A novel hyperactive antifreeze protein from R. inquisitor (RiAFP) has been overexpressed, purified and crystallized. A complete native X-ray diffraction data set was recorded to 1.3 Å resolution.
Keywords: antifreeze proteins, Rhagium inquisitor
Abstract
Antifreeze proteins (AFPs) are a specialized evolutionary adaptation of a variety of bacteria, fish, arthropods and other organisms to inhibit ice-crystal growth for survival in harsh subzero environments. The recently reported novel hyperactive AFP from Rhagium inquisitor (RiAFP) is the second distinct type of AFP in beetles and its structure could reveal important molecular insights into the evolution of AFPs. For this purpose, RiAFP was overexpressed in Escherichia coli, purified and crystallized at 293 K using a combination of 23% PEG 3350 and 0.2 M ammonium sulfate as a precipitant. X-ray diffraction data were collected to 1.3 Å resolution using a synchrotron-radiation source. The crystals belonged to the trigonal space group P3121 (or P3221), with unit-cell parameters a = b = 46.46, c = 193.21 Å.
1. Introduction
The synthesis of antifreeze proteins (AFPs) is a clever evolutionary adaptation to ensure the survival of organisms inhabiting subzero environments. It is generally accepted that AFPs inhibit ice recrystallization by adsorbing irreversibly to the ice surface, increasing the curvature of the ice–water interface and resulting in thermal hysteresis: non-colligative depression of the freezing point without affecting the solvent melting point (Raymond & DeVries, 1977 ▶; Venketesh & Dayananda, 2008 ▶).
AFPs exhibit remarkable structural diversity across the various kingdoms, phyla and species: among fish alone four distinct structural types have been identified, three of which have been determined by high-resolution crystallography (Sicheri & Yang, 1995 ▶; Gronwald et al., 1998 ▶; Jia et al., 1996 ▶; Deng & Laursen, 1998 ▶). While the most active AFPs thus far are produced in insects, they have proven to be difficult to produce in sufficient quantities for crystallographic determination. To date, only three insect AFP structures have been resolved; while they consistently contain β-sheet motifs, they have distinct tertiary architectures (Graether et al., 2000 ▶; Leinala et al., 2002 ▶).
Recently, Kristiansen and coworkers isolated and sequenced a novel hyperactive antifreeze protein from the hemolymph of the Siberian cerambycid beetle Rhagium inquisitor (RiAFP; Kristiansen et al., 2005 ▶, 2008 ▶, 2011 ▶). RiAFP exhibits thermal hysteresis activity of over 6 K, making it among the most active AFPs discovered to date (Kristiansen et al., 2011 ▶). The primary sequence of RiAFP indicates that it is a small 12.8 kDa protein with a single disulfide bond, unlike AFPs from the spruce budworm Choristoneura fumiferana (CfAFP), which forms a three-sided β-helix with five disulfide bonds spanning the core, and the mealworm beetle Tenebrio molitor (TmAFP), which forms a single-sided β-helix with seven core disulfide bonds (Graether et al., 2000 ▶; Leinala et al., 2002 ▶). A similar pattern of disulfide stapling has been suggested for the fire-colored beetle Dendroides canadensis AFP (DcAFP) based on extensive studies using mass spectrometry (Li et al., 1998 ▶).
These observations suggest that RiAFP represents the first of a new type of AFPs found in beetles. Notably, a recent structural prediction of AFP from the inchworm Campaea perlata (iwAFP) using molecular dynamics suggested that iwAFP possesses a similar overall architecture to RiAFP (Lin et al., 2011 ▶), despite belonging to a different insect order. Structural comparisons of RiAFP and other insect AFPs may provide important molecular insights into their convergent evolution. Here, we report the successful overexpression, purification and crystallization of full-length RiAFP.
