Abstract
A number of diabetogenic stimuli interact to influence insulin promoter activity, making it an attractive target for both mechanistic studies and therapeutic interventions. High-throughput screening (HTS) for insulin promoter modulators has the potential to reveal novel inputs into the control of that central element of the pancreatic β-cell. A cell line from human islets in which the expression of insulin and other β-cell-restricted genes are modulated by an inducible form of the bHLH transcription factor E47 was developed. This cell line, T6PNE, was adapted for HTS by transduction with a vector expressing green fluorescent protein under the control of the human insulin promoter. The resulting cell line was screened against a library of known drugs for those that increase insulin promoter activity. Members of the phenothiazine class of neuroleptics increased insulin gene expression upon short-term exposure. Chronic treatment, however, resulted in suppression of insulin promoter activity, consistent with the effect of phenothiazines observed clinically to induce diabetes in chronically treated patients. In addition to providing insights into previously unrecognized targets and mechanisms of action of phenothiazines, the novel cell line described here provides a broadly applicable platform for mining new molecular drug targets and central regulators of β-cell differentiated function.
Keywords: diabetes, chlorpromazine, ethopropazine
INTRODUCTION
Although both type I and type II diabetes ultimately result in β-cell destruction, β-cell dysfunction precedes outright loss of β-cell mass, contributing to disease progression.1 A number of the pathways that play a role in β-cell dysfunction affect insulin gene transcription. This includes fatty acids and glucose in type II diabetes and inflammatory mediators that are present early in the course of type I diabetes. In addition, transcription factors, including PDX-1, MafA, NeuroD1, and HNF1, which are important in insulin promoter transactivation, also act globally to maintain many aspects of β-cell function and are targets for β-cell glucolipotoxicity.2 Be cause of the central role of insulin promoter activity in β-cell function, stimulating or maintaining promoter activity in β-cell stem/progenitors or in mature β-cells has the potential to preserve and enhance β-cell function in diabetes.
To discover compounds that act on the insulin promoter, we developed a novel human cell line that mimics several important aspects of β-cell biology. TRM6, a cell line derived from human fetal islets, was previously engineered to express the homeodomain transcription factor PDX-1 and the bHLH factor NeuroD1, resulting in low levels of insulin promoter activity.3 To further augment the degree of insulin promoter activity, we expressed the NeuroD1 dimerization partner, E47. E47 activation resulted in strong upregulation of insulin gene expression and other β-cell genes. E47 induction also resulted in cell cycle arrest mediated by Kip2, a β-cell-specific cyclin-dependent kinase inhibitor (CDKI).
This cell line, termed T6PNE, was adapted for high-throughput screening (HTS) by introducing a human insulin promoter-eGFP cassette. As an initial screen, we chose to study a library of known drugs for those that activated the insulin promoter. This had the advantage that many of the compounds have known mechanisms of action, unlike commonly used large and chemically diverse compound libraries. Moreover, many commonly used drugs act on pathways that are active in β-cells, but little is known about how those drugs affect β-cell function. Surprisingly, phenothiazine neuroleptics were found to modulate the insulin promoter, inducing an acute stimulatory response followed by repression upon chronic administration. This finding demonstrates a novel mechanism by which phenothiazines can affect glucose homeostasis and may explain the propensity of patients taking these drugs to develop diabetes. T6PNE provides a simple in vitro model to study how pharmacological agents may influence β-cell function. The rapid responsiveness of the insulin promoter in this cell line to pharmacologic manipulation sets it apart from other β-cell models and makes it useful for characterization of many aspects of β-cell biology that impinge on insulin promoter activity.
