Abstract
The galectin family of glycan-binding proteins is thought to mediate many cellular processes by oligomerizing cell surface glycoproteins and glycolipids into higher-order aggregates. This hypothesis reflects the known oligomeric states of the galectins themselves and their binding properties with multivalent ligands in vitro, but direct evidence of their ability to cross-link ligands on a cell surface is lacking. A major challenge in fundamental studies of galectin–ligand interactions is that their natural ligands comprise a heterogeneous collection of glycoconjugates that share related glycan structures but disparate underlying scaffolds. Consequently, there is no obvious means to selectively monitor the behaviors of natural galectin ligands on live cell surfaces. Here we describe an approach for probing the galectin-induced multimerization of glycoconjugates on cultured cells. Using RAFT polymerization, we synthesized well-defined glycopolymers (GPs) functionalized with galectin-binding glycans along the backbone, a lipid group on one end and a fluorophore on the other. After insertion into live cell membranes, the GPs’ fluorescence lifetime and diffusion time were measured in the presence and absence of galectin-1. We observed direct evidence for galectin-1-mediated extended cross-linking on the engineered cells, a phenomenon that was dependent on glycan structure. This platform offers a new approach to exploring the “galectin lattice” hypothesis and to defining galectin ligand specificity in a physiologically relevant context.
Many cellular processes are regulated by multimerization of cell surface proteins and lipids.1 In many systems, biomolecules assemble into higher-order clusters through direct protein–protein interactions. However in some cases, auxiliary proteins provide scaffolding for oligomeric assemblies via recognition of post-translational modifications.6 The galectins, a family of secreted glycan-binding proteins, are thought to serve such a function by interacting with specific glycan structures covalently bound to cell surface proteins and lipids.8 Evidence that both the galectins as well as many of their native ligands are multivalent has led to the proposal of a “galectin lattice” model, in which galectins can segregate membrane-associated glycoproteins and glycolipids into discrete microdomains.10 Galectin-mediated assemblies have been implicated in the regulation of cell signaling, adhesion, migration, and proliferation,14 and their dysfunctions have been associated with autoimmune disease15 and cancer.17 As well, Dennis and co-workers have proposed that galectin lattices can regulate the cell surface half-lives of glycoproteins by retarding their endocytosis.18
Despite compelling evidence for the galectins’ role in modulating the behaviors of cell surface molecules, galectin-mediated ligand cross-linking has not been directly observed on live cells. A majority of studies addressing the galectins’ cross-linking ability have relied on in vitro binding assays.20 In one cell-based study, Nieminen et al. demonstrated that galectin-3 exists in a multivalent state, a requirement for cross-linking, on neutrophil and endothelial cell surfaces through Förster resonance energy transfer (FRET) imaging.23 However, the effects of galectin binding on ligand multimerization have not been directly addressed in cell-based systems, a challenge that is exacerbated by the nature of the galectins’ endogenous ligands: they comprise a heterogeneous collection of glycoconjugates that share related glycan structures but disparate underlying scaffolds. There is no straightforward means to selectively label such a complex ligand mixture with biophysical probes that would enable studies of their oligomerization. Specific glycoproteins, such as integrins,24 mucins,25 the T cell receptor,15 and EGFR,18 have been found to bind galectins in biochemical assays. In principle, the influence of galectins on these proteins’ cell-surface behavior can be monitored using GFP fusions and fluorescent antibodies. But on live cells, it is likely that only a subset of their heterogeneous glycoforms engage galectins and form oligomers, which complicates analyses focusing only on the protein component of the ligand.
