Abstract
Here we report that the transcription factor CREB-H is required for the maintenance of normal plasma triglyceride (TG) levels. CREB-H deficient mice displayed hypertriglyceridemia (HTG) secondary to inefficient TG clearance catalyzed by lipoprotein lipase (Lpl), partly due to defective expression of the Lpl coactivators, Apoc2, Apoa4, and Apoa5 and concurrent augmentation of the Lpl inhibitor, Apoc3. Multiple nonsynonymous mutations in CREB3L3 that produced hypomorphic or nonfunctional CREB-H protein were identified in patients with extreme HTG, implicating a critical role for CREB-H in human TG metabolism.
Keywords: lipid metabolism, dyslipidemia, CREB-H, transcription factor
CREB-H is an endoplasmic reticulum (ER)-bound transcription factor that is highly and selectively expressed only in the liver and the small intestine (Supplementary Fig. 1a,b)1,2. CREB-H activation requires a sequential cleavage of the precursor protein by Golgi proteases that liberate the mature N-terminal portion of the protein, which localizes to the nucleus to act as a transcriptional transactivator 3,4. It has been recently demonstrated that CREB-H mRNA is induced by metabolic cues such as fasting, fatty acids, and PPARα, suggesting that it might participate in nutrient and energy metabolism 5,6.
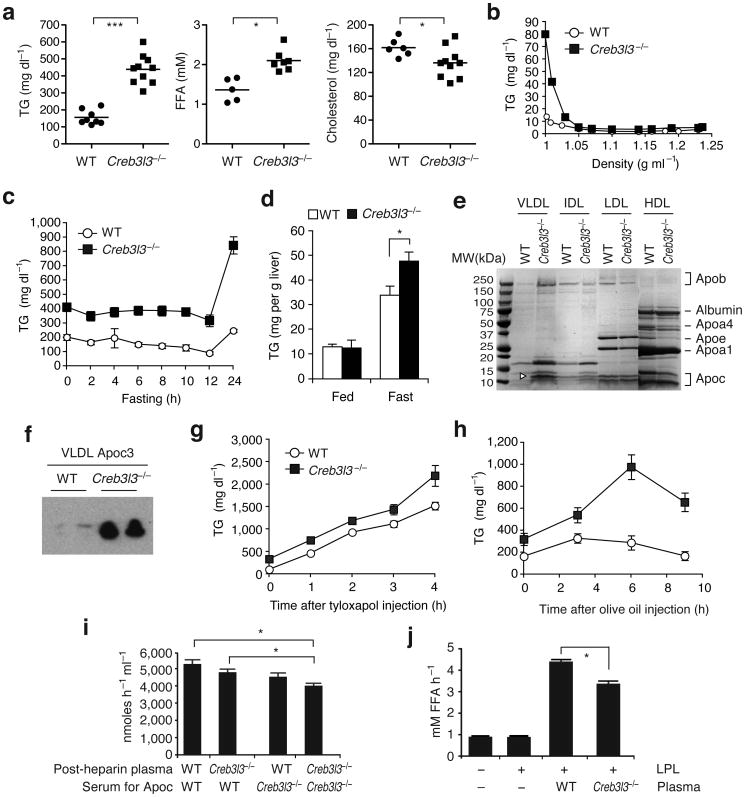
To gain insight into the role of CREB-H in nutrient metabolism, we measured metabolic parameters of CREB-H deficient Creb3l3−/− mice 1. We found that Creb3l3−/− mice had higher plasma TG levels compared to the WT mice (Fig. 1a), suggesting that CREB-H regulates TG metabolism. Plasma concentrations of free fatty acids (FFA) and cholesterol were slightly changed in Creb3l3−/− mice (Fig. 1a). Fractionation of plasma lipoproteins by density gradient ultracentrifugation demonstrated that the very low density lipoprotein (VLDL) fraction of Creb3l3−/− mice had higher TG content compared to that of WT mice (Fig. 1b). In line with the induction of the processed CREB-H(N) by fasting in the liver (Supplementary Fig. 1c,d), the HTG phenotype of Creb3l3−/− mice was exacerbated upon prolonged starvation (Fig. 1c). Fasting-induced hepatic steatosis was also more pronounced in Creb3l3−/− mice than in WT (Fig. 1d). Consistent with higher VLDL TG content in Creb3l3−/− mice versus WT, Apob level was also higher in Creb3l3−/− than in WT mice (Fig. 1e). Notably, Apoc3, an inhibitor of Lpl, was abundant in VLDL particles purified from Creb3l3−/− plasma as determined by mass spectrometry and western blot (Fig. 1e,f).