RESULTS
E47 activity is limiting for insulin gene expression in T6PN
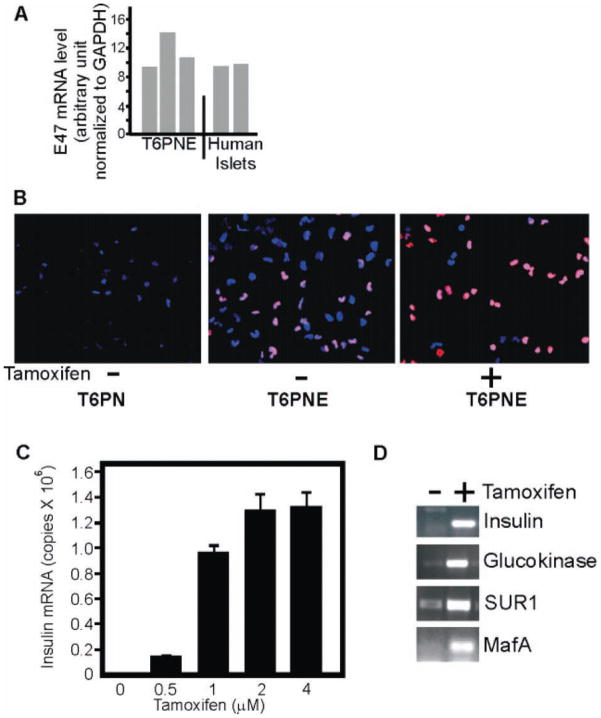
TRM-6 is a cell line derived from human fetal islets. Engineering of TRM-6 to express the homeodomain transcription factor PDX-1 and the class II bHLH factor NeuroD1 (T6PN cells) resulted in the induction of a very low level of insulin expression.3 High levels of the suppressor of E-box transactivators, Hes1, in cell lines that we have derived from human islets4 suggested that the heterodimer partner of NeuroD1 on the insulin promoter, E47,5 might be limiting insulin gene expression. As an initial test of that idea, we examined E47 expression in T6PN. Interestingly, the level of E47 mRNA was equal between T6PN and primary human islets (Fig. 1A). E47 protein was undetectable by immunohistochemistry in either T6PN (Fig. 1B, left panel) or primary islets (not shown).
FIG. 1.
E47 expression. (A) E47 mRNA level was measured by quantitative RT-PCR in 3 independent preparations of T6PNE cells and human islets from 2 different donors. (B) T6PNE and parental uninfected cells (T6PN) were assayed by immunohistochemistry for E47 (red) in the presence and absence of 4 μM tamoxifen. In the absence of tamoxifen, most cells did not have detectable E47 either in the cytoplasm or the nucleus. Some cells did have relatively weak nuclear E47, presumably due to leakiness of the modified estrogen receptor. With tamoxifen addition, nuclear E47 was greatly increased. DAPI (blue). (C) Insulin expression was responsive to the dose of tamoxifen, reaching a maximum level at 4 μM. Error bars are standard error of the mean (SEM) of 3 replicates. (D) E47 induction by tamoxifen resulted in upregulation of glucokinase, SUR-1, and MafA. Assay was done twice, and GAPDH control was equivalent between samples.
To increase the level of E47, we overexpressed its mRNA in T6PN using a retroviral vector encoding E47MER, in which E47 is fused to a modified estrogen receptor ligand binding domain (MER), rendering E47 inducible by tamoxifen.6 In the absence of tamoxifen, E47 was detectable at a very low level in these cells, termed T6PNE (Fig. 1B, middle panel). In the presence of tamoxifen, the level of nuclear E47MER increased dramatically (Fig. 1B, right panel). Induction of E47 by tamoxifen resulted in a potent and dose-dependent upregulation of insulin gene expression (Fig. 1C,D) but still approximately 1000-fold lower than in primary β-cells, in which insulin mRNA constitutes close to 10% of the mRNA.7
E47 induces multiple genes expressed in β-cells
To extend the finding that E47 plays a limiting role in insulin gene expression in T6PNE, we evaluated a subset of genes that are important for β-cell differentiation and function for E47 responsiveness. RT-PCR analysis of selected β-cell genes found that E47 induced glucokinase, SUR-1, and MafA (Fig. 1D). The myosin heavy chain gene, which is cardiac specific and highly controlled by multiple E-boxes within its promoter,8 was not expressed in T6PNE and was not induced by tamoxifen.
To probe the global pattern of gene expression in T6PNE in an unbiased fashion, we performed oligonucleotide microarray analysis on T6PNE in the presence and absence of E47 induction by tamoxifen. Transcripts detected were then compared to a list of 747 genes enriched in pancreatic islets9 and similar lists of genes expressed in other tissues in the body derived from the Neurocrine Body Atlas (GSE 3526 of the GEO database).10 Of the 17,980 transcripts detected in T6PNE (p < 0.05) and 18,482 transcripts detected in T6PNE treated with tamoxifen (p < 0.05), a total of 17,422 genes were expressed in both cell lines. Of the 747 β-cell-specific genes, 681 were detected in T6PNE induced with tamoxifen, suggesting inherently similar genetic profiling to β-cells (Suppl. Table S1).