Synthetic glycopolymers (GPs) have proven to be powerful functional surrogates for natural glycoconjugates, particularly in situations where the complexity and heterogeneity of the native biomolecules undermine experimental inquiry.27 For decades, chemists have made use of various GP architectures to study glycan–receptor interactions related to the immune response,33 viral infection,35 and neurobiology,38 and recently we employed synthetic GPs as ligands for microarray-based studies of glycan binding proteins39 and for cell-surface functionalization.41
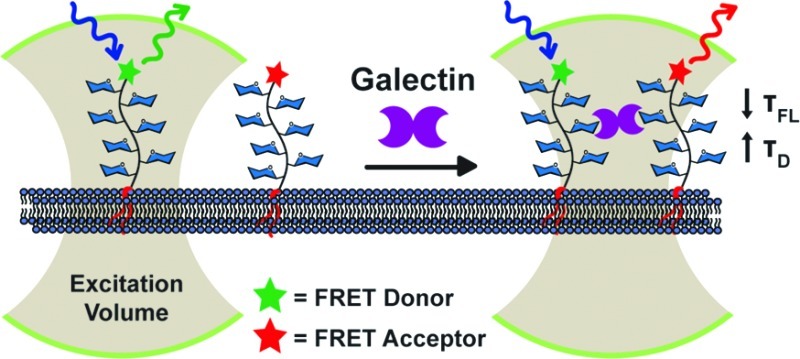
Here we present a new platform for investigating galectin-mediated cross-linking on live cell surfaces utilizing membrane-associated GPs as chemically defined ligands (Figure 1). The GPs were designed to possess the following attributes: (1) galectin-binding glycans distributed across the polymer backbone similarly to galectin-binding mucin glycoproteins,25 (2) a lipid anchor at one end, and (3) a FRET donor or acceptor fluorophore at the other end. The lipid tail enables insertion of the GP into live cell membranes and control of polymer orientation at the cell surface.41 The fluorescent dyes allow simultaneous monitoring of GP cross-linking by FRET as well as detection of higher-order assemblies by fluorescence correlation spectroscopy (FCS). Using this experimental platform we found direct evidence for the formation of cell-surface ligand clusters in the presence of galectin-1. More broadly, the method should facilitate interrogation of the galectin-lattice model in the physiologically relevant context of cell surfaces.
Figure 1.
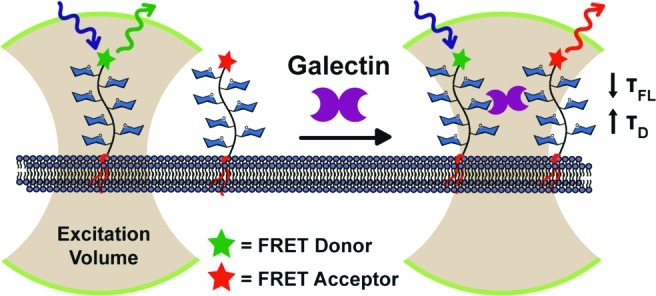
An experimental platform for probing galectin-mediated ligand cross-linking on live cell surfaces. Synthetic GPs were adorned with galectin-binding glycans (blue hexagons) and functionalized with a lipid on one end and either a FRET donor or acceptor dye on the other. The GPs were inserted into live cell membranes, and their fluorescence lifetimes (τFL) and diffusion times (τD) were monitored. The galectin-dependent decrease in τFL and increase in τD provided evidence of cell-surface GP cross-linking and oligmerization.