2. Experimental
2.1. Protein expression and purification
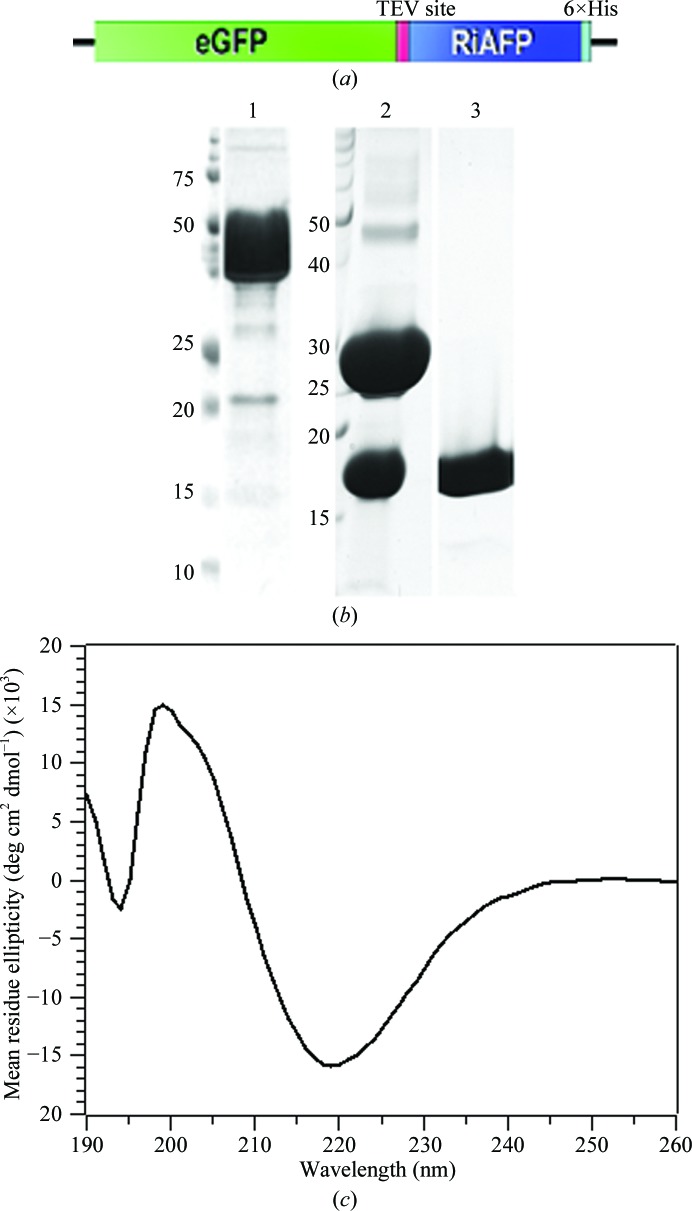
A fusion construct containing an N-terminal enhanced green fluorescent protein (eGFP), cleavable by tobacco etch virus (TEV) protease, and full-length RiAFP (gene accession HQ540314.1) was codon-optimized for Escherichia coli and synthesized (Integrated DNA Technologies). The gene was amplified by PCR for insertion into the pSB1AK8 plasmid, graciously donated by Rok Gaber (University of Ljubljana, Slovenia) for the International Genetically Engineered Machines (iGEM) competition. Briefly, the pSB1AK8 plasmid contains only a T7 promoter preceded by an SpeI cleavage site and followed by a PstI cleavage site. The forward primer, 5′-GTTTCTTCGAATTCGCGGCCGCTTCTAGAGTTAAGGAGGTAAAAAAAATGGTCTCCAAAGGTGAAGAGC-3′, contains an additional Shine–Dalgarno sequence, while the reverse primer, 5′-GTTTCTTCCTGCAGCGGCCGCTACTAGTAGCGAAAAAACCCCGCCGAAGCGGGGTTTTTTGCGTTAGTGATGATGATGATGATGGCTTGCGGTAACGGTGGTAGT-3′, contains a C-terminal His6 tag and T7 terminator. The resultant recombinant plasmid (Fig. 1 ▶ a) was transformed into E. coli strain BL21 (DE3) (Novagen) and grown at 310 K in Luria broth supplemented with 100 µg ml−1 ampicillin. Upon reaching log phase (OD600 = 0.5), cultures were cooled to 295 K, induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and grown for an additional 16 h.
Figure 1.
(a) Schematic of the eGFP-RiAFP fusion construct. (b) SDS–PAGE analysis of the GFP-RiAFP fusion construct (lane 1), TEV protease cleavage products (lane 2) and purified RiAFP after size-exclusion chromatography (lane 3). Molecular-weight markers are labeled in kDa. (c) The circular-dichroism (CD) spectrum of recombinant RiAFP reveals a minimum at 218 nm characteristic of β-strand secondary structures.
Cells were pelleted by centrifugation at 4700g for 45 min (278 K), resuspended in lysis buffer (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 1 mM PMSF) and lysed by sonication. The lysate was clarified by centrifugation at 13 000g for 60 min (278 K) and filtered through a 0.22 µm membrane. The supernatant was bound to nickel-affinity resin (GE Healthcare), washed over ten column volumes with lysis buffer supplemented with 20 mM imidazole and eluted with a gradient of increasing imidazole to 300 mM. Fractions containing RiAFP were pooled and incubated with 1:100(w:w) TEV protease overnight at 289 K. Cleaved RiAFP was purified by size-exclusion chromatography using a HiLoad 16/60 Superdex 75 column (GE Healthcare) equilibrated with 10 mM Tris–HCl pH 8.0, 100 mM NaCl. Peak fractions were concentrated in a centrifugal concentrator (Millipore) to 5 mg ml−1 as determined by SDS–PAGE against a lysozyme standard. The typical yield of purified recombinant protein was 50 mg per litre of cultured cells.
