Abstract
Our study is the first report of exceptional Mycobacterium bovis strains that have some characteristics of M. tuberculosis. The strains were isolated from 8 patients living in Kazakhstan. While molecular markers were typical for M. bovis, growth characteristics and biochemical test results were intermediate between M. bovis and M. tuberculosis.
Keywords: tuberculosis, Mycobacterium bovis, Kazakhstan, resistance, dispatch
Mycobacterium bovis causes tuberculosis (TB) mainly in cattle but has a broad host range and causes disease similar to that caused by M. tuberculosis in humans (1). It belongs to the M. tuberculosis complex (MTBC) that comprises the closely related human pathogens M. tuberculosis and M. africanum (2). Identification of M. bovis traditionally has been based on clear-cut differences in phenotypic characteristics and biochemical properties when compared to the other members of the MTBC (1,2). M. bovis shows a dysgonic colony shape on Lowenstein-Jensen medium, is negative for niacin accumulation and nitrate reduction, is susceptible to thiophene-2-carboxylic acid hydrazide (TCH), and shows microaerophilic growth on Lebek medium (1–3). A further criterion used for differentiation is the intrinsic resistance to pyrazinamide, which is found in most M. bovis isolates (1–3). In contrast, M. tuberculosis shows eugonic growth, is positive for niacin accumulation and nitrate reduction, is resistant to TCH, shows aerophilic growth on Lebek medium, and is usually not monoresistant to pyrazinamide (2,3).
More recently, several molecular methods have been developed that provide clear criteria for the identification of M. bovis. These comprise a variety of polymerase chain reaction (PCR) methods, e.g., based on DNA sequence variations in the direct repeat region of MTBC complex strains (spoligotyping [4]) or on single nucleotide polymorphisms (SNPs) in either the oxyR gene (5) or the gyrB gene (6). Furthermore, MTBC isolates can be differentiated by large sequence polymorphisms or regions of difference (RD), and according to their distribution in the genome, a new phylogenetic scenario for the different species of the MTBC has been suggested (7–9). The presence or absence of particular deletions has been proposed as being discriminative, e.g., lack of TdB1 for M. tuberculosis or lack of RD12 for M. bovis.
In routine diagnostics, the combination of phenotypic characteristics and biochemical features is sufficient to differentiate clinical M. bovis isolates, and in general, the results obtained are unambiguous. However, here we describe the characteristics of 8 strains of the MTBC that showed an unusual combination of phenotypic and biochemical attributes of both M. bovis and M. tuberculosis. Molecular analyses confirmed the strains as M. bovis, which in part have phenotypic and biochemical properties of M. tuberculosis.
The Study
During a previous investigation of 179 drug-resistant isolates from Kazakhstan (10), we determined the presence of 8 strains showing monoresistance to pyrazinamide. Kazakhstan is the largest of the central Asian republics and is divided regionally into 14 oblasts. The investigation was performed as part of a nationwide drug resistance survey conducted by the national TB program of Kazakhstan with assistance from the World Health Organization/International Union against Tuberculosis and Lung Disease Global Project in 2001. The subset of this survey investigated here (n = 158) represents 100% of strains resistant to isoniazid, rifampin, ethambutol, or streptomycin isolated in 9 of the 14 Kazakhstan oblasts during the study period; 21 samples had fungal contamination or showed no growth (10). All strains were isolated from sputum samples.
