Abstract
Minor groove hydrogen bonding (HB) interactions between DNA polymerases and N3 of purines or O2 of pyrimidines have been proposed to be essential for DNA synthesis from results obtained using various nucleoside analogues lacking the N3 or O2 contacts that interfered with primer-extension. Since there has been no direct structural evidence to support this proposal, we decided to evaluate the contribution of minor groove HB interactions with family B pols. We have used RB69 DNA pol and 3-deaza-2’-deoxyadenosine (3DA), an analog of 2-deoxyadenosine, which has the same HB pattern opposite T but with N3 replaced with a carbon atom. We then determined pre-steady state kinetic parameters for the insertion of dAMP opposite dT using primer/template (P/T) containing 3DA. We also determined three structures of ternary complexes with 3DA at various positions in the duplex DNA substrate. We found that the incorporation efficiency of dAMP opposite dT decreased 102–103 fold even when only one minor groove HB interaction was missing. Our structures show that the HB pattern and base-pair geometry of 3DA/dT is exactly the same as dA/dT, which makes 3DA an optimal analogue for probing minor groove HB interactions between a DNA polymerase and a nucleobase. In addition, our structures provide a rationale for the observed 102–103 fold decrease in nucleotide incorporation. The minor groove HB interactions between the n-2 position of the primer strand and RB69pol fixes the rotomer conformations of the K706 and D621 side chains, as well as the position of metal ion A and its coordinating ligands so that they are in the optinal orientation for DNA synthesis
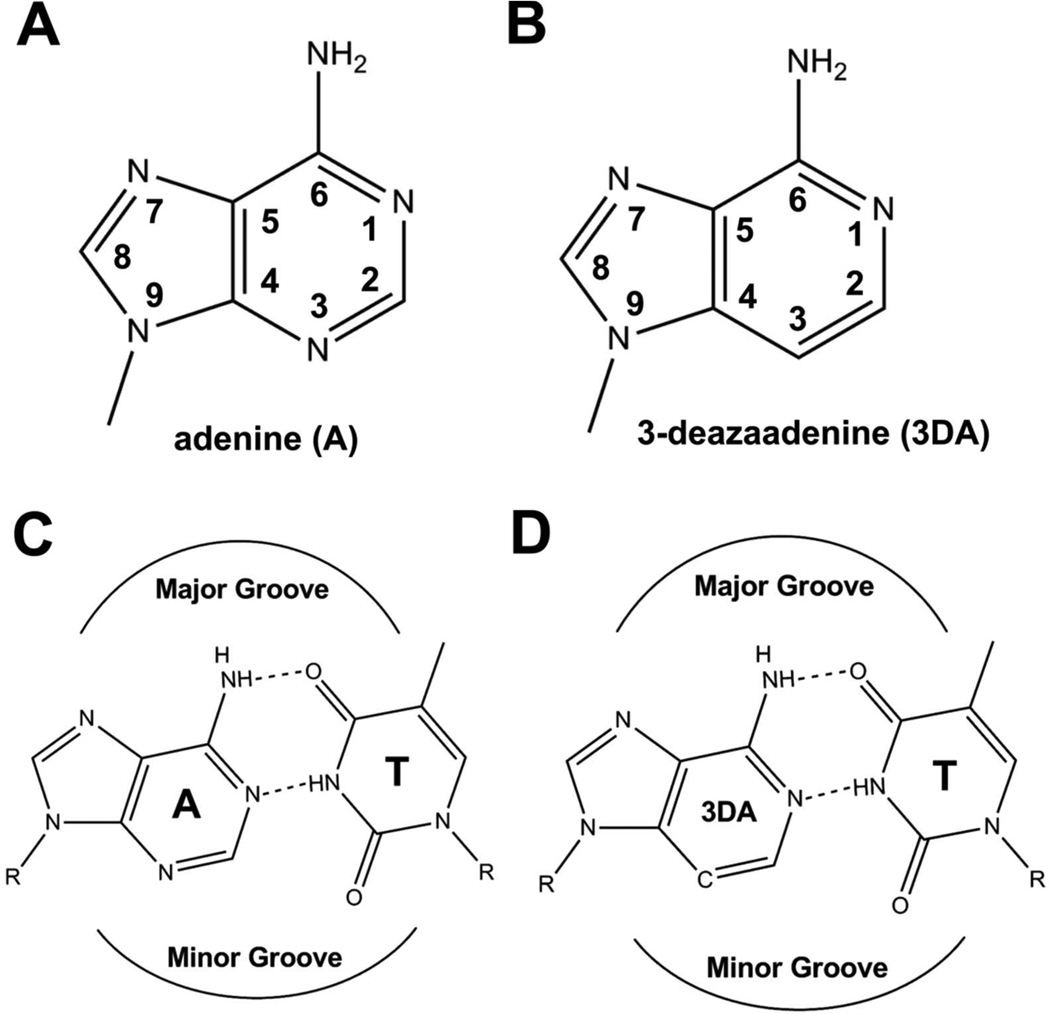
DNA polymerases are essential for the maintenance of genetic integrity with reported error frequencies for replicative DNA polymerases (pol) that are typically less than 10−6 for every insertion event.(1–5) Several factors have been identified that contribute to the high level of base selectivity; these include base stacking, base-pair (BP) geometry and inter-base hydrogen bonding (HB).(6–8) In addition minor groove HB interactions between the nucleobases in the DNA duplex and the pol contribute to the efficiency of DNA synthesis and base selectivity.(9–13) The lone pair of electrons carried by N3 in standard purines or O2 in standard pyrimidines are competent HB acceptors that can interact with HB donors presented by the pols.(14) Indeed, structures of various pols from different families have provided direct evidence for pol-minor groove HB interactions.(11, 15–19) In family A pols, a conserved Arg residue forms a HB to N3 or O2 of the nucleobase at the n-1 position (in both the primer and template strands).(18–20) Similarly, in family B pols, a conserved Tyr residue forms a HB with the nucleobase at the n-1 position of the template strand in the minor groove via a water molecule.(11, 21) Single site mutagenesis showed that substitution of certain highly conserved residues (Arg in family A pols, or Lys in family B pols) to Ala dramatically reduced catalytic efficiency.(22–25) Further evidence for the role of minor groove-pol HB interactions in enhancing the efficiency of DNA synthesis have come from experiments that employed nucleotide analogues lacking either an N3 atom in purines or an O2 atom in pyrimidines. For example, Morales et al(26) used non-HB nucleoside isosteres: 9-methyl-1H-imidazo-[4,5-b]-pyridine (Q), 4-metyl-benzimidazole (Z), and difluorotoluene (F) to probe the role of minor groove HB interactions with the Klenow Fragment (KF). They concluded that minor groove HB interactions are more important for primer extension than for insertion of base analogues.(13, 26) However there was no direct structural evidence to support this contention or to exclude other possibilities that could account for the reduction in incorporation efficiency, such as unforeseen properties of these nucleotide analogues. Our kinetic and structural studies using DNA with an abasic site in the templating position demonstrated that base stacking between incoming dNTP and BPs at the n-1 position plays an important role in the efficiency of nucleotide incorporation.(27) Introducing a nucleoside isostere incapable of HB with bases either in the primer or template strand at the n-1 position might have been expected to affect base stacking of the incoming dNTP. In addition, our recently reported structures of the ternary complex of RB69 DNA polymerase (RB69 pol) containing F in the templating position showed that F can form HBs with incoming dNTPs directly or indirectly via water molecules.(28–29) We decided to revisit this important issue regarding the role of minor groove HBs using RB69 DNA pol (RB69pol) and 3-deaza-2’-deoxyadenosine (3DA), an analogue of 2-deoxyadenosine (dA) where N3 is replaced with a carbon atom. 3DA has the same hydrogen bonding pattern with dT as does dA making it an ideal analogue to use as a probe to study these interactions (Fig. 1).
Figure 1.