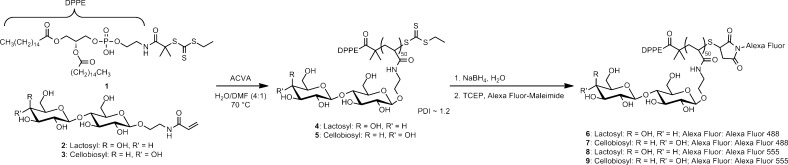
Well-defined GPs were synthesized by reversible addition–fragmentation chain transfer (RAFT) polymerization, which enables facile dual end-functionalization of the polymer chain.42 We incorporated the disaccharide lactose or cellobiose into the polymers using acrylamide monomers 2 and 3, respectively. Lactose binds a variety of galectins with KD’s in the low millimolar to high micromolar range43 and can be used at high concentrations to inhibit galectin binding to membrane-associated glycoproteins and glycolipids.44 Oligomeric forms of lactose, however, bind multimeric galectins with considerably higher avidities,47 a phenomenon that has been shown to govern natural galectin–ligand interactions as well.50 Cellobiose, the C-4′ epimer of lactose, was chosen as a structurally similar control ligand for our investigations based on its reported lack of binding to galectin-1, the family member we studied in this work.53
We synthesized lipid-functionalized trithiocarbonate 1 (Scheme 1 and Supporting Information) as a chain transfer agent that we predicted would perform well in living polymerization of acrylamide monomers 2 and 3 based on literature precedents.54 The polymerization was performed in a mixture of water and N,N-dimethylformamide (Scheme 1) and yielded GPs 4 and 5 with PDIs of ∼1.2 in high conversion (>90%). Of note, minimizing the hydrophobicity of the ‘Z’ (i.e., ethyl) group of 1 was important for optimal conversion and PDI. Long alkyl groups at this position, which are commonly employed in chain transfer agents, resulted in aggregation of the growing polymer chain during the reaction. The trithiocarbonate end groups of 4 and 5 were cleaved with sodium borohydride, and the resulting free sulfhydryl groups were conjugated with maleimide-functionalized Alexa Fluor 488 (6 and 7) or Alexa Fluor 555 (8 and 9). These fluorescent lipid-functionalized GPs were used in all subsequent experiments.
Scheme 1. RAFT Polymerization and Functionalization of GPs.
We next sought to display polymers 6 and 7 on live cells for galectin binding studies. Most cell types express endogenous glycoproteins that possess galectin-binding N-acetyllactosamine (LacNAc, Galβ1,4GlcNAc) residues.55 In this initial study, we tried to minimize the impact of endogenous ligands on galectin–GP interactions by choosing a cell line that is deficient in galactosides, the ldlD Chinese Hamster Ovary (CHO) cell mutant.56ldlD CHO cells were incubated with 6 or 7 for 50 min at rt and imaged using fluorescence microscopy to assess cell surface incorporation. Consistent with previous studies,41 both GPs produced robust fluorescence localized at the cell membrane as well as within endocytic vesicles (Figure 2B and F, and Figures S1, S6, and S7).
Figure 2.
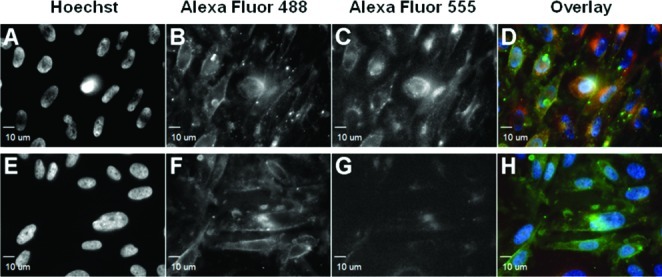
Fluorescence microscopy of ldlD CHO cells treated with GP 6 (A–D) or 7 (E–H) followed by fluorescently labeled Galectin-1, Gal-1-555 (D and G). Hoechst 33342 was used to stain the nuclei (A and E). Galectin-1 binding was observed on cells incubated with GP 6 (C) but not with GP 7 (G).
For our initial studies we chose galectin-1, a homodimer with two carbohydrate recognition domains (CRDs) at opposite poles of its 3-D structure.57 The distance between the two CRDs is ∼5 nm, well below the Förster radius, Ro, for the FRET pair Alexa Fluor 488 and 555. To explore galectin-1’s ability to bind GPs 6 and 7 when displayed on live cells, galectin-1 was fluorescently labeled with Alexa Fluor 555 (generating Gal-1-555) as previously described.58ldlD CHO cells previously treated with either 6 or 7 were incubated with labeled galectin-1, imaged using fluorescence microscopy or analyzed by flow cytometry.