To further assess the extent to which T6PNE retains a pattern of gene expression that predisposes it to endocrine differentiation in response to E47 induction, we compared data from the Illumina oligonucleotide microarray of T6PNE treated with tamoxifen to that of a number of tissues, including tongue, heart, and adipocytes. These lists were derived from the Neurocrine Body Atlas by selecting differentially expressed genes when comparing the tissue of interest to all other tissues present in the atlas. Data obtained from 50 trials of randomly selected sets of 747 genes compared with genes expressed in T6PNE treated with tamoxifen revealed a 74% average overlap by random chance. Similarly, comparisons of adipose-enriched, cardiac-enriched, and tongue-enriched gene lists to tamoxifen-treated T6PNE yielded 76%, 77%, and 76% overlap, respectively. A chi-square analysis demonstrated that T6PNE induced with tamoxifen was more consistent with the transcriptional profile of β-cells, having a 91% intersection with β-cell-enriched genes (p < 0.0005), as compared with the other tissues studied, which were not statistically similar to islets (tongue: p = 0.28; heart: p = 0.14; adipose: p = 0.41).
E47 induces growth arrest mediated by upregulation of Kip2
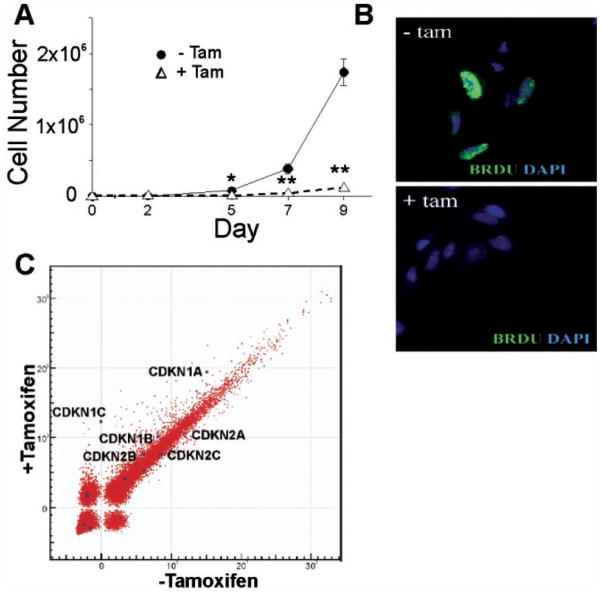
As stated above, E47 was not constitutively expressed in T6PNE but was introduced as a fusion protein with a modified estrogen receptor, rendering it inducible with tamoxifen. This was done because E47 caused growth arrest in T6PN cells, as evidenced by decreased growth rate (Fig. 2A) and BrdU incorporation (Fig. 2B).
FIG. 2.
Activation of E47 induces growth arrest in T6PNE cells. (A) Cells cultured in the absence or presence of tamoxifen (4 μM) were counted on the indicated days, demonstrating substantial growth arrest with the addition of tamoxifen *p < 0.05, **p < 0.001. (B) BrdU (green) and DAPI (blue) staining of T6PNE cells, demonstrating the absence of BrdU incorporation in the presence of tamoxifen (4 μM). (C) Microarray analysis was used to determine genes in T6PNE cells that were affected by E47, with particular attention to CDKIs. T6PNE were treated with tamoxifen (4 μM) and assayed for the expression of 24,000 human genes using Illumina BeadArray microarrays. Each dot on the scatter plot (power function) represents a single gene. The values on the x- and y-axes represent the level of hybridization to the oligonucleotide on the array, which is an indirect measure of the level of mRNA in the sample. p57Kip2 was undetectable in the absence of tamoxifen and was expressed at an approximately 2000-fold higher level in cells in which E47 was induced by tamoxifen. p21Cip1 was induced 2-fold.