Chemical structure and hydrogen bonding pattern of 3DA. (A) Chemical structure of adenine. (B) Chemical structure of 3DA. (C) Complementary dA/dT base pair. (D) 3DA/dT base pair.
We chose RB69pol as a representative of B family pols and because it has extensive sequence similarity to human and other eukaryotic replicative DNA pols.(30) In addition, there is a wealth of structural and kinetic data available that would be expected to aid in the interpretation of results obtained with this enzyme.(11, 31–37) The structure of the dCTP/dG containing ternary complex of RB69 pol at 1.8 Å resolution has shown that: (i) Y567 forms a HB with N3 of G at the n-1 position of the template strand via a water molecule and; (ii) K706 forms a HB with N3 of A at the n-2 position of the primer strand.(35) Both Y567 and K706 are highly conserved residues in family B pols.(30) To assess whether RB69pol employs minor groove hydrogen bond interactions as one of the ways that RB69pol, and likely other pols, optimize substrate binding and reactive center alignment, we examined the incorporation efficiency of correct incoming nucleotides into DNA that had 3DA at the n-1, n-2 and n-3 position in the primer strand and at the n-1 position in the template strand. Our pre-steady-state kinetic data showed that minor groove HB interactions are essential for DNA synthesis. Missing just one minor groove HB interaction resulted in nearly a 103 fold decrease in catalytic efficiency for dAMP insertion opposite dT. We also determined three structures of dATP/dT-containing ternary complexes; one with wt RB69pol and control DNA without 3DA; a second with a P/T that had 3DA at the n-1 position in the template strand and a third with a P/T that had 3DA at the n-3 position in the primer strand. These structures provide a basis for understanding why the absence of one minor groove HB interaction with a pol resulted in a 102–103 fold decrease in efficiency of nucleotide incorporation.
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals were the highest quality commercially available; dNTPs were purchased from Roche (Roche Applied Science, Indianapolis, IN). T4 polynucleotide kinase was purchased from New England BioLabs (Boston, MA). [γ-32P]ATP was purchased from MP Biomedicals (Irvine, CA).
Enzymes
wt RB69pol and the Y567A mutant RB69pol, in an exonuclease-deficient background (D222A and D327A), were over-expressed in E. coli strain BL21(DE), then purified, and stored as previously described.(35, 37–38)
DNA Substrates
All oligonucleotides were synthesized at the Keck facilities (Yale University) and purified via polyacrylamide gel electrophoresis. The sequence of the primer-template (P/T) used in this study was shown in Fig 2. For kinetic studies, the primer was labeled on the 5’-end with 32P using T4 polynucleotide kinase and [γ-32P]-ATP and annealed to the corresponding templates as previously described.(34–35, 37) For crystallization, ddT or ddC terminated primers were used to prevent phosphoryl transfer.
Figure 2.
P/T sequences used in this study. (A) P/T sequences used in chemical quench experiments. (B) P/T sequences used in fluorescence lifetime experiments. (C) P/T sequences used in fluorescence anisotropy experiments.
Chemical Quench Experiments
Rapid chemical quench experiments were performed at 23°C using a KinTek RFQ-3 instrument (KinTek Corp., University Park, PA). Each reaction mixture contained 66 mM Tris-HCl (pH 7.4) and 10 mM MgSO4. All experiments were performed under single-turnover conditions, with a 10-fold excess of RB69pol over the P/T. The concentrations of enzyme after mixing were 1 µM and the [P/T] was 83 nM. Products were separated by 19:1% (w/v)
Determination of dissociation constants for RB69pol-P/T binary complexes
Fluorescence anisotropy experiments were performed as previously described.(37) The binding buffer contained 50mM Tris-HCl (pH 7.4) and 10 mM MgSO4. All experiments were performed in duplicate.
Fluorescence Lifetime Determinations
These experiments were carried as described previously except that tCo was used in place of 2AP.(39) The excitation and emission wavelengths for tCo are 364 nm and 450 nm respectively. The final reaction mixtures consisted of 50 mM Tris (pH7.4), 10 mM MgSO4, 200 nM P/T and 4 µM RB69pol. The methods and data analysis were the same as described previously.(39)
Crystallization of dATP/dT-containing wt RB69pol ternary complexes
Wild type RB9pol was mixed in an equimolar ratio with a dideoxy terminated P/T to give a final protein concentration of 120 µM. dATP was then added to give a final concentration of 2 mM. Crystals was grown under silicon oil using a microbatch procedure.(28–29, 35–38) A solution containing 120 mM CaCl2, 12% (w/v) PEG 350 monomethyl ether (MME), and 100 mM sodium cacodylate pH 6.5 was mixed with an equal volume of the protein complex. The square rod-shaped crystals grew in 2 days at 20°C and had typical dimensions of about 150 µm × 70 µm × 70 µm. Crystals were transferred from the mother liquor to a cryoprotectant solution with the same components but containing a high concentration of PEG 350 MME (30% w/v) prior to freezing in liquid nitrogen.
Data collection, structure determination, and refinement
X-ray diffraction data were collected using the synchrotron radiation sources at beam line 24ID-E, Northeast Collaborative Access Team (NECAT), Advanced Photon Source, Argonne National Laboratory (APS, ANL, Chicago, IL). The data were processed using HKL2000 program suite (Table 1 and Table S1).(40) The structures were determined by molecular replacement using Phaser,(41) starting with the wt RB69pol structure of the ternary complex (3NCI), and refined using REFMAC5.(42) The P/T duplex and dNTP were built using the program COOT.(43) Structure refinement statistics are summarized in Table 1 and Table S1. All figures were made using the program Pymol.(44)
Table 1.
Crystallographic Statistics for Data Collection and Structure Refinement of dATP/dT-Containing Ternary Complexes of wt RB69pol
| RB69pol Ternary Complex P/T duplex DNA |
wt/dATP/dT control |
wt/dATP/dT 3DA at n-1 (T) |
wt/dATP/dT 3DA at n-3 (P) |
|---|---|---|---|
| Space group | P212121 | P212121 | P212121 |
| Unit cell dimensions | 74.98, 119.83, 129.95 | 74.64, 120.18, 130.67 | 78.14, 119.50, 130.54 |
| [a, b, c (Å) ] | |||
| Resolution range (Å) | 50.0-2.15 (2.23-2.15) | 50.0-2.02 (2.09-2.02) | 50.0-2.31 (2.39-2.31) |
| Number of reflections | |||
| Unique | 63,686 | 76,820 | 55,522 |
| Redundancy | 4.4 (3.8) | 4.0 (4.0) | 4.8 (4.9) |
| Completeness (%) | 99.2 (99.7) | 99.4 (99.8) | 99.9 (100.0) |
| Rmerge (%) | 13.5 (95.8) | 10.5 (93.2) | 7.6 (96.0) |
| I/σ | 11.3 (1.2) | 12.7 (1.1) | 16.8 (1.3) |
| Final model | |||
| Amino acid residues | 903 | 903 | 903 |
| Water molecules | 354 | 537 | 187 |
| Metal ions | 5 | 5 | 5 |
| Template nucleotides | 18 | 18 | 18 |
| Primer nucleotides | 13 | 13 | 13 |
| dNTP molecules | 1 | 1 | 1 |
| Refinement Statistics | |||
| Reflections | 60,252 | 72,694 | 52,490 |
| Rwork (%) | 19.7 (25.7) | 19.0 (32.4) | 20.8 (26.5) |
| Rfree (%) | 24.4 (31.6) | 22.6 (35.9) | 25.9 (33.5) |
| r.m.s.d. | |||
| Bond length (Å) | 0.009 | 0.008 | 0.007 |
| Bond angle (° ) | 1.190 | 1.165 | 1.149 |
| PDB access code | 4DU1 | 4DU3 | 4DU4 |
Footnotes:
, Statistics for the highest resolution shell are in parenthesis.
