The Crimean–Congo haemorrhagic fever virus nucleocapsid protein was expressed in E. coli, purified and crystallized. X-ray diffraction data were collected to 2.1 Å resolution.
Keywords: Crimean–Congo haemorrhagic fever virus, nucleocapsid proteins
Abstract
Crimean–Congo haemorrhagic fever virus (CCHFV) is a member of the Nairovirus genus within the Bunyaviridae family of segmented negative-sense RNA viruses. This paper describes the expression, purification and crystallization of full-length CCHFV nucleocapsid (N) protein and the collection of a 2.1 Å resolution X-ray diffraction data set using synchrotron radiation. Crystals of the CCHFV N protein belonged to space group C2, with unit-cell parameters a = 150.38, b = 72.06, c = 101.23 Å, β = 110.70° and two molecules in the asymmetric unit. Circular-dichroism analysis provided insight into the secondary structure, whilst gel-filtration analysis revealed possible oligomeric states of the N protein. Structural determination is ongoing.
1. Introduction
The Bunyaviridae family contains at least 350 named species and is divided into the Orthobunyavirus, Tospovirus, Nairovirus, Phlebovirus and Hantavirus genera. All five genera contain species that impact on human health or welfare, either directly by causing disease or indirectly by impinging on the productivity of livestock or crop plants (Elliott, 2009 ▶). Classification into this family is based on the possession of a genome comprising three single-stranded RNA segments of either negative-sense or ambisense polarity, designated small (S), medium (M) and large (L) (Walter & Barr, 2011 ▶). Bunyavirus replication is confined to the cytoplasm of infected cells and virus particles are enveloped, with a diameter of approximately 80–120 nm.
Crimean–Congo haemorrhagic fever (CCHF) is a potentially fatal tick-borne viral zoonotic haemorrhagic disease within one of seven serogroups of the Nairovirus genus. CCHF virus (CCHFV) has been classified as an ‘emerging virus’ owing to increasing numbers of outbreaks associated with anthropogenic and climate factors which have the potential to alter host tick–virus dynamics, such as changes in global land use, agriculture and movement of livestock (Whitehouse, 2004 ▶; Maltezou et al., 2010 ▶). Similar to other tick-borne zoonotic agents, CCHFV normally circulates in nature unnoticed, causing symptomless infection in both ticks and animal vertebrates, both of which the virus life cycle relies upon (Ergönül, 2006 ▶). Human infection is characterized by a sudden febrile illness and in severe cases can progress to haemorrhagic fever manifestations with up to a 70% mortality rate (Schwarz et al., 1997 ▶). Despite the medical importance and severity of the disease, the molecular biology of CCHFV remains poorly understood, primarily because outbreaks are sporadic and its handling requires the highest containment facilities: biosafety level 4 (BSL-4). There is currently no vaccine or efficacious antiviral treatment for CCHFV infection (Vorou et al., 2007 ▶).
As is the case for other members of the Bunyaviridae family, the pathogenesis of CCHFV depends on formation of the ribonucleoprotein (RNP) complex, in which the CCHFV genome and positive-sense antigenome are entirely encapsidated by the S-segment-encoded nucleocapsid (N) protein. Only in the form of the RNP is the genome replicated, transcribed and packaged into new progeny particles. Furthermore, as all bunyaviruses lack a matrix protein, the RNP is also expected to directly interact with the viral glycoproteins, allowing packaging of the RNA segments into mature infectious particles (Rodgers et al., 2006 ▶; Estrada & De Guzman, 2011 ▶).
The only high-resolution structural information for a Bunyavirus N protein is for Rift Valley fever virus (RVFV), a member of the Phlebovirus genus. These data showed that the RVFV N protein forms a ring-shaped hexamer with sixfold symmetry. These hexamers reveal the structural basis for multimerization, as well as providing evidence for an RNA-binding groove populated with many positively charged amino acids postulated to bind the viral RNA within the RNP (Ferron et al., 2011 ▶; Raymond et al., 2010 ▶).