To determine the mechanism responsible for the growth arrest, we examined the microarray data for genes involved in cell cycle control that were induced by tamoxifen. Most prominently, the cyclin-dependent kinase inhibitor p57Kip2 was strongly induced in T6PNE by tamoxifen (Fig. 2C). This was confirmed by RT-PCR, demonstrating that tamoxifen induction of E47 resulted in an increase in the level of Kip2 mRNA from almost undetectable to a level similar to that in islets. Interestingly, p57Kip2 is expressed in a β-cell-restricted manner in the adult human (but not rodent) pancreas and is deleted in the β-cell hyperproliferative disorder focal PHHI (persistent hyperinsulinemia and hypoglycemia of infancy).11
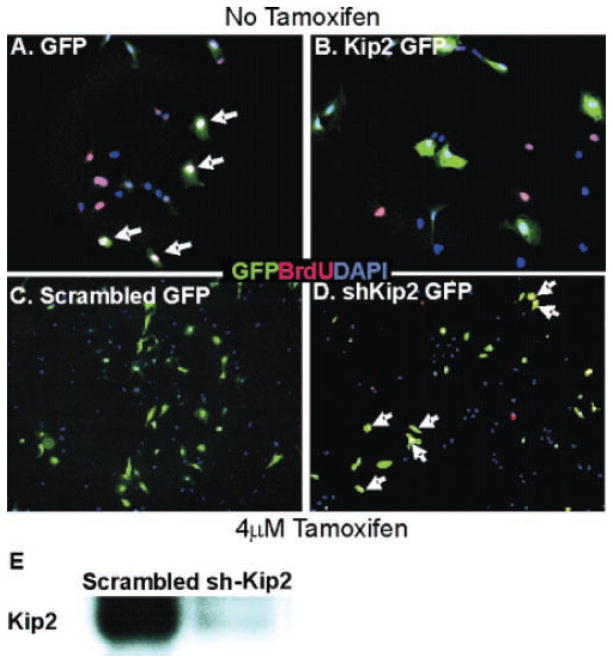
To determine whether Kip2 was sufficient to induce growth arrest in T6PNE cells, it was overexpressed using a bicistronic retroviral vector also expressing green fluorescent protein (GFP). In the absence of tamoxifen (no E47 induction), 59% of the cells infected with a control virus expressing GFP alone incorporated BrdU. In contrast, no cells infected with the Kip2 and GFP-expressing virus were positive for BrdU (Fig. 3A).
FIG. 3.
Kip2 controls proliferation in T6PNE cells. A retroviral vector expressing green fluorescent protein (GFP) alone (A) or expressing GFP and Kip2 (B) was used to infect T6PNE cells in the absence of tamoxifen. Cell cycle status of GFP-positive cells was assessed by BrdU staining (red). No GFP-BrdU double-positive cells were found in wells infected with the bicistronic GFP-Kip2 vector (B), whereas many cells infected with the GFP control vector did take up BrdU (arrows in A). DAPI is shown in blue. To inhibit Kip2 expression, T6PNE cells were transfected with a plasmid encoding an shKip2 (D) or scrambled (C) RNA along with GFP. The efficacy of the Kip2 shRNA construct was assessed by Western blot analysis (E). In T6PNE cells in which growth was inhibited by tamoxifen (4 μM)–mediated induction of E47, transfection of the shKip2 but not control shRNA (visualized by co-coexpression of GFP) caused cell cycle reentry as determined by colocalization of BrdU (red) and GFP (these are green cells with yellow nuclei; 6 such cells are marked by white arrows in D).
To determine whether Kip2 was sufficient to account for the growth arrest observed in T6PNE cells when E47 activity was induced, we used a bicistronic lentiviral vector expressing both GFP and a Kip2 shRNA. The specific shRNA but not control, scrambled shRNA suppressed Kip2 expression in tamoxifen-treated cells (Fig. 3E). Even though knockdown of Kip2 by shRNA was incomplete and it is unknown how much Kip2 expression is required to inhibit proliferation, the degree of inhibition that was achieved resulted in significant reversal of the inhibitory effects of 4 μM tamoxifen on growth, increasing the percentage of cells incorporating BrdU from 22% to 31% (p = 0.04; Fig. 3).
To investigate the mechanism by which E47 induced Kip2 gene expression, we used cotransfection, ChIP assays, and site-directed mutagenesis, finding that E47 directly activated the p57Kip2 promoter through a specific E-box between −3 and −4 kb. Seven E47 binding motifs within this region were identified, but deletion analysis revealed that the second E-box (E2) site was critical for promoter activity (Suppl. Fig. S1).