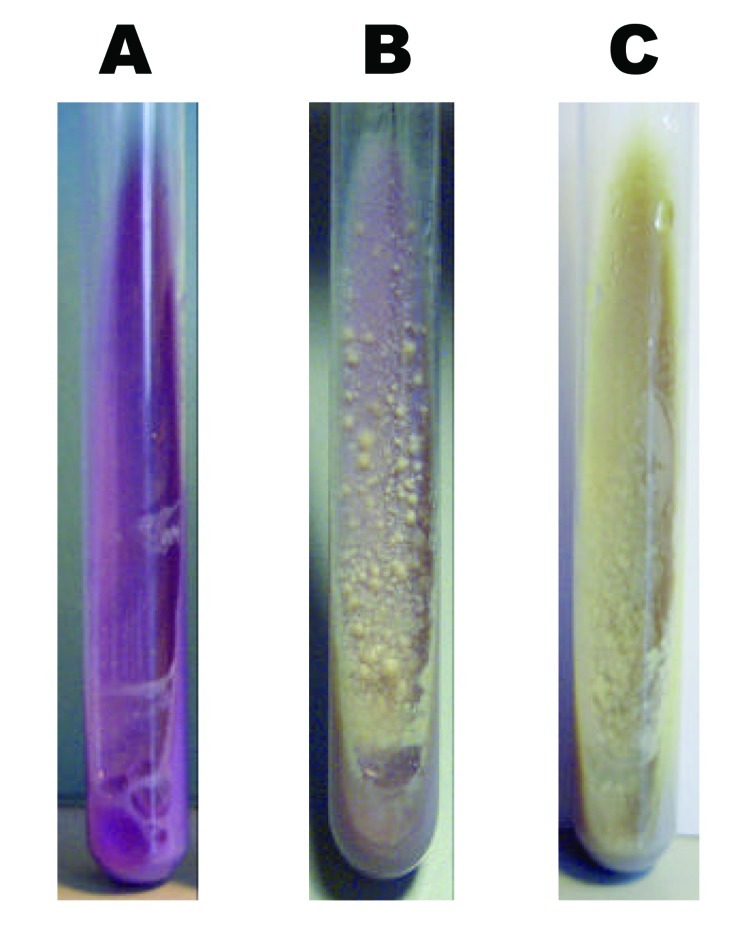
To further clarify if these strains were monoresistant M. tuberculosis or M. bovis isolates intrinsically resistant to pyrazinamide, we performed several routine diagnostic tests traditionally used for species differentiation (6). All strains showed eugonic growth characteristics on Lowenstein-Jensen slants and on bromcresol purple medium (Figure), which in general is typical for M. tuberculosis. However, on bromcresol purple medium, classic M. tuberculosis isolates induce a pH-dependent change of color from blue to yellow, which was not observed in these cases (Figure). Furthermore, all 8 isolates were positive for niacin accumulation, negative for nitrate reduction, susceptible to TCH, and showed aerophilic growth on Lebek medium. Considering all results, the 8 strains showed a combination of test results that did not allow a clear differentiation as M. bovis or as M. tuberculosis (Table 1). Such a combination of test results would apply best to M. africanum, a species from which more variable test results have been reported (3). However, this species was probably was not isolated because M. africanum strains are usually not monoresistant to pyrazinamide (3).
Figure.
Growth morphology on bromcresol purple medium of Mycobacterium bovis (A), M. tuberculosis (C), and 1 of the strains analyzed (B).
Table 1. Phenotypic characteristics of type strains Mycobacterium tuberculosis H37 (ATCC 27294), M. bovis (ATCC19210), M. africanum (ATCC25420), and the strains analyzed*.
| Strains | Colony morphology† | Test result |
|||||
|---|---|---|---|---|---|---|---|
| TCH‡ | PZA | Niacin accumulation | Nitrate reduction | Change of color of bromcresol medium | Growth on Lebek medium | ||
| Kazakhstan (n = 8)§ | Eugonic | S | R | + | – | – | Aerophilic |
| ATCC M. bovis | Dysgonic | S | R | – | – | – | Microaerophilic |
| ATCC H37 | Eugonic | R | S | + | + | + | Aerophilic |
| ATCC M. africanum | Dysgonic | S | S | + | + | – | Microaerophilic |
*TCH, thiophene-2-carboxylic acid hydrazide; PZA, pyrazinamide; +, positive test result; –, negative test result; S, susceptible; R, resistant. †Growth characteristics on Lowenstein-Jensen and bromcresol purple slants. ‡Growth in presence of TCH. §All 8 strains showed identical test results.