2.2. Circular-dichroism spectroscopy
In order to assess the secondary structure of recombinant RiAFP, circular-dichroism (CD) measurements were recorded for RiAFP in 50 mM sodium phosphate buffer pH 7.4, 100 mM NaCl. Each measurement was an average of three scans in steps of 1 nm at 293 K. The differential spectrum was obtained by subtracting the buffer spectrum before conversion to molar ellipticity. The characteristic minimum at 218 nm is suggestive of a folded protein consisting predominately of β-strands or β-sheets.
2.3. Crystallization
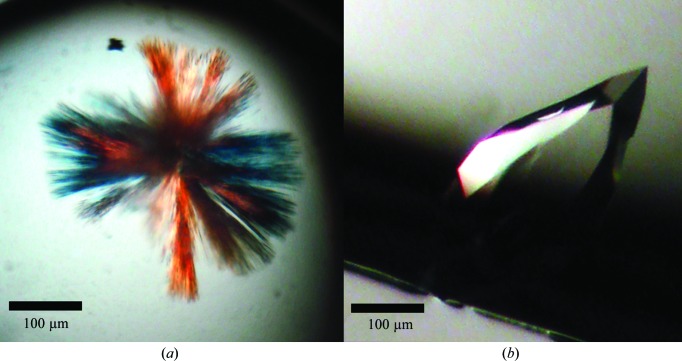
Initial crystallization conditions were established via sitting-drop vapor diffusion using the Wizard screens (Emerald BioSystems) and the JCSG+ Suite (Qiagen) in 96-well plates (Greiner) using a high-throughput nanolitre Mosquito robot (TTP LabTech). Sitting drops were prepared by mixing equal volumes (200 nl each) of the protein and reservoir solutions. Thin needle clusters emerged from a condition consisting of 25%(w/v) polyethylene glycol 3350 (PEG 3350), 0.2 M lithium sulfate, 0.1 M bis-Tris pH 5.5 (Fig. 2 ▶ a) within 5 d. Despite multiple rounds of optimization of pH, precipitant and salts, the crystals did not grow thicker and were not suitable for diffraction. Additional screening around polyethylene glycol (PEGs and PEGs II Suites; Qiagen) and ammonium sulfate (AmSO4 Suite; Qiagen) yielded thick rectangular crystals from 25% PEG 3350, 0.2 M ammonium sulfate. The best crystals grew after one week of incubation at 293 K in reservoir solution consisting of 23–25% PEG 3350, 0.2 M ammonium sulfate, 0.1 M sodium acetate pH 4.0 (Fig. 2 ▶ b). Incubations for longer than one week without crystal growth resulted in needle clusters. Both hanging and sitting drops were used for optimization, with no apparent effect on crystal morphology and diffraction.
Figure 2.
Native crystals of full-length RiAFP. (a) Initial needle clusters obtained using 25%(w/v) polyethylene glycol 3350, 0.1 M bis-tris pH 5.5, 0.2 M lithium sulfate. (b) Optimized crystals obtained using 23%(w/v) polyethylene glycol 3350, 0.1 M sodium acetate pH 4.0, 0.2 M ammonium sulfate.