, Rmerge = ΣhklΣj |Ij(hkl)-<Ij(hkl)>|/Σhkl Σj <I(hkl)>, statistics for merging all observations for given reflections.
, R = Σhkl |Fobs(hkl)-Fcalc(hkl)|/Σhkl Fobs(hkl), statistics for crystallographic agreement between the measured and model-calculated amplitudes. Rfree is the agreement for cross-validation data set.
, Root mean squares deviations (rmsd) to ideal values.
PDB accession numbers
Coordinates and structure factors for the dATP/dT-containing wt RB69pol ternary complex structure have been deposited in the Protein Data Bank under accession code 4DU1 (with control DNA), 4DU3 (with 3DA at the n-1 position of template strand), and 4DU4 (with 3DA at the n-3 position of primer strand). In addition, coordinates and structure factor for the dQTP/dT-containing ternary complex of RB69pol triple mutant was deposited under accession code 4E3S.
RESULTS AND DISSCUSION
Incorporation of dAMP into DNA Containing 3DA at Different Positions in the P/T
To determine the precise role of HB interactions between RB69pol and the minor groove in a P/T duplex, we designed P/Ts with 3DA at the n-1, n-2 or n-3 position in the primer strand or at the n-1 position in the template strand (Fig. 2A). As shown in Figure 1, substitution of nitrogen with carbon eliminates the possibility of HB between the 3-position of adenine and the pol while the overall geometry and W-C HB capability between the incoming dATP and the templating thymine remain unchanged. Thus, any variation in the kinetic parameters, e.g kpol or Kd,app for insertion of dAMP opposite dT, resulting from the dA to 3DA substitution can be ascribed to the disruption of the critical HB interaction between pol and the 3-position of the adenine residue in the P/T duplex. Under pre-steady-state conditions, the incorporation efficiency of dAMP opposite dT by wt RB69pol varies markedly depending on the position of 3DA in the duplex. As shown in Table 2, substitution of dA with 3DA at the n-1 position of the template strand results in a 5 fold decrease in kpol and an 18 fold increase in Kd,app. The change is even more dramatic when the 3DA substitution is in the primer strand. Replacing dA with 3DA at the n-1 position of the primer strand causes the catalytic efficiency to decrease by 144 fold. When the 3DA is at the n-2 position in the primer strand, the kpol and Kd,app values for incorporation dAMP opposite dT were 10 s−1 and 1100 µM respectively, which are almost 30 fold lower for kpol and 30 fold higher for Kd,app resulting in a 900 fold decrease in catalytic efficiency compared to the kinetic parameters obtained with control DNA. Interestingly, replacing dA with 3DA at the n-3 position of the template strand did not affect kpol and Kd,app values for incorporation dAMP opposite dT. Therefore, some minor groove-pol HB interactions are not essential for nucleotide incorporation.
Table 2.
Pre-Steady-State Kinetic Parameters for Incorporation of dAMP Opposite dT by wt and Y567A RB69pol.a,c
| RB69 pol | P/T b | P/T Details | kpol (s−1) | Kd,app (µM) | kpol / Kd (µM−1 s−1) | Ratio |
|---|---|---|---|---|---|---|
| wt | SeqA1 | Control | 296 | 36 | 8.2 | |
| wt | SeqA2 | 3DA at n-1 (T) | 61 | 665 | 9.1 × 10−2 | 1/90 |
| wt | SeqA3 | 3DA at n-1 (P) | 38 | 670 | 5.7 × 10−2 | 1/144 |
| wt | SeqA4 | 3DA at n-2 (P) | 10 | 1100 | 9.1 × 10−3 | 1/901 |
| wt | SeqA5 | 3DA at n-3 (P) | 293 | 38 | 7.7 | 1/1.1 |
| Y567A | SeqA1 | Control | 290 | 40 | 7.2 | |
| Y567A | SeqA2 | 3DA at n-1 (T) | 283 | 42 | 6.7 | 1.0 |
The structure that we have reported for a dATP/dT-containing ternary complex of Y567A RB69pol shows that the minor groove HB network at the n-1 position of the template strand and the pol was disrupted by replacing Y567 with Ala.(37, 39) It is very likely that the Y567A RB69pol mutant was not able to sense the dA to 3DA substitution at this position. As predicted, the kinetic parameters for incorporation of dAMP opposite dT by the Y567A RB69pol mutant were almost identical for both control DNA and DNA with 3DA at the n-1 position of the template strand (Table 2).
Determination of Fluorescence Lifetimes
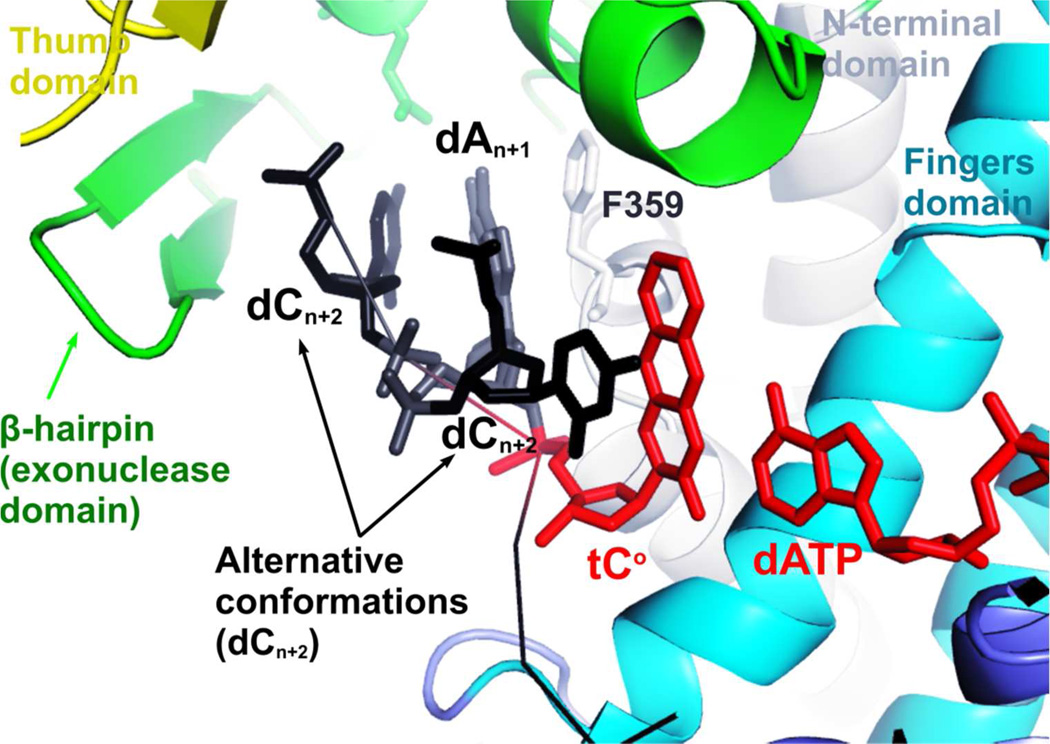
To determine the effect of HB interactions between RB69pol and the minor groove of the P/T duplex in partitioning the DNA substrate between the pol and exo domains, we designed three duplex DNAs with tCo, an analogue of dC, at the n position of the template strand (Fig. 3). At the n-1 position, the first duplex (Fig.2B: SeqB1) has a normal W-C BP dT/dA; the second (Fig.2B: SeqB2) has a dT/3DA BP; and the third (Fig.2B: SeqB3) has an dA/dA mismatched BP. The lifetime of tCo fluorescence depends largely on its local environment.(45–46) We assume that, in a pol-P/T binary complex, the majority of SeqB1 will be in the pol domain, and the majority of SeqB3 will be in the exo domain. If minor-groove HB interactions are important for keeping a P/T in the pol domain, then the local environment of tCo in SeqB2 will resemble that of tCo in SeqB1, otherwise, it will be closer to that of tCo in SeqB3. As shown in Table 3, a single fluorescent species with a 3.8 to 3.9 ps fluorescence lifetime was observed for duplex DNA SeqB1 and SeqB2. Upon binding RB69pol, the tCo fluorescence intensity increases, as does the fluorescence lifetime. Three fluorescent species with 4.3 ns, 6.4 ns and 39 ps fluorescence lifetimes, respectively, were observed for the pol-SeqB1 (control DNA) binary complex. The existence of multiple fluorescent species is consistent with our recently reported dATP/tCo-containing ternary complex of Y567A RB69pol.(47) The 5’ overhang of the template strand has two alternative conformations; the base of dC at the n+2 position of the template strand can either stack between the β-hairpin and the dA base at the n+1 position in the template strand or flip backward forming a Hoogsteen base-pair with dG at the n-1 position of the template strand (Fig. 3). The β-hairpin in the exonuclease domain of RB69pol acts as a wedge to separate the end of the primer strand from the template. Thus, a conformational change involving the β-hairpin will affect the local environment of tCo at the n position when the 3’-end of the primer partitions into the exonuclease domain. Replacing dA with 3DA at the n-1 position of template strand affects fluorescence lifetimes as follows: (i) the amplitudes of the two nano-second lifetime species decreases by 30% (Table 3); (ii) the amplitude of the pico-second lifetime species increases by 50% (Table 3); (iii) the value of lifetime for the pico-second lifetime species decreases by 40% (Table 3). Thus, the local environment of tCo in SeqB2 is not exactly the same as that of tCo in SeqB1. By replacing a normal WC BP with a dA/dA mismatch, the amplitude for the pico-second species increases by 75%, and the lifetime of the pico-second species decreases by 50%. By comparison, the values and amplitudes of fluorescence lifetimes of tCo in SeqB2 are closer to those of SeqB3 than SeqB1. Clearly, pol-minor groove HB interactions help to stabilize the P/T in the pol domain.