Cells displaying lactosyl GP 6 showed significant Gal-1-555 binding (Figure 2C), whereas cells displaying cellobiosyl GP 7 did not (Figure 2G), mirroring the known monomeric ligand preference of galectin-1. Flow cytometry analysis of cells treated similarly gave comparable results (Figure S2), although a low amount of Gal-1-555 binding to cell-associated GP 7 was observed. It is likely that galectin-1’s interaction with cellobiose, though too weak to detect at the monomer level, becomes discernible with multivalent polymers. Overall, these results show that cells deficient in endogenous ligands can be engineered using synthetic GPs to bind galectin-1.
The impact of galectin-1 binding on the oligomerization state and mobility of bound GPs was assessed using fluorescence lifetime measurements by time-correlated single-photon counting (TCSPC) and FCS. The principle of the experiment is as follows. GPs 6 and 8 possess identical backbone and glycan structures but disparate dyes that constitute a FRET pair. The GP pair can be codisplayed on cell membranes in a 1:1 ratio. In the ns regime, fluorescence lifetime measurements can probe for galectin-1-mediated cross-linking since the depletion of excited states of donor 6 by proximal acceptor 8 (d < Ro) decreases the overall fluorescence lifetime (τFL) of 6.59 Further, FCS operates by performing an autocorrelation analysis on the fluorescence fluctuations in an ∼1 fL excitation volume over many time scales, ns to s.60 A diffusion time (τD) parameter can be extracted from the autocorrelation function, ultimately quantifying the relative mobility of the GPs on the cell surface. The formation of GP clusters would be implied by observed increases in diffusion time.
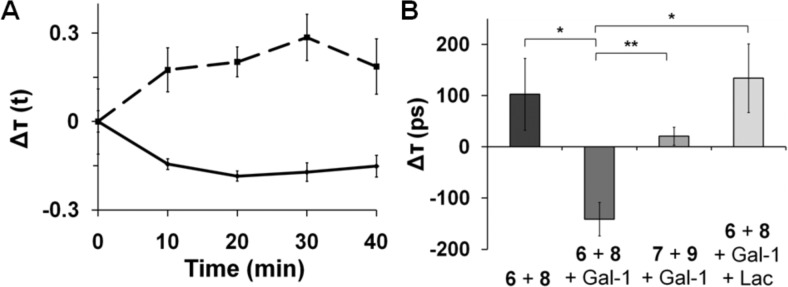
ldlD CHO cells were first incubated with GPs 6 and 8 (1:1 ratio), and the time-resolved fluorescence intensity of donor 6 was monitored on single live cells with a ps laser pulse at 10 MHz in the presence or absence of unlabeled galectin-1.61 Fluorescence lifetime and diffusion time were calculated at 10-min intervals. In the absence of galectin-1, donor 6’s fluorescence lifetime increased with time (Figure 3A). We attribute this phenomenon to endocytosis of the GPs, as evident in our microscopy images. As a consequence, the GPs’ density on the cell surface decreases over time, which would reduce the background level of FRET among unclustered GP molecules. Indeed, at lower temperatures at which endocytosis is slower (11–13 °C), the fluorescence lifetime of donor 6 increased at a slower rate (Figure S3).
Figure 3.
Fluorescence lifetime measurements of GPs on ldlD CHO cells. (A) Fluorescence lifetime, Δτ(t), measurements of donor 6 in the absence (dotted) or presence (solid) of galectin-1 as a function of time. Δτ(t) represents the difference between τ(t) and τ(0). Error bars indicate the standard deviation. (B) The difference in fluorescence lifetime (Δτ) between t = 20 and t = 0 min, averaged over six different cells. *P < 0.01; **P < 0.005. Gal-1: unlabeled galectin-1; Lac: 200 mM lactose. Error bars indicate the standard error.
