Abstract
Dental tissue engineering efforts have yet to identify scaffolds that instruct the formation of bioengineered teeth of predetermined size and shape. Here we investigated whether extracellular matrix (ECM) molecules present in natural tooth scaffolds can provide insight on how to achieve this goal. We describe methods to effectively decellularize and demineralize porcine molar tooth buds, while preserving natural ECM protein gradients. Natural tooth ECM composition was assessed using histological and immunohistochemical (IHC) analyses of fibrillar and basement membrane proteins. Our results showed that Collagen I, Fibronectin, Collagen IV, and Laminin gradients were detected in natural tooth tissues, and retained in decellularized samples. Second harmonic generation (SHG) image analysis and 3D reconstructions were used to show that natural tooth tissue exhibited higher collagen fiber density, and less oriented and less organized collagen fibers, as compared to decellularized tooth tissue. We also found that reseeded decellularized tooth scaffolds exhibited distinctive collagen content and organization as compared to decelluarized scaffolds. Our results show that SHG allows for quantitative assessment of ECM features that are not easily characterized using traditional histological analyses. In summary, our results demonstrate the potential for natural decellularized molar tooth ECM to instruct dental cell matrix synthesis, and lay the foundation for future use of biomimetic scaffolds for dental tissue engineering applications.
Keywords: Decellularization, second harmonic generation (SHG), tooth bud, extracellular matrix
1. Introduction
Current efforts in whole tooth tissue engineering focus on identifying methods to accurately control bioengineered tooth size and shape, create functional tooth roots, and eliminate ectopic mineralized tissue formation in in vivo implanted bioengineered tooth and bone constructs. To date, strategies for tooth tissue engineering have utilized a variety of scaffold materials, growth factors, and cell sources, achieving some level of success [1–4]. We hypothesized that detailed characterizations of extracellular matrix (ECM) composition and organization in natural tooth development could facilitate human tooth tissue engineering efforts. Evidence in support of this includes the fact that amelogenin and its associated natural cleavage products have been shown to direct the proper self-assembly of enamel crystals into microribbons [5], and that biglycan decorated nanofiber scaffolds can induce amelogenin expression and subsequent enamel formation and maturation [6–8]. These and other reports indicate that functional characterizations of tooth expressed ECM molecules, including their respective developmental and spatial organization, may facilitate the design of effective scaffolds for tooth regeneration.
Based on the fact that the ECM provides morphogenetic cues that guide proper cellular interactions during natural and bioengineered organogenesis, recent reports have focused on elucidating roles for natural ECM molecules and gradients in craniofacial tissues and organs [9–11]. In the tooth bud, dental epithelial and dental mesenchymal cell layers develop into enamel and pulp organs, respectively. As the tooth matures, dental mesenchymal cells differentiate into odontoblasts and secrete a matrix that eventually mineralizes to form dentin, and dental epithelial cells differentiate into ameloblasts, which secrete an enamel matrix. To date, the fabrication of biomimetic scaffolds that support robust dentin and enamel formation in a predictable manner remains an elusive goal. Recently, tissue decellularization methods have been used to preserve natural tissue-specific ECM composition and spatial organization, creating acellular scaffolds for a variety of tissue and organ engineering applications [12–17]. In this study, we first devised nondestructive decellularization and demineralization methods to process natural porcine tooth buds, and then compared ECM protein expression patterns present in natural and processed tooth buds using histological and immunohistochemical (IHC) approaches.
In addition, we employed second harmonic generation (SHG) imaging to obtain quantitative information about collagen content, organization, and remodeling in natural and processed tooth scaffolds. SHG is a non-linear scattering process that monitors the interaction of two photons with molecules that lack centrosymmetry, resulting in the scattering of a single photon at half the wavelength [18]. This process requires a high density photon beam that is typically available only at the focal point emanating from a microscope objective [18]. Thus, it offers intrinsic optical sectioning capabilities in three dimensions, and enables micron-level resolution imaging of tissues extending over a few hundred microns in depth. Based on the fact that fibrillar collagens are non-centrosymmetric structures that provide intrinsic SHG contrast, avoiding the need to stain or process specimens prior to imaging, numerous studies have used SHG imaging to assess collagen organization and structure in vivo, ex vivo and in vitro [19–22]. SHG microscopy is often performed simultaneously with two-photon excited fluorescence (TPEF) imaging [21–23], another non-linear imaging process involving the simultaneous absorption of two low energy photons, resulting in the excitation and emission of a single higher energy fluorescent photon upon decay to the ground state [24]. Certain chromophores within cells such as NADH and FAD, and proteins including collagen and elastin, are natural fluorophores. The combined use of SHG and TPEF allows for non-invasive evaluation of cellular and ECM components of tissues, and can be used to assess their interactions during normal or diseased tissue development [21–26].
Here we report the characterization of natural ECM molecules and fibrillar proteins present in natural and processed tooth bud tissues. IHC, SHG, and TPEF were used to define ECM molecule gradients, collagen fiber content, and 3D organization in natural tooth tissues, processed decellularized and demineralized samples, and in processed tooth scaffolds reseeded and cultured with dental mesenchymal cells. Our long term goal is to apply knowledge gained from these studies to fabricate instructive biomimetic tooth scaffolds that promote the formation of engineered tooth and bone constructs of specified size and shape, for future applications in craniofacial tissue engineering [27–29].
2. Materials and Methods
2.1 Decellularization and decalcification of porcine molar tooth buds
All reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless otherwise specified. Recently discarded 5 ½ month old pig jaws were obtained following USDA guidelines. Second and third molar tooth buds (M2 and M3, respectively) were harvested from hemi-split jaws, rinsed thoroughly in PBS, and fixed in 10% neutral buffered formalin (NBF) overnight, and decellularized based on published formulations [30, 31]. Briefly, harvested M2 and M3 tooth buds were rinsed in Hank’s Balanced Salt Solution (HBSS) (Gibco, Carlsbad, CA, USA) supplemented with 2x Penicillin Streptomycin Amphotericin (PSA) (Gibco), and weighed prior to decellularization (see Table 1). Tooth buds were placed in buffered solution (0.01 M Tris-HCl, 1 mM EDTA, pH 8.2) for 48 hours at 4°C with constant stirring. All subsequent steps were performed with constant rocking at 24°C unless otherwise specified. For the 1 and 2 cycle decellularization procedure, Protease Inhibitor Cocktail Set III (EMD Chemicals, Gibbstown, NJ) was added to the Tris-HCl Buffer and detergent solutions. For the 5 cycle decellularization procedure, Aprotinin (10 KIU per gram wet weight tissue, Worthington Biochemical, Lakewood, NJ) and 10 µM EDTA were added to the Tris-HCl Buffer and detergent solutions at a ratio of 25:1 [32]. Decellularized tooth buds were exposed to one of three detergent cycle treatments hereafter referred to as Methods I, II, and III. Method I consisted of incubating tooth buds first in 1% SDS (w/v) with protease inhibitors for 24 hours, followed by three, 30 minute rinse cycles in 25 mM Tris Buffered Saline (TBS) (pH 7.6) at 24°C, followed by one cycle in 1% TritonX-100 (w/v) for 24 hours at 24°C and three subsequent rinse cycles for 30 min at 24°C. These SDS/rinse/TritonX-100/rinse treatment cycles were repeated an additional 4 times. Method II consisted of an initial 5% SDS/rinse/TritonX-100 treatment cycle, followed by 4 additional cycles of 1% SDS/rinse/TritonX-100 treatments. Method III was performed in a similar fashion using 5% SDS (w/v) for all treatments in place of 1% SDS. All samples were next treated with 3.6 U/mL DNase I, RNase-free (Roche Diagnostics Corporation, Indianapolis, IN, USA) and 1.2 U/mL RNase, DNase-free (Sigma-Aldrich, St. Louis, MO, USA) for 3 hours at 37°C (Hybaid/Thermo Fisher Scientific, Waltham, MA, USA), followed by heat-inactivation at 75°C for up to 20 minutes, and a 1x TE buffer (100 µM EDTA, pH 8.0) wash as per manufacturer specifications. Tooth tissues were then rinsed three times in TBS prior to the decalcification process.
Table 1.