Adaptation of T6PNE for HTS
The ability to precisely modulate insulin gene expression with tamoxifen suggested that T6PNE cells could be used as a platform for HTS to detect compounds that affect insulin promoter activity. To adapt the cell line for HTS assays, we infected T6PNE cells with a lentiviral vector expressing the eGFP gene from a 1.4-kb fragment containing the human insulin promoter,12,13 providing for direct visualization of changes in insulin promoter activity. The specificity of the insulin promoter-eGFP transgene has been demonstrated previously.13
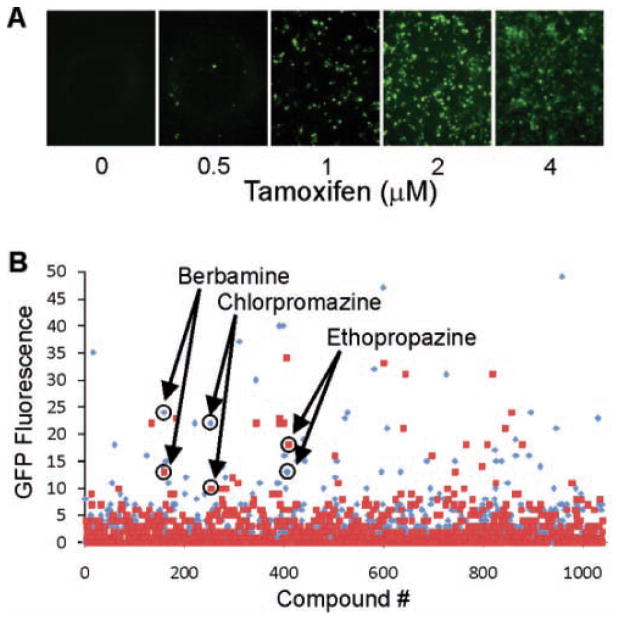
Tamoxifen dose-dependent GFP fluorescence, precisely in concert with endogenous insulin mRNA level, was observed in T6PNE cells infected with the transgene construct (Fig. 4A). Therefore, we were able to prime T6PNE with an intermediate level of insulin promoter activity by administering low levels of tamoxifen, allowing for detection of insulin promoter activators and inhibitors. Assay optimization of T6PNE cells in 384-well plates was performed using increasing doses of tamoxifen from 0.5 to 4 μM, yielding Z′ scores that ranged between 0.2 and 0.6. The variation in the Z′ occurred because the extent to which GFP expression was induced by tamoxifen varied in different pilot studies for reasons that are unclear but appears to be related to growth characteristics of the T6PNE cells.
FIG. 4.
High-throughput screening for insulin promoter modulators. (A) T6PNE cells infected with a lentiviral vector expressing an insulin promoter-eGFP cassette13 exhibited a tamoxifen dose-dependent increase in green fluorescence similar to that seen with endogenous insulin mRNA levels (Fig. 1C). (B) Scatter plot of a high-throughput screen with T6PNE expressing the insulin promoter–GFP cassette. In total, 1040 known drugs from the NIH/JDRF Custom Collection were screened in duplicate. Each set of replicates is depicted as either red squares or blue triangles. The duplicates for each compound were assayed on separate 384-well plates, but the values for each compound are shown consecutively for ease of visualization. Duplicate values for the 3 compounds that passed the primary confirmatory and secondary assays (berbamine, chlorpromazine, and ethopropazine) are circled.
High-throughput screening
Despite the variability in the Z′ values, we proceeded with a screen of a library of 1040 known drugs from the NIH/JDRF Custom Collection (NJCC), from MicroSource Discovery Systems, Inc. (Gaylordsville, CT). Because of the variability, the primary screen was done in duplicate in separate plates. A number of compounds, including merbromin, acriflavinium hydrochloride, calcein, aklavine hydrochloride, isoreserpine, and pyrvinium pamoate, were found to exhibit autofluorescence and thus were not pursued further. Others were cytotoxic and increased GFP by creating fluorescent cell aggregates. As expected, known estrogenic compounds such as estrone, clomiphene citrate, estradiol diacetate, estrone acetate, diethylstilbestrol, estradiol cypionate, estriol, and estradiol acetate were positive in the assay through their ability to activate the E47MER transgene.
Of the remaining compounds, 3 compounds were identified as increasing the number of GFP-positive cells in both wells from the primary screen. These compounds—chlorpromazine, ethopropazine, and berbamine—were subjected to a primary confirmatory assay, consisting of a repeat of the initial assay but with a dose response and 8 replicates for each dose. Image acquisition and analysis was performed as for the primary screen. All 3 of these compounds demonstrated dose-increases in the percentage of GFP-positive cells.