Figure 3.
Two alternative conformations of 5’ overhang of template strand DNA in the dATP/tCocontaining ternary complex of Y567A RB69pol. Chemical structure of tCo was shown in Fig S3.
Table 3.
| RB69 pol | P/T b | P/T Details | Fluorescence lifetimes |
|||
|---|---|---|---|---|---|---|
| τ1 (ns) | τ2 (ns) | τ3 (ps) | chi2 | |||
| SeqB1 | Control | 3.8 (1.0) | 1.04 | |||
| SeqB2 | 3DA at n-1 (T) | 3.9 (1.0) | 0.98 | |||
| wt | SeqB1 | Control | 4.3 (0.29) | 6.4 (0.34) | 39 (0.37) | 1.05 |
| wt | SeqB2 | 3DA at n-1 (T) | 4.3 (0.19) | 7.6 (0.25) | 23 (0.56) | 1.03 |
| wt | SeqB3 | dA/dA at n-1 | 4.3 (0.19) | 7.0 (0.16) | 19 (0.65) | 1.07 |
Determination of RB69pol-P/T Dissociation Constants
A concern raised by the results of the fluorescence lifetime measurements is whether minor groove HB interactions affect the dissociation of pol-P/T binary complexes. We tested this using fluorescence anisotropy. As shown in Table 4, the Kd for the wt RB69pol with control DNA is 152 nM. Substitution of dA with 3DA at the n-1 position of the template strand in the DNA duplex did not affect the corresponding binding affinity to RB69pol as the two Kds are almost identical (Table 4). Thus the increase in Kd,app values observed with 3DA at the n-1 position, determined using single turnover conditions, was mainly due to substrate misalignment and a sparser population of pol-P/T binary complex in the pol mode. In addition, replacing dA/dT with dA/dA at the n-1 position of P/T duplex did not affect the binding affinity of pol to P/T duplex (Table 4). Therefore, the change of fluorescence lifetime of tCo in SeqB3 is due to the partitioning of the primer strand from the pol to the exo domain.
Table 4.
Ground-state Dissociation Constants of RB69pol-P/T Binary Complexes.a
| P/T b | P/T Details | Kd (nM) |
|---|---|---|
| SeqC1 | Control | 152 |
| SeqC2 | 3DA at n-1 (T) | 156 |
| SeqC3 | dA/dA at n-1 | 150 |
The standard deviation for the Kd values was ±25%.
Sequences of P/Ts are shown in Fig 2C.
Structural Studies of the dATP/dT-containing Ternary Complex of Wild Type RB69pol with 3DA at Various Positions in the DNA Duplex
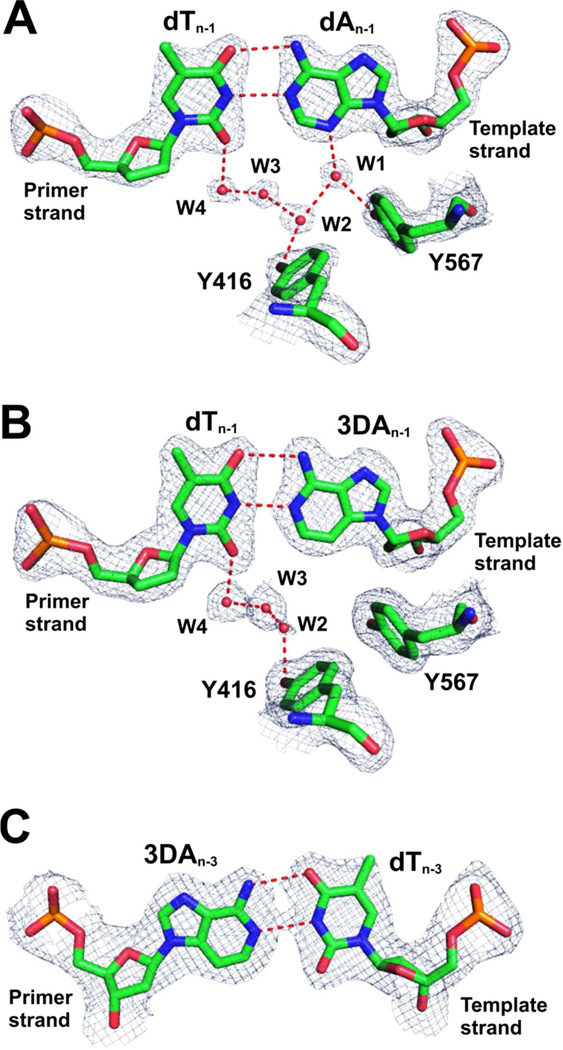
In an attempt to provide a structural basis that could account for the 102 to 103 fold difference in incorporation efficiency when 3DA was substituted for dA in various position of the P/T, we determined the crystal structures of three dATP/dT-containing ternary complexes of wt RB69pol that differed as follows: one contained control duplex DNA; the second had 3DA at the n-1 position in the template strand of the P/T; and the third had 3DA at the n-3 position in the primer strand of the P/T. These three structures were determined with resolutions ranging from 2.02 to 2.31 Å and free R factors ranging from 22.6% to 25.9%. The overall structures of all three ternary complexes were identical to our reported 1.8 Å dCTP/dG-containing wt RB69pol ternary complex with root-mean-squares Cα deviations varying between 0.18 Å to 0.21 Å.(35) The electron densities for the P/T duplex DNA and the surrounding network of ordered water molecules were well-defined in the structures reported here (Fig. 4).
Figure 4.
Structure of base-pairs at the n-1 and n-3 position of the P/T. (A) Final 2Fo-Fc electron density map for the wt RB69pol with control DNA contoured at 1.8 σ. (B) Final 2Fo-Fc electron density map for the wt RB69pol with 3DA at the n-1 position of the template strand contoured at 1.2 σ. (C) Final 2Fo-Fc electron density map for the wt RB69pol with 3DA at the n-3 position of the primer strand contoured at 1.2 σ.