In contrast, no structural information has been reported to date for the CCHFV N protein. We have initiated the process of determining the three-dimensional structure of the CCHFV N protein in order to obtain insight into its functions. Here, we report the expression of CCHFV N in Escherichia coli, together with its purification and crystallization and initial analysis of the X-ray diffraction data.
2. Experimental procedures and results
2.1. Protein expression and purification
The cDNA encoding the full-length CCHFV N protein was cloned into pET28(a) downstream of the coding sequence for both SUMO (type 3) and a His6 epitope. The cDNA encoded an unpublished N-protein sequence with three conserved amino-acid substitutions (T111I, R195H and H445D) from the published Baghdad 12 strain (protein accession CAD61342.1). The His6 tag allowed metal-affinity chromatography of the protein; the His-SUMO fusion tag was cleavable using U1p protease. The pET28(a)-SUMO-N construct was transformed into E. coli BL21 Rosetta2 (Merck) and grown in Luria–Bertani broth (Fisher Scientific, Germany) supplemented with 50 µg ml−1 kanamycin and 34 µg ml−1 chloramphenicol at 310 K until the OD600 reached 0.7. Expression of the recombinant fusion protein was then induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 16 h at 291 K. Typically, 1.5 l cell culture was pelleted by centrifugation at 7660g for 12 min and resuspended in 25 ml lysis buffer [500 mM NaCl, 20 mM Tris–HCl pH 7, 0.1% Triton X-100, 5% glycerol (Sigma), 1 U deoxyribonuclease 1 (DNase I) (1 U µl−1; Invitrogen), 25 U Benzonase (25 U µl−1; Novagen), 7000 U chicken egg-white lysozyme solution (7000 U mg−1; Sigma), 1 mM MgCl2] and stored in a 50 ml Falcon tube on ice.
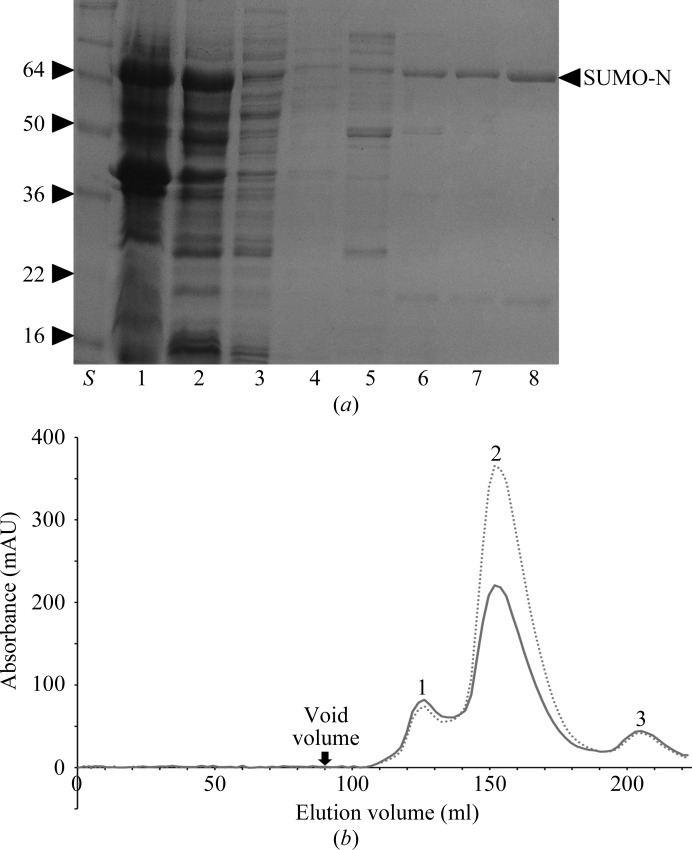
The cell suspension was lysed by sonication on ice and one slow freeze–thaw cycle. Soluble and insoluble fractions were separated by centrifugation at 23 488g for 15 min. Unless described otherwise, all subsequent steps were performed at 277 K. Typically, 7 ml nickel Superflow Plus resin (Generon) was added to the supernatant and the suspension was washed twice consecutively with wash buffer 1 (500 mM NaCl, 20 mM Tris–HCl pH 7), wash buffer 2 (500 mM NaCl, 50 mM imidazole, 20 mM Tris–HCl pH 7), wash buffer 3 (500 mM NaCl, 90 mM imidazole, 20 mM Tris–HCl pH 7) and wash buffer 4 (500 mM NaCl, 120 mM imidazole, 20 mM Tris–HCl pH 7). The bound protein was typically eluted using one column volume of elution buffer (300 mM imidazole, 500 mM NaCl, 20 mM Tris–HCl pH 7) and 50 µl from the elution was collected and resuspended in SDS–PAGE sample buffer for analysis (Fig. 1 ▶ a). The N-terminal His6-SUMO moiety tag was removed by digestion with SUMO protease (U1p), which cleaves between a glycine and a serine (QIG/SMENK), leaving a single non-native serine residue at the N-terminus of the CCHFV N protein. The cleaved protein was subject to size-exclusion chromatography (SEC) using an ÄKTApurifier (GE Healthcare) and a Superdex S75 16/60 gel-filtration preparative column equilibrated with 300 mM NaCl, 20 mM Tris–HCl, 1 mM EDTA pH 7 at a flow rate of 2.5 ml min−1. The UNICORN program (GE Healthcare) was used to operate all steps of the purification. Selected fractions were then combined and concentrated to 40 mg ml−1, and aliquots were stored at 277 K. Approximately 10 mg purified CCHFV N protein was obtained per 1.5 l of cell culture.
