Abstract
Pathogenic microorganisms are persistently expressing resistance towards present generation antibiotics and are on the verge of joining the superbug family. Recent studies revealed that, notorious pathogens such as Salmonella typhi, Shigella dysenteriae and Vibrio cholerae have acquired multiple drug resistance and the treatment became a serious concern. This necessitates an alternative therapeutic solution. Present study investigates the utility of computer aided method to study the mechanism of receptor-ligand interactions and thereby inhibition of virulence factors (shiga toxin of Shigella dysenteriae, cholera toxin of Vibrio cholerae and hemolysin-E of Salmonella typhi) by novel phytoligands. The rational designs of improved therapeutics require the crystal structure for the drug targets. The structures of the virulent toxins were identified as probable drug targets. However, out of the three virulent factors, the structure for hemolysin-E is not yet available in its native form. Thus, we tried to model the structure by homology modeling using Modeller 9v9. After extensive literature survey, we selected 50 phytoligands based on their medicinal significance and drug likenesses. The receptor-ligands interactions between selected leads and toxins were studied by molecular docking using Auto Dock 4.0. We have identified two novel sesquiterpenes, Cadinane [(1S, 4S, 4aS, 6S, 8aS)- 4- Isopropyl- 1, 6- dimethyldecahydronaphthalene] and Cedrol [(8α)-Cedran-8-ol] against Shiga (binding energy -5.56 kcal/mol) and cholera toxins (binding energy -5.33 kcal/mol) respectively which have good inhibitory properties. Similarly, a natural Xanthophyll, Violaxanthin [3S, 3'S, 5R, 5'R, 6S, 6'S)-5, 5', 6, 6'-Tetrahydro-5, 6:5', 6'-diepoxy-β, β-carotene-3, 3'-diol] was identified as novel therapeutic lead for hemolysin-E (binding energy of –5.99 kcal/mol). This data provide an insight for populating the pool of novel inhibitors against various drug targets of superbugs when all current generation drugs seem to have failed.
Keywords: Multiple drug resistance, Shiga toxin, Phytoligands, Drug likenesses, Sesquiterpenes, Superbugs
Background
The discovery and introduction of treatments by antibiotics after World War I resulted in a dramatic decrease in number of deaths due to bacterial infections. However, in the present era, these antibiotics have lost their status as the “miracle drug” and “treatment failure” is a new and often seen situation. The antibiotic resistance became sustainable in the environment as already resistant bacteria emerged as new dominant population and evolve as superbugs [1].
The use, overuse and misuse of antibiotics in human and veterinary medicine are major promoters for the development and spread of multidrug resistant bacteria world-wide. However, large usage of antibiotics results in environmental releases. Even though, levels of antibiotics found in the environment are low, minimum inhibitory concentration for most bacteria, they may still result in an increased selection pressure at some sites [2]. In general practice, there are concerns that some common infections are becoming increasingly difficult to treat and that illnesses due to antibiotic resistant bacteria may take longer to resolve. With the evolution of new antibiotic resistant microorganisms, there are reports indicating the end of era of antibiotics treatment and the need for new methodologies to target the pathogenic microorganisms [3].
Cholera toxin is a protein complex secreted by the bacterium Vibrio cholerae which is responsible for the massive, watery diarrhea (cholera). It is a self-limiting illness. However, antibiotics are commonly administered as part of the treatment regimen. In recent days, the organism is becoming resistant for the multiple antibiotics [4]. Shiga toxin produced by Shigella dysenteriae is responsible for shigellosis (bacillary dysentery). The disease has epidemic potential and threat in Central Africa, India and other developing countries in the world [5]. Many strains of S. dysenteriae were multidrug resistant, resistance to commonly used antimicrobials such as Ampicillin, Tetracycline, Cotrimoxazole, Amoxicillin, Nalidixic acid and Fluoroquinolones (such as Ciprofloxacin and Norfloxacin), which had unusually high minimum inhibitory concentrations [6]. Similarly, Hemolysin-E of Salmonella typhi act as virulent factors for enteric fever and food borne illness. In developed countries many strains were found to be zoonotic in origin and acquire their resistance in the food-animal host before onward transmission to humans through the food chain. The multi-drug resistant (MDR) strains of Salmonella typhi display resistance to most of the antimicrobials and exhibit decreased susceptibility to Ciprofloxacin and other current therapies [7].