A subset of phenothiazines activates the insulin promoter
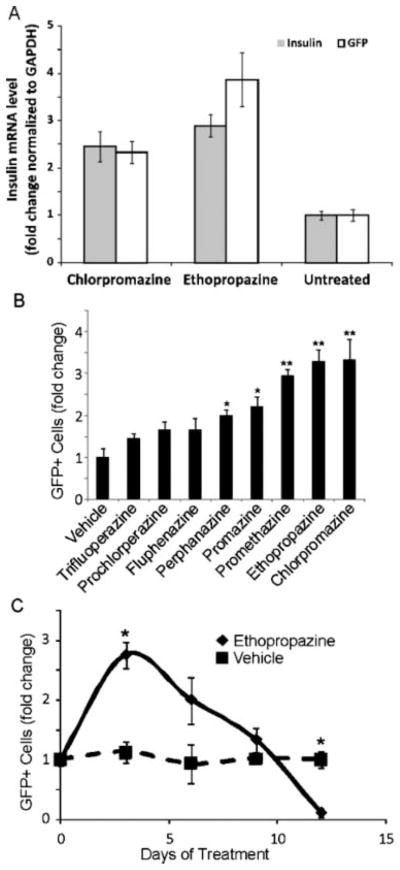
The 3 compounds that passed the primary counterscreens—ethopropazine, chlorpromazine, and berbamine (Fig. 4B)—were subjected to a secondary assay consisting of quantitative RT-PCR for GFP and endogenous insulin mRNA. Berbamine increased GFP but not endogenous insulin mRNA and thus was not studied further (Fig. 5A). However, ethopropazine stimulated an increase in both GFP and insulin mRNA above 0.1% DMSO by 3.86 ± 0.33–fold and 2.89 ± 0.13–fold, respectively (Fig. 5A). Chlorpromazine acted similarly, stimulating an increase in GFP and insulin mRNA of 2.33 ± 0.14–fold and 2.45 ± 0.19–fold, respectively (Fig. 5A).
FIG. 5.
Effect of phenothiazines on insulin promoter activity. (A) Quantitative RT-PCR of green fluorescent protein (GFP) mRNA and endogenous insulin mRNA. Ethopropazine and chlorpromazine increased endogenous insulin mRNA and eGFP mRNA relative to untreated cells. Insulin mRNA levels were normalized to GAPDH. Error bars are SEM of 3 independent experiments. (B) Retesting of all phenothiazines in the NIH/JDRF library for effect on the insulin promoter. Values are expressed as fold change in GFP-positive cells relative to vehicle-treated cells. Each compound is shown at its maximal tolerated dose, which was 20 μM for all compounds except fluphenazine, perphenazine, and trifluoperazine, which were used at 10 μM because of toxicity at higher doses (n = 6; experiment was performed 3 times; error bars are standard error; *p < 0.05, **p < 0.01). (C) Effect of chronic administration of ethopropazine on insulin promoter activity. Ethopropazine (15 μM) was administered to T6PNE cells for 12 days. Media were changed every 3 days. A lower concentration of drug was used than in acute treatment to minimize toxic effects on the cells. Values are expressed as fold change in GFP-positive cells relative to vehicle-treated cells (n = 3; error bars are SEM; *p < 0.05).
To ensure that the phenothiazines were not acting in a manner similar to the estrogenic compounds that were true but biologically uninteresting positives in the initial screen, we tested their ability to activate the insulin promoter in the absence of added tamoxifen, finding that there was no effect under that condition (Suppl. Fig. S2).
Structure-activity relationship
Given that the NJCC library contained many phenothiazines, it was interesting that the only members of the class that were detected as hits in the primary screen were chlorpromazine and ethopropazine. To examine the structural specificity of phenothiazines on insulin gene expression further, we retested all of the phenothiazines using the methodology employed for the primary confirmatory screen described above. Results of this inquiry indicated that many phenothiazines in the NJCC library were weakly positive but fell below the threshold value used in the high-throughput screen to detect hits. This analysis also revealed a false negative in the assay, promethazine, which indeed turned out to activate insulin promoter activity (Fig. 5B). Examining the structures of the positive and negative phenothiazines revealed that the most active compound was chlorpromazine, which differs from promazine only by the addition of a chlorine onto the carbon at the 3 position. This suggests that adding an electron-withdrawing group at that position increases activity. The increasing activity from promazine to ethopropazine suggests that increasing the hydrophobicity at the terminal amine by adding alkyl substituents also increases activity. The presence of a piperazine rather than an alkyl group at the terminal amine had a strong negative effect on activity. This suggests the possibility that this side group is limiting interaction with a specific receptor by steric hindrance (Suppl. Fig. S3).
