The expression, purification and crystallization of the alginate lyase AlgL from P. aeruginosa is described. The crystals diffracted to a resolution of 1.64 Å.
Keywords: AlgL, Pseudomonas aeruginosa, alginate biosynthetic complex, biofilms, exopolysaccharides, alginate lyases
Abstract
The periplasmic alginate lyase AlgL is essential for the synthesis and export of the exopolysaccharide alginate in Pseudomonas sp. and also plays a role in its depolymerization. P. aeruginosa PAO1 AlgL has been overexpressed and purified and diffraction-quality crystals were grown using the hanging-drop vapour-diffusion method. The crystals grew as thin plates, with unit-cell parameters a = 56.4, b = 59.6, c = 102.1 Å, α = β = γ = 90°. The AlgL crystals exhibited the symmetry of space group P212121 and diffracted to a minimum d-spacing of 1.64 Å. Based on the Matthews coefficient (V M = 2.20 Å3 Da−1), one molecule is estimated to be present in the asymmetric unit.
1. Introduction
Pseudomonas aeruginosa is an opportunistic pathogen that forms chronic biofilm infections in the lungs of cystic fibrosis (CF) patients. Following initial colonization, P. aeruginosa converts over time in the oxidative environment of the CF lung to a mucoid phenotype, which is characterized by the overproduction of the exopolysaccharide alginate (Govan & Deretic, 1996 ▶; Breidenstein et al., 2011 ▶). Alginate is a random linear polysaccharide composed of β-(1–4)-linked d-mannuronate and its C5 epimer α-l-guluronate (Evans & Linker, 1973 ▶).
Alginate is synthesized by the 12 proteins encoded on the algD operon, AlgD/8/44/K/E/G/X/L/I/J/F/A, and AlgC. AlgC is also involved in rhamnolipid and LPS metabolism and is not linked to the algD operon. The biosynthesis of alginate begins with d-fructose 6-phosphate, which is converted to GDP-d-mannuronic acid in the cytoplasm by the enzymes AlgA, AlgC and AlgD (Franklin et al., 2011 ▶). Polymerization occurs on the cytoplasmic face of the inner membrane and both the putative glycosyl transferase Alg8 and the bis-(3′,5′)-cyclic dimeric guanosine monophosphate-binding Alg44 are required (Oglesby et al., 2008 ▶; Merighi et al., 2007 ▶; Remminghorst & Rehm, 2006a ▶,b ▶; Remminghorst et al., 2009 ▶). Within the periplasm, residues in the polymannuronate polymer can be modified by addition of acetyl groups to their O2′ and/or O3′ hydroxyls through the concerted action of AlgI, AlgJ and AlgF (Franklin & Ohman, 2002 ▶), epimerized to α-l-guluronate by AlgG (Franklin et al., 1994 ▶) or remain unaltered. As the polysaccharide passages through the periplasm to AlgE, the outer membrane β-barrel porin, it is believed to be protected by a multi-protein complex consisting of Alg44, AlgG, AlgK, AlgX and AlgL (Franklin et al., 2011 ▶). While the function of AlgX is unknown, the presence of multiple copies of the tetratricopeptide-like protein–protein interaction motif in AlgK has led to the proposal that it is a scaffold protein that assembles the periplasmic biosynthetic complex (Keiski et al., 2010 ▶). AlgL is an alginate lyase which appears to have two roles in alginate biosynthesis: (i) as part of the multi-protein alginate-secretion complex required for export of the polymer (Jain & Ohman, 2005 ▶; Albrecht & Schiller, 2005 ▶) and (ii) depolymerizing alginate that escapes into the periplasm (Bakkevig et al., 2005 ▶; Jain & Ohman, 2005 ▶).