Figure 1. Creb3l3−/− mice display HTG secondary to inefficient TG clearance catalyzed by Lpl.
(a)Plasma TG, FFA, and cholesterol levels measured after a 16 h fast. Each dot represents an individual mouse. (b) We separated lipoproteins by density gradient ultracentrifugation of pooled plasma (n = 3 per each) after a 24 h fast. TG levels in and density of fractions (1 ml each) sequentially collected from top to bottom. (c) Plasma TG levels in WT (n = 9) and Creb3l3−/− (n = 7) male mice deprived of food, starting at 8 AM. (d) Hepatic TG levels measured at fed state or after a 24h fast. n = 5 per group. (e) After concentration, fractions from (b) were separated on a 4–20% gradient SDS-polyacrylamide gels. The gel was stained by Coomassie Brilliant Blue G-250. Arrowhead indicates Apoc3 identified by mass spectrometry. VLDL, d < 1.006 g ml−1; IDL d = 1.006–1.019 g ml−1; LDL, d = 1.019–1.063 g ml−1; HDL, d = 1.063–1.21 g ml−1. (f) Apoc3 western blot of VLDL fractions. (g) Plasma TG concentrations measured after a 4h fasting followed by i.v. injection with tyloxapol (500 mg kg−1). n = 4 per group. (h) Plasma TG concentrations measure after a 16 h fasting followed by oral gavage of olive oil (10 ml kg−1). n = 6 per group. (i) Post-heparin Lpl activity (n = 6 per group) measured in the presence of heat inactivated serum pooled from WT or Creb3l3−/− mice as Apoc sources. (j) Recombinant LPL was incubated with triolein substrate in the presence of plasma collected from WT or Creb3l3−/− mice. Values represent FFA concentration released from triolein. n = 8 per group. *P < 0.05, ***P < 0.0001, compared to WT mice.
Fasting plasma TG concentration reflects a balance between the production of VLDL from the liver and the clearance of TG-rich lipoproteins from the circulation, governed predominantly by Lpl present on the surface of vascular endothelium in various organs 7. To determine if CREB-H plays a role in VLDL secretion from the liver, we measured the rate of TG accumulation in plasma after administration of an Lpl inhibitor, tyloxapol. The VLDL secretion rate was not significantly different between WT and Creb3l3−/− mice, arguing against a role for CREB-H in VLDL secretion (Fig. 1g). We next examined if CREB-H deficiency led to decreased Lpl-mediated TG clearance. Olive oil gavage resulted in more pronounced HTG in Creb3l3−/− mice than in WT (Fig. 1h), suggesting a defect in TG clearance in the absence of CREB-H. Creb3l3−/− mice exhibited lower postheparin Lpl activity compared to WT mice (Fig. 1i), despite normal Lpl mRNA levels in adipose tissue and skeletal muscle (Supplementary Fig. 2), suggesting that plasma cofactors for Lpl might be limiting in Creb3l3−/− mice. Consistent with this idea, the addition of WT serum as a source of Apoc proteins significantly increased the Lpl activity retained in post-heparin plasma of Creb3l3−/− mice (Fig 1i). The potency of Creb3l3−/− serum in augmenting Lpl-mediated hydrolysis of triolein substrate was also attenuated compared to that of WT serum (Fig. 1i). Further, transfusion of WT but not Creb3l3−/− plasma efficiently reduced TG levels in Creb3l3−/− mice (Supplementary Fig. 3a). Collectively, these data establish that the HTG observed in Creb3l3−/− mice is a consequence primarily of decreased TG clearance from plasma, rather than increased TG output from the liver.