The spread of the MDR superbugs urges the need for an alternative and promising therapy. Computer aided approach is a novel platform to screen and select better therapeutic substances from wide varieties of lead molecules. Many herbal derived compounds have significant inhibitory and antimicrobial properties against a broad range of pathogenic microorganisms [8]. Our previous studies reported the applications of novel lead molecules against multidrug resistant Clostridium perfringens [9] and Staphylococcus aureus [10]. This study aims the selection of ligands from medicinal herbs and their utility as potential inhibitors against virulent toxins. There are many molecular studies indicated the scope of shiga toxin [11], cholera toxin [12] and hemolysin-E [13] of Shigella dysenteriae, Vibrio cholerae and Salmonella typhi respectively as the probable drug targets for drug discovery. The 3D structures of these toxins are very essential for computer aided drug discovery and the structure of shiga toxin and cholerae toxin are available in their native form. Since there is no 3D crystal structure of hemolysin-E of Salmonella typhi, there is a need for a homology model. Furthermore, we have demonstrated the effective interactions between selected phytoligands and the drug targets by docking simulations.
Methodology
Selection of the probable drug targets:
The cholera toxin of Vibrio cholerae, shiga toxin of Shigella dysenteriae and hemolysin-E of Salmonella typhi were identified as probable drug targets based on their virulent function in the diseases. The 3D structures of proteins are the fundamental requirement for structure based drug designing. The crystal structures of shiga toxin, PDB: 1DM0 [14] and cholera toxin, PDB: 1XEZ [15] are available in their native form. But, the 3D structure of hemolysin-E is not available in native state. Hence, our preliminarily aim in this study was to focus on the hypothetical modeling of hemolysin-E by computer aided approach.
Target retrieval and template selection:
The amino acid sequence of hemolysin-E, accession number Q8Z727 [16], was retrieved from UniProt database. The FASTA sequence of the hemolysin-E was subjected to PSI – BLAST [17] analysis against PDB database for the selection of best homologous templates. The best template was selected based on percentage of identity, similarity, expectation value, bit scores and query coverage. A multiple sequence analysis was performed by T-COFFEE [18] and the phylogenetic characterization was carried out by NJ plot [19]. These steps ensured the accurate selection of the template for homology modeling.
Homology modeling of hemolysin-E:
A 3D model of hemolysin-E was generated by Modeller 9v9 [20]. It is a UNIX based programme based on satisfaction of spatial restraints derived from the alignment and probability density functions (PDFs). The chain-A of hemolysin from E. coli, PDB: 1QOY [21], was identified as the best template for comparative modeling. The template consists of 69% α-helices, 14% coils, 12% of β-sheets and 5% of turns. The 3D structure was solved by X-ray diffraction studies and resolution was 2.0Ao. The R-factor of the structure was 0.198 and R-free value was 0.252. The modeled protein was visualized using Chimera [22]. The model was energy minimized by CHARMM [23] and the stereo-chemistry was validated by Procheck [24]. The overall quality factor of non-bonded interactions between different atoms was calculated by ERRAT [25]. The backbone of the modeled protein was threaded against the template chain by DaliLite [26] and root mean square deviation (RMSD) between the model and the template was calculated. The hypothetical model was deposited to Protein model data base [27]. The modeled structure of hemolysin-E of Salmonella typhi, crystal structures of cholera toxin (1XEZ) from Vibrio cholerae and shiga toxin (1DM0) from Shigella dysenteriae were considered as the probable drug targets.