Decellularization and demineralization treatments.
| Incubation time |
Treatment | Temperature |
|---|---|---|
| 48 hrs | Hypotonic TE Buffer | 4°C |
| 24 hrs | SDS (1% or 5%) + Protease Inhibitors* | 24°C |
| 24 hrs | TritonX-100 (1%) + Protease Inhibitors* | 24°C |
| 3 hrs | Nuclease Treatment | 37°C |
| 16 hrs | Normal Buffered Formalin (10%) | 24°C |
| 3 months | EDTA Demineralization (10%) | 24°C |
Hypotonic TE Buffer, 10 mM Tris-HCl Buffer, 1 mM EDTA, pH 8.2; Protease Inhibitors: Aprotinin (10 KIU) and EDTA (10 µM); Nuclease Treatment (pH 7.5), 50 mM Tris-HCl, 10 mM magnesium chloride; 50 µg/mL bovine serum albumin (pH 7.5), 3.6 U/mL DNase I (RNase-free) and 1.2 U/mL RNase (DNase-free); pH of detergent solutions: 7.6–7.8
Alternated for 5 cycles (with TBS rinses in between), prior to nuclease treatment.
To facilitate subsequent paraffin sectioning, decellularized decellularized and natural tooth buds were decalcified in 10% (v/v) EDTA, pH 7.4, at 24°C with constant gentle rocking, with solution changes 1–2 times a week. Demineralization was continued until calcified precipitates were no longer detected using an endpoint detection solution consisting of 5:1 sample demineralization (pH 3.2–3.6) and saturated ammonia oxalate solution [33, 34]. Demineralized samples were rinsed thoroughly in distilled water, and stored in 50% ethanol at 4°C for subsequent ECM molecule characterizations, or in 75% ethanol until subsequent use in reseeding studies.
2.2 Dissociation, isolation, and culture of porcine dental mesenchymal cells
Tooth buds were harvested as follows. Briefly, the jaw outer surface was scraped to remove soft tissue, and sanitized using iodine and 75% ethanol. Tooth buds were extracted using a hammer and chisel, as previously described by us [27]. Next, enamel organ (EO) and pulp organ (PO) tissues were mechanically separated, minced, and washed three times in HBSS supplemented with 2% PSA. The tissues were digested for 30 minutes at 37°C with gentle agitation in HBSS (0.4 mg/mL collagenase II and 0.2 mg/mL dispase, Worthington), triturated for 10 minutes using a 25 mL pipette and 10 minutes using a 10 mL pipette, strained (Cell Strainer, 40µm, BD Biosciences, San Jose, CA), and pelleted via centrifugation. PO-derived dental mesenchymal cells were plated at 2–3 ×106 cells (26,000–40,000 cells/cm2) and cultured in DMEM/F12 media containing 10% FBS, 1% PSA, 1% GlutaMAX (Gibco) and 25 µg/mL L-ascorbic acid.
2.3 Reseeding acellular porcine tooth bud scaffolds
Decellularized/demineralized M2 tooth buds were sectioned into 1.5 mm slices, which were then quartered. Cultured porcine dental mesenchymal cells (Passage 6) were suspended in a 1 mg/mL rat tail collagen type I gel (BD Biosciences), and seeded drop-wise onto decellularized tooth bud scaffold slices at a density of 1×107 cells/cm3. Unseeded control decellularized scaffolds were coated with 1 mg/mL collagen gel alone. Cell culture media consisted of a 50:50 mix of high glucose DMEM:LHC-8 Media with supplements 10% FBS, 1x PSA, 0.5 µg/mL epinephrine, 0.05 mM ascorbic acid, 100 nM dexamethasone, and 10 mM β-glycerolphosphate. Dental mesenchymal cell-seeded and unseeded scaffolds were cultured at 5% CO2, 37°C for up to 30 days.
2.4 Histological and immunohistochemical analyses
Decellularized/demineralized porcine tooth bud specimens were dehydrated in an ethanol series, cleared in acetone and chloroform, paraffin-embedded, and serial sectioned at 6 microns. Selected sections were stained with Hematoxylin and Eosin (H&E), Masson’s Trichrome (American MasterTech Scientific, Lodi, CA) or Movat’s Pentachrome (Rowley Biochemical Inc., Danvers, MA). IHC analyses were performed as previously described [35]. Briefly, endogenous peroxidase activity was quenched using 3% hydrogen peroxidase in methanol. Mounted sections were unmasked in 1 mg/mL hyaluronidase solution in PBS (Sigma H3884; Type IV-S) for 1 hour at 37°C, and blocked with 10% normal donkey serum for 15 min. Sections were incubated for 1 hour at 24°C with the following primary antibodies diluted in 2% normal donkey serum: Mouse Monoclonal Anti-Laminin (Clone LAM-89, Abcam, ab49726, Cambridge, MA, 1:100); Mouse Monoclonal Anti-Collagen Type I (Clone COL-1, Sigma, C2456, 1:1000); Rabbit Polyclonal Anti-Collagen Type IV (Abcam, ab6586, 1:700); and Rabbit Polyclonal Anti-Fibronectin (Abcam, ab23751, 1:800). After washing, sections were incubated for 45 minutes at 24°C with the secondary antibodies, Biotin-SP Donkey Anti-Rabbit IgG (H+L) (Jackson Laboratories, 1:500) or Biotin-SP Donkey Anti-Mouse IgG (H+L) (Jackson Laboratories, 1:500), followed by washing. VECTASTAIN ABC Kit (Vector Laboratories NC9206402, Burlingame, CA) and DAB Peroxidase Substrate (Sigma D0426; SIGMAFast DAB with Metal Enhancer) were used for antibody detection. Sections were counterstained in 0.2% Fast Green, dehydrated, fixed in Xylene, and cover-slipped with Permount (Fisher Scientific). Bright-field images were acquired using a compound microscope (Zeiss Axiophot Imager Z.1, Zeiss, Germany) and Zeiss Plan- Apochromat 20×/0.8, and EC Plan-NEOFLUAR 40×/0.75 objectives. Digital images were acquired using a Zeiss AxioCamHRc digital camera (Stuttgart, Germany) and Axiovision software (Carl Zeiss Imaging, AxioVS40 v4.7.2.0).
2.5 DNA quantification
Third molar tooth halves were used to quantitate DNA content in control and decellularized samples. Triplicate decellularized (Methods I, II, III) and duplicate control (Natural) tooth buds were cut in half. Triplicate decellularized and duplicate half samples were then pooled, providing two samples per condition. Tooth samples were placed in 50% ethanol overnight, rinsed in sterile DI water, and frozen at −80°C. The samples were next lyophilized (Labconco FreeZone Plus Freeze Dry Systems, 7960042, Kansas City, MO), at 0.018–0.060 Torr and 76°C. Lyophilized tooth bud halves were cut into thin strips and weighed. Sample digestion and DNA isolation was then performed using established protocols [36, 37]. Briefly, tooth sections were placed in digestion buffer [0.1 mg/ml Proteinase K (Worthington, Lakewood, NJ), 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, 25 mM EDTA (pH 8.0), 0.5% SDS (Sigma), and UltraPure water (Invitrogen)], and incubated at 55°C for up to 144 hours. DNA was then extracted using 25:24:10 (v/v/v) phenol/chloroform/isoamyl alcohol (Invitrogen). After centrifugation at 10,000g for 10 min at 4°C the supernatant was collected, the DNA was precipitated using 3M sodium acetate (pH 5.2) and ethanol, and pelleted via centrifugation at 10,000g for 10 min, followed by a 70% ethanol wash. The DNA pellets were resuspended in 1xTE Buffer and frozen at −20°C until quantification using the Quant-It PicoGreen dsDNA Quantitation Kit (Molecular Probes, Eugene, OR) as per manufacturer specifications. Briefly, Quant-It Pico Green reagent was added 1:1 with serially diluted samples, and aliquots were quantified (ex: 480 nm; em: 520 nm; cutoff: 515nm) using a top-reader spectrophotometer (SpectraMax M2, Molecular Devices, Sunnyvale, CA). DNA content was expressed as ng/mg dry weight.
2.6 Non-linear microscopy and image analysis
Processed Natural M2 porcine control and decellularized Method I-treated tooth samples were used for non-linear SHG and TPEF imaging and reseeding studies as follows. Samples were hand-sectioned into 1–1.5 mm slices (Stainless Steel Tissue Matrix, Electron Microscopy Sciences, Hatfield, PA). SHG and TPEF images were acquired using a Leica TCS SP2 laser scanning confocal microscope (Wetzlar, Germany) equipped with a tunable Mai Tai (710–920 nm) titanium sapphire laser emitting 100 fs pulses at 80 MHz (Spectra Physics, Mountain View, Calif., USA). Tissue autofluorescence was excited at 755 nm and captured with a non-descanned PMT with a filter cube containing a 700 nm short pass filter (Chroma SPC700bp) a dichroic mirror (Chroma 495dcxr), and an emitter bandpass filter centered at 460 nm (Chroma 460bp40). Backward scattered SHG signal was collected at 800 nm excitation through a filter centered at 400 nm (Chroma hq400/20m-2) for decellularized scaffolds. Power at the level of the sample was 43.5 mW (755 nm) and 34.5 mW (800 nm). Forward scattered signal was collected at 800nm excitation through a filter centered at 400nm for reseeded scaffolds. Images were merged in ImageJ (v1.37) or rendered in OsiriX (v3.0.2).