Effect of chronic exposure to ethopropazine
A puzzling aspect of the finding that some phenothiazines increase insulin expression in our assay is that this class of drugs has been reported to be diabetogenic in chronically treated patients.14 To address that discrepancy, we investigated the effect of chronic exposure of T6PNE cells to ethopropazine. Although the positive effect on insulin expression was confirmed early in the course of treatment, chronic exposure to the drug led not only to the loss of stimulatory activity but also to repression of insulin promoter activity (Fig. 5C).
DISCUSSION
This study describes the generation of a new model of human β-cell growth and differentiation and its application to HTS for compounds that affect the activation state of the insulin promoter, a key feature of the differentiated β-cell. The principal finding from this small screen of known drugs was that a subset of phenothiazine neuroleptics was able to acutely activate the insulin promoter, and chronic treatment led to downregulation of the insulin promoter.
A key to the development of the T6PNE β-cell line model described here was the finding that E47 controls both proliferation, through direct binding to the Kip2 promoter, and β-cell differentiation. Thus, selection for growth, as inevitably occurs in cells cultured in vitro, resulted in downregulation of E47 activity and loss of β-cell differentiation, including insulin expression. Of note, the effect of E47 on insulin and other β-cell genes was replicated in another cell line that we have studied, blox5, which also loses differentiated gene expression over time.15 The low levelE47 activity observed of in T6PNE compared with T6PN produces a small effect on proliferation, but this is sufficient to result in selection against maintenance of E47MER expression over time. Thus, it was important to start periodically with earlier passage or cloned T6PNE cells.
Although glucose-responsive insulin secretion is not seen with induction of E47 activity in T6PNE, the upregulation of many β-cell genes in this system provides a model to study some aspects of β-cell differentiation and modulation of the insulin promoter. The microarray analysis revealed that T6PNE cells express a large number of the genes that are preferentially expressed in islets. However, it is clear that the level of expression is much lower in T6PNE than in primary β-cells.
For most purposes, the low level of expression of β-cell genes, particularly insulin, in T6PNE would be regarded as a drawback. However, it was precisely this low-level expression that made it possible to use HTS for insulin promoter modulators. Detecting differences in expression from the insulin promoter–GFP transgene is straightforward in T6PNE because the level of GFP can be titrated by adjusting the tamoxifen concentration. However, in primary β-cells or even conventional insulinoma cell lines such as MIN6, where the insulin gene is expressed at an enormous level, detecting the rapid change in GFP expression that is required for a practical high-throughput screen in primary islets is problematic.
It was surprising that the only class of drugs within the 1040-compound library that influenced insulin gene expression was the phenothiazines, a class of drugs known to predispose to diabetes upon chronic treatment.14 It should be noted that the screen was of short duration, with only 2 days of exposure to compound. Prolonged treatment with ethopropazine in T6PNE led to repression of insulin promoter activity. This could result from desensitization of a pathway that plays an important role in β-cell function and could contribute to the deleterious effects on β-cells that are found in most published studies, in which phenothiazines have been shown to inhibit glucose-responsive insulin secretion.16 Thus, as with other diabetogenic molecules such as fatty acids, acute exposure can stimulate aspects of β-cell function, whereas chronic exposure is harmful.2 Unfortunately, it was not possible to perform such chronic exposure studies with primary islets due to toxicity of the compounds. Using lower doses that are not toxic would require much longer times of exposure, exceeding the time in which islets can be maintained in vitro. However, longer term in vivo studies of the effects of phenothiazines on islet function would be of value.
The targets of phenothiazine neuroleptics are diverse, having been shown to affect a variety of proteins, including dopamine receptors, protein kinase C, calmodulin, and others.17 We have not as yet identified the target in T6PNE that signals to the insulin promoter. However, it is interesting to note that only a subset of phenothiazines was able to increase insulin promoter activity and that a specific structural feature of the terminal amine determined the level of activity. This suggests not only that a specific molecule is being targeted but that it is also likely to be a molecule that is different from those relevant to the psychiatric actions of the phenothiazines. There is poor concordance between the phenothiazines that we have found to act on the insulin promoter with those that have been reported to inhibit insulin secretion.16 With additional understanding of the target that is involved in insulin promoter modulation, it may be possible to design compounds that are more specific for the psychiatrically relevant target, with fewer effects on glucose homeostasis.
Supplementary Material
Acknowledgments
We thank Kees Murre, Michael German, and Sam Okret for providing materials. Steve Vasile and the Burnham Institute High Content Screening Core provided support with HTS. Roy Williams (Informatics Core) and Kang Liu (Genomics Core) provided support for the microarray studies.