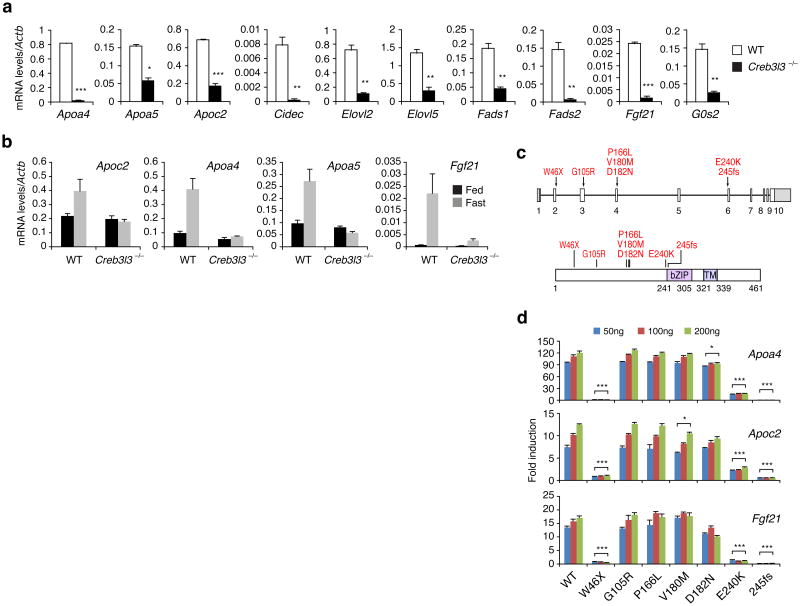
To identify CREB-H target genes that might contribute to the HTG phenotype of Creb3l3−/− mice, we performed microarray analysis of RNAs isolated from WT and Creb3l3−/− mouse liver (Supplementary Fig. 2b). qRT-PCR confirmed lower levels in Creb3l3−/− than in WT liver of Fads1, Fads2, Elovl2, Cidec, Apoc2 and Apoa5, which have been associated with human TG metabolism (Fig. 2a) 8. Hepatic Fgf21, Apoa4, and Elov5, and G0s2 levels were also lower in Creb3l3−/− than in WT mice. Fgf21 is induced by fasting and reduces plasma TG level 9. Fads and Elovl genes encode a fatty acid desaturase and elongase, respectively, which play important roles in the synthesis of long chain polyunsaturated fatty acids and the regulation of fatty acid metabolism 10. G0s2 and Cidec are induced in the liver by fasting, and are known to regulate TG hydrolysis, and lipid droplet formation, respectively 11,12. Apoa4, Apoa5, Apoc2 and Fgf21 were induced in WT liver by fasting, an effect which was abrogated in Creb3l3−/− mice (Fig. 2b). Apoa4 and Apoc2 are normally highly expressed in mouse small intestine. Intestinal mRNA levels of these genes were also lower in Creb3l3−/− compared to WT mice (Supplementary Fig. 2c). Lower plasma Apoa4 protein level in Creb3l3−/− than in WT mice correlated well with Apoa4 mRNA levels in the liver and the intestine (Supplementary Fig. 2d). Apoa5 and Apoc2 activate Lpl, whose activity is further augmented by Apoa4, to facilitate the delivery of hydrolyzed fatty acids to peripheral cells and hence to lower plasma TG levels 13,14. Despite markedly higher VLDL associated-Apoc3 level in Creb3l3−/− than in WT mice, its mRNA levels were not significantly different between these two groups (Supplementary Fig. 2e), suggesting post-transcriptional control of Apoc3 by CREB-H. We postulated that the concomitant reduction of Lpl activators, Apoa4, Apoa5, and Apoc2, and induction of Lpl inhibitor, Apoc3 impaired TG clearance, resulting in higher plasma TG levels in Creb3l3−/− than in WT mice. Indeed, injection of recombinant Apoc2 effectively reduced plasma TG levels in Creb3l3−/− mice (Supplementary Fig. 3b). In contrast, Apoc2 injection only modestly decreased plasma TG levels in WT mice.