Image analysis was conducted on one scaffold per condition in multiple fields of view. Z-stacks containing 32 images were acquired at 2 µm steps for both natural and decellularized scaffolds. For the reseeded samples, 10 sequential optical sections were acquired at 1.5 µm steps. The acquired data was analyzed using algorithms developed in-house on the Matlab software platform to quantify collagen fiber density, orientation index (OI), and entropy [38]. Fiber density was calculated as the percentage of pixels with nonzero intensity after initial thresholding, as compared to the total number of pixels within the field. OI represents the percentage of fibrils within the image parallel to a dominant fiber direction and is calculated with Fourier-based methods [38]. Entropy is a measure of the level of organization of dominant fibers in the image determined by a Hough transform-based approach [38], where a lower entropy value correlates to more organized fibers.
2.7 Statistics
Two-tailed, unpaired Student T-tests of significance (P<0.05) were conducted in Excel for the DNA content studies and SHG image analysis.
3. Results
3.1. Decellularized Sample Analyses
3.1.1. Morphology and gross anatomy of natural and decellularized M2 tooth buds
Decellularization efficiency was evaluated in dental pulp tissues of decellularized and control samples (Fig. 1A, boxed region). Reduced numbers of nuclei were apparent in H&E stained specimens after 1, 2, and 5 cycles of detergent treatment (Fig. 1B–D) as compared to untreated natural control tissue (Fig. 1E). After two cycles, nuclei were still detected in Method I and II treated samples (Fig. 1B–C), while no nuclei were detected in Method III treated samples. Five detergent cycles removed the majority of nuclei in all three decellularized Methods, while preserving the tissue matrix and vascular tissues (Fig. 1F–H). Both the fragile and transparent dental sac tissues and sturdy demineralized cusp tissues appeared intact after treatment. (Fig. 1I–L).
Figure 1. Comparison of natural and decellularized tooth buds.
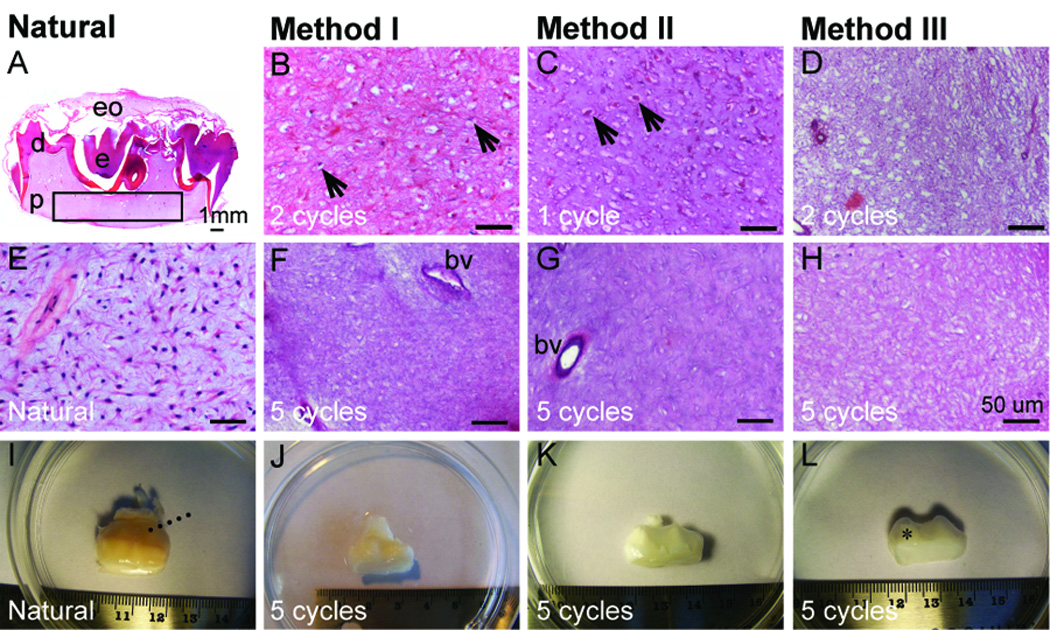
(A, E) H&E stained natural M2 molar tooth section reveals highly cellularized dental pulp (p) (boxed region). H&E stained sectioned pulp after various Method I (B, F), Method II (C, G), and Method III (D, H) treatments. (I–L) Low mag view of teeth after various treatments. Black arrows indicate nuclei, dashed lines indicate the dental sac, and * denotes tooth cusps. Abbreviations: bv, blood vessel; d, dentin, e, enamel; eo, enamel organ. Scale bars: (A) 1.0 mm; (B–H) 50 microns.
3.1.2. DNA content of natural and decellularized tissues (M3)
Decellularization efficiency was evaluated via DNA content in decellularized and control M3 molar teeth (Fig. 2). Surprisingly, although Method III treatment appeared to efficiently remove cell nuclei, no statistically significant difference in DNA content was observed between control and Method III treated decellularized samples (Fig. 2F, J, and N), (p < 0.05). It is possible that the high SDS concentration used in Method III may have interfered with the absorbance values of these samples. A significant 2.5- fold reduced DNA content was observed using either Method I or Method II treatment, as compared to Natural tooth bud controls (p < 0.0001). No significant difference was observed in DNA content between Method I and Method II treated samples (p < 0.05).
Figure 2. Decellularized tooth samples.
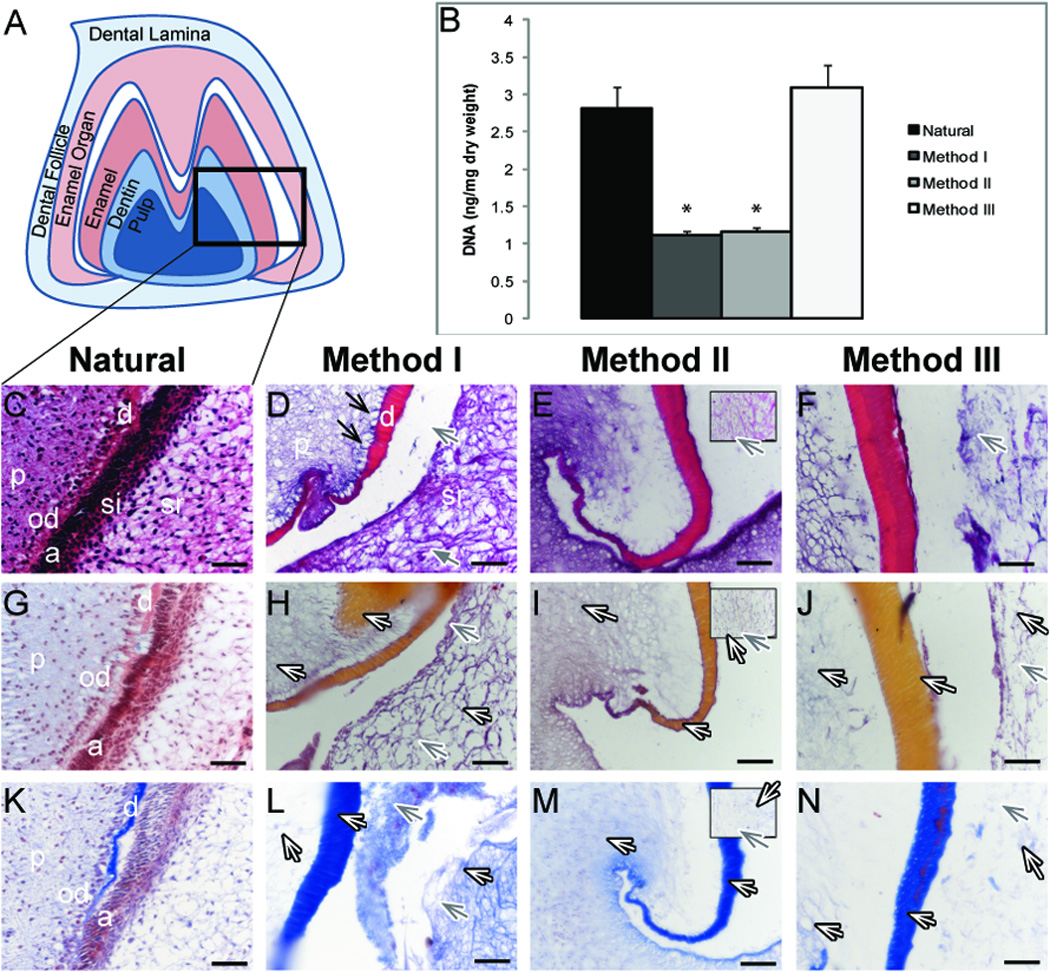
(A). Schematic of M3 tooth. B) DNA content before and after decellularization. H&E (C–F), Movat’s Pentachrome (G–J) and Masson’s Trichrome (K–N) stained paraffin sections. Method II boxed inlays display stellate reticulum (E, I, M). Efficiently removed odontoblasts (D, black arrows) and ameloblasts (D, gray arrows), and preserved collagen (white arrows). Abbreviations: a ameloblasts; d, dentin; p, pulp; od, odontoblasts; si, stratum intermedium; sr, stellate reticulum. * = p < 0.0001. Scale bar = 50 microns.