Therefore, we investigated all strains with several molecular techniques previously used for differentiation within the MTBC (Table 2). They all had identical spoligotype patterns (hexcode 6B-57-5F-7F-FF-60, performed according to the methods of Kamerbeek et al. [4]), that lacked spacers 39–43 and identical IS6110 DNA fingerprint patterns with 2 IS6110 copies (data not shown, performed according to the methods of van Embden et al. [11]). All isolates carried the M. bovis–specific polymorphism in the oxyR gene (5), and none of them had the M. bovis BCG– specific deletion in the RD1 region (12). PCR analysis of other RDs (RD3, RD4, RD5, RD9, RD10, RD12, TbD1, and IS1541) showed results typical for M. bovis when compared with the RD signatures of the American Type Culture Collection strains of M. tuberculosis, M. africanum, and M. bovis (Table 2) and with previously published data (7,8). The intrinsic resistance to pyrazinamide was confirmed by DNA sequence analysis as all strains carried the M. bovis–specific single point mutation at nucleotide position 169 of the pncA gene.
Table 2. Genetic characteristics of type strains Mycobacterium tuberculosis H37 (ATCC 27294), M. bovis (ATCC19210), M. africanum (ATCC25420), and the strains analyzed*.
| Strains | Test result |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TbD1 | RD1 | RD3 | RD4 | RD5 | RD9 | RD10 | RD12 | IS1561 | oxyR† | gyrB‡ | |
| Kazakhstan (n = 8)§ | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | M. bovis |
| M. bovis ATCC | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | M. bovis |
| M. tuberculosis H37 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | M. tuberculosis |
| M. africanum ATCC | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | M. africanum |
*RD, region of difference; 0, region deleted; 1, region present. †Presence of oxyR mutation G to A at position 285; 1, polymorphism present; 0, polymorphism not present. ‡Classification according to gyrb-polymerase chain reaction restriction fragment length polymorphism analysis (Niemann et al. [3]). §All 8 strains showed identical test results.
Seven of the 8 strains were isolated from 30- to 55-year-old men, and 1 strain was from a 72-year-old woman. All but 1 patient had a history of previous antituberculosis treatment, but none of the strains showed any further resistance (data not shown). The patients originated from the oblast of Kostanajskaya in north Kazakhstan. Among all patients, no direct epidemiologic links could be established. However, 3 of the patients lived in the city of Kostanaj, while 5 came from rural areas. Before 1950, the Kazakh Steppe was a broad, continuous belt of grassland that stretched from the Ural River to the Altai foothills, covering large parts of Kostanajskaya; after the 1950s, the region was used extensively for agriculture. Information on contact with animals is not available, since cattle herds are only kept privately.
Conclusions
We describe 8 strains of M. bovis with exceptional phenotypic characteristics that are intermediate between M. tuberculosis and M. bovis. This fact initially complicated a clear species differentiation; however, the battery of molecular tests performed clearly confirmed all strains as M. bovis. These tests included the presence of characteristic single nucleotide polymorphisms as well as an RD profile that is typical for the M. bovis lineage of the MTBC (6,7). To our knowledge, this is the first report describing M. bovis isolates with phenotypic characteristics and biochemical properties of M. tuberculosis. In our previous investigation of 176 M. bovis strains from Germany, all strains had phenotypic characteristics typical of M. bovis, and no strains similar to the isolates from Kazakhstan could be identified (13). The same result applies for the spoligotype patterns, as none of the strains in our database had an identical spoligotype pattern (data not shown). A further comparison with the international M. bovis spoligotype database (available from http://www.mbovis.org/spoligodatabase) identified 1 strain isolated in Argentina with an identical spoligotype pattern; however, no further information about his strain is available.
