Abstract
Despite advances toward understanding the prevention and treatment of many cancers, patients who suffer from oral squamous cell carcinoma (OSCC) confront a survival rate that has remained unimproved for more than two decades indicating our ability to treat them pharmacologically has reached a plateau. In an ongoing effort to improve the clinical outlook for this disease, we previously reported that an essential component of the mechanism by which the proteasome inhibitor bortezomib (PS-341, Velcade) induced apoptosis in OSCC required the activation of a terminal unfolded protein response (UPR). Predicated on these studies, we hypothesized that high throughput screening (HTS) of large diverse chemical libraries might identify more potent or selective small molecule activators of the apoptotic arm of the UPR to control or kill OSCC. We have developed complementary cell-based assays using stably transfected CHO-K1 cell lines that individually assess the PERK/eIF2α/CHOP (apoptotic) or the IRE1/XBP1 (adaptive) UPR sub-pathways. A ~66K compound collection was screened at the University of Michigan Center for Chemical Genomics that included a unique library of pre-fractionated natural product extracts. The mycotoxin methoxycitrinin was isolated from a natural extract and found to selectively activate the CHOP-luciferase reporter at 80μM. A series of citrinin derivatives were isolated from these extracts, including a unique congener that has not been previously described. In an effort to identify more potent compounds we examined the ability of citrinin and the structurally related mycotoxins ochratoxin A and patulin to activate the UPR. Strikingly, we found that patulin at 2.5 – 10μM induced a terminal UPR in a panel of OSCC cells that was characterized by an increase in CHOP, GADD34 and ATF3 gene expression and XBP1 splicing. A luminescent caspase assay and the induction of several BH3-only genes indicated that patulin could induce apoptosis in OSCC cells. These data support the use of this complementary HTS strategy to identify novel modulators of UPR signaling and tumor cell death.
Keywords: unfolded protein response, endoplasmic reticulum stress, cell-based assay, luciferase reporter, natural products
Introduction
Oral squamous cell carcinoma (OSCC) accounts for approximately 90% of malignant lesions affecting the oral cavity, oropharynx, hypopharynx and larynx and represents the sixth most common cancer world-wide with an estimated 650,000 new cases and 350,000 deaths each year 1. Despite advances in our understanding in the prevention and treatment of many cancers, aggressive surgical management remains the preeminent approach offered to patients even with early disease (stage I–II). Unfortunately, neither recently introduced monoclonal antibodies nor the addition of radiation to the standard chemotherapy regimen consisting of cisplatin, taxane or 5-fluorouracil, have significantly improved event-free survival. We previously reported that an essential component of the mechanism by which the proteasome inhibitor bortezomib (PS-341, Velcade) induced apoptosis in a panel of OSCC cell lines was the activation of a terminal unfolded protein response (UPR) 2, 3. These studies motivated us to conduct a high throughput screen (HTS) developed to identify more potent UPR-exacerbating small molecules that could induce cell death in OSCC cells.
The UPR is orchestrated through activation of the protein kinases IRE1 and PERK and cleavage of the transcription factor ATF6 4, 5. These three endoplasmic reticulum (ER) resident transmembrane stress sensors are maintained in an inactive state through interaction with the ER lumen chaperone BiP/GRP78. Upon accumulation of misfolded protein, BiP dissociates from the sensors in an attempt to facilitate the restoration of homeostatic polypeptide folding. BiP release permits IRE1 and PERK homodimerization, autophosphorylation and activation. IRE1 and PERK activation initiates parallel yet genetically distinct signaling cascades that culminate in the transcriptional activation of genes encoding ER chaperones and the endoplasmic reticulum-associated degradation (ERAD) machinery and the attenuation of global protein synthesis 6. IRE1 activation elicits an endoribonuclease function that mediates a non-conventional splicing reaction on XBP1 mRNA to activate genes encoding ER protein chaperones, lipid biosynthetic enzymes, and multiple factors that function in ERAD 7, 8. Activated PERK phosphorylates eIF2α at Ser 51 to inhibit the initial step of polypeptide biosynthesis leading to general translation attenuation which has become a hallmark of ER stress 9, 10. Recent studies indicate that induction of CHOP, a C/EBP homologous transcription factor that forms heterodimers with other C/EBP family members, following eIF2α phosphorylation, is essential for the apoptotic response that occurs when defects with ER protein folding are not resolved 11–13. Although many mechanistic details of CHOP-induced apoptosis remain unknown, it is required for induction of the pro-apoptotic factors DR5 and TRB3 and can repress the transcription of anti-apoptotic BCL2 14, 15. Small molecules that could selectively induce pro-apoptotic CHOP but not adaptive XBP1 signaling are therefore, extremely attractive as chemotherapeutics. By enhancing stress, such small molecules might directly induce apoptosis in highly secretory malignant cells or sensitize them to conventional chemotherapies.
The mechanisms by which microenvironmental stresses modulate growth, vascularization and metastasis in solid tumors remain an avid area of research interest. Recent in vivo studies using luciferase reporters have revealed that ATF4 and XBP1 levels are elevated in the hypoxic regions of xenografts with human fibrosarcoma and primary mammary tumors, respectively, and may predict tumor growth 16, 17. Several intriguing reports have together examined approximately 50 OSCC cell lines and observed increased rates of protein synthesis that the authors independently attributed to the overexpression of the translation factors eIF-4E and eIF2α 18–21. Considered together, and predicated on our discovery that bortezomib-induced death of OSCC cell lines was dependent on the induction of a terminal UPR, these studies strongly support our hypothesis that small molecules that induce ER stress and overwhelm the hyperactive secretory pathway of OSCC cells will be an effective therapeutic approach to reduce tumor burden. Importantly, targeting the translation machinery of OSCC cells and not a specific signaling cascade or intermediate provides a unique approach where the events leading to cell death cannot be short circuited by any redundant signaling mechanism.