2.4. X-ray data collection and processing
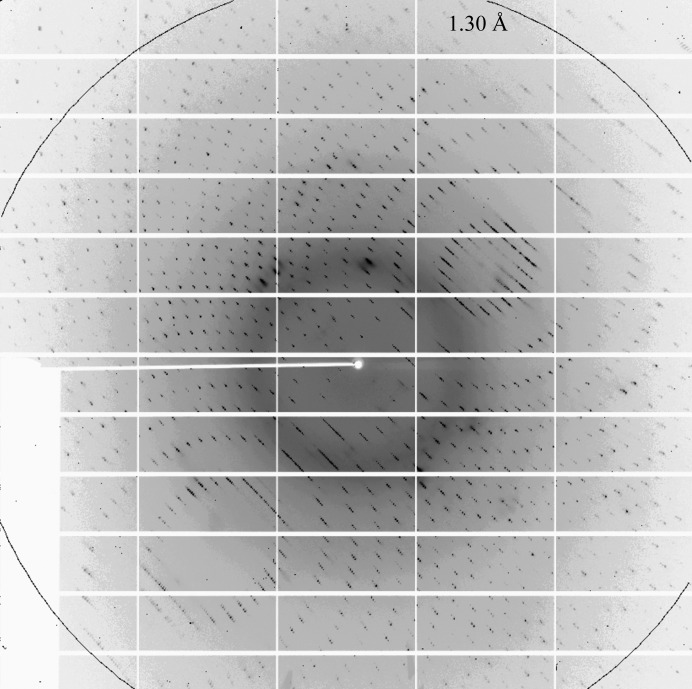
Native crystals were transferred from the mother liquor into cryoprotectant solution consisting of the reservoir solution supplemented with 25%(v/v) glycerol and were flash-frozen in liquid nitrogen. Native data were collected under a stream of nitrogen gas (100 K) on beamline X25 of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory. Owing to the problems of a relatively large unit cell and poor spot separation (Fig. 3 ▶), despite low mosaicity (Table 1 ▶) we opted to sacrifice resolution and collected data sets to various resolution cutoffs using a 0.3° oscillation width. Diffraction images were indexed, integrated and scaled using the XDS package (Kabsch, 2010 ▶).
Figure 3.
(a) A typical 0.3° oscillation diffraction image of the full-length RiAFP crystal, collected on a PILATUS 6M detector.
Table 1. Data-collection and reduction statistics for RiAFP.
Values in parentheses are for the highest resolution shell.
| Space group | P3121 or P3221 |
| Wavelength (Å) | 1.100 |
| Beamline | NSLS X25 |
| Resolution (Å) | 50–1.15 (1.18–1.15) |
| Unit-cell parameters (Å) | a = b = 46.46, c = 193.21 |
| No. of measured reflections | 541809 |
| No. of unique reflections | 136527 |
| Mosaicity (°) | 0.546 |
| Multiplicity | 6.4 (2.8) |
| 〈I/σ(I)〉 | 10.93 (1.58) |
| Completeness (%) | 81.8 (8.3) |
| Rmerge† (%) | 6.8 (20.1) |
| No. of molecules in asymmetric unit | 2 |
| Matthews coefficient (Å3 Da−1) | 2.15 |
| Solvent content (%) | 42.78 |
R
merge = 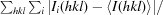
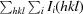 , where Ii(hkl) and 〈I(hkl)〉 are the intensity of the ith measurement and the mean intensity of the reflection with indices hkl, respectively.
, where Ii(hkl) and 〈I(hkl)〉 are the intensity of the ith measurement and the mean intensity of the reflection with indices hkl, respectively.
3. Results and discussion
Full-length AFP from R. inquisitor was cloned, overexpressed and purified to homogeneity from a cleavable N-terminal GFP fusion construct (Figs. 1 ▶ a and 1b). Notably, we observed little to no expression in E. coli from a construct of RiAFP without the GFP tag, suggesting that the presence of GFP promoted expression. These results corroborated a previous observation that AFPs, particularly those containing disulfide bonds, are notoriously difficult to express in E. coli (Bar et al., 2006 ▶).
Initial crystallization screening resulted in several hits for the native protein as needle clusters which were thin and not suitable for X-ray diffraction (Fig. 2 ▶ a). Follow-up screening and optimization yielded robust single crystals from a 1:1 mixture of protein and precipitant solution containing a combination of 23–25% PEG 3350 and 0.2 M ammonium sulfate. These crystals often had a rectangular prismatic morphology but tended to have poor-quality highly mosaic diffraction patterns. The best diffracting crystals had a teardrop-like morphology with average dimensions of 150 × 150 × 300 µm (Fig. 2 ▶ b). The crystals diffracted to 1.3 Å resolution (Fig. 3 ▶) and belonged to space group P3121 (or its enantiomorph P3221), with unit-cell parameters a = b = 46.46, c = 193.21 Å. The calculated Matthews coefficient V M (Matthews, 1968 ▶) of 2.15 A3 Da−1, corresponding to a solvent content of 42.78%, suggests the presence of two molecules per asymmetric unit.
Because RiAFP has no sequence similarity to any other proteins according to a PSI-BLAST search, we were interested in determining its secondary structure to find potential molecular-replacement models. Because AFPs within the same family tend to have similar structures, RiAFP was predicted to adopt a β-sheet structure similar to but distinct from CfAFP or TmAFP (Lin et al., 2011 ▶). Consistent with this hypothesis, CD measurements on the purified protein suggest that RiAFP is a well folded protein with significant β-strand content (Fig. 1 ▶ b).