Figure 2. CREB-H controls genes involved in TG metabolism and mutations of CREB3L3 are associated with human HTG.
(a)Hepatic mRNA levels determined by qRT-PCR. Mice were fasted for 24h before sacrifice. n = 3 mice per group. Error bars indicate standard deviation. (b) Apoc2, Apoa4, Apoa5 and Fgf21 mRNA levels at fed state or after a 24 h fast in WT and Creb3l3−/− mice as determined by qRT-PCR. n = 4 mice per group. (c) Locations of the nonsynonymous mutations found in individuals with HTG, and predicted amino acid changes. bZIP: basic leucine zipper; TM: transmembrane domain. (d) Hepa1.6 cells were co-transfected with Apoa4, Apoc2, or Fgf21 promoterdriven luciferase reporters and the indicated CREB-H(N) constructs. Values represent fold induction of luciferase activities compared to the reporter only transfection. *P < 0.05, **P < 0.01, ***P < 0.0001.
To further explore the identity of CREB-H target genes involved in TG metabolism, we generated transgenic mice that over-expressed a constitutively active CREB-H(N) in the liver (Supplementary Fig. 4a). Both the endogenously processed and the transgenic CREB-H(N) were expressed as 42 kDa proteins in the liver (Supplementary Fig. 4b). Hepatic CREB-H(N) protein levels were 14-fold higher in the transgenic mice compared with WT littermates, despite that the transgenic mRNA level was only 1.25-fold higher than the endogenous Creb3l3 (Supplementary Fig. 4c), suggesting that ∼10% of CREB-H protein is basally processed to the mature form in the liver of chow diet fed WT mice. CREB-H(N) overexpression strongly induced Apoc2, Apoa4, Fgf21, and Cidec mRNAs (Supplementary Fig. 4d), mirroring the downregulation of these genes in Creb3l3−/− mice. Other apolipoprotein genes located within the same gene clusters as Apoc2 and Apoa4 on chromosomes 7 and 9, respectively, were unaffected by CREB-H(N) overexpression in the transgenic liver. Notably, plasma TG levels were significantly lower in CREB-H(N) transgenic mice than in WT littermates, in contrast to the hypertriglyceridemic phenotype of Creb3l3−/− mice (Supplementary Fig. 4f). CREB-H(N) overexpression by adenoviral infection also decreased plasma TG levels in mice, concomitantly increasing the potency of the serum to stimulate LPL (Supplementary Fig. 4g,h). Transient transfection assays showed that CREB-H strongly transactivated the Apoa4, Apoc2 and Fgf21 promoters (Supplementary Fig. 4i), indicating transcriptional activation of these genes by CREB-H.
Emerging evidence suggests that complex traits such as cholesterol and TG are contributed by both common and rare genetic variants 8,15. Common variants in CREB3L3 were not observed to be associated with TG in recent large genome wide association studies 16. Since CREB-H deficiency caused a dramatic increase in plasma TG with no other overt abnormalities in mice, we asked whether mutations in CREB3L3 could similarly contribute to HTG in humans. Hence, we searched for rare mutations in the CREB3L3 gene in an established clinical cohort with HTG 17. We sequenced CREB3L3 coding regions in 449 unrelated HTG individuals of European Caucasian ancestry (TG > 3.37 mM) and normal LPL, APOC2, and APOA5 genes, as well as in 238 matched normolipidemic controls (TG < 2.3 mM) 17. Remarkably, heterozygous nonsynonymous or insertional mutations in CREB3L3 were identified in 12/449 HTG individuals, but were completely absent from 238 controls (P = 0.01, 2-sided Fisher's exact test, Supplementary Table 1). In contrast, synonymous variants had similar frequency in HTG and control groups (13/238 control versus 12/449 HTG, P = 0.22). All mutations localize to the N-terminal region of the protein preceding the bZIP domain (Fig. 2c). Three unrelated HTG individuals were heterozygous for the same complex mutation (designated 245fs), which consisted of a G insertion in the first nucleotide of codon 245 together with a A>T point mutation 7 nucleotides downstream (Supplementary Fig. 5a). One HTG individual was heterozygous for the nonsense CREB3L3 W46X mutation. Eight HTG probands had heterozygous missense mutations, which included two probands with G105R, two with P166L, one with V180M, one with D182N, and two with E240K mutations (clinical features summarized in Supplementary Table 1). Lipoprotein profiles of the nuclear families of the 4 individuals with CREB3L3 nonsense mutations showed a significantly elevated mean plasma TG level in 11 mutation carriers compared with 5 non-carrier first-degree relatives (9.67 ± 4.70 vs. 1.66 ± 0.55 mM, P = 0.021, Wilcoxon test) (Supplementary Fig. 5b). Finally, low-coverage resequencing of 179 additional multiethnic genomes in the 1000 Genomes project only identified the G105R variant from our sample and one additional variant not observed in our sample 18, suggesting the occurrence of nonsynonymous variants in CREB3L3 is extremely rare in normolipidemic populations.