3.1.3 Histology of the pulp and surrounding tissues (M3)
Histological analyses of paraffin serial sectioned specimens were used to characterize molar tooth bud tissues before and after decellularization/demineralization treatments. In the labeled schematic (Fig. 2A), ameloblasts, stratum intermedium, and stellate reticulum were collectively referred to as the enamel organ. Polarized odontoblasts and ameloblasts were identifiable in natural tooth bud control tissue sections using H&E, Movat’s Pentachrome, and Masson’s Trichrome stains (Fig. 2C, G, and K, respectively). All three decellularization Methods appeared to remove the majority of cells from the normally cellularized tissues (Fig. 2C–F). Interestingly, while odontoblasts appeared to be efficiently removed (Fig. 2D, black arrows), the pulp itself remained intact. Distinct areas of polarized ameloblasts in the enamel organ were also removed by the decellularization treatment (Fig. 2D–F, H–J, L–N, gray arrows). The stratum intermedium (Fig. 2C) was absent in decellularized sections, while collagens were preserved in multiple dental tissues, as evidenced by the (pink-to-orange) Movat’s and (blue) Masson’s staining, present in the pulp, dentin, and stellate reticulum (Fig. 2HJ, L–N, white arrows).
3.2. ECM proteins present in natural and decellularized tooth bud tissues
3.2.1. Immunohistochemical (IHC) analyses (M3 and M2 molar teeth)
ECM molecule expression patterns in Method I and II treated decellularized, and control M3 tooth buds, were analyzed using IHC (Fig. 3). Decellularized samples exhibited somewhat higher background levels than were observed in Natural control samples. Fibronectin appeared diffusely expressed in all dental tissues (Fig. 3A) and in enamel organ tissues (data not shown). Collagen I gradients were detected in the dental lamina of decellularized samples (Fig. 3B), consistent with published reports [27]. Collagen I appeared highly expressed in pre-dentin, and more diffuse in dental pulp tissue (Fig. 3B, bottom panels).
Figure 3. IHC analyses of fibrillar ECM protein expression.
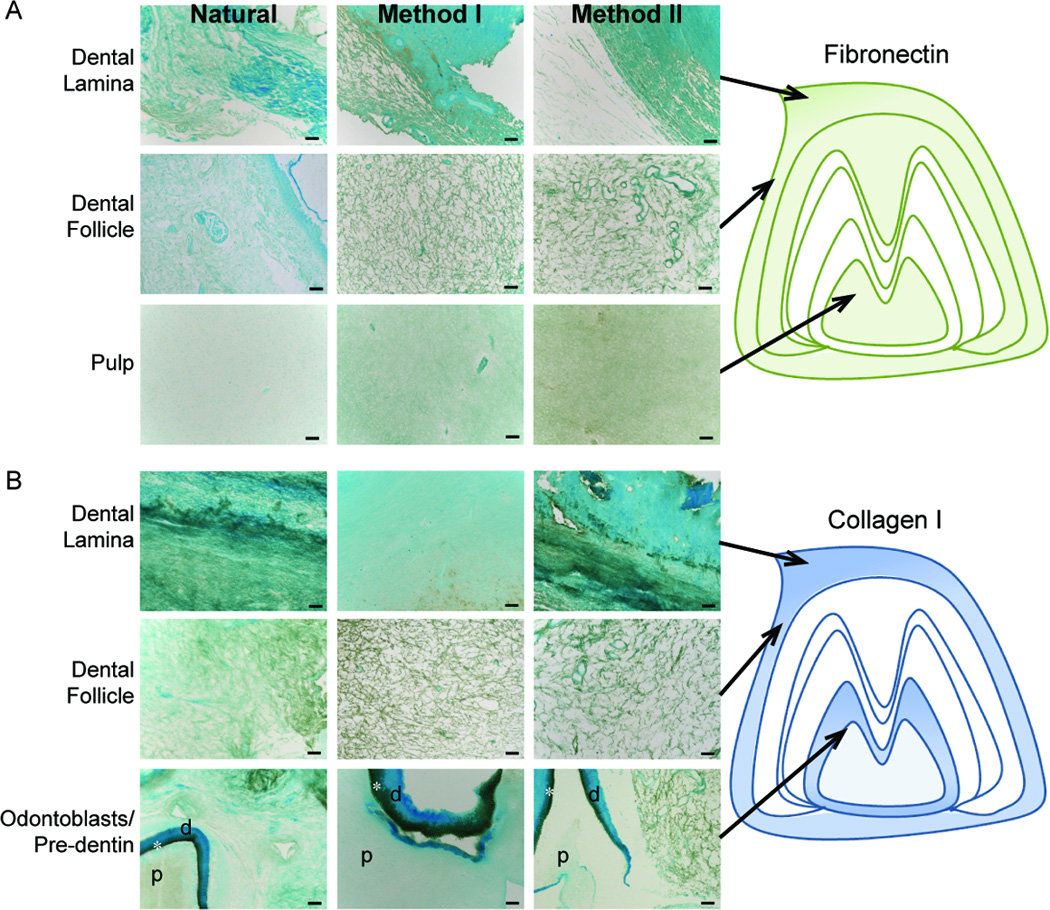
(A) Fibronectin and (B) Collagen I expression in Natural, Method I, or Method II treated teeth. Schematic representations are shown to the right. Strong collagen I expression was observed in the pre-dentin region (*) between the pulp (p) and dentin (d). Scale = 50 microns.
The basement membrane proteins Collagen IV and Laminin were detected primarily in nerves and blood vessels present in dental epithelial and mesenchymal tissues (Fig. 4). In general, ECM proteins present in blood vessels and nerves of enamel organ and pulp tissues were well preserved in decellularized samples. The weak Laminin and Collagen IV expression detected in enamel organ tissues of Natural tooth bud controls was not detectable in any decellularized samples (Fig. 4A and B, middle panels). Similar ECM expression patterns were detected in both M2 and M3 molar scaffolds (Supplemental Fig. 1).
Figure 4. IHC analyses of basement membrane ECM protein expression.
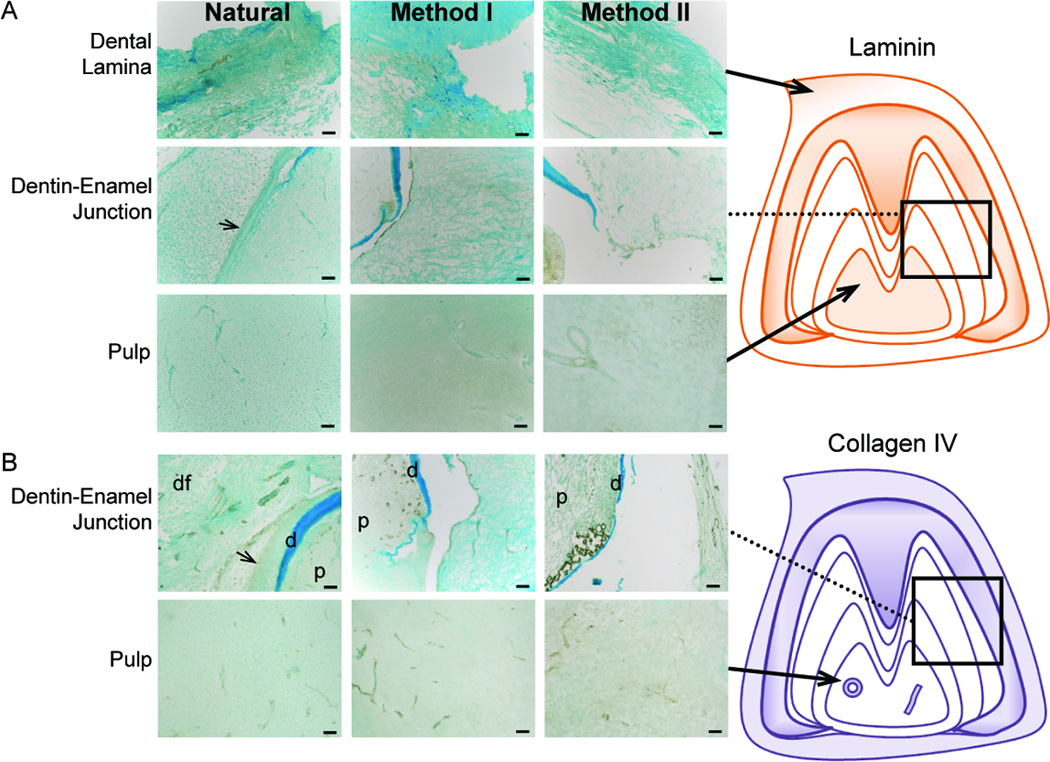
(A) Laminin and (B) Collagen IV expression in Natural, Method I, or Method II treated teeth. Schematic representations are shown to the right. Vascular and nerve tissues expressed both basement membrane proteins. Abbreviations: d, dentin; p, pulp; dental follicle, df. Scale = 50 microns.