Bioactive natural products isolated from plants and microorganisms such as bacteria and fungi have been a rich source of pharmaceuticals with an estimated 60% of drugs currently on the market deriving from natural sources 22. In terms of cancer therapies, rapamycin, the vinca alkaloids vincristine and vinblastine, anthracyclines such as daunorubicin, doxorubicin, the quinoline alkaloid camptothecin and the cyclic diterpene paclitaxel are all naturally-derived. Although historically underrepresented in terms of therapeutic use, many studies are currently underway examining the benefit of marine-derived natural products for diverse clinical applications ranging from analgesics to cancer and inflammation. Importantly, the number of compounds isolated from living organisms undergoing preclinical and clinical evaluation continues to expand with many promising candidates 23, and the recent FDA granting of “orphan drug” status to ET-743 (Trabectedin) for soft tissue sarcomas and ovarian cancer 24.
A collection of 5036 bacterial and fungal-derived pre-fractionated natural extracts was screened for the ability to activate the CHOP and XBP1 luciferase reporters. Using bioassay-guided fractionation we identified and isolated methoxycitrinin from an extract that preferentially activated the CHOP reporter. Purified methoxycitrinin, its analog citrinin, and patulin and ochratoxin A, two compounds with similar structural features, were examined for their ability to induce UPR signaling in reporter and gene expression assays. Patulin was found to be a robust UPR activator, whereas citrinin demonstrated activity similar to methoxycitrinin and ochratoxin A was unable to activate either UPR reporter. Patulin is a secondary metabolite produced by Aspergillus and Penicillium species and has been demonstrated to have a cytotoxic affect in human promyelocytic leukemia and hepatoma cell lines via a mechanism that involves perturbation of redox status and inhibition of amino acid uptake, respectively 25, 26. Further analyses with a panel of OSCC cell lines revealed activation of caspases and induction of BH3-only pro-apoptotic genes following low-dose patulin exposure. This study demonstrates the value of high throughput screening of natural product extracts to identify novel modulators of the unfolded protein response and cell death in human oral squamous cell carcinoma cell lines.
Materials and Methods
Cell lines, reporters and reagents
For screening, single high-expressing clones that provided Z′ values >0.5 were selected from stably transfected CHO-K1 cells that individually report on either the PERK/eIF2α/CHOP or the IRE1/XBP1 pathway. The CHO-CHOP-luciferase reporter cell line was screened previously 27. In this cell line, approximately 8kb of the murine CHOP promoter is upstream of the luciferase coding region and can be induced by ER stress. The CHO-XBP1-luciferase cell line was made by transfection and G-418 selection of CHO-K1 cells with pcDNA3.1-XBP1-luc. pcDNA3.1-XBP1-luc contains a 26 base intron from XBP1 placed into the 5′ end of the luciferase mRNA; luciferase is not translated unless the intron is removed via IRE1-mediated splicing that occurs upon activation of the UPR 28. The human floor of mouth (FOM) squamous cell carcinoma lines UMSCC1, UMSCC14A and laryngeal squamous cell carcinoma cell line UMSCC23 were kindly provided by Dr. Thomas Carey at the University of Michigan. The tongue carcinoma cell line CAL27 (CRL-2095), the pharyngeal carcinoma cell line FaDu (HTB-43) and the epidermoid carcinoma cell line A-253 (HTB-41) were from ATCC (Manassas, VA). OSCC cell lines were grown in DMEM supplemented with penicillin and streptomycin and 10% fetal bovine serum. Normal human epidermal keratinocytes (nHEK)(ScienCell Research Laboratories Carlsbad, CA) were grown in keratinocyte medium (#2101) with keratinocyte growth supplement (#2152) (ScienCell Research Laboratories, Carlsbad, CA). Citrinin (ALX-380-058), patulin (ALX-270-111), and ochratoxin A (ALX-630-089) were purchased Enzo Life Sciences (San Diego, CA).
High Throughput Screening
Reporter cells were plated onto white 384-well white tissue culture plates (5000 cells/well) using a Multidrop (Thermo Fisher Scientific, Waltham, MA). Plates were incubated for 16 hours at 37°C, 5% CO2 prior to compound addition. Compounds were added using a high-density replication tool on a Biomek FX liquid handler (Beckman Coulter, Brea, CA) in a 0.2μL volume (DMSO concentration = 0.4% in all wells). All compounds were tested as singletons. 2.5μg/ml tunicamycin (Tm), or an equal volume of DMSO was each added to one row of wells on each plate for assay controls. Six to eight hours later the medium was aspirated to 10μl with an Elx 405 plate washer (Bio Tek U.S., Winooski, VT) and 10μl of Steady-Glo (Promega Corp., Madison, WI) was added to each well. Luminescence was measured on a Pherastar multi-mode plate reader (BMG Labtech, Cary, NC). A hit was defined as a compound able to induce luciferase to a level > 50% of the tunicamycin control. For the CHOP assay this definition produced 1064 hits for an overall hit rate of 1.6%. Within the natural products collection this definition produced 39 hits (0.8%). The 1025 hits (excluding the natural products) were further scrutinized by the Vahlteich Medicinal Chemistry Core synthesis laboratory (VMCC) at the University of Michigan. Compounds were triaged based on their ability to activate reporters in other cell-based assays (including our XBP1 reporter) or if they had molecular weights, total polar surface areas, AlogP values or known structural alerts that might preclude them from being reasonably considered for therapeutic development. Compounds surviving this level of scrutiny were subjected to confirmatory dose-response assays performed in duplicate with both the CHOP and XBP1 reporter cell lines. The initial triage and subsequent dose-response assays identified 368 bona fide CHOP-selective hits, reducing the overall hit rate for the screen to 0.6%. The 368 compounds were clustered using SARNavigator software at 65%+ similarity which produced 278 clusters. Signposts for the 278 clusters were provided to the VMCC where the compounds were prioritized as High (222), Medium (75) or Low (71) for further study based on their predicted potential for therapeutic development.
Fermentation and extraction
Fungal strain 7792-2H was isolated from marine sediment, collected in Las Baulas Marine National Park in Costa Rica in 2006. The strain was identified based on a phylogenetic sequence analysis of 18s rRNA using the primers ITS1 and ITS4. The rRNA gene sequence data (553 bp) was submitted to a match search using BLAST (http://www.ncbi.nlm.nih.gov/blast/), showing 99% similarity with Penicillium sp. The fungus was cultured for three weeks at 28°C in two 6.0L Fernbach flasks containing 2.5 – 5.0L of ISP2 medium (1L of deionized water, 10g of malt extract, 4g yeast extract, 4g dextrose and 30g NaCl) with shaking at 160 rpm. Cultures were centrifuged at 7000 rpm and the fermentation medium was extracted with amberlite XAD-16 resin. After shaking overnight the resin was washed with 1L of deionized H2O and sequentially extracted with MeOH, acetone and EtOAc. The resulting extracts were concentrated by rotary evaporation and re-dissolved in DMSO at 15mg/ml for storage at −28°C.