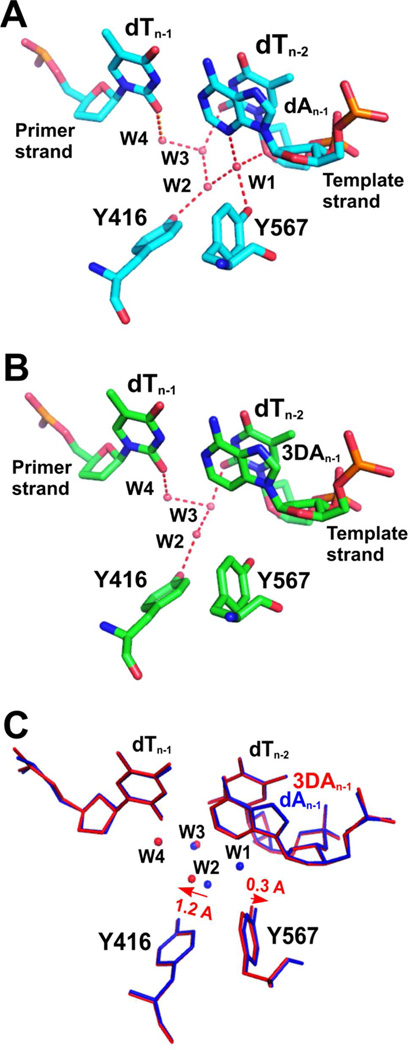
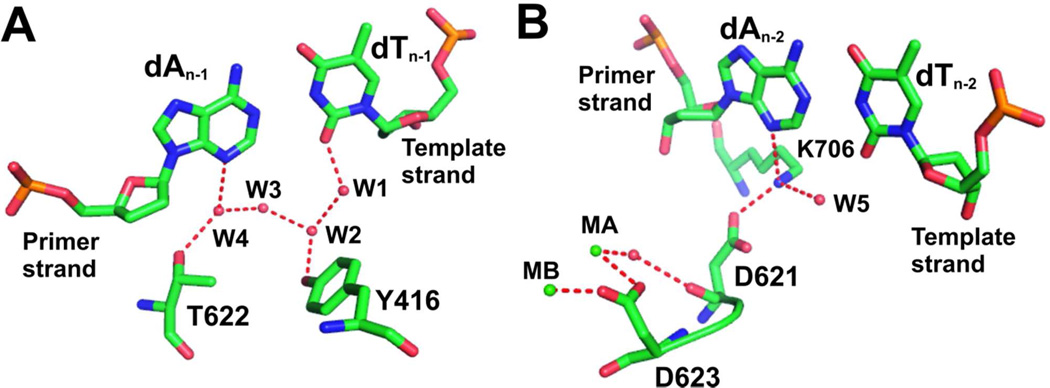
Consistent with our previous findings, a rigid HB network involving the γ-OH groups of Y567, Y416, and five ordered water molecules (four of them shown in Fig. 5A) was observed in the minor groove of the nascent P/T duplex in our structure containing control DNA. As shown in Fig 5A, the W1 water molecule was in perfect tetrahedral coordination. It formed a HB with N3 of dA at the n-1 position of the template strand. The HB distance between W1 and N3 was 2.69 Å. The adjacent W2 water molecule was in an unusual planar trigonal geometry, a feature that was consistently observed in structures of RB69pol ternary complexes.(28–29, 35–38) All four water molecules are polarized and serve as extensions of protein side chains to mediate pol-DNA interactions. In contrast, the W1 water molecule was missing in the structure where dA was replaced with 3DA at the n-1 position of the template strand (Fig. 5B). Least squares superposition of these two structures shows that: (i) the dT/3DA BP is completely superimposable with the dT/dA BP at the n-1 position and the HB pattern and geometry of the two BPs are almost identical (Fig. 5C); (ii) the γ-hydroxyl of Y567 tilted 0.3 Å toward the main chain amide of residue G568 (Fig. 5C); (iii) the W2 water molecule shifted 1.2 Å laterally toward the Y416 side chain (Fig. 5C). Thus, the 90 fold difference in the efficiency of nucleotide incorporation using different duplex DNAs (control DNA vs DNA with 3DA at the n-1 position of template strand) was due to the disruption of this rigid HB network at the n-1 position of the template strand of the P/T duplex.
Figure 5.
Minor groove HB at the n-1 position. (A) Overview of the NBP of wt RB69pol with control DNA. A rigid HB network with ordered water molecules labeled as W1 through W4 contacts the minor groove of the P/T DNA duplex. (B) Overview of the NBP of wt RB69pol with 3DA at the n-1 position of the template strand. (C) Superposition of A (shown in blue sticks) and B (shown in red sticks).
Substitution of dA with 3DA at the n-3 position of the primer strand did not disrupt the HB network in the minor groove of the nascent P/T duplex, as all four water molecules shown in Fig 5A can be unambiguously identified. This is consistent with the kinetics which showed that replacement of dA with 3DA at the n-3 position of the template strand had no effect on the kinetic parameters for nucleotide incorporation (Table 2). There is no water molecule within HB distance to the C3 of 3DA at the n-3 position of the primer strand (Fig. 6A), whereas one water molecule formed a HB with O2 of dT at the n-3 position of the primer strand in the structure of ternary complex with the control DNA (Fig. 6B). Clearly, this minor groove HB interaction at the n-3 position is not essential, and does not affect the kinetic behavior of wt RB69pol.
Figure 6.
Minor groove HB at the n-3 position. (A) 3DA/dT base-pairs at the n-3 position of the P/T duplex. (B) dA/dT base-pairs at the n-3 position of the P/T duplex in the wt RB69pol with control DNA.
Role of Minor Groove HB Interactions at the n-1 and n-2 Positions of the Primer Strand
Since we could not obtain a dideoxy-3DA terminated primer and because of the high Kd,app for the incoming dNTP, we were unable to obtain a structure with 3DA at the n-1 or n-2 positions of the primer strand. However the structure of the ternary complex with control DNA provides insight into why the dA to 3DA substitution at the n-1 or n-2 position of primer strand has greater impact on incorporation efficiency than when the 3DA for dA substitution is at the n-1 position of the template strand. As shown in Fig 7A, the W4 water molecule formed a HB with N3 of dA at the n-1 position of the template strand. The orientation of the hydrogen atom from W4 is determined by the side chain of T622 and by another water molecule, W3. Replacement of dA with 3DA at the n-1 position of the primer strand creates a steric clash between the C3-H of 3DA and the W4 water molecule. It’s very likely that W4 will be missing just as W1 is absent in the structure where dA was replaced with 3DA at the n-1 position of the template strand. Because 3DA is at the primer-terminus where its 3’ OH group is in a position to attack the Pα of an incoming dNTP, disruption of the HB network involving the minor groove of the primer strand will clearly have a greater impact on catalysis than the case where disruption of the HB network occurs at the n-1 position of the template strand. This could explain why there is 144 fold decrease in kpol/Kd for dAMP incorporation opposite dT when 3DA is at the n-1 position in the primer strand, and only a 90 fold decrease in kpol/Kd when 3DA is at the n-1 position in the template strand.
Figure 7.
Minor groove HB at the n-1 and n-2 positions. (A) HB network and dA/dT base-pair at the n-1 position of the P/T duplex. (B) HB interactions between K706 and dA at the n-2 position of the primer strand, and between K706 and D621.