AlgL is a 40 kDa poly-(β-d-mannuronate) lyase that preferentially degrades deacetylated polymannuronate via a β-elimination reaction, resulting in an unsaturated uronic acid at the nonreducing end of the molecule (Linker & Evans, 1984 ▶). AlgL is an endolytic enzyme that cleaves the 1–4 glycosidic linkage, resulting in disaccharides and trisaccharides as its major products (Wong et al., 2000 ▶; Linker & Evans, 1984 ▶). The Phyre 2 structure-prediction server (Kelley & Sternberg, 2009 ▶) suggests that AlgL has an α6/α5-barrel fold and is similar to Sphingomonas spp. alginate lyase A1-III (Yoon et al., 1999 ▶). This structure prediction places AlgL in polysaccharide family 5 (Lombard et al., 2010 ▶). Using site-directed mutagenesis and residue-conservation analysis, several residues, including His202 and Tyr256, have been suggested to play a role in AlgL activity; however, the exact mechanism of catalysis and the molecular determinants of substrate specificity are not yet known (Albrecht & Schiller, 2005 ▶; Preston et al., 2000 ▶, 2001 ▶).
In addition to its enzymatic function, AlgL appears to be required for alginate export, as alginate accumulates in the periplasm of algL deletion strains (Jain & Ohman, 2005 ▶; Bakkevig et al., 2005 ▶). In vivo, point mutants that disrupt lyase activity result in a non-mucoid phenotype, indicating that AlgL activity is crucial for alginate export and/or biosynthesis (Bakkevig et al., 2005 ▶; Albrecht & Schiller, 2005 ▶). Thus, the scaffolding complex responsible for the export of alginate across the periplasm is hypothesized to include AlgL (Franklin et al., 2011 ▶).
To understand the catalytic mechanism of AlgL and the role that it plays in the overall process of alginate biosynthesis and export, we have undertaken structural studies of this protein. Here, we describe the overexpression, purification and crystallization of AlgL.
2. Materials and methods
2.1. Cloning and expression
The nucleotide sequence of algL (PA3547) from P. aeruginosa PAO1 was obtained from the Pseudomonas Genome Database (Stover et al., 2000 ▶; Winsor et al., 2011 ▶) and used to design gene-specific primers. AlgL minus its signal sequence was amplified from genomic DNA using the forward and reverse primers 5′-TTGTATTTCCAGGGCGCCGACCTGGTACCCCCG-3′ and 5′-CAAGCTTCGTCATCAACTTCCCCCTTCGCGGCTG-3′. The amplified PCR product was treated with BD In-Fusion enzyme (BD Biosciences) and cloned into BseRI-linearized pET28-MHL vector (Structural Genomics Consortium, Toronto) using ligation-independent cloning (LIC). The fidelity of the algL nucleotide sequence was confirmed by DNA sequencing (ACGT DNA Technologies Corporation). The resulting expression vector (pNMAlgL28–367) encodes residues 28–367 of AlgL fused to a TEV-cleavable N-terminal His6 tag (His6AlgL28–367) for purification purposes. As repeated attempts to crystallize the full-length protein failed to yield any crystallization hits, a new construct was made based on limited proteolysis experiments and mass-spectrometric analysis (data not shown). The new construct, pNMAlgL28–362, removes the last five residues from the C-terminal end of the protein. This construct was generated by amplification from genomic DNA as described above using the same forward primer and the reverse primer 5′-CAAGCTTCGTCATCAGCTGAACACCCGCGTCACTTC-3′. The resulting vector enables the expression of N-terminally His6-tagged AlgL28–362 under the control of an isopropyl β-d-1-thiogalactopyranoside (IPTG) inducible promoter.
Prior to its expression, the pNMAlgL28–362 expression vector was transformed into Escherichia coli Origami 2 (DE3) competent cells. The cells were grown in 1 l Luria–Bertani (LB) broth supplemented with 0.05 mg ml−1 kanamycin and 0.0125 mg ml−1 tetracycline at 310 K. The cells were induced with IPTG at a final concentration of 1.0 mM when the OD600 of the cell culture reached 0.6. The induced cells were then incubated for 20 h at 292 K and harvested via centrifugation at 6260g for 30 min at 277 K. The resulting cell pellet was stored at 193 K until required.