Next, we monitored the fluorescence lifetime of donor 6 after adding galectin-1 to cells labeled with 6 and 8. We observed a marked decrease in fluorescence lifetime of donor 6 over a period of 40 min (Figure 3A). We repeated the experiment on six different cells and observed similar results; the average changes in fluorescence lifetime are shown in Figure 3B. These observations suggest that cross-linking of donor 6 and acceptor 8 by galectin-1 enhanced FRET and, consequently, decreased the fluorescence lifetime of donor 6. We performed a comparable experiment using the cellobiosyl GP FRET pair 7 and 9. Despite the presence of galectin-1, the fluorescence lifetime of donor 7 increased with time, consistent with endocytosis and little cross-linking. The observed increase was not as dramatic as that observed in the absence of galectin-1, probably reflecting GP 7’s weak interaction with the protein as previously observed by flow cytometry (Figure S2). Importantly, the galectin-1-dependent decrease in fluorescence lifetime of GP 6 was entirely inhibitable by soluble lactose (200 mM) (Figure 3B). In the presence of this galectin-1 competitor, no significant galectin-1-mediated cross-linking was observed. Additional evidence for a direct interaction between galectin-1 and donor 6 was demonstrated through a FRET experiment with fluorescently labeled galectin-1 (Figure S3).
Using the same data acquired for fluorescence lifetime measurements, diffusion times were calculated for donor 6 over the 40-min time course in the presence or absence of galectin-1. Examples of autocorrelation functions (A and B) and diffusion time values (C) are shown in Figure 4. In the absence of galectin-1, diffusion times for donor 6 were relatively stable (Figure 4C), indicating that the mobility of 6 on living cells does not change significantly over time. Notably, the diffusion time for donor 6 noticeably increased over time after addition of galectin-1. This dramatic galectin-1-dependent decrease in donor 6’s mobility provides evidence for oligomerization on the cell surface. As before, we averaged the diffusion time values from six different cells and calculated the difference between those values at t = 20 and 0 min (Figure 4D). The data confirm that galectin-1 increased the diffusion time of donor 6, which is indicative of reduced mobility and oligomerization of the GP. Free lactose abrogated galectin-1’s influence on mobility, consistent with a glycan-binding mechanism of oligomerization. In the presence of 200 mM lactose, the change in diffusion time mirrored that of donor 6 in the absence of galectin-1.
Figure 4.
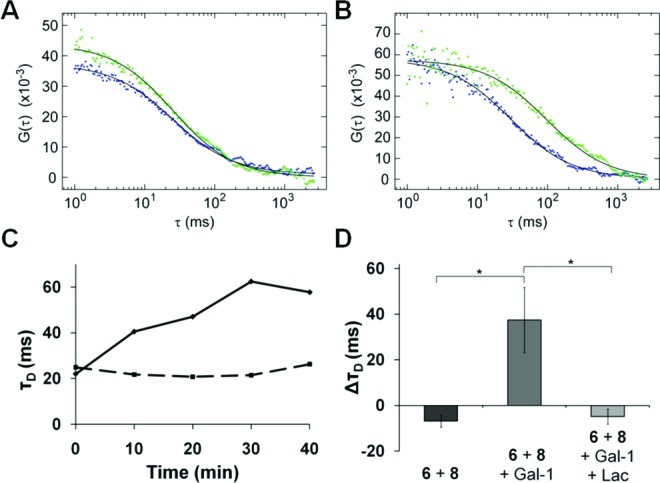
FCS of GP 6 on ldlD CHO cells. Autocorrelation curves for donor 6 in the absence (A) or presence (B) of galectin-1 at 0 min (blue) and 20 min (green). (C) Diffusion time, τD, for donor 6 in the absence (dotted) or presence (solid) of galectin-1. (D) The difference in diffusion time (ΔτD) of donor 6 at t = 20 and 0 min averaged over six different cells. *P < 0.05. Gal-1: unlabeled galectin-1; Lac: 200 mM lactose. Error bars indicate the standard error.