Isolation of compounds 1 – 6
The methanol and acetone extracts (3.2g and 2.2g) were first fractionated by C18 LOBAR B (Ø=25 mm × 310 mm) LiChroprep column eluting with MeOH/H2O 7:3. The active fractions were further subjected to HPLC purification on a XBridge Prep C18 column (Ø=10 mm × 250 mm) with MeOH/H2O gradient to afford citrinin 29 together with five more non-active compounds 30, 31, including one that has not been previously described. These structures were determined by a combination of spectroscopic and spectrometric experiments. IR spectra were recorded with a Perkin Elmer BX FT-IR infrared spectrometer using NaCl plates. Mass spectra were carried out with a Micromass AutoSpec Yltima Magnetica mass spectrometer. 1H, 13C and 2D NMR spectra were recorded in CD3OD on a Varian INOVA 600 MHz NMR spectrometer at 600 MHz for 1H NMR and 150 MHz for 13C, using TMS as internal standard.
PCR
Total mRNA was harvested with Trizol (Invitrogen Corp., Carlsbad, CA) and cDNA was made with iScript cDNA kits (BioRad, Hercules, CA). Semi-quantitative reverse transcription PCR (RT-PCR) analysis of spliced and un-spliced XBP1 was performed with a single human-specific primer pair ACA CGC TTG GGA ATG GAC AC (forward) and CCA TGG GAA GAT GTT CTG GG (reverse); amplicons were visualized with a Qiagen Qiaxcel automated nucleic acid fragment analyzer using a high resolution cartridge on the M500 setting using a 15bp - 1kb alignment marker and a 50bp – 800bp size marker. Quantitative real-time reverse transcription PCR (qRT-PCR) was performed with following Life Technologies Taqman primer/probe sets: CHOP/DDIT3 (Hs01090850_m1), 18S (Hs99999901_s1), GADD34 (Hs00169585_m1), ATF3 (Hs00910173_m1), ATF4 (Hs00909569_g1), BiP/GRP78 (Hs99999174_m1), Bax (Hs99999001_m1), Bak (Hs00832876_g1), Bim (Hs00708019_s1), Bid (Hs00609632_m1), NOXA (Hs00560402_m1), PUMA (Hs00248075_m1) and NBK (Hs00154189_m1), as described 32.
Viability and Caspase Assays
Cell Titer-Glo Luminescent Cell Viability Assay (G7570) and ). Caspase-Glo 3/7 Assay (G8090) from Promega Corp., Madison, WI were performed with 12,500 cells (in 50μl) per well in 96 well white opaque cell culture plate, as described 32.
Results
Construction of Reporter Cell Lines
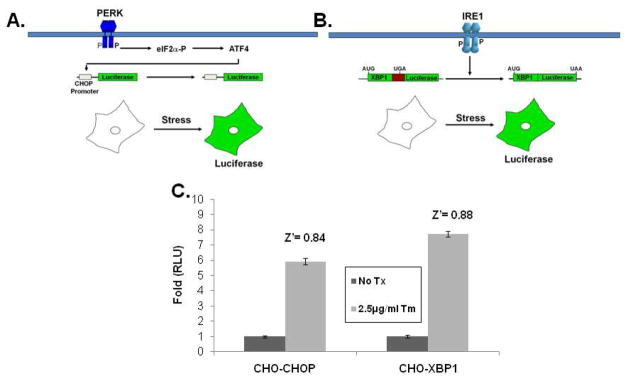
To identify small molecules that modulate specific sub-pathways of the UPR we developed two (complementary) cell-based assays using stably transfected CHO-K1 cells that individually report on the PERK/eIF2α/CHOP or the IRE1/XBP1 sub-pathways of the unfolded protein response (UPR). Counterscreening with these two cell lines has allowed us to identify compounds that specifically perturb only one pathway and has allowed us to rule out many compounds that generally perturb properties of protein synthesis or folding such as ER lumenal Ca2+ concentration or cellular redox status. The CHO-CHOP-luciferase reporter cell line was previously described and used successfully for a chemical genomic screen 27. The CHOP reporter construct consists of approximately 8kb of the murine CHOP promoter cloned immediately upstream of the luciferase coding region (Fig. 1A). The CHO-XBP1-luciferase cell line was created by the stable transfection of a reporter that contains a 26 base intron from XBP1 placed into the 5′ end of the luciferase mRNA; luciferase is not translated unless the intron is removed via IRE1-mediated splicing that occurs upon activation of the UPR (Fig. 1B) 28. Stable high-expressing clones from each cell line were chosen that provided Z′ values >0.5 for HTS (Fig. 1C) 33. The assay was optimized for a 384 well format and screened with a 66,000 compound library that included a collection of 5036 pre-fractionated natural product extracts. Hit criteria, rates and chemoinformatic analysis are described under Methods.
Figure 1. Reporter construct and cell lines.
Illustration of the CHOP-luciferase (A) and XBP1-luciferase (B) reporters used to make stable CHO-K1 cell lines for high throughput screening. Single high expressing clones providing Z′ values > 0.5 (C) were chosen for screening. Error bars represent standard deviation.