Whether the 8 strains analyzed represent strains of an ancestral phylogenetic lineage of M. bovis that might have been conserved because of the geographic isolation of that region of Kazakhstan or whether they gained their special characteristics by new mutations is a question that cannot be answered by the data obtained in this study. All strains have been isolated from humans. We cannot say if we have found an exceptional outbreak of a particular M. bovis strain or if the patients were infected directly by wildlife, livestock, or food, and the disease developed by chance during the study. However, an overall percentage of ≈5% of all resistant strains investigated in this study indicates that these isolates may be important in Kazakhstan. This also poses the question of whether these strains might become more virulent in humans if they acquired phenotypic/biochemical characteristics usually observed exclusively in M. tuberculosis. However, to address this question more precisely, longitudinal studies on the population structure of MTBC isolates in Kazakhstan obtained from humans and animals, in combination with experiments in virulence model systems, will be necessary. In any case, these strains represent ideal model organisms for analyzing the nature of the biologic differences observed between M. bovis and M. tuberculosis. To ensure a clear differentiation from other M. bovis strains, we suggest the name M. bovis subtype "Almaty" for this genotype. Almaty is the former capital and largest city of Kazakhstan.
Biography
Dr Kubica is a physician and working in a postdoctoral position at the German National Research Center for Mycobacteria, Research Center Borstel, Borstel, Germany. Her major research interests include molecular epidemiology and the diagnosis and treatment of mycobacterial infections.
Footnotes
Suggested citation for this article: Kubica T, Agzamova R, Wright A, Rakishev G, Rüsch-Gerdes S, Niemann S. Mycobacterium bovis isolates with intermediate M. tuberculosis characteristics. Emerg Infect Dis [serial on the Internet]. 2006 May [date cited]. http://dx.doi.org/10.3201/eid1205.050200
References
- 1.Ayele WY, Neill SD, Zinsstag J, Weiss MG, Pavlik I. Bovine tuberculosis: an old disease but a new threat to Africa. Int J Tuberc Lung Dis. 2004;8:924–37. [PubMed] [Google Scholar]
- 2.Wayne LG, Kubica GP. The mycobacteria. In: Sneath PHA and Holt JG, editors. Bergeys manual of systematic bacteriology, vol. 2. Baltimore: Williams & Wilkins Co.; 1986. p. 1435–57. [Google Scholar]
- 3.Niemann S, Richter E, Rüsch-Gerdes S. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J Clin Microbiol. 2000;38:152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strains differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sreevatsan S, Escalante P, Pan X, Gillies DA II, Siddiqui S, Khalaf CN, et al. Identification of a polymorphic nucleotide in oxyR specific for Mycobacterium bovis. J Clin Microbiol. 1996;34:2007–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niemann S, Harmsen D, Rüsch-Gerdes S, Richter E. Differentiation of clinical Mycobacterium tuberculosis complex strains by gyrB DNA sequence polymorphism analysis. J Clin Microbiol. 2000;38:3231–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99:3684–9. 10.1073/pnas.052548299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huard RC, de Oliveira Lazzarini LC, Butler WR, van Soolingen D, Ho JL. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J Clin Microbiol. 2003;41:1637–50. 10.1128/JCM.41.4.1637-1650.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J Infect Dis. 2002;186:74–80. 10.1086/341068 [DOI] [PubMed] [Google Scholar]
- 10.Kubica T, Agzamova R, Wright A, Aziz MS, Rakishev G, Bismilda V, et al. The Beijing genotype is a major cause of drug resistant tuberculosis in Kazakhstan. Int J Tuberc Lung Dis. 2005;9:646–53. [PubMed] [Google Scholar]
- 11.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talbot EA, Williams DL, Frothingham R. PCR identification of Mycobacterium bovis BCG. J Clin Microbiol. 1997;35:566–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubica T, Rüsch-Gerdes S, Niemann S. Mycobacterium bovis subsp. caprae caused one-third of human M. bovis–associated tuberculosis cases reported in Germany between 1999 and 2001. J Clin Microbiol. 2003;41:3070–7. 10.1128/JCM.41.7.3070-3077.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]