In conclusion, we have established a new methodology for investigating galectin–glycan interactions on live cell membranes using fluorescently labeled GPs in conjunction with FRET and FCS. To our knowledge, we have also provided the first experimental evidence for galectin-1-mediated cross-linking from the perspective of the bound ligand. Studies are underway addressing the ability of different members of the galectin family to induce such cross-linking. The information should shed light on the dimensions and dynamics of putative galectin lattices on the cell surface as well as the effects of the glycan structure and presentation on galectin recognition and cross-linking. This approach could ultimately provide insight into how the various galectin members exert different signaling and organizational functions through cell surface interactions. Beyond galectin biology, we envision applications in the study of glycan–receptor interactions between two cells, wherein changes in oligomerization of receptor-bound GPs might reveal the preferred cluster size of the associated glycan-binding protein. As well, the platform can be extended to cell-surface interactions not involving glycans, as the polymers are wholly synthetic and can be adorned with any ligand type.
Acknowledgments
We thank Dr. Linda Baum for providing the plasmid encoding human galectin-1 and Jason Hudak and Ramray Bhat for helpful discussions and critical reading of the manuscript. This work was funded by a grant from the NIH to C.R.B. (GM59907) and in part by the Director, Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division under Contract No. DE-AC02-05CH11231. B.B. was supported by an NSF predoctoral fellowship.
Supporting Information Available
Experimental procedures, characterization data, supporting figures, and schemes. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Groves J. T.; Kuriyan J. Nat. Struct. Mol. Biol. 2010, 17, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg A. J.; Li H.; Patel D. J.; Allis C. D. Nat. Rev. Mol. Cell Biol. 2007, 8, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N. W.; Barondes S. H. Glycobiology 1999, 9, 979. [DOI] [PubMed] [Google Scholar]
- Rabinovich G. A.; Toscano M. A.; Jackson S. S.; Vasta G. R. Curr. Opin. Struct. Biol. 2007, 17, 513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lella S.; Sundblad V.; Cerliani J. P.; Guardia C. M. A.; Estrin D. A.; Vasta G. R.; Rabinovich G. A. Biochemistry 2011, 50, 7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriou M.; Granovsky M.; Quaggin S.; Dennis J. W. Nature 2001, 409, 733. [DOI] [PubMed] [Google Scholar]
- Liu F. T.; Rabinovich G. A. Nat. Rev. Cancer 2005, 5, 29. [DOI] [PubMed] [Google Scholar]
- Partridge E. A.; Le Roy C.; Di Guglielmo G. M.; Pawling J.; Cheung P.; Granovsky M.; Nabi I. R.; Wrana J. L.; Dennis J. W. Science 2004, 306, 120. [DOI] [PubMed] [Google Scholar]
- a Ahmad N.; Gabius H.-J.; André S.; Kaltner H.; Sabesan S.; Roy R.; Liu B.; Macaluso F.; Brewer C. F. J. Biol. Chem. 2004, 279, 10841. [DOI] [PubMed] [Google Scholar]; b Bourne Y.; Bolgiano B.; Liao D.-I.; Strecker G.; Cantau P.; Herzberg O.; Feizi T.; Cambillau C. Nat. Struct. Biol. 1994, 1, 863. [DOI] [PubMed] [Google Scholar]
- Nieminen J.; Kuno A.; Hirabayashi J.; Sato S. J. Biol. Chem. 2006, 282, 1374. [DOI] [PubMed] [Google Scholar]