Complementary screens identify active natural product extracts
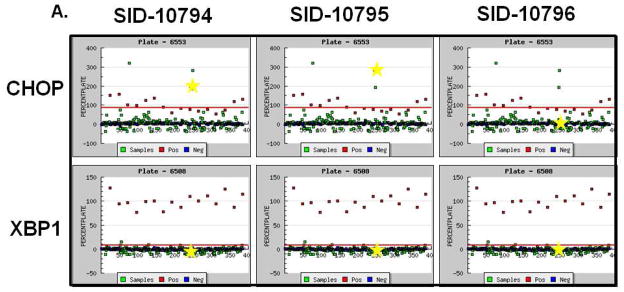
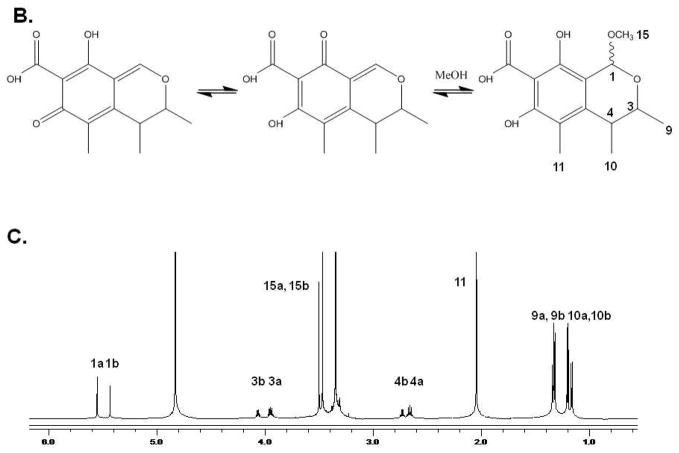
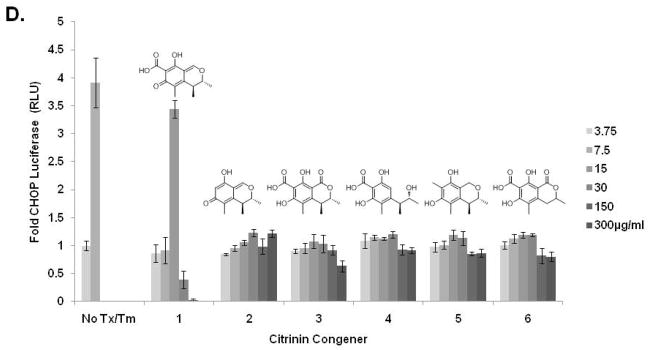
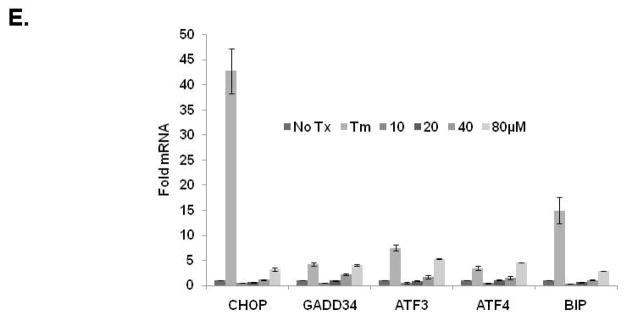
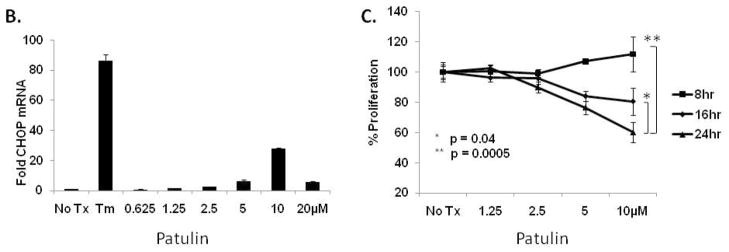
To support our effort to identify bioactive compounds derived from natural products we focused our attention toward the pre-fractionated natural products that demonstrated selectivity for the CHOP reporter. The methanol (SID-10794) and acetone (SID-10795) extracts from strain 7792-2 potently activated the CHOP-luciferase but not the XBP1-luciferase reporter, suggesting that active metabolites able to selectively induce the apoptotic arm of the UPR might be present (Fig. 2A). Subsequent dose-resposnse assays performed with aliquots of all three extracts used in the primary screen confirmed this activity (data not shown). In an attempt to isolate and identify the active metabolite(s) in SID’s 10794 and 10795, archived spore stocks from the original fungal strain were re-cultured in 5L of ISP2 medium, and methanol and acetone extracts were prepared as described. The extracts were separated using a medium pressure column into eight fractions and each was re-tested with triplicate samples in a dose-response fashion (3.75μg/ml – 300μg/ml) for the ability to activate the CHOP-luciferase reporter. Active fractions were further purified by HPLC and a MeOH adduct of citrinin, which was present as an isomer in equilibrium with the parent compound (Fig. 2B) 34, was identified by NMR and mass spectrometry analysis (Fig. 2C). Citrinin (compound 1) was able to activate the CHOP luciferase reporter only at 15μg/ml (~60μM) and then appeared to be toxic (120μM+) as evidenced by lower than baseline luminescence. Additional citrinin derivatives that failed to activate the CHOP-luciferase reporter (at the same concentrations) were purified in adjacent fractions, including one that had not been previously described (structure 6) (Fig. 2D). The structure of this new citrinin congener was determined by a combination of HRMS and 1D and 2D NMR experiments including 1HNMR, 13CNMR, COSY, HSQC, HMBC and NOESY (see supporting information). Purified methoxycitrinin was found to induce the CHOP reporter at 80μM; and experiments performed with the human OSCC cell line UMSCC1 revealed that methoxycitrinin could induce the expression of the UPR genes CHOP, GADD34, ATF3, ATF4, and BiP after six hours (Fig. 2E). Conventional PCR analysis of the same cDNA failed to detect XBP1 splicing (data not shown) suggesting methoxycitrinin might preferentially activate the apoptotic arm of the UPR.
Figure 2. Complementary screens identify CHOP-selective natural extracts.
Scatter diagram of 384 well plates of CHO-CHOP and CHO-XBP1 cells treated with the same parent compounds; SID-10794, SID-10795 and SID-10796 (stars) (A). Blue = DMSO (neg) controls; Red = Tunicamycin (pos) controls; Green = natural product extract. Tautomeric equilibrium and methanol adduct of citrinin (B). 1HNMR spectrum of two isomers of methoxycitrinin adduct (C). Confirmatory CHO-CHOP-luciferase assays with purified citrinin and inactive congeners (D). UMSCC1 cells were treated with methoxycitrinin for 6 hours and UPR-related transcripts were identified using qRT-PCR (E). Tm = tunicamycin. All error bars represent standard deviation.