3.2.2. Non-linear microscopy and image analysis of natural and decellularized M2 molar tooth pulp
SHG analyses of natural and decellularized samples confirmed the presence of fibrillar collagen in the treated dental pulp sample (shown in green, Fig. 5), consistent with our histological and IHC analyses. TPEF excited at 755 nm and detected in the 440–480 nm region (shown in red, Fig. 5), generated a signal that appeared to be distributed throughout both natural and Method I decellularized tissue samples, but TPEF was much more prominent in the natural scaffolds. Two possible sources of this signal include elastin fibers and cells [25]. However, the morphology of this signal, along with the fact that its presence is significantly decreased in the decellularized scaffolds suggests that it is emanating from cells. Three-dimensional reconstructions revealed SHG visualized collagen and TPEF visualized elastin and cell signals distributed through a depth of approximately 60 microns (Fig. 5A, 5D and Movies 1–2.). Single images from the z-series displayed typical SHG signal for natural and decellularized scaffolds (Fig. 5B, 5E, respectively). Thresholding the image allowed for quantification of collagen fiber density (Fig. 5C and 5F), showing that natural tooth scaffolds displayed a much higher collagen fiber density as compared to decellularized scaffolds. In addition, Natural tooth scaffold collagen fibers exhibited more random orientation (as exhibited by the significantly lower orientation index or OI; P<0.05), and were less organized (higher entropy) as compared to the decellularized scaffold fibers (Table 2).
Figure 5. Non-linear microscopy of natural and decellularized M2 molar pulp.
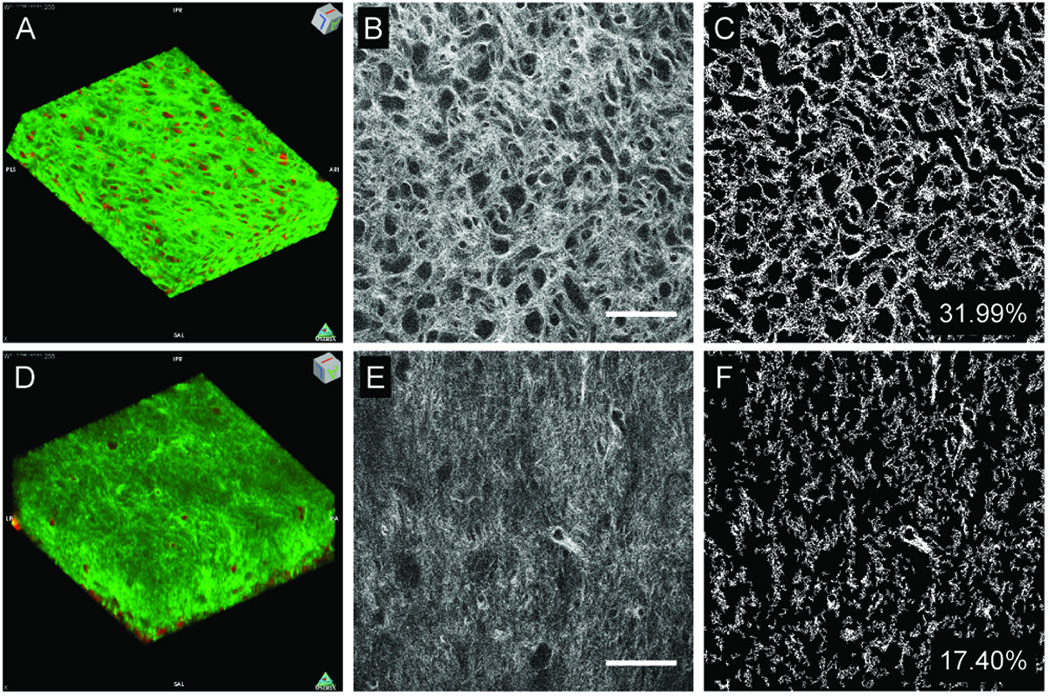
TPEF (red pseudocolor) and SHG (green pseudocolor) reconstructions of natural (A) and Method I treated (D) tooth scaffolds (238×238×64 µm3 tissue volumes). Representative natural (B) and decellularized (E) tooth scaffolds. Thresholded natural (C) and decellularized (H) images were used to assess collagen fiber density and organization. Scale = 50 microns.
Table 2.
SHG analyses of Natural and Method I treated tooth scaffolds.
| Fiber Density |
OI | Entropy | |
|---|---|---|---|
| Natural | 31.68 ± 1.58 | 4.65 ± 4.29 | 6.74 ± 0.06 |
| Method I | 17.78 ± 3.20 | 18.00 ± 4.47 | 6.68 ± 0.10 |
| P-values | 9.21E-26 | 4.17E-18 | 0.003 |
3.3. Reseeded M2 tooth bud samples
3.3.1. Non-linear microscopy and image analysis of reseeded tooth tissues after 4 weeks in vitro culture
SHG and TPEF were used to characterize collagen fibers and cells in decellularized treated and control samples without the need for fixation or exogenous fluorophores. Although some degree of autofluorescence was observed in the unseeded controls, bright autofluorescence was observed in dental mesenchymal cell-seeded samples (Fig. 6A versus 6D, and Movies 3–4), likely due to the presence of cells. As can be observed in representative images, collagen fiber density was significantly higher (P<0.05) in the dental mesenchymal cell-seeded scaffold as compared to the unseeded control (Fig. 6B–C, versus 6E–F). Cell-seeded scaffolds also exhibited more organized collagen, as reflected in the lower entropy reading compared to unseeded controls (Table 3). The OI was also higher in dental cell-seeded scaffolds as compared to unseeded controls, further supporting that the collagen fibers were more highly aligned.
Figure 6. Non-linear microscopy of re-seeded tooth scaffolds after four weeks of culture.
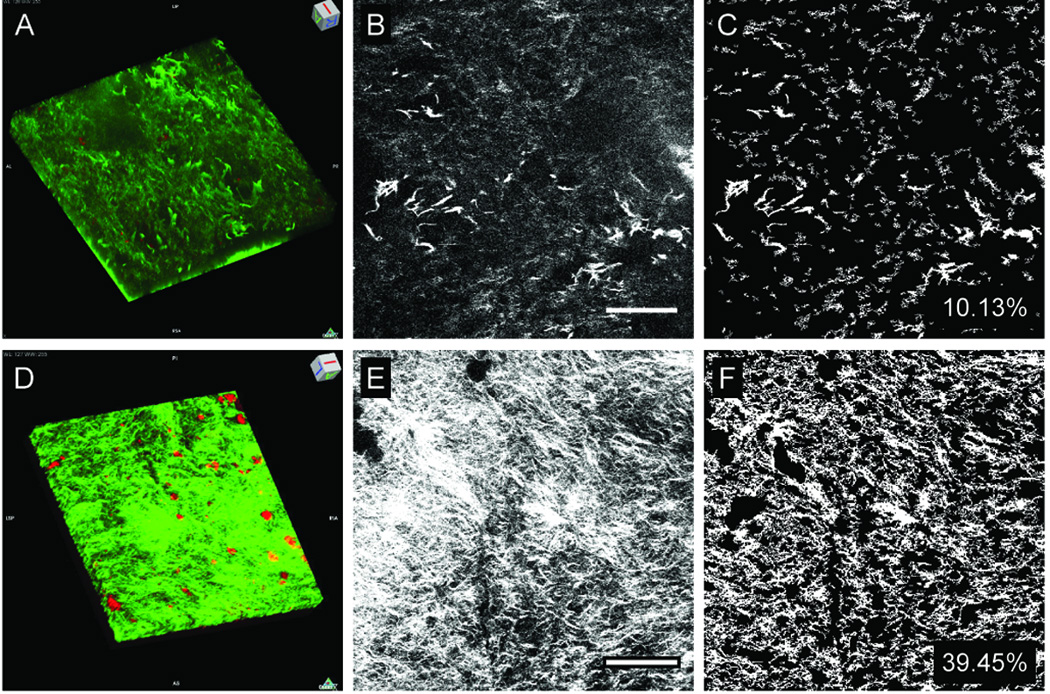
SHG (green pseudocolor) and TPEF (red pseudocolor) reconstructions of cultured unseeded (A), and cultured dental cell seeded (D) decellularized scaffolds (238×238×33 µm3 tissue volumes). Representative images of cultured unseeded (B) and mes-seeded (E) scaffolds. Collagen fiber densities were determined from thresholded images (C, H). Scale bars = 50 microns.