2.2. Purification
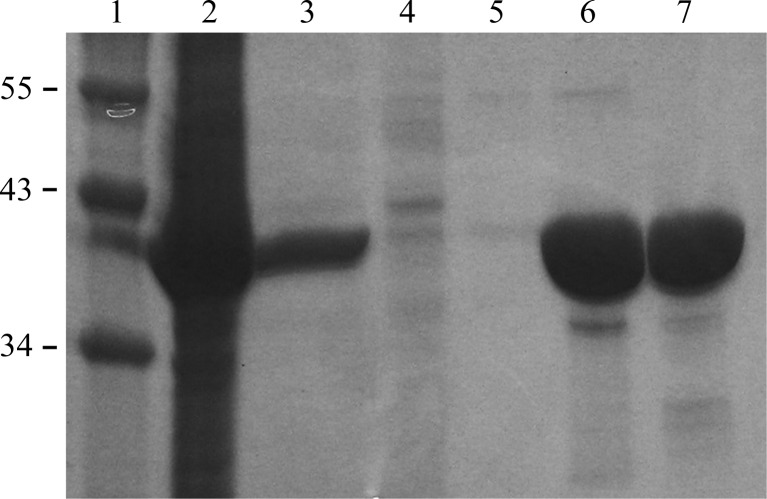
To purify the His6-AlgL28–362 protein, the cell pellet from a 2 l bacterial culture was thawed and resuspended in 40 ml buffer A (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 10 mM imidazole) containing one tablet of SIGMAFAST EDTA-free protease-inhibitor cocktail (Sigma). The resuspended medium was lysed by sonication (Misonix Sonicator 3000) while cooling on ice for 2 min (5 s on/10 s off) or until the lysate was translucent. The insoluble cell lysate was removed by centrifugation at 34 000g for 30 min at 277 K. The supernatant was then loaded onto a 5 ml Ni2+–NTA Superflow Cartridge (Qiagen) which had been pre-equilibrated with buffer A. The column was washed with at least five column volumes of buffer A to remove any contaminants. Bound AlgL was eluted with five column volumes of buffer B (50 mM Tris–HCl pH 7.5, 300 mM NaCl, 200 mM imidazole) at a flow rate of 5 ml min−1. The elution peak was pooled and concentrated by centrifugation (1240g at 277 K) using a Millipore concentrator with a 10 kDa molecular-weight cutoff. AlgL was then further purified and buffer-exchanged into buffer C (20 mM Tris–HCl pH 7.5, 150 mM NaCl) by size-exclusion chromatography using a HiLoad 16/60 Superdex 200 prep-grade gel-filtration column (GE Healthcare). Fractions corresponding to AlgL were collected and concentrated to 8 mg ml−1 before flash-cooling and could be stored at 193 K for up to one month without compromising the ability to form crystals. The purity of the protein was visualized at all stages using SDS–PAGE (Fig. 1 ▶).
Figure 1.
SDS–PAGE analysis of AlgL expression and purification. Lane 1, molecular-weight markers (labelled in kDa); lane 2, cell pellet; lane 3, soluble cell lysate; lane 4, Ni–NTA flowthrough; lane 5, Ni–NTA column wash; lane 6, purified His6-AlgL28–362 following Ni–NTA chromatography; lane 7, after size-exclusion chromatography.
2.3. Crystallization
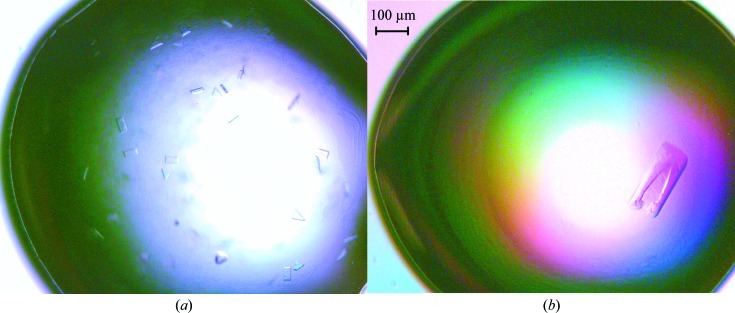
Initial crystallization trials were set up with six different sparse-matrix screens from Emerald BioSystems and Hampton Research. The trials were set up in 48-well VDX plates (Hampton Research) by hand with 2 µl protein solution (8 mg ml−1) and mother liquor in a 1:1 ratio over a reservoir containing 0.2 ml mother liquor. The crystal trays were stored at 293 K. These trials yielded a number of hits in various conditions, most of which contained a low-molecular-weight polyethylene glycol (PEG) as the precipitant. The best crystals were obtained from condition No. 9 [0.2 M ammonium acetate, 0.1 M sodium citrate tribasic dihydrate pH 5.6, 30%(w/v) PEG 4000] from Crystal Screen (Hampton Research; Fig. 2 ▶ a). Optimization of the crystallization conditions was performed by varying the precipitant concentration and the buffer pH and carrying out an additive screen (ADDit, Emerald BioSystems). The optimized condition [0.2 M ammonium acetate, 0.1 M sodium citrate tribasic dihydrate pH 4.6, 28%(w/v) PEG 4000, 0.3 M PIPES] yielded diffraction-quality crystals that grew as thin plates to maximum dimensions of 275 × 100 × 10 µm and took approximately 4 d to grow (Fig. 2 ▶ b).