Figure 1.
Bacterially expressed CCHFV N protein fused to an N-terminal His6-SUMO tag was affinity-purified using Ni-Superflow Plus resin (Generon), after which the His6-SUMO moiety was cleaved by SUMO protease, and applied onto a Superdex S75 gel-filtration column at a flow rate of 2.5 ml min−1. (a) Coomassie Brilliant Blue R-250-stained 15% polyacrylamide-gel analysis of affinity purification of soluble SUMO-CCHFV N protein. Lane S, molecular-weight markers (labelled in kDa). Lane 1, insoluble fraction; Lane 2, soluble fraction. Lane 3, flowthrough. Lane 4, wash. Lane 5, 50 mM imidazole wash. Lane 6, 90 mM imidazole wash. Lane 7, 120 mM imidazole wash. Lane 8, 300 mM imidazole elution (SUMO-CCHFV N protein is marked by an arrow). (b) The SEC elution profile of SUMO-cleaved full-length CCHFV N protein is shown as a dotted line and a solid line at 280 and 260 nm wavelength, respectively. Peaks are labelled as follows: 1, CCHFV N dimer; 2, CCHFV N monomer; 3, His6-SUMO.
2.2. Biochemical analysis of purified CCHFV N protein
During purification, the CCHFV N protein eluted from the size-exclusion column in two forms: a highly abundant slow-eluting species and a less abundant form that eluted more rapidly. Comparison of these two species with molecular-weight standards indicated that the slower eluting peak 2 had an apparent mass of 55 kDa according to comparisons with calibrated standards and was likely to be N-protein monomer, which has a predicted mass of 54 kDa. In contrast, the faster eluting species in peak 1 had an apparent mass of 110 kDa and was likely to be N-protein dimer (Fig. 1 ▶ b). The OD260nm/OD280nm ratios of the two peaks were measured to determine the presence of bound nucleic acid. The OD260nm/OD280nm ratio of peak 1 (1.10) was considerably higher than that of peak 2 (0.6). The monomeric and nucleic acid-free CCHFV N protein from peak 2 was chosen for crystallization trials owing to its increased abundance and homogeneity.
The CCHFV N protein purified from peak 2 was subjected to circular dichroism to obtain insight into its secondary structure. Recombinant CCHFV N protein was diluted to a concentration of 0.5 mg ml−1 in 50 mM sodium phosphate buffer pH 7.5; 200 µl filtered protein solution was placed in a 1 mm path-length far- and near-UV CD cuvette (Hellma precision cells, synthetic far-UV quartz). CD spectra were measured between 260 and 180 nm at a scanning speed of 1 nm s−1 using a Chirascan CD spectrometer (Applied Photophysics). Two spectra were recorded and averaged, followed by baseline correction via subtraction of the buffer spectrum. The mean residue ellipticity was calculated and expressed in units of deg cm2 dmol−1.