Table 3.
SHG analyses of cultured unseeded and dental cell-seeded tooth scaffolds.
| Incubation time |
Treatment | Temperature |
|---|---|---|
| 48 hrs | Hypotonic TE Buffer | 4°C |
| 24 hrs | SDS (1% or 5%) + Protease Inhibitors* | 24°C |
| 24 hrs | TritonX-100 (1%) + Protease Inhibitors* | 24°C |
| 3 hrs | Nuclease Treatment | 37°C |
| 16 hrs | Normal Buffered Formalin (10%) | 24°C |
| 3 months | EDTA Demineralization (10%) | 24°C |
Hypotonic TE Buffer, 10 mM Tris-HCl Buffer, 1 mM EDTA, pH 8.2; Protease Inhibitors: Aprotinin (10 KIU) and EDTA (10 µM); Nuclease Treatment (pH 7.5), 50 mM Tris-HCl, 10 mM magnesium chloride; 50 µg/mL bovine serum albumin (pH 7.5), 3.6 U/mL DNase I (RNase-free) and 1.2 U/mL RNase (DNase-free); pH of detergent solutions: 7.6–7.8
Alternated for 5 cycles (with TBS rinses in between), prior to nuclease treatment.
4. Discussion
Recent efforts in tooth tissue engineering have focused on creating bioengineered teeth and supporting structures, using post natal cells harvested from dental pulp and periodontal tissues [2, 29]. We have previously demonstrated the successful generation of bioengineered tooth and bone tissues, generated from porcine and rat tooth bud cells seeded onto PGA/PLGA [27–29, 39], and silk fibroin scaffolds [40]. Our currently used bioengineered tooth scaffolds lack ECM molecule gradients present in naturally formed teeth that provide essential cues for proper tooth development, periodontal tissues, and surrounding alveolar bone [32]. In the present study, we have compared ECM protein gradients present in natural and decellularized tooth buds, and have tested whether re-seeded acellular tooth bud scaffolds can support dental mesenchymal cell matrix deposition. First, using traditional IHC, and non-linear SHG and TPEF analyses, we identified decellularization methods that preserved the ECM protein gradients present in naturally formed teeth. Next, we examined Method I decellularized samples using SHG imaging analyses.
In support of this approach, recent publications report the successful viability of cells reseeded onto 1% SDS/1%TritonX-100 treated scaffolds [16, 41]. As evident in both histological and optical sections, Method I also appeared to effectively remove cell layers in the tooth buds. Interestingly, although 5% SDS treatment appeared to remove all cellular material as evidenced by lack of cell nuclei, we found that the DNA content was not decreased Method III treated samples. This unexpected result could be attributed to: a) removing tissue, but not removing actual DNA from the samples despite DNAse treatment; b) high residual amounts of detergent interfering with PicoGreen excitation; or c) our inability to remove both the concentrated detergent and nucleic debris from the tissue even with extensive washing.
Our characterizations of decellularized and control scaffolds using both IHC and SHG/TPEF exploit the strengths of both techniques. Although other methods to quantify collagen content exist, including hydroxyproline assays [42], they cannot provide detailed information regarding collagen protein localization or fiber structure, characteristics that may prove to be quite important for biomimetic scaffold design. And although IHC can be used to detect Collagen I, it is not a quantitative method for collagen protein detection, or of collagen fiber structure. While IHC is subject to proper antibody recognition of the epitope, and to variations in antigen retrieval and tissue processing, SHG generates a highly specific signal for fibrillar collagen with minimal background, allowing for sensitive measurements without the need for additional fixation, fluorophores, or processing [22]. Still, the value of a combined approach, using IHC and SHG/TPEF, or immunofluorescence (IF) combined with SHG acquisition, is the ability to distinguish between various types of collagens, since SHG signal alone can be generated from a variety of fibrillar collagens including collagen I, III and V, all of which are present in dental pulp [43, 44].
Having first confirmed differences in Type I collagen expression in our decellularized and control samples via IHC, we then used SHG to quantify these differences. Non-invasive imaging via SHG provided a quantitative analysis not only of collagen fiber content, but also revealed differences in collagen fiber distribution and alignment. Notably, decellularized scaffolds exhibited lower collagen fiber densities than natural tissue, likely the result of the decellularization procedure itself. Also, decellularized tooth scaffolds exhibited higher collagen alignment and organization, as compared to natural tooth tissue. It is possible that the rocking used during the detergent treatment cycles induced this higher level of alignment. The re-seeded acellular scaffolds exhibited significantly increased collagen fiber density relative to the corresponding unseeded scaffolds (Table 3), with levels that were similar to the collagen fiber density of the natural teeth (Table 2). However, both the entropy and the OI values indicate that the collagen present in the reseeded scaffolds is even more organized and aligned than the decellularized scaffolds (Table 3). Thus, it appears that aligned collagen fibers in decellularized scaffolds directed cell matrix production of enhanced alignment and organization in reseeded scaffolds. In support of our results, recent reports show that aligned nanofiber orientation can facilitate increased cell matrix secretion and mineralization as compared to random nanofiber orientation [45–47]. When comparing the fiber characteristics of the decellularized scaffolds of Table 2 and 3, there are differences in the detected fiber density and OI. These could be real differences as the scaffolds in Table 3 were coated with collagen and cultured for 30 days. Alternatively, the SHG measurements of the reseeded scaffolds (Table 3) were performed in the forward direction, while those of the natural and decellularized ones (Table 2) were acquired in the backward direction. This could affect our signal to noise ratio, which could in turn affect the extracted fiber density. Previous studies have demonstrated that similar information in terms of the fiber orientation is typically contained in the forward and backward direction [48, 49]. Future analyses of larger sample numbers should be performed in order to assess sample to sample variation and identify the origins of the observed differences.
Both the decellularization and reseeding processes were monitored using non-linear microscopy capable of analyzing a tissue depth of 64 µm and 33 µm, respectively, to reveal 2D and 3D collagen structures in processed natural tooth scaffolds, and to monitor matrix remodeling of dental cell seeded scaffolds after 30 days in cell culture. The most significant differences we found were in the collagen fiber density of natural versus decellularized tooth scaffolds, and in unseeded versus reseeded scaffolds. Interestingly, more dense patches of collagen were detected in areas of the reseeded scaffold, as compared to the unseeded control scaffolds, consistent with the fact that denser collagen fibers were generated by de novo cell matrix production by repopulated dental cells. It is noteworthy that one study reported increased SHG signal due to seeded cell contraction of existing collagen fibers [50]. In contrast, our observed increase in collagen fiber density combined with a heightened autofluorescent signal in the reseeded scaffold suggests that viable dental cells established residence in the decellularized scaffold, and remodeled the matrix. Additional studies are underway to confirm and expand upon these observations. Since non-linear microscopy allows for repeatable, non-destructive imaging, future studies will use SHG/TPEF to characterize dental cell repopulation, and changes in collagen fiber density, orientation, and organization over time.
Since natural tooth tissues exhibit variable cell density, collagen fiber density, and ECM scaffold porosity, future studies will focus on detailed 3D characterizations of tooth pulp tissue. Our results using SHG to evaluate collagen in natural, decellularized, and reseeded tooth bud matrices raise many intriguing questions regarding fibrillar collagen remodeling. The decellularized scaffolds, in addition to exhibiting a lower collagen fiber density than that of natural tooth bud tissue, also exhibited lower entropy, and appeared more aligned. It is possible that the decellularization procedure, in addition to removing cells, may also have removed weaker, more disorganized collagen fibers, revealing a dominant directionality present in the remaining, sturdier collagen fibers. And based on the fact that the reseeded scaffold exhibited increased collagen fiber density and alignment as compared to unseeded controls, it is possible that the reseeded cells preferentially secreted new matrix in a manner that reinforced the existing and dominant collagen fiber matrix. A preference to adopt an existing dominant fiber direction is also consistent with the fact that entropy remained low after the reseeded cells elaborated new ECM. Further examination of collagen fiber destruction and remodeling as a consequence of decellularization and reseeding will be performed to explore this hypothesis.