Figure 2.
Crystals of AlgL. (a) Initial crystals grown from Hampton Research Crystal Screen condition No. 9; (b) crystals after optimization. The optimized crystals with dimensions of 275 × 100 × 10 µm were grown in 28%(w/v) PEG 4000, 0.1 M sodium citrate tribasic dihydrate pH 4.6, 0.2 M ammonium acetate, 0.3 M PIPES.
2.4. Data collection
To prepare them for data collection, the crystals were cyroprotected in well solution with the addition of 20%(v/v) glycerol. The cryoprotectant was added stepwise to the drop and the crystals were soaked for 25–30 s prior to vitrification in liquid nitrogen. The crystals were screened in-house using the X-ray diffraction facilities at The Hospital for Sick Children (Cu Kα X-ray radiation from an RU-H3R rotating-anode generator with an R-AXIS IV++ image-plate detector) prior to shipping the crystals to the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory. X-ray diffraction data were collected on beamline X29. 360 images of 1° ϕ oscillation were collected on an ADSC Q315 CCD detector with a 240 mm crystal-to-detector distance and an exposure time of 0.5 s per image. The data were processed using DENZO and integrated and scaled using SCALEPACK from the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
3. Results
The periplasmic alginate lyase AlgL has been expressed and purified to near-homogeneity (∼98%; Fig. 1 ▶). Approximately 8 mg purified His6-AlgL protein could be obtained per litre of cell culture. The crystals diffracted to 1.64 Å resolution on average. The crystals belonged to space group P212121, with unit-cell parameters a = 56.4, b = 59.6, c = 102.1 Å, α = β = γ = 90°. The data-collection statistics are summarized in Table 1 ▶. Density calculations (Matthews, 1968 ▶) predict that each asymmetric unit consists of one monomer of AlgL (V M = 2.20 Å3 Da−1). Currently, we are in the process of determining the structure of AlgL using selenomethionine incorporation and the anomalous dispersion technique (Hendrickson, 1991 ▶).
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.075 |
| Temperature (K) | 100 |
| Space group | P212121 |
| Unit-cell parameters (Å, °) | a = 56.4, b = 59.6, c = 102.1, α = β = γ = 90 |
| Resolution (Å) | 50.00–1.64 (1.70–1.64) |
| Total No. of reflections | 542308 |
| No. of unique reflections | 43027 |
| Multiplicity | 13.2 (10.6) |
| Completeness (%) | 95.2 (71.2) |
| Average I/σ(I) | 25.8 (3.2) |
| Rmerge† (%) | 8.9 (55.6) |
R
merge = 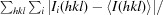
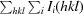 , where Ii(hkl) and 〈I(hkl)〉 represent the diffraction-intensity values of the individual measurements and the corresponding mean values, respectively.
, where Ii(hkl) and 〈I(hkl)〉 represent the diffraction-intensity values of the individual measurements and the corresponding mean values, respectively.
Acknowledgments
The authors thank Dr Dante Neculai and SGC Toronto for the gift of the pET28-MHL plasmid. This work was supported by a research grant from the Canadian Institutes of Health Research (CIHR #13337) to PLH. Beamline X29 at the National Synchrotron Light Source is supported by the US Department of Energy and the NIH National Center for Research Resources. PLH is the recipient of a Canada Research Chair; AMN was supported by a postdoctoral fellowship from CIHR.