This work was supported by the Sanford Children’s Health Research Center (FL) and grants from the NIH (1R21NS057001 and R01 DK055283-08S1 to FL), Juvenile Diabetes Research Foundation (FL, PI-A), the UCSD Genetics Training Grant (AK, TC), a California Institute for Regenerative Medicine (CIRM) fellowship (S-HL), and the UC Systemwide Biotechnology Research and Education Program Graduate Research and Education in Adaptive Bio-Technology (GREAT) Training Program (AK).
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental. Journal of Biomolecular Screening 15(6); 2010 DOI: 10.1177/1087057110372257
References
- 1.Porte D, Jr, Kahn SE. The key role of islet dysfunction in type II diabetes mellitus. Clin Invest Med. 1995;18:247–254. [PubMed] [Google Scholar]
- 2.Poitout V. Glucolipotoxicity of the pancreatic beta-cell: myth or reality? Biochem Soc Trans. 2008;36:901–904. doi: 10.1042/BST0360901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Itkin-Ansari P, Marcora E, Geron I, Tyrberg B, Demeterco C, Hao E, et al. NeuroD1 in the endocrine pancreas: localization and dual function as an activator and repressor. Dev Dyn. 2005;233:946–953. doi: 10.1002/dvdy.20443. [DOI] [PubMed] [Google Scholar]
- 4.Ball AJ, Abrahamsson AE, Tyrberg B, Itkin-Ansari P, Levine F. HES6 reverses nuclear reprogramming of insulin-producing cells following cell fusion. Biochem Biophys Res Commun. 2007;355:331–337. doi: 10.1016/j.bbrc.2007.01.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol Cell Biol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 7.Tillmar L, Carlsson C, Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3′-untranslated region pyrimidine-rich sequence. J Biol Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- 8.Molkentin JD, Brogan RS, Jobe SM, Markham BE. Expression of the alpha-myosin heavy chain gene in the heart is regulated in part by an E-box-dependent mechanism. J Biol Chem. 1993;268:2602–2609. [PubMed] [Google Scholar]
- 9.Kutlu B, Burdick D, Baxter D, Rasschaert J, Flamez D, Eizirik DL, et al. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kassem SA, Ariel I, Thornton PS, Hussain K, Smith V, Lindley KJ, et al. p57(KIP2) expression in normal islet cells and in hyperinsulinism of infancy. Diabetes. 2001;50:2763–2769. doi: 10.2337/diabetes.50.12.2763. [DOI] [PubMed] [Google Scholar]
- 12.Odagiri H, Wang J, German MS. Function of the human insulin promoter in primary cultured islet cells. J Biol Chem. 1996;271:1909–1915. doi: 10.1074/jbc.271.4.1909. [DOI] [PubMed] [Google Scholar]
- 13.Hao E, Tyrberg B, Itkin-Ansari P, Lakey JR, Geron I, Monosov EZ, et al. Beta-cell differentiation from nonendocrine epithelial cells of the adult human pancreas. Nat Med. 2006;12:310–316. doi: 10.1038/nm1367. [DOI] [PubMed] [Google Scholar]
- 14.Kato MM, Goodnick PJ. Antipsychotic medication: effects on regulation of glucose and lipids. Expert Opin Pharmacother. 2001;2:1571–1582. doi: 10.1517/14656566.2.10.1571. [DOI] [PubMed] [Google Scholar]
- 15.Dufayet de la Tour D, Halvorsen T, Demeterco C, Tyrberg B, Itkin-Ansari P, Loy M, et al. b-cell differentiation from a human pancreatic cell line in vitro and in vivo. Mol Endocrinol. 2001;15:476–483. doi: 10.1210/mend.15.3.0604. [DOI] [PubMed] [Google Scholar]
- 16.Krausz Y, Eylon L, Cerasi E. Calcium-binding proteins and insulin release: differential effects of phenothiazines on first- and second-phase secretion and on islet cAMP response to glucose. Acta Endocrinologica. 1987;116:241–246. [PubMed] [Google Scholar]
- 17.Palmer GC, Blosser JC, McCreedy SA, Barrantes MA, Manian AA. Correlation of activity of chlorpromazine and respective hydroxy, dimethoxy and sulphoxide analogues on dopamine, muscarinic, histamine and calmodulin sites of action. Xenobiotica. 1988;18:277–289. doi: 10.3109/00498258809041664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.