Computer aided lead discovery
Screening and selection of phytoligands:
Many antibiotics are available against the pathogens in contention of our study. Since the resistance towards these antibiotics is increasing, there is a need for the identification of natural substances which can act as potential drug candidates. Various phytoligands against the drug targets were identified by extensive literature studies. The 3D structures of all identified ligands were retrieved from PubChem [28] and ChemSpider [29] databases. The pharmacokinetic and druggish activities were studied by Lipinski's Rule of Five [30]. The rule is one of the major landmarks for selecting the pharmacophoric pattern and biological behavior of the drug molecules. In our study, we selected 50 molecules were of herbal origin and that none of them showed any violation towards Lipinski's rule.
Docking studies and structure based drug design:
The selected phytoligands were docked against three toxins by Auto Dock 4.0 [31] using Lamarckian genetic algorithm. The catalytic and binding site of the target was identified and selected by AutoGrid. The structure and chemical properties of the active site allows the recognition and binding of the ligand. The electrostatic interactions were evaluated by interpolating the values of the electrostatic potential and multiplying by the charge on the atom. Around 2,500,000 conformations were generated by ten iterations and the best conformations were screened in terms of lowest binding energy generated in the clustering histogram. The AutoDock program was executed to simulate the real time molecular interactions of the receptor and ligands. The function scoring, energy calculations and ranking of the best conformations were carried out as per energy minimization and stabilizing interactions.
Disscussion
The shiga toxin of Shigella dysenteriae consists of two subunits. The subunit-A act as major virulent factor in most of the shigella infections. Crystal structure of the toxin (PDB: 1DM0) has 267 amino acids in which 34 % alpha helical structures (12 helices constitutes 99 residues) and 23% beta sheet (15 strands; 67 residues). This polypeptide is inhibiting protein synthesis through the catalytic inactivation of 60s ribosomal subunits. The subunit-B is 69 amino acids long (17% helical- 1 helices; 12 residues; 36% beta sheet - 8 strands; 25 residues) and is responsible for the binding of the holotoxin to specific receptors on the target cell surface, such as globotriaosylceramide (Gb3) in human intestinal microvilli.
The cholera toxin is a cytolysin which consists of 741 amino acids (PDB: 1XEZ) and has significant role in the pathogenesis of Vibrio cholerae. The toxin has one polypetide chain, which constitutes 6% helices (10 helices; 50 residues) and 45 % beta sheets (56 strands; 327 residues.). The molecular weight of the toxin is 799.43 Kda and it act as potential virulent factor. During infection it causes cytolysis by forming heptameric pores in target host membranes. By considering the functional aspects of the toxins in pathogenesis, the crystal structures of toxin were used as probable drug targets.
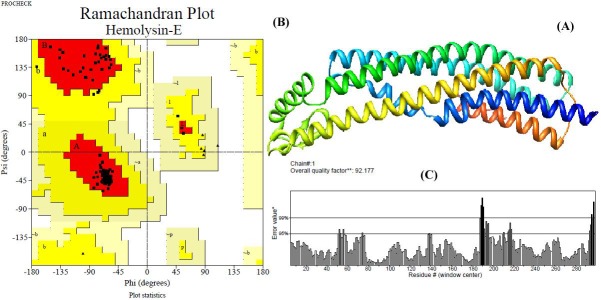
Salmonella typhi is another multiple drug resistance bacteria responsible for severe health hazards all over the world. Most strains of Salmonella secrete powerful toxin called hemolysin-E which act as major virulent factor. Hemolysin-E lyses erythrocytes and mammalian cells, forming transmembrane pores with a minimum internal diameter of 25 Ao. The three dimensional structure of toxin is not available in native form. Hence, we have modeled the structure of the toxin from its basic sequences. The sequence consists of 303 amino acids. The template selected for the modeling was chain-A of E. coli hemolysin with the length of 318 amino acids. The modeled protein has six alpha helical domains and it was visualized by Chimera (Figure 1A). The structure was energy minimized by CHARMM which yielded the energy value -2.14 kcal/mol from the previous energy value of -1.04 kcal/mol. The backbone structure of modeled protein is threaded with chain-A of template by DaliLite. The superimposition showed RMSD value of 0.2Ao with 298 aligned residue and 91% identity in their alignment. The modeled structure was steriochemically validated by Procheck. Ramachandran plot of the model indicated 98.2% of the residues in the allowed region, 1.2% of the residues in the additional allowed regions and no residues in the generously allowed and disallowed regions (Figure 1B).