Preliminary electron-density maps from molecular-replacement experiments using CfAFP (PDB entry 1m8n; Leinala et al., 2002 ▶) as a search model were promising and revealed a single six-stranded β-sheet. However, electron density connecting the individual β-strands was missing, and despite the high-resolution data the protein sequence could not be traced by automated model-building software such as Buccaneer (Cowtan, 2006 ▶) or ARP/wARP (Perrakis et al., 1999 ▶). Further attempts to solve the high-resolution structure using direct methods are currently under way.
Acknowledgments
The authors acknowledge N. Grindley and C. Joyce for use of laboratory space, Keck Biotechnology Resource Laboratory for sponsoring primers and sequencing and Integrated DNA Technologies for sponsorship of the eGFP-RiAFP fusion gene. We would also like to thank the personnel at beamline X25 of NSLS, Brookhaven. We acknowledge F. Isaacs and the Yale International Genetically Engineered Machines Team, C. Akusobi, K. Brower, A. Li, A. Lewis and S. Zhu, as well as W. Meng, B. Le, D. Spakowicz, T. Craggs, G. Blaha, C. A. Innis, T. Padukkavidana, A. Trexler, M. Heisig and D. Blaho and the Y. Modis, T. A. Steitz, R. H. G. Baxter and S. A. Strobel laboratories for helpful discussions and advice. This work was supported by Yale College Dean’s Fellowships to AH and DFZ, the Arnold and Mabel Beckman Foundation Scholarship awarded to DT, and NIH grant P01 GM022778, as well as the Raymond and Beverly Sackler Institute for Biological, Physical and Engineering Sciences, the Yale Science and Engineering Association (YSEA) and Yale Departments of Molecular Biophysics and Biochemistry (MB&B), Biomedical Engineering (BME), and Physics.
References
- Bar, M., Bar-Ziv, R., Scherf, T. & Fass, D. (2006). Protein Expr. Purif. 48, 243–252. [DOI] [PubMed]
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Deng, G. & Laursen, R. A. (1998). Biochim. Biophys. Acta, 1388, 305–314. [DOI] [PubMed]
- Graether, S. P., Kuiper, M. J., Gagné, S. M., Walker, V. K., Jia, Z., Sykes, B. D. & Davies, P. L. (2000). Nature (London), 406, 325–328. [DOI] [PubMed]
- Gronwald, W., Loewen, M. C., Lix, B., Daugulis, A. J., Sönnichsen, F. D., Davies, P. L. & Sykes, B. D. (1998). Biochemistry, 37, 4712–4721. [DOI] [PubMed]
- Jia, Z., DeLuca, C. I., Chao, H. & Davies, P. L. (1996). Nature (London), 384, 285–288. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kristiansen, E., Pedersen, S. A. & Zachariassen, K. E. (2008). Cryobiology, 57, 122–129. [DOI] [PubMed]
- Kristiansen, E., Ramløv, H., Hagen, L., Pedersen, S. A., Andersen, R. A. & Zachariassen, K. E. (2005). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 142, 90–97. [DOI] [PubMed]
- Kristiansen, E., Ramløv, H., Højrup, P., Pedersen, S. A., Hagen, L. & Zachariassen, K. E. (2011). Insect Biochem. Mol. Biol. 41, 109–117. [DOI] [PubMed]
- Leinala, E. K., Davies, P. L., Doucet, D., Tyshenko, M. G., Walker, V. K. & Jia, Z. (2002). J. Biol. Chem. 277, 33349–33352. [DOI] [PubMed]
- Li, N., Kendrick, B. S., Manning, M. C., Carpenter, J. F. & Duman, J. G. (1998). Arch. Biochem. Biophys. 360, 25–32. [DOI] [PubMed]
- Lin, F. H., Davies, P. L. & Graham, L. A. (2011). Biochemistry, 50, 4467–4478. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Perrakis, A., Morris, R. & Lamzin, V. S. (1999). Nature Struct. Biol. 6, 458–463. [DOI] [PubMed]
- Raymond, J. A. & DeVries, A. L. (1977). Proc. Natl Acad. Sci. USA, 74, 2589–2593. [DOI] [PMC free article] [PubMed]
- Sicheri, F. & Yang, D. S. C. (1995). Nature (London), 375, 427–431. [DOI] [PubMed]
- Venketesh, S. & Dayananda, C. (2008). Crit. Rev. Biotechnol. 28, 57–82. [DOI] [PubMed]