2.3. Crystallization and data collection
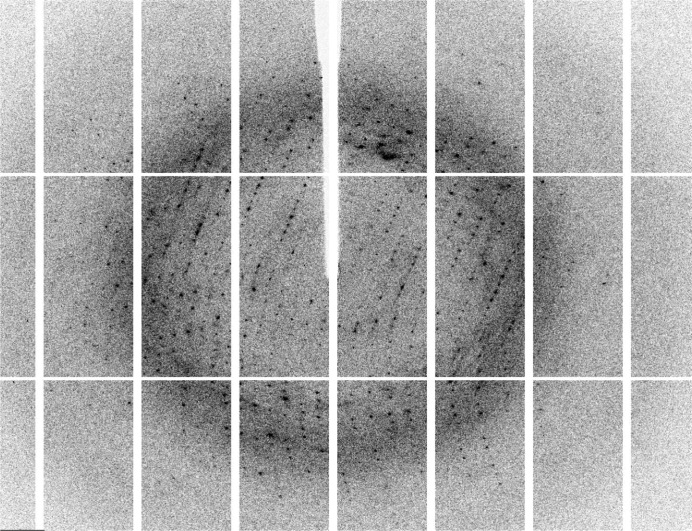
The concentration of the purified CCHFV N monomeric protein was estimated by the Bradford assay (Bio-Rad Protein Dye Reagent; Bio-Rad) using BSA as a standard and concentrated to 40 mg ml−1. Crystallization trials using the monomeric protein purified from SEC were carried out at 298 K using the sitting-drop vapour-diffusion method in MRC plates (Molecular Dimensions). Crystallization was performed with Crystal Screen, Crystal Screen 2, Index, SaltRx (Hampton Research), Wizard 1 and Wizard 2 (Emerald BioSystems) using an Oryx 6 crystallization robot (Douglas Instruments). Initial crystal hits were seen in 0.2 M magnesium acetate tetrahydrate, 0.1 M sodium cacodylate trihydrate pH 6.5, 20%(w/v) polyethylene glycol (PEG) 8000. Sequential rounds of optimization were carried out using a range of PEGs, different buffers (MOPS, MES and BTP) and salts; ultimately, diffraction-quality crystals were grown in 0.2 M NaCl, 25%(v/v) PEG 600, 0.1 M bis-Tris propane (BTP) pH 6.5 using the hanging-drop vapour-diffusion method (Fig. 2 ▶). X-ray diffraction data were collected from crystals cryocooled straight from the drop and data were collected at 100 K on beamline I04-1 at Diamond Light Source (UK) using 13.516 keV X-rays with a Pilatus 2M detector, a crystal-to-detector distance of 267.7 mm and 0.2° oscillations (Fig. 3 ▶). The best data were collected to a maximum Bragg spacing of 2.1 Å with an average mosaicity of 0.46°. Data were indexed and integrated using the MOSFLM package (Leslie, 2006 ▶) and scaled using SCALA (Evans, 2006 ▶) from the CCP4 suite (Winn et al., 2011 ▶).
Figure 2.

Monoclinic crystals of the CCHFV N protein grown in 0.2 M NaCl, 25%(v/v) PEG 600, 0.1 M bis-Tris propane (BTP) pH 6.5 at 298 K. The crystals were approximately 75 × 75 × 75 µm in size.
Figure 3.
Diffraction image of a CCHFV N crystal. The oscillation was 0.2° and reflections were observed to beyond 2.1 Å.
The CCHFV N protein crystals belonged to the monoclinic system; the space group was identified as C2. Diffraction data statistics are summarized in Table 1 ▶. The Matthews coefficient (V M) was calculated and revealed that there are likely to be two copies of the CCHFV N protein in the asymmetric unit with a V M of 2.47 Å3 Da−1, corresponding to a solvent content of 50.17%.