Structure-based analog selection identifies more potent compounds
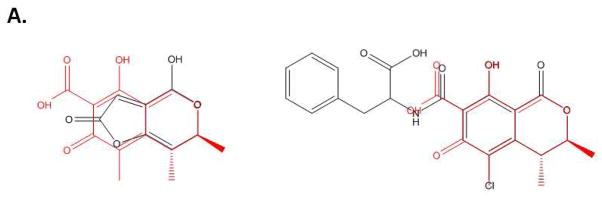
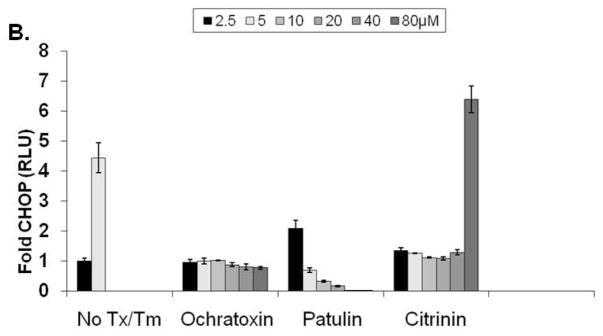
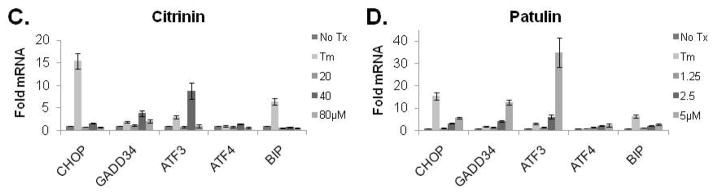
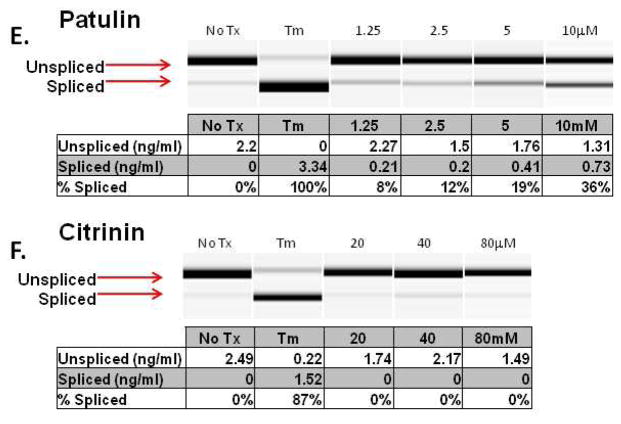
In an attempt to identify more potent UPR-inducing natural products (than methoxycitrinin), we obtained citrinin (Penicillium citrinum), patulin (Penicillium expansum) and ochratoxin A (Aspergillus ochraceus) (Fig. 3A), three mycotoxins that have previously been studied together for their ability to inhibit lymphocyte growth 35. Similar to purified methoxycitrinin, citrinin could activate the CHOP-luciferase reporter at 80μM and ochratoxin A was unable to induce luciferase expression at any concentration tested (Fig. 3B). Patulin at 2.5μM induced a 2-fold increase in luciferase expression and then appeared to be toxic as shown by lower than baseline luminescence suggesting patulin might be a more potent UPR activator than any citrinin congener (Fig. 3B). To determine the relative ability of citrinin and patulin to modulate the expression of CHOP and other UPR-associated genes, dose-response assays were performed with oral squamous cell carcinoma (OSCC) cells. UMSCC14A cells were treated with citrinin (20 – 80μM) or patulin (1.25 – 5μM) for six hours. Quantitative real-time gene expression analysis revealed that patulin treatment led to a more robust accumulation of mRNA transcripts of CHOP, GADD34, and ATF3 than citrinin (Fig. 3C and D). Additionally, comparatively low doses of patulin led to an accumulation of the spliced form of XBP1 indicating that this natural product, unlike any citrinin congener, could activate the UPR (Fig. 3E and F).
Figure 3. Structure-based analog selection identifies similar mycotoxins.
Visual analysis of the structures of patulin and ochratoxin A revealed structural similarity to citrinin (super imposed black) (A). CHO-CHOP-luciferase cells were treated with ochratoxin A, patulin or citrinin (2.5 – 80μM) for 6 hours (B). UMSCC1 cells were treated for 6 hours with citrinin or patulin as indicated; qRT-PCR analysis of total mRNAs for accumulation of UPR-related transcripts (C and D). Error bars represent standard deviation. In a separate experiment UMSCC1 cells were treated with 1.25 – 5.0μM patulin (E) or 20 – 80μM citrinin (F) for 6 hours and conventional RT-PCR analysis with primers that amplify spliced and un-spliced XBP1 mRNA was performed; data were quantified with Qiaxcel software, as described.
Patulin inhibits proliferation of OSCC cells via oxidative stress
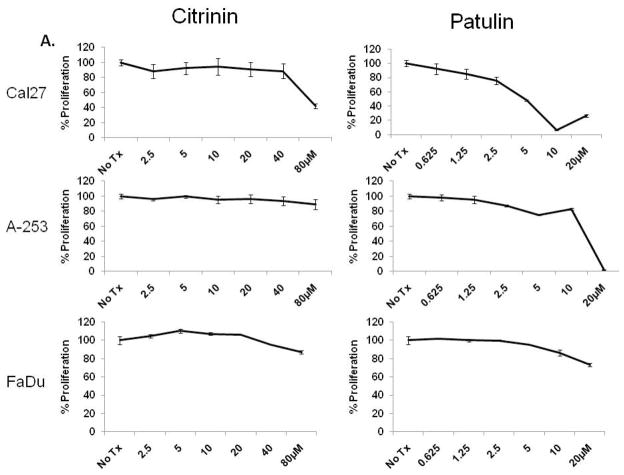
Since patulin and citrinin were shown to induce markers of UPR signaling, we next examined their ability to alter the proliferative capacity of a panel of OSCC cells in vitro. CAL 27, A-253 and FaDu cells were cultured with 2.5 – 80μM citrinin or 0.625 – 20μM patulin for 24 hours. An ATP-based (luciferase) proliferation assay revealed that patulin could substantially inhibit the growth of CAL 27 and A-253 cells, but provided only modest inhibition of FaDu (Fig. 4A). Additionally, UMSCC14A cells were found to be sensitive at a concentration of patulin similar to CAL-27 and A-253 (data not shown). In contrast, these experiments revealed that citrinin could only reduce proliferation in CAL 27 cells at a dose of 80μM (the same concentration required to activate the CHOP reporter). To determine whether or not patulin was toxic in non-malignant cells, normal human epidermal keratinocytes (nHEK) were cultured with 1.25 – 10μM patulin and proliferation was determined after 8, 16 and 24 hours. Modest but significant inhibition of proliferation occurred after 16 and 24 hours at 10μM, a concentration that correlated with CHOP mRNA induction (Fig. 4B and C). However, the fact that FaDu cells were able to tolerate higher concentrations of patulin, via an unknown mechanism, suggests this natural product is not generally cytotoxic.