The incorporation efficiency decreases by almost three orders of magnitude when dA is replaced with 3DA at the n-2 position of the primer strand. As shown in Fig 7B, the ε-amino group of K706 forms a HB with N3 of dA at the n-2 position of the primer strand. This is the only case that has been observed in RB69pol ternary complexes where a protein side interacts directly with the N3 of a purine or the O2 of a pyrimidine (which are hydrogen bond acceptors in the P/T duplex) that does not involve a water molecule. The epsilon amino nitrogen of K706 is sp3-hybridized and is in perfect tetrahedral geometry (Fig. 7B). It forms one HB with the carboxyl group of D621 and another with the W5 water molecule (Fig. 7B). The carbonyl oxygen of D621 interacts with metal ion A via a water molecule. The adjacent residue, D623, interacts with both metal ions, and is essential for catalysis. Therefore, K706 and D621 are indirectly involved in metal ion coordination, consistent with our previous mutagenesis studies.(33) The D621 to Ala replacement decreases incorporation efficiency of dGMP opposite dC by 103 fold. The K706A mutation almost eliminated pol activity. When dA was replaced with 3DA at the n-2 position of the primer strand, the side chain of K706 had to be repositioned to avoid a steric clash with the C3-H of 3DA and the epsilon amino group of K706. This repositioning of K706 affects the rotomer conformation of D621 and further interferes with the coordination of the metal ion A which has been proposed to activate the attacking 3’-OH of the primer-terminus. Thus, any alteration of metal ion A’s coordination would be expected to have a strong negative effect on catalysis. This could explain why the dA to 3DA substitution at the n-2 position of the primer strand resulted in a decrease in incorporation efficiency of three orders of magnitude.
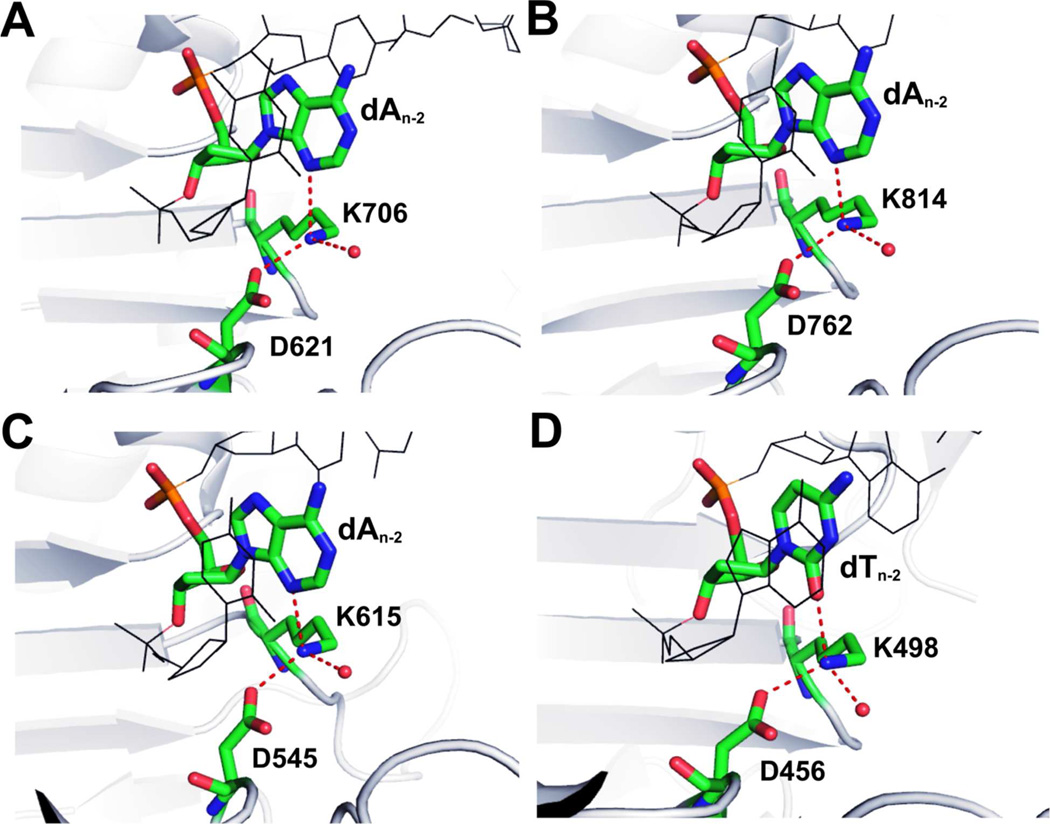
In addition, both K706 and D621 are highly conserved in the family B pols. For example, the corresponding residues are: K814 and D762 in Polδ;(48) K615 and D545 in PolII;(49) K498 and D456 in Phi29 pol.(50) Interestingly, the tetrahedral geometry of the Lys ε-amino nitrogen is also conserved. As shown in Fig 8, the ε-amino group of Lys forms one HB with the carboxyl group of Asp, one HB with a water molecule, and one HB with either N3 of a purine or O2 of a pyrimidine. In contrast, no HB is observed between K706 and D621 in the the apo structure of RB69pol.(11) For these reasons, we propose that the minor groove HB interaction at the n-2 position of the primer strand in RB69pol is essential for the correct orientation of K706 and D621 side chains and for metal ion A coordination. All these features are conserved in family B pols.
Figure 8.
The conserved tetrahedral geometry of the Lys ε-amino nitrogen in (A) RB69pol; (B) polδ; (C) polII; (D) phi29 DNA pol.
Probing minor groove HB interactions with nucleotide analogues
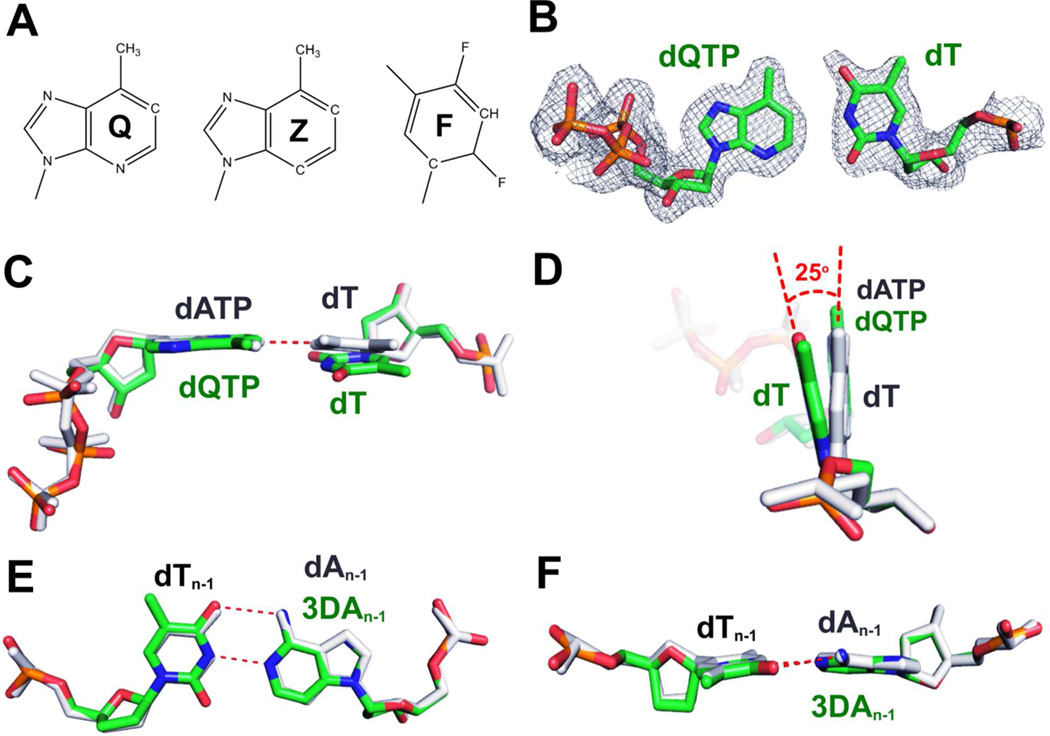
Since Steitz(51) pointed out that the minor groove HB interactions between protein side chains and the N3 of purines or the O2 of pyrimidines are important for replication fidelity, numerous kinetic and structural studies have been conducted on DNA pols from different families.(26, 52–56) In particular, various purine and pyrimidine analogues have been used to address the crucial role of minor groove HB in DNA replication. Morales et al were the first to use Q, Z and F (Fig. 9A) to study minor groove interactions between pol and DNA.(26, 55, 57) Q and Z are nonpolar purine analogues, and F was assumed to be a nonpolar pyrimidine analogue. According to Morales’s steady-state kinetics studies,(26) Klenow Fragment (KF) was able to bypass a dQ/dF pair 300 fold more efficiently than a dZ/dF pair. Since dZ lacks a minor groove HB acceptor at the C3 position of the purine ring, Morales et al concluded that a single minor groove interaction in the primer strand with KF was responsible for the 300 fold decrease in primer extension efficiency.(26) Nevertheless, a question remained as to whether Q, Z or F were appropriate nucleotide analogues to study minor groove HB interactions. We raise this issue because our recent report on dF-containing RB69 ternary complexes showed that dF can actually form HBs and thus F can’t be classified as a strictly non-polar isostere of dT.(28–29)
Figure 9.