Table 1. Crystal parameters, diffraction data and processing statistics for the CCHFV N protein.
Values in parentheses are for the highest resolution shell.
| Space group | C2 |
| No. of molecules in asymmetric unit | 2 |
| Unit-cell parameters | |
| a (Å) | 150.38 |
| b (Å) | 72.06 |
| c (Å) | 101.23 |
| β (°) | 110.70 |
| Wavelength (Å) | 0.9173 |
| Resolution (Å) | 48.79–2.10 (2.21–2.10) |
| Rmerge† | 0.044 (0.467) |
| Rp.i.m.‡ | 0.028 (0.301) |
| No. of unique reflections | 56441 (8347) |
| Wilson B factor (Å2) | 35.5 |
| Completeness (%) | 95.5 (97.2) |
| Multiplicity | 3.3 (3.3) |
| 〈I/σ(I)〉 | 13.9 (2.5) |
R
merge = 
 .
.
R
p.i.m. = 
 .
.
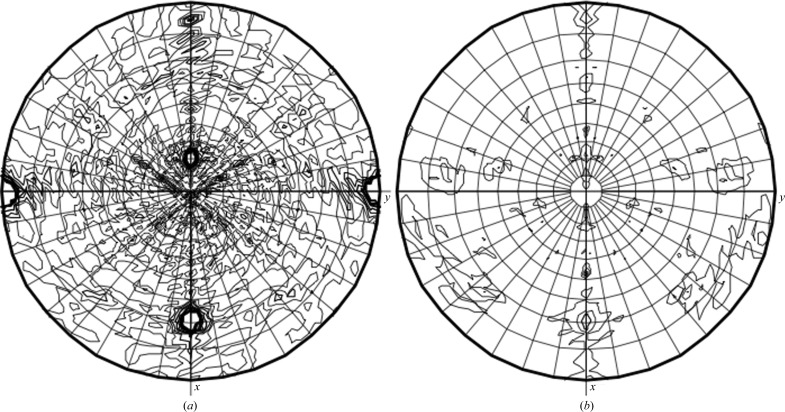
Self-rotation functions were calculated with the MOLREP program (Vagin & Teplyakov, 2010 ▶). The two strongest peaks for κ = 180° at ϕ = 180° and ϕ = 0° are derived from the crystallographic twofold-symmetry axes. The two additional peaks observed on the κ = 180° plot correspond to noncrystallographic twofold symmetry (Fig. 4 ▶). Considering that a potential dimer was observed during SEC, in conjunction with the twofold noncrystallographic symmetry, a dimeric form is likely to be present in the asymmetric unit. There was no evidence of fourold or sixfold crystallographic symmetry for κ = 90° (Fig. 4 ▶) or κ = 60° (data not shown). Tetrameric or hexameric complexes similar to those seen for other bunyaviruses do not appear to be present, unless a combination of asymmetric units results in, for instance, a tetramer (Mohl & Barr, 2009 ▶). It is also possible that apo CCHFV N does not oligomerize, or that the twofold NCS is unrelated to higher order RNP assemblies.
Figure 4.
Self-rotation function calculated by MOLREP. Self-rotation searches with (a) κ = 180° and (b) κ = 90° were used to identify twofold and fourfold NCS axes, respectively.
The molecular weight of the CCHFV N protein (54 kDa) is significantly greater than that of the RVFV N protein (22 kDa) and is similar to that of the Lassa fever virus N protein (63 kDa), which contains an extra domain with exonuclease activity. Analysis of the CD spectrum suggested predominately α-helical content (Fig. 5 ▶). The percentage of each type of secondary structure was predicted to be 66% α-helix, 0.005% β-sheet and 29% random coil by the k2d neural-network program. The high proportion of α-helices in the CCHFV N protein is common to many negative-sense RNA virus nucleocapsid proteins and is consistent with the crystal structure of the RVFV N protein (Ferron et al., 2011 ▶; Raymond et al., 2010 ▶). Furthermore, the distinctive α-helical spectrum suggests that very little β-sheet secondary structure was present and suggests that the CCHFV N protein is unlikely to possess domains homologous to the Lassa fever virus N protein exonuclease C-terminal domain, which is predominantly composed of β-sheet. However, additional functional domains cannot be ruled out.