Figure 4. Patulin inhibits proliferation of OSCC cells in an oxidative stress-dependent manner.
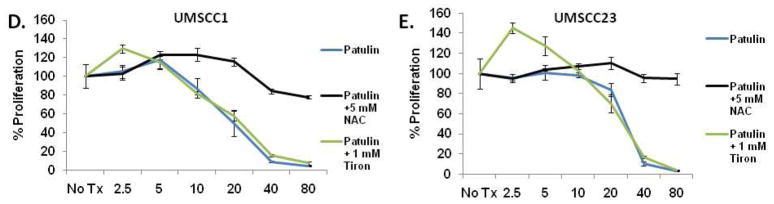
Luminescent proliferation assays were performed after CAL27, A-253 and FaDu were treated as indicated for 24 hours (A). Normal human epidermal keratinocytes were treated with patulin (0.625 – 20μM) for 6 hours and qRT-PCR analysis for CHOP was performed (B). In parallel cultures cells were analyzed with a luminescent proliferation assay after 24 hour patulin exposure (C). UMSCC1 and UMSCC23 cells were treated with 2.5 – 80μM patulin for 24 hours in the presence or absence of 1mM tiron or 5mM N-acetyl cysteine and subject to a luminescent proliferation assay (D and E).
Previous studies have revealed that patulin can generate reactive oxygen species (ROS) in Chinese hamster ovary (CHO) cells, peripheral blood lymphocytes and several human cancer cell lines including HL60 and HEK293 36, 37. In light of our previous studies demonstrating that ROS can induce ER stress and the UPR 2, we sought to determine whether patulin could induce oxidative stress in OSCC. UMSCC1 and UMSCC23 cells were treated with increasing doses of patulin in the presence or absence of the cell permeable antioxidants tiron or N-acetylcysteine (NAC) for 24hrs. Whereas tiron is well-regarded as a specific inhibitor of superoxide, NAC is a precursor to glutathione and can increase cellular pools of glutathione and scavenge a variety of ROS, including superoxide. Both cell lines continued to proliferate after 24 hours even at very high concentrations of patulin when cultured with NAC but not tiron (Fig. 4D and E). Importantly, supplementing culture medium with essential and or non-essential amino acids could not protect UMSCC1 or UMSCC23 from celastrol treatment (data not shown) indicating that the protection provided by N-acetylcysteine was likely the result of its broad free radical scavenging ability and not amino acid repletion.
Patulin induces apoptosis in OSCC cells
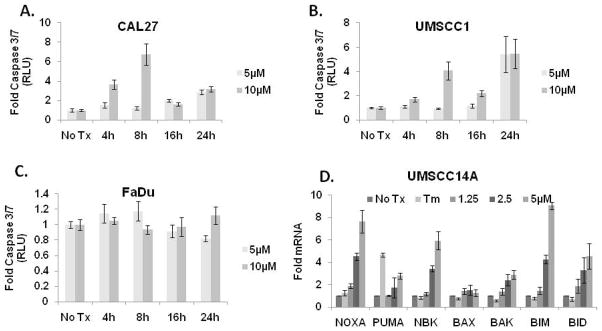
The ability of patulin to inhibit the proliferation of OSCC cells in culture prompted us to examine whether or not it was cytotoxic. CAL 27 and UMSCC1 (patulin-sensitive) and FaDu (patulin-resistant) cells were treated with 5 and 10μM patulin and the activation of the initiator caspases 3 and 7 was examined after 4, 8 and 16 hours with a luminescent assay. Consistent with the notion that patulin-mediated reduction in proliferation was due to increased cell death, CAL 27 and UMSCC1, but not FaDu cells displayed a 4- to 7-fold increase in caspase 3/7 activation after 8 hours (Fig. 5A – C). To better understand the mechanism of patulin-induced apoptosis, the expression profile of a panel of “BH3-only” proapoptotic BCL2 family members was examined in UMSCC14A cells. Cultures were treated with 2.5μM patulin for 6 hrs. Quantitative real-time PCR (q-RT-PCR) analysis of Trizol harvested mRNAs revealed an upregulation of NOXA, PUMA Bim, Bid and NBK which are consistent with apoptotic genes associated with UPR-mediated apoptosis (Fig 5D).
Figure 5. Patulin induces apoptosis in OSCC cells.
CAL27 (A), UMSCC1 (B) and FaDu (C) cells were treated with 5 and 10μM patulin for 4, 8, 16 and 24 hours and subject to a luminescent caspase 3/7 activity assay. UMSCC14A cells were treated with 1.25 – 5μM patulin for 6 hours and qRT-PCR analysis was performed with TaqMan primer probe sets specific for pro-apoptotic BCL2 family members. All errors bars represent standard deviation.
Discussion
The recent introduction of affordable user-friendly screening technologies has facilitated the development of sensitive cell-based and biochemical high throughput assays within the realm of academia. The application of HTS in an academic setting offers promise for the development of novel therapeutics and encourages the continued investigation of compounds that are not likely to succeed in the clinic as molecular tools to address fundamental biological questions. In the current study, a cell-based HTS strategy was designed to identify natural products that could specifically activate the apoptotic arm of the UPR from pre-fractionated extracts. Using two cell lines that individually reported on the apoptotic or adaptive stress responses facilitated the rapid identification of bona fide UPR sub-pathway-specific hits. An additional advantage provided by the use of such an assay (cell-based) is the mitigation of challenges associated with hit compound permeability and ATP-dependent efflux once it passes the membrane. We identified methoxycitrinin and five additional congeners, including one not previously described, however, when purified, very high concentrations were required to induce the UPR and they were not appreciably toxic.