Figure 1.
Three dimensional structure of hemolysin-E of Salmonella typhi generated by homology modeling; (A) The secondary structure of the protein displayed in Chimera shown that the toxin has seven alpha helical domains; (B) The Ramachandran plot of the model indicate 98.2% residues (A, B, L in the plot) are most favored region and 1.8 % (a, b, l, p regions) residues are additional allowed regions revealed the stupendous quality of the model; (C) The overall quality factor of non-bonded interactions between different atoms types estimated by ERRAT is 92.17 implies good validity of our structure.
The number of non-glycine and non-proline residues were identified as 282 and the number of glycine and proline residues present in the model was 15 and 4 respectively. The overall quality factor of the hemolysin-E identified by ERRAT was 92.17 % (Figure 1C). The modeled structures further validated by PROVE indicated the Z-score mean as 0.96, Zscore standard deviation as 29.87 and Z-score RMS as 29.873 which indicate the reliable quality of the model. The model was deposited to Protein model data base which can be downloaded by the accession number PM0077415.
Since, most strains of Vibrio cholerae, Shigella dysentriae and Salmonella typhi showed multiple drug resistance, therapeutic significance of these drugs became limited. Hence, there is a need for a novel approach. The present approach mainly focused to inhibit the key virulent toxins of the pathogens. Many studies revealed the utilities of phytoligands as potential inhibitors against wide varieties of drug resistant bacteria [8]. Therefore, we also tried to screen 50 best phytoligands from various medicinal plant sources by extensive literature studies. The selected ligands were screened for better pharmacological features by Lipinski's rule of five. All molecules showed very good physiochemical properties and drug likeness, enabling the molecule to exhibit good pharmacological properties.
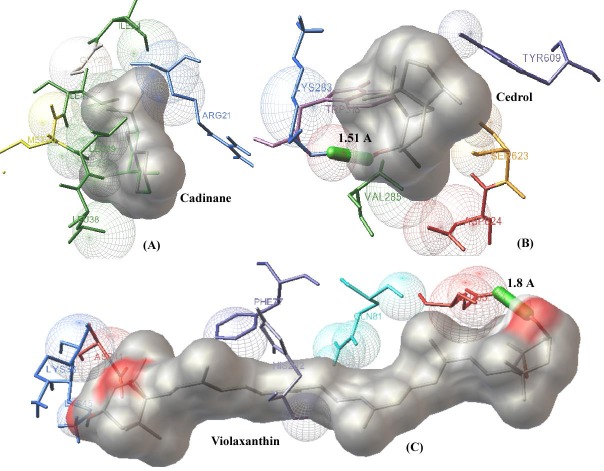
The fundamental principle behind computer aided lead designing is the study of receptor ligand interactions. The inhibitory properties and best binding conformations of novel ligands can be studied by molecular docking. In the present study, docking was preceded by the addition of polar hydrogen charges to the target proteins. All phytoligands were docked against cholera toxin of Vibrio cholerae, shiga toxin of Shigella dysenteriae and hemolysin-E of Salmonella typhi, Table 1 (see supplementary material). The best binding poses and interactions were screened by minimum energy conformers. Moreover, other factors influencing the stability of the ligand– receptor complexes, number of hydrogen bonds and Vander waals interactions, were also taken into consideration. Based on all these theoretical principles employed in docking studies, we identified the best phytolead against shiga toxin to be Cadinane, a medicinal compound of Gum myrhh (Commiphora myrrha), with minimum energy value of -5.56 kcal/mol and the interacting residues are Ile 24, Gly 25, Leu 38, Leu 39, Met 40 and Arg 21, Ile 41 (Figure 2A). The best inhibitor against cholera toxin identified from our study is Cedrol, a phytoligand present in Italian cypress (Cupressus sempervirens). The binding energy was found to be -5.33 kcal/mol and the interaction is stabilized by a hydrogen bond. The interacting residues are Lys 283, Val 285, Trp 318, Asp 624, Ser 623 and Tyr 609 (Figure 2B). Similarly, the active compound found in varieties of squash, gourd, and pumpkin (Cucurbita pepo) known as Violaxanthin was found to have significant inhibitory property against hemolysin-E. The binding energy was –5.99 kcal/mol and the interactions are stabilized by a hydrogen bond (Figure 2C). The amino acids present in the binding cavity of the toxin are Lys 38, Asp 41, Lys 45, Phe 71, Glu 85, Gln 81 and His 292 Table 2 (see supplementary material).