- Moiseeva E. P.; Williams B.; Goodall A. H.; Samani N. J. Biochem. Biophys. Res. Commun. 2003, 310, 1010. [DOI] [PubMed] [Google Scholar]
- Argüeso P.; Guzman-Aranguez A.; Mantelli F.; Cao Z.; Ricciuto J.; Panjwani N. J. Biol. Chem. 2009, 284, 23037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kiessling L. L.; Gestwicki J. E.; Strong L. E. Curr. Opin. Chem. Biol. 2000, 4, 696. [DOI] [PubMed] [Google Scholar]; b Coullerez G.; Seeberger P. H.; Textor M. Macromol. Biosci. 2006, 6, 634. [DOI] [PubMed] [Google Scholar]
- Courtney A. H.; Puffer E. B.; Pontrello J. K.; Yang Z. Q.; Kiessling L. L. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal G. B.; Mammen M.; Dahmann G.; Whitesides G. M. J. Am. Chem. Soc. 1996, 118, 3789. [Google Scholar]
- Rawat M.; Gama C. I.; Matson J. B.; Hsieh-Wilson L. C. J. Am. Chem. Soc. 2008, 130, 2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godula K.; Rabuka D.; Nam K. T.; Bertozzi C. R. Angew. Chem., Int. Ed. 2009, 48, 4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabuka D.; Forstner M. B.; Groves J. T.; Bertozzi C. R. J. Am. Chem. Soc. 2008, 130, 5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moad G.; Rizzardo E.; Thang S. H. Aust. J. Chem. 2005, 58, 379. [Google Scholar]
- Hirabayashi J.; Hashidate T.; Arata Y.; Nishi N.; Nakamura T.; Hirashima M.; Urashima T.; Oka T.; Futai M.; Muller W. E.; Yagi F.; Kasai K. Biochim. Biophys. Acta, Gen. Subj. 2002, 1572, 232. [DOI] [PubMed] [Google Scholar]
- Grigorian A.; Demetriou M. In Methods in Enzymology; Elsevier: 2010; Vol. 480, pp 245–266. [DOI] [PubMed] [Google Scholar]
- a Gouin S. G.; García Fernández J. M.; Vanquelef E.; Dupradeau F.-Y.; Salomonsson E.; Leffler H.; Ortega-Muñoz M.; Nilsson U. J.; Kovensky J. ChemBioChem 2010, 11, 1430. [DOI] [PubMed] [Google Scholar]; b Vrasidas I.; Andre S.; Valentini P.; Bock C.; Lensch M.; Kaltner H.; Liskamp R. M. J.; Gabius H.-J.; Pieters R. J. Org. Biomol. Chem. 2003, 1, 803. [DOI] [PubMed] [Google Scholar]
- a Lundquist J. J.; Toone E. J. Chem. Rev. 2002, 102, 555. [DOI] [PubMed] [Google Scholar]; b Collins B. E.; Paulson J. C. Curr. Opin. Chem. Biol. 2004, 8, 617. [DOI] [PubMed] [Google Scholar]
- Lantéri M.; Giordanengo V.; Hiraoka N.; Fuzibet J.-G.; Auberger P.; Fukuda M.; Baum L. G.; Lefebvre J.-C. Glycobiology 2003, 13, 909. [DOI] [PubMed] [Google Scholar]
- Lowe A. B.; McCormick C. L. Prog. Polym. Sci. 2007, 32, 283. [Google Scholar]
- Varki A.; Cummings R. D.; Esko J. D.; Freeze H. H.; Stanley P.; Bertozzi C. R.; Hart G. W.; Etzler M. E.. Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2008. [PubMed] [Google Scholar]
- Kingsley D. M.; Kozarsky K. F.; Hobble L.; Krieger M. Cell 1986, 44, 749. [DOI] [PubMed] [Google Scholar]
- López-Lucendo M. F.; Solís D.; André S.; Hirabayashi J.; Kasai K.; Kaltner H.; Gabius H.-J.; Romero A. J. Mol. Biol. 2004, 343, 957. [DOI] [PubMed] [Google Scholar]
- Stowell S. R.; Dias-Baruffi M.; Penttila L.; Renkonen O.; Nyame A. K.; Cummings R. D. Glycobiology 2003, 14, 157. [DOI] [PubMed] [Google Scholar]
- Levitt J. A.; Matthews D. R.; Ameer-Beg S. M.; Suhling K. Curr. Opin. Biotechnol. 2009, 20, 28. [DOI] [PubMed] [Google Scholar]
- Kim S. A.; Heinze K. G.; Schwille P. Nat. Methods 2007, 4, 963. [DOI] [PubMed] [Google Scholar]
- ldlD CHO cells were washed with 200 mM lactose prior to incubation with GPs in order to remove endogenous galectins.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.