In an attempt to find more potent mycotoxins, we obtained the structurally related natural products ochratoxin A and patulin to determine their corresponding biological activity profile in this assay system. Patulin treatment increased the expression of CHOP, GADD34, ATF3, ATF4 and BiP, and led to XBP1 splicing in a dose-dependent fashion indicating that it could induce the UPR in OSCC cells. Low-dose patulin-induced stress was robust enough in five of the six OSCC cell lines examined to overwhelm the adaptive capacity of the UPR and induce apoptosis. Patulin was also observed to increase transcripts for CHOP, and induce significant cell death in normal human epidermal keratinocytes after 24 hours at a rate that was similar to malignant cells. Although these experiments indicated that patulin did not exhibit selectivity for malignant cells, the fact that FaDu and UMSCC23 could tolerate higher doses indicates that patulin is not generally cytotoxic. The mechanism by which these two cell lines were resistant to patulin treatment is not known. It is possible that a variety of mutational events in genetically unstable cancer cells could alter the ability of CHOP or ATF4 protein to be expressed following stress, or that lower than average basal levels of stress or even aneuploidy contributed to the resistance. Additionally, it is plausible that slower doubling times and reduced protein synthesis rates in culture could limit their dependence on the UPR for survival and narrow the therapeutic window of any apoptosis-inducing compounds.
The constant pressure on cancer cells to meet enhanced oxygen and nutrient demands, likely depends on enhanced basal activation of the UPR for vascularization and proliferation. Oral squamous cell carcinoma cells (OSCC) were selected for the present work due to the fact many highly secretory OSCC cell lines and fresh surgical biopsies have been shown to express increased levels of the eukaryotic translation factors eIf4E and eIf2α. This important finding and our own work demonstrating the ability of the proteasome inhibitor bortezomib to induce UPR-dependent cell death in a panel of OSCC cells has driven the hypothesis that malignant cell populations might be preferentially sensitive to UPR-inducing compounds. Additionally, epithelial cell-derived tumors may be more susceptible to UPR-inducing compounds due to increased rates of protein secretion. Therapies that target oncogene-driven growth signals are often rendered ineffective due to redundant molecular networks for these intermediates. The continued identification and investigation of agents that interfere with protein folding homeostasis will continue to shed light on whether they may be leveraged against the reliance of cancer cells (vs non-malignant adjacent cells) on UPR, and escape the need for neoplastic cell targeting. In the present work we have shown that following patulin treatment the UPR is activated in numerous OSCC cell lines.
Mycotoxins are metabolites or toxins secreted from filamentous fungi such as Penicillium, Aspergillus, Fusarium and associated genera 38. Patulin is a common mycotoxin, contaminating apples and other commercial fruit crops, that has been studied for more than seven decades, including early work that examined patulin as a potential cancer therapy and a large multicenter trial using it as a nasal rinse to fight the common cold 39. Patulin has been shown to interfere with glutathiones and induce oxidative damage resulting in the formation of DNA adducts and strand breaks and is toxic to cancer cells 40, 41. Consistent with this mechanism we found that N-acetyl cysteine, a precursor to glutathione, protected OSCC cells from patulin while tiron, a superoxide-specific scavenger, was not protective, suggesting a specific pattern of oxidative stress is required for patulin-induced toxicity. We have previously shown that tiron could protect OSCC from bortezomib 2 suggesting varied oxidative stress can induce CHOP dependent cell death and additionally that CHOP is also involved in the production of oxidative stress suggesting a complex interplay between ER and oxidative stress 42. It is likely that fungi developed the ability to secrete generally toxic oxidative stress-inducing agents in response to environmental stress as a survival mechanism 43. Patulin has also been shown to interfere with uptake of amino acids and protein production 26. Taken together, our results indicate that patulin likely induces an oxidative stress that culminates in the activation of ER stress, the UPR and eventually cell death.
Despite its interesting effects on the UPR, we do not believe that patulin is a promising lead for anti-cancer drug development in an un-modified form. Previous work on the mechanism of action of patulin has indicated that it is an efficient and non-specific trap for cellular thiols, including glutathione 40, 41. Close structural analogs of patulin, lacking its inherent electrophilicity, are much less active as cytotoxins 44 suggesting that the mechanism of action is largely, if not entirely, dependent upon reaction with cellular thiols. In general, thiol-reactivity is unlikely to be a mechanism of action that will be highly selective for cancer cells. In fact, patulin has been reported to be a general genotoxin and carcinogen, as well as having negative effects on multiple organs and inducing DNA damage and apoptosis upon contact with skin 45.
The results reported here provide an example of the power of high throughput screening to identify natural products that target specific cellular pathways involved in UPR signaling and malignancy. Additional efforts to identify molecular probes and potential new therapeutic agents will undoubtedly be uncovered by further efforts to harness the broad chemical diversity from unique sources of microbial-derived secondary metabolites.
Supplementary Material
Acknowledgments
Portions of this work were supported by NIH grants DK042394, HL052173, and HL057346, as well as a MH084182 and MH089782 (RJK) and DE019678 (AMF). PGC gratefully acknowledges the Spanish Foundation of Science and Technology (FECYT) for a postdoctoral fellowship. Additional aspects of this work were supported by NIH grant U01 TW007404 as part of the International Cooperative Biodiversity Group initiative at the Fogarty International Center (DHS and GT-C), and the H. W. Vahlteich Professorship (DHS). The authors wish to thank Luis Guillermo Acosta and Frank Gonzalez (Bioprospecting Unit, INBio). The marine sediment sample was obtained under the permit R-CM-INBio-03-2006-OT.