Mutant CREB-H proteins encoded by these human variants were functionally evaluated in transactivation assays using luciferase reporters driven by the proximal Apoa4, Apoc2 or Fgf21 promoters. The W46X and 245fs mutations precluded translation of the DNA binding and bZIP domains, and as expected, the resulting mutant proteins failed to transactivate any of the reporters (Fig. 2d). The E240K mutation also severely abrogated the induction of all reporters by CREB-H(N) cotransfection. The D182N mutation exhibited modest but reproducible decreases in fold inductions on all reporters, while V180M did so only on the Apoc2 reporter (Fig. 2d). In contrast, the G105R and P166L mutations did not significantly affect CREB-H transactivation of the promoters tested. The precise effects of these mutations on the function of CREB-H and their contribution, if any, to the elevated TG levels in the carriers remain to be investigated. In summary, we demonstrated that rare heterozygous variants in CREB3L3 are significantly associated with the polygenic trait of HTG, similar to other established HTG-associated genes such as LPL and APOA5 17.
In this report, we identify CREB-H as a novel transcription factor that plays a crucial role in TG metabolism. CREB-H is induced by fasting in the liver and required for the expression of a series of genes that play important roles in TG metabolism, which include Apoa4, Apoa5, Apoc2, Cidec, Elovl2, Elovl5, Fads1, Fads2, Fgf21, and G0s2. Creb3l3−/− mice displayed HTG and a defect in Lpl-mediated TG clearance, which might be ascribed to defective expression of Apoc2, Apoa4 and Apoa5 coupled with the augmentation of VLDL-associated Apoc3 with contributions from other yet-to-be-identified CREB-H targets. We further discovered hypomorphic or null mutations in CREB3L3 that were associated with HTG in humans, implicating a critical role for CREB-H in human TG metabolism. The causal relationship between CREB3L3 variations and HTG remains to be determined. It will be informative to test if mutant mice harboring any CREB3L3 mutations found in HTG individuals are predisposed to develop HTG. It will also be of interest to investigate the correlation between plasma TG levels and the frequency of CREB3L3 sequence variations and their effects on CREB-H function in larger and more diverse populations.
CREB-H is structurally related to SREBPs which play major roles in TG and cholesterol biosynthesis in the liver, as they are synthesized as ER-localizing precursor proteins and activated by S1P and S2P proteases in the Golgi apparatus 3,19. CREB-H is also functionally related to PPARα which plays an important role in FA oxidation in the liver 20, as both genes are induced by fasting and have common targets such as Fgf21, Apoa5 and G0s2. Future studies will address the molecular mechanisms by which CREB-H is activated and regulates TG homeostasis in concert with other transcriptional regulatory proteins. Elucidation of the signaling pathways that activate CREB-H may lead to novel therapeutic targets to treat human dyslipidemias.