The dQTP/dT-containing ternary complex of RB69pol triple mutant. (A) Chemical structure of Q, Z and F. (B) Final 2Fo-Fc electron density map for the dQTP/dT contoured at 1.6 σ. (C) Superposition of the dQTP/dT-containing ternary complex of RB69pol triple mutant with the dATP/dT-containing ternary complex of wt RB69pol. (D) An orthogonal view of panel C. (E) Superposition of the structure with control DNA to the structure with 3DA at the n-1 position of the template strand. (F) An orthogonal view of panel E.
In addition, we determined the structure of a dQTP/dT-containing ternary complex of RB69pol at 2.0 Å (data collection and structure refinement statistics of this structure is provided in the supplement). As shown in Fig 9B, there is no direct interbase HB between dQTP and dT. Superimposition of this structure with the dATP/dT-containing ternary complex of wt RB69pol shows that dT tilts 25° toward residue S565 to avoid a steric clash between dQTP’s C1-H and dT’s N3-H (Fig. 9C and 9D). As a result, the dQTP and dT are not in the same plane due to the interbase repulsive interaction. We also expect that dQ and dT will not be in the same plane when they are at the n-1 position in a P/T duplex, nor will the BPs of dQ/dF or dZ/dF. The resulting distortion of the DNA duplex at the P/T junction would be expected to cause misalignment between the 3’ hydroxyl group at the primer-terminus and the α–phosphorus atom of the incoming dNTP. Thus, the decrease in incorporation efficiency is caused not only by the absence of minor groove HB interactions but also by distortion of the BP at the n-1 position of the P/T duplex. Since N3 was replaced with C3 in dZ, the dZ/dF BP may be more tilted or distorted at the P/T junction compared to dQ/dF. Thus, it is very likely that lack of minor groove-pol HB interactions at the primer-template junction is not the sole reason for the 300 fold difference in bypass efficiency between dQ/dF and dZ/dF. In contrast, the structure of the ternary complex with dT/3DA at the n-1 position of template strand can be superimposed perfectly on the ternary complex containing dT/dA at the n-1 position of the template strand in the P/T duplex (Fig. 9E). 3DA and dT are in the same plane just like dT/dA (Fig. 9F). This illustrates why 3DA is a better purine analogue than Z or Q for probing minor groove HB interactions between the N3 of a purine and a pol. The observed 90 fold difference in the incorporation of correct dNMP past P/Ts containing dT/dA or dT/3DA at the primer-termini was due solely to the disruption of the minor groove HB network involving the n-1 position of the template strand.
Hendrickson et al(56) used 3-deaza-2-deoxyadnosine triphosphate (c3dATP) to probe minor groove interactions in three family A pols and three family B pols. However their primer-extension assays did not provide results that allowed quantitative evaluation of minor groove HB interactions with the pols. In addition, the three family B pols chosen were all thermostable pols with optimum temperatures for extension of about 72°C. Thus, it is not clear that their findings would apply to the majority of family B pols. In addition, pyrimidine analogues lacking the 2-keto group, such as 2-amino-5-(2’-deoxy-β-D-ribofuranosyl)-pyridine-5’-triphosphate (d*CTP) and 5-(2’-deoxy-β-D-ribofuranosyl)-3-methyl-2-pyridone-5’-triphosphate (d*TTP), were found to inhibit DNA pol activity.(54) It should be noted that no study has been conducted on the influence of the pyrimidine 2-keto group at the n-1 position of a DNA duplex where it has been shown that pol-minor groove HB interactions are conserved in family A pols.(15–17) McCain et al(58) determined steady-state and pre-steady-state kinetic parameters for DNA synthesis by KF using 10 different duplexes in which 3-deazaguanine (3DG) was placed at different positions. They found that R668 forms a HB fork between the minor groove of the primer-terminus and the deoxyribose ring oxygen of the incoming dNTP. Since KF is a model family A pol, our study on minor groove HB interactions using 3DA and RB69pol, a model family B pol complements the results of McCain et al.(58)
In summary, we have shown that minor groove HB interactions are essential for efficient nucleotide incorporation. For example the minor groove HB interaction mediated by one water molecule between the n-1 position of the template strand and Y567 is critical for efficient nucleotide incorporation; and the minor groove HB interaction at the n-2 position of the primer strand in RB69pol is essential for correctly orienting the K706 and D621 side chains and optimizing metal ion A coordination with D623 and D411. The absence of a single minor groove HB interaction can result in up to a 103 fold decrease in incorporation efficiency. In addition, our fluorescence lifetime measurements have shown that minor groove HBs help to stabilize primer/template (P/T) duplex in the pol domain. Our structures show that 3DA is an ideal analogue for probing minor groove HB interactions between pol and nucleobases in duplex DNA.
Supplementary Material
ACKNOWLEDGMENT
We thank the staff of the NE-CAT beamline 24-ID-E at the Advanced Photon Source of Argonne National Laboratory.
ABBREVIATIONS
- pol
polymerase
- exo
exonuclease
- RB69pol
RB69 DNA polymerase
- NBP
Nascent Base-pair binding Pocket
- P/T
primer/template
- W-C
Watson-Crick
- wt
wild type
- BP
Base-pair
- HB
hydrogen bonding or hydrogen bond
- KF
Klenow Fragment
- tCo
1,3,-diaza-2 oxophenoxazine
- 3DA
3-deaza-2’-deoxyadenosine
- 3DG
3-deaza-2’-deoxyguanisine
- 2AP
2-aminopurine
- F
2,4-difluorotoluene deoxynucleoside
- Z
4-methylbenzimidazole deoxynucleoside
- Q
9-methyl-1H-imidazo-[4,5-b]-pyridine
- dQTP
9-methyl-1H-imidazo-[4,5-b]-pyridine triphosphate
- c3dATP
3-deaza-2-deoxyadnosine triphosphate
- PEG 350 MME
polyethylene glycol 350 monomethyl ether
- Kd
dissociation constant for the P/T-polymerase binary complex
- Kd,app
the [dNTP] that gives half of the maximal rate
Footnotes
This work was supported by NIH RO1-GM063276-09 (to W.H.K.) and by SCSB-GIST (to J.W.).