Figure 2.
The docking simulations of novel phytoligands against probable drug targets of multidrug resistant pathogens. The ligands are displayed in the form of molecular surfaces and interacting residues and hydrogen bonds are shown in stick figures. (A) The interaction of shiga toxin (virulent factor of Shigella dysenteriae) and the best phytoligand, Cadinane is stabilized by weak interactions (residues Ile 24, Gly 25, Leu 39, Leu 38, Arg 21 Met 40 and Ile 41) with a minimum energy of -5.56 kcal/mol; (B) The docked conformation of Cholera toxin (virulent factor of Vibrio cholerae) and herbal ligand Cadinane is stabilized by a hydrogen bond with an energy minimum of -5.33 kcal/mol. The interacting residues are Lys 283, Val 285, Trp 318, Asp 624, Ser 623 and Tyr 609; (C) Hemolysin-E (virulent factor of Salmonella typhi) interacted with Violaxanthin by a hydrogen bond and other interactions (Lys 38, Asp41, Lys 45, Phe 71, Glu85, Gln 81 and His 292) by the free energy minimum of –5.99 kcal/mol. Present docking studies revealed that herbal leads have good binding affinities against the drug targets and the data has crucial applications in further studies.
Our studies concluded that, phytoligands such as Cadinane, Cedrol and Violaxanthin have significant inhibitory properties against shiga, cholera and hemolysin-E toxins respectively. These compounds can be used to design novel therapy against multidrug resistant bacteria when all current generation antibiotics seem to have failed. From our study it is evident that present data find significant applications for further experimental studies. Moreover, the applied methods pave a new therapeutic insight against multiple drug resistant pathogens.
Conclusion
The increase of multiple drug resistance has lead to the evolution of many superbugs including notorious pathogens such as Shigella dysenteriae, Vibrio cholerae and Salmonella typhi. Our present study concluded that computer aided approach may serve as an effective method to screen novel therapeutic leads when all current generation drugs seems to have failed. We could identify novel drug targets for multidrug resistant bacteria. In addition, we also studied the utility of structure based virtual screening to select new therapeutic substances. Phytoligands such as Cedrol, Cadinane and Violaxanthin can be used as potential inhibitors against virulent toxins of Shigella dysenteriae, Vibrio cholerae and Salmonella typhi respectively. However, experimental studies need to be performed to confirm the efficiency of the applied approach and present data finds applications for such studies.
Supplementary material
Acknowledgments
The authors thankfully acknowledge the R & D Centre of Life Sciences and Engineering, Dayananda Sagar Institutions for providing all necessary facilities and also grateful to Dr. P.S Rao, Vice President, (R & D) in Life Sciences and Dr. G S Jagannatha Rao, Senior Professor and Head, Department of Biotechnology, Dayananda Sagar College of Engineering for their constant support and encouragement throughout the study.
