [DOI] [PubMed] [Google Scholar]
- 55.Karp NA, Lilley KS. Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteomics. 2009;9:388–397. doi: 10.1002/pmic.200800485. [DOI] [PubMed] [Google Scholar]
- 56.Thresher WC, Swaisgood HE. Characterization of specific interactions of coenzymes, regulatory nucleotides and cibacron blue with nucleotide binding domains of enzymes by analytical affinity chromatography. J Mol Recognit. 1990;3:220–228. doi: 10.1002/jmr.300030509. [DOI] [PubMed] [Google Scholar]
- 57.Bjorhall K, Miliotis T, Davidsson P. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics. 2005;5:307–317. doi: 10.1002/pmic.200400900. [DOI] [PubMed] [Google Scholar]
- 58.Zhou M, Lucas DA, Chan KC, Issaq HJ, Petricoin EF, 3rd, Liotta LA, Veenstra TD, Conrads TP. An investigation into the human serum"interactome". Electrophoresis. 2004;25:1289–1298. doi: 10.1002/elps.200405866. [DOI] [PubMed] [Google Scholar]
- 59.Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 60.Tannu NS, Hemby SE. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat Protoc. 2006;1:1732–1742. doi: 10.1038/nprot.2006.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen R, Brentnall TA, Pan S, Cooke K, Moyes KW, Lane Z, Crispin DA, Goodlett DR, Aebersold R, Bronner MP. Quantitative proteomics analysis reveals that proteins differentially expressed in chronic pancreatitis are also frequently involved in pancreatic cancer. Mol Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.Chen R, Pan S, Yi EC, Donohoe S, Bronner MP, Potter JD, Goodlett DR, Aebersold R, Brentnall TA. Quantitative proteomic profiling of pancreatic cancer juice. Proteomics. 2006;6:3871–3879. doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- 63.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 64.Ke E, Patel BB, Liu T, Li XM, Haluszka O, Hoffman JP, Ehya H, Young NA, Watson JC, Weinberg DS, Nguyen MT, Cohen SJ, Meropol NJ, Litwin S, Tokar JL, Yeung AT. Proteomic Analyses of Pancreatic Cyst Fluids. Pancreas. 2009;38:33–42. doi: 10.1097/MPA.0b013e318193a08f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian M, Cui YZ, Song GH, Zong MJ, Zhou XY, Chen Y, Han JX. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer. 2008;8:241. doi: 10.1186/1471-2407-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou L, Lu Z, Yang A, Deng R, Mai C, Sang X, Faber KN, Lu X. Comparative proteomic analysis of human pancreatic juice: methodological study. Proteomics. 2007;7:1345–1355. doi: 10.1002/pmic.200600086. [DOI] [PubMed] [Google Scholar]