SUPPORTING INFORMATION
Additional one table and three figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kunkel TA, Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim Biophys Acta. 1988;951:1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- 2.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 4.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 5.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 6.Loeb LA, Kunkel TA. Fidelity of DNA synthesis. Annu Rev Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 7.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 8.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 9.Schweitzer BA, Kool ET. Aromatic Nonpolar Nucleosides as Hydrophobic Isosteres of Pyrimidine and Purine Nucleosides. J Org Chem. 1994;59:7238–7242. doi: 10.1021/jo00103a013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 11.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 12.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 13.Kool ET, Sintim HO. The difluorotoluene debate--a decade later. Chem Commun (Camb) 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 14.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eom SH, Wang J, Steitz TA. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 16.Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spratt TE. Identification of hydrogen bonds between Escherichia coli DNA polymerase I (Klenow fragment) and the minor groove of DNA by amino acid substitution of the polymerase and atomic substitution of the DNA. Biochemistry. 2001;40:2647–2652. doi: 10.1021/bi002641c. [DOI] [PubMed] [Google Scholar]
- 19.Singh K, Modak MJ. Presence of 18-A long hydrogen bond track in the active site of Escherichia coli DNA polymerase I (Klenow fragment). Its requirement in the stabilization of enzyme-template-primer complex. J Biol Chem. 2003;278:11289–11302. doi: 10.1074/jbc.M211496200. [DOI] [PubMed] [Google Scholar]
- 20.Thompson EH, Bailey MF, van der Schans EJ, Joyce CM, Millar DP. Determinants of DNA mismatch recognition within the polymerase domain of the Klenow fragment. Biochemistry. 2002;41:713–722. doi: 10.1021/bi0114271. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 22.Polesky AH, Steitz TA, Grindley ND, Joyce CM. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 23.Polesky AH, Dahlberg ME, Benkovic SJ, Grindley ND, Joyce CM. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J Biol Chem. 1992;267:8417–8428. [PubMed] [Google Scholar]
- 24.Joyce CM, Steitz TA. Function and structure relationships in DNA polymerases. Annu Rev Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 25.Yang G, Lin T, Karam J, Konigsberg WH. Steady-state kinetic characterization of RB69 DNA polymerase mutants that affect dNTP incorporation. Biochemistry. 1999;38:8094–8101. doi: 10.1021/bi990653w. [DOI] [PubMed] [Google Scholar]
- 26.Morales JC, Kool ET. Minor Groove Interactions between Polymerase and DNA: More Essential to Replication than Watson-Crick Hydrogen Bonds? J Am Chem Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia S, Vashishtha A, Eom SH, Wang J, Konigsberg W. Contribution of partial chargeinteractions and base-stacking to the efficiency of primer-extension at and beyond abasic sites in DNA. Biochemistry. 2012 doi: 10.1021/bi300296q. (Under review). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia S, Konigsberg WH, Wang J. Hydrogen-bonding capability of a templating difluorotoluene nucleotide residue in an RB69 DNA polymerase ternary complex. J Am Chem Soc. 2011;133:10003–10005. doi: 10.1021/ja2021735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia S, Eom SH, Konigsberg WH, Wang J. Structural Basis for Differential Insertion Kinetics of dNMPs Opposite a Difluorotoluene Nucleotide Residue. Biochemistry. 2012;51:1476–1485. doi: 10.1021/bi2016487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braithwaite DK, Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry. 2002;41:2526–2534. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- 32.Yang G, Franklin M, Li J, Lin TC, Konigsberg W. A conserved Tyr residue is required for sugar selectivity in a Pol alpha DNA polymerase. Biochemistry. 2002;41:10256–10261. doi: 10.1021/bi0202171. [DOI] [PubMed] [Google Scholar]
- 33.Zakharova E, Wang J, Konigsberg W. The activity of selected RB69 DNA polymerase mutants can be restored by manganese ions: the existence of alternative metal ion ligands used during the polymerization cycle. Biochemistry. 2004;43:6587–6595. doi: 10.1021/bi049615p. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Beckman J, Wang J, Konigsberg W. RB69 DNA polymerase mutants with expanded nascent base-pair-binding pockets are highly efficient but have reduced base selectivity. Biochemistry. 2009;48:6940–6950. doi: 10.1021/bi900422b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, Xia S, Blaha G, Steitz TA, Konigsberg WH, Wang J. Insights into base selectivity from the 1.8 A resolution structure of an RB69 DNA polymerase ternary complex. Biochemistry. 2011;50:581–590. doi: 10.1021/bi101192f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia S, Wang M, Blaha G, Konigsberg WH, Wang J. Structural Insights into Complete Metal Ion Coordination from Ternary Complexes of B Family RB69 DNA Polymerase. Biochemistry. 2011;50:9114–9124. doi: 10.1021/bi201260h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia S, Wang M, Lee HR, Sinha A, Blaha G, Christian T, Wang J, Konigsberg W. Variation in mutation rates caused by RB69pol fidelity mutants can be rationalized on the basis of their kinetic behavior and crystal structures. J Mol Biol. 2011;406:558–570. doi: 10.1016/j.jmb.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia S, Eom SH, Konigsberg WH, Wang J. Bidentate and tridentate metal-ion coordination states within ternary complexes of RB69 DNA polymerase. Protein Sci. 2012;21:447–451. doi: 10.1002/pro.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reha-Krantz LJ, Hariharan C, Subuddhi U, Xia S, Zhao C, Beckman J, Christian T, Konigsberg W. Structure of the 2-aminopurine-cytosine base pair formed in the polymerase active site of the RB69 Y567A-DNA polymerase. Biochemistry. 2011;50:10136–10149. doi: 10.1021/bi2014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymology. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.The PyMOL Molecular Graphics System, Version 1.2r3pre. Schrodinger, LLC; [Google Scholar]
- 45.Borjesson K, Sandin P, Wilhelmsson LM. Nucleic acid structure and sequence probing using fluorescent base analogue tC(O) Biophys Chem. 2009;139:24–28. doi: 10.1016/j.bpc.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 46.Preus S, Borjesson K, Kilsa K, Albinsson B, Wilhelmsson LM. Characterization of nucleobase analogue FRET acceptor tCnitro. J Phys Chem B. 2010;114:1050–1056. doi: 10.1021/jp909471b. [DOI] [PubMed] [Google Scholar]
- 47.Xia S, Beckman J, Wang J, Konigsberg W. Using fluorescent cytosine analogue tCo to probe the effect of Y567 to Ala substitution on the pre-insertion steps of dNMP incorporation by RB69 DNA polymerase. Biochemistry. 2012 doi: 10.1021/bi300241m. (Under review). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat Struct Mol Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F, Yang W. Structural insight into translesion synthesis by DNA Pol II. Cell. 2009;139:1279–1289. doi: 10.1016/j.cell.2009.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berman AJ, Kamtekar S, Goodman JL, Lazaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steitz TA. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990;23:205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- 52.Cosstick R, Li X, Tuli DK, Williams DM, Connolly BA, Newman PC. Molecular recognition in the minor groove of the DNA helix. Studies on the synthesis of oligonucleotides and polynucleotides containing 3-deaza-2'-deoxyadenosine. Interaction of the oligonucleotides with the restriction endonuclease EcoRV. Nucleic Acids Res. 1990;18:4771–4778. [PMC free article] [PubMed] [Google Scholar]
- 53.Spratt TE. Klenow fragment-DNA interaction required for the incorporation of nucleotides opposite guanine and O6-methylguanine. Biochemistry. 1997;36:13292–13297. doi: 10.1021/bi971253g. [DOI] [PubMed] [Google Scholar]
- 54.Guo MJ, Hildbrand S, Leumann CJ, McLaughlin LW, Waring MJ. Inhibition of DNA polymerase reactions by pyrimidine nucleotide analogues lacking the 2-keto group. Nucleic Acids Res. 1998;26:1863–1869. doi: 10.1093/nar/26.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morales JC, Kool ET. Varied Molecular Interactions at the Active Sites of Several DNA Polymerases: Nonpolar Nucleoside Isosteres as Probes. J Am Chem Soc. 2000;122:1001–1007. doi: 10.1021/ja993464+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hendrickson CL, Devine KG, Benner SA. Probing minor groove recognition contacts by DNA polymerases and reverse transcriptases using 3-deaza-2'-deoxyadenosine. Nucleic Acids Res. 2004;32:2241–2250. doi: 10.1093/nar/gkh542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morales JC, Kool ET. Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat Struct Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 58.McCain MD, Meyer AS, Schultz SS, Glekas A, Spratt TE. Fidelity of mispair formation and mispair extension is dependent on the interaction between the minor groove of the primer terminus and Arg668 of DNA polymerase I of Escherichia coli. Biochemistry. 2005;44:5647–5659. doi: 10.1021/bi047460f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.