Supplementary Material
Acknowledgments
We thank. B. Zhai and S. Gygi for mass spectrometry analysis, K. Sigrist for assistance with the development of transgenic mice, R. Hassell for assistance with genomic DNA sequencing, B. Monia for Apoc3 antibody (Isis Innovation Ltd) and D. Cohen for invaluable suggestions and comments on the manuscript. The research was supported by the Harvard University Accelerator Fund (L.H.G), an HHMI fellowship (P. G), the Harvard Digestive Disease Center, NIH Grant P30 DK34845 (A.-H.L), NIH grant DK082448 (L.H.G.) and grants from the American Heart Association (A.-H.L. and J.H.L.), Canadian Institutes for Health Researc,h and Genome Canada through the Ontario Genomics Institute.
Footnotes
Author contributions: A.-H.L. designed the experiments. J.H.L. performed in vivo experiments using Creb3l3−/− mice, and mutational analysis of CREB-H protein. A.-H.L. and P.G. generated and characterized CREB-H(N) transgenic mice. J.D.B. performed post-heparin LPL assays. J.W. and C.T.J. performed sequencing experiments. S.A.D. provided Creb3l3−/− mice. A.H.L., L.H.G. R.A.H. and J.P. analyzed the data. A.-H.L., L.H.G. and R.A.H. wrote the manuscript.
References
- 1.Luebke-Wheeler J, et al. Hepatocyte nuclear factor 4alpha is implicated in endoplasmic reticulum stress-induced acute phase response by regulating expression of cyclic adenosine monophosphate responsive element binding protein H. Hepatology. 2008;48:1242–1250. doi: 10.1002/hep.22439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omori Y, et al. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liverspecific expression. Nucleic Acids Res. 2001;29:2154–2162. doi: 10.1093/nar/29.10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 4.Bailey D, Barreca C, O'Hare P. Trafficking of the bZIP transmembrane transcription factor CREB-H into alternate pathways of ERAD and stressregulated intramembrane proteolysis. Traffic. 2007;8:1796–1814. doi: 10.1111/j.1600-0854.2007.00654.x. [DOI] [PubMed] [Google Scholar]
- 5.Danno H, et al. The liver-enriched transcription factor CREBH is nutritionally regulated and activated by fatty acids and PPARalpha. Biochem Biophys Res Commun. 2010;391:1222–1227. doi: 10.1016/j.bbrc.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 6.Lee MW, et al. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 2010;11:331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;43:1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- 8.Hegele RA. Plasma lipoproteins: genetic influences and clinical implications. Nat Rev Genet. 2009;10:109–121. doi: 10.1038/nrg2481. [DOI] [PubMed] [Google Scholar]
- 9.Kliewer SA, Mangelsdorf DJ. Fibroblast growth factor 21: from pharmacology to physiology. Am J Clin Nutr. 2010;91:254S–257S. doi: 10.3945/ajcn.2009.28449B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillou H, Zadravec D, Martin PG, Jacobsson A. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog Lipid Res. 2010;49:186–199. doi: 10.1016/j.plipres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Gong J, Sun Z, Li P. CIDE proteins and metabolic disorders. Curr Opin Lipidol. 2009;20:121–126. doi: 10.1097/MOL.0b013e328328d0bb. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, et al. The G(0)/G(1) switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010;11:194–205. doi: 10.1016/j.cmet.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg IJ, Scheraldi CA, Yacoub LK, Saxena U, Bisgaier CL. Lipoprotein ApoC-II activation of lipoprotein lipase. Modulation by apolipoprotein A-IV. J Biol Chem. 1990;265:4266–4272. [PubMed] [Google Scholar]
- 14.Jong MC, Hofker MH, Havekes LM. Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler Thromb Vasc Biol. 1999;19:472–484. doi: 10.1161/01.atv.19.3.472. [DOI] [PubMed] [Google Scholar]
- 15.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansen CT, Kathiresan S, Hegele RA. Genetic determinants of plasma triglycerides. J Lipid Res. 2011;52:189–206. doi: 10.1194/jlr.R009720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genomes Project C, et al. A map of human genome variation from populationscale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duval C, Muller M, Kersten S. PPARalpha and dyslipidemia. Biochim Biophys Acta. 2007;1771:961–971. doi: 10.1016/j.bbalip.2007.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.