Additional improvements to our method may include analyzing scaffold rinse solutions for hydroxyproline content over time to measure the rate of collagen removal [51, 52], and to monitor reseeded scaffolds over time to quantitate increased collagen content [53]. Knowledge of the rate and timing of collagen remodeling could provide informative data to supplement SHG analyses. For example, it is possible that reseeded dental cells initially elaborate collagen fibers that adopt an existing dominant orientation, and eventually reach a threshold density that allows for the subsequent elaboration of intermediate fibers.
5. Conclusions
Here, we have described reliable methods to decellularize and demineralize composite hard and soft dental tissues, and to characterize these tissues using both traditional histological, immunohistochemical and state-of-the-art non-linear microscopic methods. These studies describe, for the first time, distinct extracellular matrix molecular gradients in early stage tooth structures We also demonstrate the use of SHG imaging to analyze heterogeneous tooth organ tissues, and describe a systematic quantification of tooth tissues, using the parameters of OI, entropy, and collagen density. Future studies will build upon these results by performing real-time monitoring of dental cell matrix elaboration and remodeling of dental and other progenitor cell reseeded decellularized/demineralized tooth scaffolds.
Supplementary Material
Collagen (SHG, green) and elastin/cells (TPEF, red) visualized from molar pulp.
Collagen (SHG, green) and elastin/cells (TPEF, red) visualized from molar pulp.
Collagen (SHG, green) and elastin/cells (TPEF, red) visualized from molar pulp.
Collagen (SHG, green) and elastin/cells (TPEF, red) visualized from molar pulp.
(A) Fibronectin, (B) Collagen I, (C) Laminin, and (D) Collagen IV expression in Natural, Method I, or Method II treated tooth samples. Fibronectin, Collagen I and Laminin were highly expressed in the dental follicle, while Collagen IV was predominantly expressed in blood vessels. Scale bars = 50 microns.
Acknowledgements
The authors wish to acknowledge Leah Bellas for lyophilizing the samples prior to Pico Green analysis and Weibo Zhang for expert histological training and advice. We also thank Stephen Badylak and Steven Tottey for expert advice on DNA isolation and quantification. This work was funded by NIH R01DE016132 (PCY), NSF BES0547292 (IG, NF), NIH RO1EB007542 (IG, NF), and NIH P41 EB002520 (Tissue Engineering Resource Center).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Honda MJ, Fong H, Iwatsuki S, Sumita Y, Sarikaya M. Tooth-forming potential in embryonic and postnatal tooth bud cells. Med Mol Morphol. 2008;41:183–192. doi: 10.1007/s00795-008-0416-9. [DOI] [PubMed] [Google Scholar]
- 2.Li YC, Jin F, Du Y, Ma ZW, Li F, Wu G, et al. Cementum and periodontal ligament-like tissue formation induced using bioengineered dentin. Tissue Eng Part A. 2008;14:1731–1742. doi: 10.1089/ten.tea.2007.0268. [DOI] [PubMed] [Google Scholar]
- 3.Nakao K, Morita R, Saji Y, Ishida K, Tomita Y, Ogawa M, et al. The development of a bioengineered organ germ method. Nat Methods. 2007;4:227–230. doi: 10.1038/nmeth1012. [DOI] [PubMed] [Google Scholar]
- 4.Scheller EL, Krebsbach PH, Kohn DH. Tissue engineering: state of the art in oral rehabilitation. J Oral Rehabil. 2009;36:368–389. doi: 10.1111/j.1365-2842.2009.01939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du C, Fan DM, Sun Z, Fan YW, Lakshminarayanan R, Moradian-Oldak J. Immunogold labeling of amelogenin in developing porcine enamel revealed by field emission scanning electron microscopy. 9th International Conference on the Chemistry and Biology of Mineralized Tissues; Karger; Austin, TX. 2007. pp. 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Z, Sargeant TD, Hulvat JF, Mata A, Bringas P, Koh CY, et al. Bioactive nanofibers instruct cells to proliferate and differentiate during enamel regeneration. J Bone Miner Res. 2008;23:1995–2006. doi: 10.1359/JBMR.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenorio DMH, Santos MF, Zorn TMT. Distribution of biglycan and decorin in rat dental tissue. International Symposium on Extracellular Matrix (SIMEC 2002); Assoc Bras Divulg Cientifica; Angra Reis, Brazil. 2002. pp. 1061–1065. [DOI] [PubMed] [Google Scholar]
- 8.Wen X, Zou YM, Luo W, Goldberg M, Moats R, Conti PS, et al. Biglycan overexpression on tooth enamel formation in transgenic mice. Anat Rec. 2008;291:1246–1253. doi: 10.1002/ar.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci. 2006;119:3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- 10.Fukumoto S, Miner JH, Ida H, Fukumoto E, Yuasa K, Miyazaki H, et al. Laminin alpha 5 is required for dental epithelium growth and polarity and the development of tooth bud and shape. J Biol Chem. 2006;281:5008–5016. doi: 10.1074/jbc.M509295200. [DOI] [PubMed] [Google Scholar]
- 11.Manabe RI, Tsutsui K, Yamada T, Kimura M, Nakano I, Shimono C, et al. Transcriptome-based systematic identification of extracellular matrix proteins. Proc Natl Acad Sci U S A. 2008;105:12849–12854. doi: 10.1073/pnas.0803640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, et al. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–933. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 13.Bolland F, Korossis S, Wilshaw SP, Ingham E, Fisher J, Kearney JN, et al. Development and characterisation of a full-thickness acellular porcine bladder matrix for tissue engineering. Biomaterials. 2007;28:1061–1070. doi: 10.1016/j.biomaterials.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Grayson WL, Bhumiratana S, Cannizzaro C, Chao PHG, Lennon DP, Caplan AI, et al. Effects of initial seeding density and fluid perfusion rate on formation of tissue-engineered bone. Tissue Eng Part A. 2008;14:1809–1820. doi: 10.1089/ten.tea.2007.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macchiarini P, Jungebluth P, Go T, Asnaghi MA, Rees LE, Cogan TA, et al. Clinical transplantation of a tissue-engineered airway. Lancet. 2008;372:2023–2030. doi: 10.1016/S0140-6736(08)61598-6. [DOI] [PubMed] [Google Scholar]
- 16.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, et al. Perfusion-decellularized matrix: using nature's platform to engineer a bioartificial heart. Nat Med. 2008;14:213–221. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 17.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329:538–541. doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campagnola PJ, Wei M, Lewis A, Loew LM. High resolution nonlinear optical imaging of live cells by second harmonic generation. Biophys J. 1999;77:3341–3349. doi: 10.1016/S0006-3495(99)77165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown E, McKee T, diTomaso E, Pluen A, Seed B, Boucher Y, et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med. 2003;9:796–800. doi: 10.1038/nm879. [DOI] [PubMed] [Google Scholar]
- 20.Campagnola PJ, Millard AC, Terasaki M, Hoppe PI, Malone CJ, Ohler WA. Three-dimensional high resolution second-harmonic generation imaging of endogenous structural proteins in biological tissues. Biophys J. 2002;82:493–508. doi: 10.1016/S0006-3495(02)75414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zoumi A, Yeh A, Tromberg B. Imaging cells and extracellular matrix in vivo by using second-harmonic generation and two-photon excited fluorescence. Proc Natl Acad Sci U S A. 2002;99:11014–11019. doi: 10.1073/pnas.172368799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strupler M, Pena AM, Hernest M, Tharaux PL, Martin JL, Beaurepaire E, et al. Second harmonic imaging and scoring of collagen in fibrotic tissues. Opt Express. 2007;15:4054–4065. doi: 10.1364/oe.15.004054. [DOI] [PubMed] [Google Scholar]