Footnotes
Citation:Skariyachan et al, Bioinformation 8(9): 420-425 (2012)
References
- 1.S Schjorring, AK Karen. Int J Microbiol. 2011;2011:312956. doi: 10.1155/2011/312956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.E Kristiansson, et al. PLoS One. 2011;6:e17038. doi: 10.1371/journal.pone.0017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.P Nordmann, et al. J Antimicrob Chemother. 2011;66:689. doi: 10.1093/jac/dkq520. [DOI] [PubMed] [Google Scholar]
- 4.M Kitaoka, et al. J Med Microbiol. 2011;60:397. doi: 10.1099/jmm.0.023051-0. [DOI] [PubMed] [Google Scholar]
- 5.R Bercion, et al. Trans R Soc Trop Med Hyg. 2006;100:1151. doi: 10.1016/j.trstmh.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 6.S Dutta, et al. J Clin Microbiol. 2003;41:5833. doi: 10.1128/JCM.41.12.5833-5834.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.EJ Threlfall. FEMS Microbiol Rev. 2002;26:141. doi: 10.1111/j.1574-6976.2002.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 8.TB Machado, et al. Int J Antimicrob Agents. 2003;21:279. doi: 10.1016/s0924-8579(02)00349-7. [DOI] [PubMed] [Google Scholar]
- 9.S Skariyachan, et al. Bioinformation. 2011;7:222. [Google Scholar]
- 10.S Skariyachan, et al. Bioinformation. 2011;6:375. [Google Scholar]
- 11.TJ Rogers, JC Paton. Expert Rev Anti Infect Ther. 2009;7:683. doi: 10.1586/eri.09.51. [DOI] [PubMed] [Google Scholar]
- 12.MH Fazil, DV Singh. Future Microbiol. 2011;6:1199. doi: 10.2217/fmb.11.93. [DOI] [PubMed] [Google Scholar]
- 13.S Hunt, et al. Adv Exp Med Biol. 2010;677:116. doi: 10.1007/978-1-4419-6327-7_10. [DOI] [PubMed] [Google Scholar]
- 14.ME Fraser, et al. Nat Struct Biol. 1994;1:59. doi: 10.1038/nsb0194-59. [DOI] [PubMed] [Google Scholar]
- 15.R Olson, E Gouaux. J Mol Biol. 2005;350:997. doi: 10.1016/j.jmb.2005.05.045. [DOI] [PubMed] [Google Scholar]
- 16.J Oscarsson, et al. Infect Immun. 2002;70:5759. doi: 10.1128/IAI.70.10.5759-5769.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M Johnson, et al. Nucleic Acids Res. 2008;36:W5. [Google Scholar]
- 18.C Notredame, et al. J Mol Biol. 2000;302:205. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 19.G Perrière, M Gouy. Biochimie. 1996;78:364. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 20.H Fan, et al. Methods Mol Biol. 2012;819:105. doi: 10.1007/978-1-61779-465-0_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.AJ Wallace, et al. Cell. 2000;100:265. [Google Scholar]
- 22.EF Pettersen, et al. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 23.BR Brooks, et al. J Comput Chem. 2009;30:1545. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.RA Laskowski, et al. J Appl Cryst. 1993;26:283. [Google Scholar]
- 25.C Colovos, TO Yeates. Protein Sci. 1993;2:1511. [Google Scholar]
- 26.L Holm, J Park. Bioinformatics. 2000;16:566. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 27.T Castrignanò, et al. Nucleic Acids Res. 2006;34:D306. doi: 10.1093/nar/gkj105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Y Wang, et al. Nucleic Acids Res. 2012;40:D400. doi: 10.1093/nar/gkr1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.AJ Williams. Curr Opin Drug Discov Devel. 2008;11:393. [PubMed] [Google Scholar]
- 30.CA Lipinski, et al. Adv Drug Deliv Rev. 2001;46:3. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 31.GM Morris, et al. J Comput Aided Mol Des. 1996;10:293. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.