References
- Albrecht, M. T. & Schiller, N. L. (2005). J. Bacteriol. 187, 3869–3872. [DOI] [PMC free article] [PubMed]
- Bakkevig, K., Sletta, H., Gimmestad, M., Aune, R., Ertesvåg, H., Degnes, K., Christensen, B. E., Ellingsen, T. E. & Valla, S. (2005). J. Bacteriol. 187, 8375–8384. [DOI] [PMC free article] [PubMed]
- Breidenstein, E. B., de la Fuente-Núñez, C. & Hancock, R. E. (2011). Trends Microbiol. 19, 419–426. [DOI] [PubMed]
- Evans, L. R. & Linker, A. (1973). J. Bacteriol. 116, 915–924. [DOI] [PMC free article] [PubMed]
- Franklin, M. J., Chitnis, C. E., Gacesa, P., Sonesson, A., White, D. C. & Ohman, D. E. (1994). J. Bacteriol. 176, 1821–1830. [DOI] [PMC free article] [PubMed]
- Franklin, M. J., Nivens, D. E., Weadge, J. T. & Howell, P. L. (2011). Front. Microbiol. 2, 167. [DOI] [PMC free article] [PubMed]
- Franklin, M. J. & Ohman, D. E. (2002). J. Bacteriol. 184, 3000–3007. [DOI] [PMC free article] [PubMed]
- Govan, J. R. & Deretic, V. (1996). Microbiol. Rev. 60, 539–574. [DOI] [PMC free article] [PubMed]
- Hendrickson, W. A. (1991). Science, 254, 51–58. [DOI] [PubMed]
- Jain, S. & Ohman, D. E. (2005). Infect. Immun. 73, 6429–6436. [DOI] [PMC free article] [PubMed]
- Keiski, C. L., Harwich, M., Jain, S., Neculai, A. M., Yip, P., Robinson, H., Whitney, J. C., Riley, L., Burrows, L. L., Ohman, D. E. & Howell, P. L. (2010). Structure, 18, 265–273. [DOI] [PMC free article] [PubMed]
- Kelley, L. A. & Sternberg, M. J. (2009). Nature Protoc. 4, 363–371. [DOI] [PubMed]
- Linker, A. & Evans, L. R. (1984). J. Bacteriol. 159, 958–964. [DOI] [PMC free article] [PubMed]
- Lombard, V., Bernard, T., Rancurel, C., Brumer, H., Coutinho, P. M. & Henrissat, B. (2010). Biochem. J. 432, 437–444. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Merighi, M., Lee, V. T., Hyodo, M., Hayakawa, Y. & Lory, S. (2007). Mol. Microbiol. 65, 876–895. [DOI] [PubMed]
- Oglesby, L. L., Jain, S. & Ohman, D. E. (2008). Microbiology, 154, 1605–1615. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Preston, L. A., Bender, C. L. & Schiller, N. L. (2001). DNA Seq. 12, 455–461. [DOI] [PubMed]
- Preston, L. A., Wong, T. Y., Bender, C. L. & Schiller, N. L. (2000). J. Bacteriol. 182, 6268–6271. [DOI] [PMC free article] [PubMed]
- Remminghorst, U., Hay, I. D. & Rehm, B. H. A. (2009). J. Biotechnol. 140, 176–183. [DOI] [PubMed]
- Remminghorst, U. & Rehm, B. H. A. (2006a). Appl. Environ. Microbiol. 72, 298–305. [DOI] [PMC free article] [PubMed]
- Remminghorst, U. & Rehm, B. H. A. (2006b). FEBS Lett. 580, 3883–3888. [DOI] [PubMed]
- Stover, C. K. et al. (2000). Nature (London), 406, 959–964.
- Winsor, G. L., Lam, D. K. W., Fleming, L., Lo, R., Whiteside, M. D., Yu, N. Y., Hancock, R. E. W. & Brinkman, F. S. L. (2011). Nucleic Acids Res. 39, D596–D600. [DOI] [PMC free article] [PubMed]
- Wong, T. Y., Preston, L. A. & Schiller, N. L. (2000). Annu. Rev. Microbiol. 54, 289–340. [DOI] [PubMed]
- Yoon, H.-J., Mikami, B., Hashimoto, W. & Murata, K. (1999). J. Mol. Biol. 290, 505–514. [DOI] [PubMed]