- 23.Palero JA, de Bruijn HS, van den Heuvel AV, Sterenborg H, Gerritsen HC. Spectrally resolved multiphoton imaging of in vivo and excised mouse skin tissues. Biophys J. 2007;93:992–1007. doi: 10.1529/biophysj.106.099457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 25.Opitz F, Schenke-Layland K, Cohnert TU, Starcher B, Halbhuber KJ, Martin DP, et al. Tissue engineering of aortic tissue: dire consequence of suboptimal elastic fiber synthesis in vivo. Cardiovasc Res. 2004;63:719–730. doi: 10.1016/j.cardiores.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Rice WL, Kaplan DL, Georgakoudi I. Two-photon microscopy for non-invasive, quantitative monitoring of stem cell differentiation. PLoS One. 2010;5(4):13. doi: 10.1371/journal.pone.0010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young CS, Terada S, Vacanti JP, Honda M, Bartlett JD, Yelick PC. Tissue engineering of complex tooth structures on biodegradable polymer scaffolds. J Dent Res. 2002;81:695–700. doi: 10.1177/154405910208101008. [DOI] [PubMed] [Google Scholar]
- 28.Young CS, Abukawa H, Asrican R, Ravens M, Troulis MJ, Kaban LB, et al. Tissue-engineered hybrid tooth and bone. Tissue Eng. 2005;11:1599–1610. doi: 10.1089/ten.2005.11.1599. [DOI] [PubMed] [Google Scholar]
- 29.Zhang WB, Abukawa H, Troulis MJ, Kaban LB, Vacanti JP, Yelick PC. Tissue engineered hybrid tooth-bone constructs. Methods. 2009;47:122–128. doi: 10.1016/j.ymeth.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Mirsadraee S, Wilcox HE, Korossis SA, Kearney JN, Watterson KG, Fisher J, et al. Development and characterization of an acellular human pericardial matrix for tissue engineering. Tissue Eng. 2006;12:763–773. doi: 10.1089/ten.2006.12.763. [DOI] [PubMed] [Google Scholar]
- 31.Wilshaw SP, Kearney J, Fisher J, Ingham E. Biocompatibility and potential of acellular human amniotic membrane to support the attachment and proliferation of allogeneic cells. Tissue Eng Part A. 2008;14:463–472. doi: 10.1089/tea.2007.0145. [DOI] [PubMed] [Google Scholar]
- 32.Marquez SP, Martinez VS, Ambrose WM, Wang J, Gantxegui NG, Schein O, et al. Decellularization of bovine corneas for tissue engineering applications. Acta Biomater. 2009;5:1839–1847. doi: 10.1016/j.actbio.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 33.Kiernan JA. Histological and histochemical methods: Theory and practice. 4th ed. Oxford, UK: Scion Publishing; 2008. [Google Scholar]
- 34.Rosen AD. Endpoint determination in edta decalcification using ammonium oxalate. Stain Technol. 1981;56:48–49. doi: 10.3109/10520298109067275. [DOI] [PubMed] [Google Scholar]
- 35.Zhang W, Ahluwalia IP, Yelick PC. Three dimensional dental epithelial-mesenchymal constructs of predetermined size and shape for tooth regeneration. Biomaterials. 2010;31:7995–8003. doi: 10.1016/j.biomaterials.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wainwright JM, Czajka CA, Patel UB, Freytes DO, Tobita K, Gilbert TW, et al. Preparation of cardiac extracellular matrix from an intact porcine heart. Tissue Eng Part C Methods. 2010;16:525–532. doi: 10.1089/ten.tec.2009.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert TW, Freund JM, Badylak SF. Quantification of DNA in biologic scaffold materials. J Surg Res. 2009;152(1):135–139. doi: 10.1016/j.jss.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayan C, Levitt JM, Miller E, Kaplan D, Georgakoudi I. Fully automated, quantitative, noninvasive assessment of collagen fiber content and organization in thick collagen gels. J Appl Phys. 2009;105:11. doi: 10.1063/1.3116626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duailibi MT, Duailibi SE, Young CS, Bartlett JD, Vacanti JP, Yelick PC. Bioengineered teeth from cultured rat tooth bud cells. J Dent Res. 2004;83:523–528. doi: 10.1177/154405910408300703. [DOI] [PubMed] [Google Scholar]
- 40.Xu WP, Zhang W, Asrican R, Kim HJ, Kaplan DL, Yelick PC. Accurately shaped tooth bud cell-derived mineralized tissue formation on silk scaffolds. Tissue Eng Part A. 2008;14:549–557. doi: 10.1089/tea.2007.0227. [DOI] [PubMed] [Google Scholar]
- 41.Uygun BE, Soto-Gutierrez A, Yagi H, Izamis ML, Guzzardi MA, Shulman C, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16 doi: 10.1038/nm.2170. 814-U120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy GK, Enwemeka CS. A simplified method for the analysis of hydroxyproline in biological tissues. Clin Biochem. 1996;29:225–229. doi: 10.1016/0009-9120(96)00003-6. [DOI] [PubMed] [Google Scholar]
- 43.Tung PS, Domenicucci C, Wasi S, Sodek J. Specific immunohistochemical localization of osteonectin and collagen type-i and type-iii in fetal and adult porcine dental-tissues. J Histochem Cytochem. 1985;33:531–540. doi: 10.1177/33.6.3889139. [DOI] [PubMed] [Google Scholar]
- 44.Wang HM, Nanda V, Rao LG, Melcher AH, Heersche JNM, Sodek J. Specific immunohistochemical localization of type-iii collagen in porcine periodontal tissues using the peroxidase-antiperoxidase method. J Histochem Cytochem. 1980;28:1215–1223. doi: 10.1177/28.11.7000890. [DOI] [PubMed] [Google Scholar]
- 45.Vaquette C, Kahn C, Frochot C, Nouvel C, Six J-L, De Isla N, et al. Aligned poly(l-lactic-co-e-caprolactone) electrospun microfibers and knitted structure: a novel composite scaffold for ligament tissue engineering. J Biomed Mater Res A. 2010;94A:1270–1282. doi: 10.1002/jbm.a.32801. [DOI] [PubMed] [Google Scholar]
- 46.Lee CH, Shin HJ, Cho IH, Kang Y-M, Kim IA, Park K-D, et al. Nanofiber alignment and direction of mechanical strain affect the ecm production of human acl fibroblast. Biomaterials. 2005;26:1261–1270. doi: 10.1016/j.biomaterials.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 47.Ma JY, He XZ, Jabbari E. Osteogenic differentiation of marrow stromal cells on random and aligned electrospun poly(l-lactide) nanofibers. Ann Biomed Eng. 2010;39:14–25. doi: 10.1007/s10439-010-0106-3. [DOI] [PubMed] [Google Scholar]
- 48.Williams RM, Zipfel WR, Webb WW. Interpreting second-harmonic generation images of collagen i fibrils. Biophys J. 2005;88:1377–1386. doi: 10.1529/biophysj.104.047308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao RAR, Mehta MR, Leithem S, Toussaint JKC. Quantitative analysis of forward and backward second-harmonic images of collagen fibers using fourier transform second-harmonic-generation microscopy. Opt Lett. 2009;34:3779–3781. doi: 10.1364/OL.34.003779. [DOI] [PubMed] [Google Scholar]
- 50.Pena A-M, Fagot D, Olive C, Michelet J-F, Galey J-B, Leroy F, et al. Multiphoton microscopy of engineered dermal substitutes: assessment of 3-d collagen matrix remodeling induced by fibroblast contraction. J Biomed Opt. 2010;15 doi: 10.1117/1.3503411. 056018. [DOI] [PubMed] [Google Scholar]
- 51.Wu Z, Zhou Y, Li NY, Huang MH, Duan HY, Ge J, et al. The use of phospholipase a(2) to prepare acellular porcine corneal stroma as a tissue engineering scaffold. Biomaterials. 2009;30:3513–3522. doi: 10.1016/j.biomaterials.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 52.Wilshaw SP, Kearney JN, Fisher J, Ingham E. Production of an acellular amniotic membrane matrix for use in tissue engineering. Tissue Eng. 2006;12:2117–2129. doi: 10.1089/ten.2006.12.2117. [DOI] [PubMed] [Google Scholar]
- 53.Schenke-Layland K, Opitz F, Gross M, Doring C, Halbhuber KJ, Schirrmeister F, et al. Complete dynamic repopulation of decellularized heart valves by application of defined physical signals - an in vitro study. Cardiovasc Res. 2003;60:497–509. doi: 10.1016/j.cardiores.2003.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Collagen (SHG, green) and elastin/cells (TPEF, red) visualized from molar pulp.
Collagen (SHG, green) and elastin/cells (TPEF, red) visualized from molar pulp.
Collagen (SHG, green) and elastin/cells (TPEF, red) visualized from molar pulp.
Collagen (SHG, green) and elastin/cells (TPEF, red) visualized from molar pulp.
(A) Fibronectin, (B) Collagen I, (C) Laminin, and (D) Collagen IV expression in Natural, Method I, or Method II treated tooth samples. Fibronectin, Collagen I and Laminin were highly expressed in the dental follicle, while Collagen IV was predominantly expressed in blood vessels. Scale bars = 50 microns.