Figure 5.
Circular-dichroism spectrum of the CCHFV N protein.
3. Discussion
We report the first crystallization of an N protein from the Nairovirus genus of the Bunyaviridae family. Attempts to determine initial phase information by molecular-replacement approaches using a monomer and a hexamer of the recently solved RVFV N protein (Raymond et al., 2010 ▶; Ferron et al., 2011 ▶) as a search model failed. This may reflect significant differences in secondary, tertiary and quaternary structure between these two nucleocapsids. These differences are also highlighted by comparison with the similar-sized nucleocapsid of the hantaviruses, which has been proposed to form trimers as intermediates in RNP assembly (Kaukinen et al., 2004 ▶). Our data suggest that the CCHFV N protein may potentially utilize a dimeric intermediate for RNP formation and provides further evidence that bunyaviruses may not utilize a common strategy of RNP formation (Walter & Barr, 2011 ▶). From this structure, we hope to obtain insight into RNA binding, RNP formation and virus assembly.
Acknowledgments
We thank Dr Roger Hewson at the HPA for supplying the CCHFV N cDNA. We would also like to thank the beamline scientists at the Diamond Light Source and DORIS (HASYLAB, Hamburg) for their excellent support. Circular dichroism was performed with the help of G. Nasir Khan. Additional thanks is given to Drs Arwen Pearson and Chi Trinh for support with data collection and X-ray beam time provided by Diamond Light Source (UK).
References
- Elliott, R. M. (2009). Clin. Microbiol. Infect. 15, 510–517.
- Ergönül, O. (2006). Lancet Infect. Dis. 6, 203–214. [DOI] [PMC free article] [PubMed]
- Estrada, D. F. & De Guzman, R. N. (2011). J. Biol. Chem. 286, 21678–21686. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Ferron, F., Li, Z., Danek, E. I., Luo, D., Wong, Y., Coutard, B., Lantez, V., Charrel, R., Canard, B., Walz, T. & Lescar, J. (2011). PLoS Pathog. 7, e1002030. [DOI] [PMC free article] [PubMed]
- Kaukinen, P., Kumar, V., Tulimäki, K., Engelhardt, P., Vaheri, A. & Plyusnin, A. (2004). J. Virol. 78, 13669–13677. [DOI] [PMC free article] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Maltezou, H. C., Andonova, L., Andraghetti, R., Bouloy, M., Ergonul, O., Jongejan, F., Kalvatchev, N., Nichol, S., Niedrig, M., Platonov, A., Thomson, G., Leitmeyer, K. & Zeller, H. (2010). Euro Surveill. 15, 19504. [PubMed]
- Mohl, B.-P. & Barr, J. N. (2009). RNA, 15, 391–399. [DOI] [PMC free article] [PubMed]
- Raymond, D. D., Piper, M. E., Gerrard, S. R. & Smith, J. L. (2010). Proc. Natl Acad. Sci. USA, 107, 11769–11774. [DOI] [PMC free article] [PubMed]
- Rodgers, J. W., Zhou, Q., Green, T. J., Barr, J. N. & Luo, M. (2006). Acta Cryst. F62, 361–364. [DOI] [PMC free article] [PubMed]
- Schwarz, T. F., Nsanze, H. & Ameen, A. M. (1997). Infection, 25, 364–367. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Vorou, R., Pierroutsakos, I. N. & Maltezou, H. C. (2007). Curr. Opin. Infect. Dis. 20, 495–500. [DOI] [PubMed]
- Walter, C. T. & Barr, J. N. (2011). J. Gen. Virol. 92, 2467–2484. [DOI] [PubMed]
- Whitehouse, C. A. (2004). Antiviral Res. 64, 145–160. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.