Contributor Information
Andrew M. Fribley, Wayne State University, Carmen and Ann Adams Department of Pediatrics, Division of Hematology/Oncology, 2228 Elliman Building, 421 E. Canfield, Detroit, MI 48201, 313-577-8510
Patricia G. Cruz, University of Michigan, Life Sciences Institute
Justin R. Miller, Wayne State University, Carmen and Ann Adams Department of Pediatrics, Division of Hematology/Oncology
Michael U. Callaghan, Wayne State University, Carmen and Ann Adams Department of Pediatrics, Division of Hematology/Oncology
Peter Cai, Wayne State University School of Medicine
Neha Narula, Wayne State University School of Medicine
Richard R. Neubig, University of Michigan, Department of Pharmacology, Center for Chemical Genomics
Hollis D. Showalter, University of Michigan, College of Pharmacy, Department of Medicinal Chemistry
Scott D. Larsen, University of Michigan, College of Pharmacy, Department of Medicinal Chemistry
Paul D. Kirchhoff, University of Michigan, College of Pharmacy, Department of Medicinal Chemistry
Martha J. Larsen, University of Michigan, Life Sciences Institute, Center for Chemical Genomics
Douglas A. Burr, Perkin Elmer, Inc
Pamela J. Schultz, University of Michigan, Life Sciences Institute
Renju R. Jacobs, University of Michigan, Life Sciences Institute, Center for Chemical Genomics
Giselle Tamayo-Castillo, University of Costa Rica, National Institute for Biodiversity (INBio)
David Ron, University of Cambridge Institute of Metabolic Science
David H. Sherman, University of Michigan, Life Sciences Institute, Center for Chemical Genomics and Department Medicinal Chemistry
Randal J. Kaufman, University of Michigan, Departments of Biological Chemistry and Internal Medicine
References
- 1.Parkin DM, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Fribley A, Zeng Q, Wang CY. Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol Cell Biol. 2004;24(22):9695–704. doi: 10.1128/MCB.24.22.9695-9704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fribley AM, et al. Proteasome inhibitor PS-341 induces apoptosis in cisplatin-resistant squamous cell carcinoma cells by induction of Noxa. J Biol Chem. 2006;281(42):31440–7. doi: 10.1074/jbc.M604356200. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13(10):1211–33. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 5.Ron D. Translational control in the endoplasmic reticulum stress response. J Clin Invest. 2002;110(10):1383–8. doi: 10.1172/JCI16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 7.Iwakoshi NN, Lee AH, Glimcher LH. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev. 2003;194:29–38. doi: 10.1034/j.1600-065x.2003.00057.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23(21):7448–59. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397(6716):271–4. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 10.Scheuner D, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7(6):1165–76. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 11.Oyadomari S, Mori M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 2004;11(4):381–9. doi: 10.1038/sj.cdd.4401373. [DOI] [PubMed] [Google Scholar]
- 12.Scheuner D, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11(7):757–64. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi H, Wang HG. CHOP is involved in endoplasmic reticulum stress-induced apoptosis by enhancing DR5 expression in human carcinoma cells. J Biol Chem. 2004;279(44):45495–502. doi: 10.1074/jbc.M406933200. [DOI] [PubMed] [Google Scholar]
- 14.McCullough KD, et al. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21(4):1249–59. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohoka N, et al. TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. Embo J. 2005;24(6):1243–55. doi: 10.1038/sj.emboj.7600596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spiotto MT, et al. Imaging the unfolded protein response in primary tumors reveals microenvironments with metabolic variations that predict tumor growth. Cancer Res. 70(1):78–88. doi: 10.1158/0008-5472.CAN-09-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fels DR, et al. Preferential cytotoxicity of bortezomib toward hypoxic tumor cells via overactivation of endoplasmic reticulum stress pathways. Cancer Res. 2008;68(22):9323–30. doi: 10.1158/0008-5472.CAN-08-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franklin S, et al. Detection of the proto-oncogene eIF4E in larynx and hypopharynx cancers. Arch Otolaryngol Head Neck Surg. 1999;125(2):177–82. doi: 10.1001/archotol.125.2.177. [DOI] [PubMed] [Google Scholar]
- 19.Nathan CO, et al. Detection of the proto-oncogene eIF4E in surgical margins may predict recurrence in head and neck cancer. Oncogene. 1997;15(5):579–84. doi: 10.1038/sj.onc.1201216. [DOI] [PubMed] [Google Scholar]
- 20.Sorrells DL, et al. Pattern of amplification and overexpression of the eukaryotic initiation factor 4E gene in solid tumor. J Surg Res. 1999;85(1):37–42. doi: 10.1006/jsre.1999.5653. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, et al. Expression of eukaryotic translation initiation factors 4E and 2alpha correlates with the progression of thyroid carcinoma. Thyroid. 2001;11(12):1101–7. doi: 10.1089/10507250152740939. [DOI] [PubMed] [Google Scholar]
- 22.Molinari G. Natural products in drug discovery: present status and perspectives. Adv Exp Med Biol. 2009;655:13–27. doi: 10.1007/978-1-4419-1132-2_2. [DOI] [PubMed] [Google Scholar]
- 23.Schwartsmann G, et al. Marine-derived anticancer agents in clinical trials. Expert Opin Investig Drugs. 2003;12(8):1367–83. doi: 10.1517/13543784.12.8.1367. [DOI] [PubMed] [Google Scholar]
- 24.Pommier Y, et al. DNA sequence- and structure-selective alkylation of guanine N2 in the DNA minor groove by ecteinascidin 743, a potent antitumor compound from the Caribbean tunicate Ecteinascidia turbinata. Biochemistry. 1996;35(41):13303–9. doi: 10.1021/bi960306b. [DOI] [PubMed] [Google Scholar]
- 25.Wu TS, et al. Mechanism of patulin-induced apoptosis in human leukemia cells (HL-60) Toxicol Lett. 2008;183(1–3):105–11. doi: 10.1016/j.toxlet.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 26.Arafat W, Musa MN. Patulin-induced inhibition of protein synthesis in hepatoma tissue culture. Res Commun Mol Pathol Pharmacol. 1995;87(2):177–86. [PubMed] [Google Scholar]
- 27.Harding HP, et al. Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab. 2005;2(6):361–71. doi: 10.1016/j.cmet.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Back SH, et al. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J Biol Chem. 2006;281(27):18691–706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- 29.Cartwright NJ, Robertson A, Whalley WB. A synthesis of citrinin. Nature. 1948;162(4133):94. doi: 10.1038/163094b0. [DOI] [PubMed] [Google Scholar]
- 30.Lu ZY, et al. Citrinin dimers from the halotolerant fungus Penicillium citrinum B-57. J Nat Prod. 2008;71(4):543–6. doi: 10.1021/np0704708. [DOI] [PubMed] [Google Scholar]
- 31.Li DH, et al. Two new metabolites with cytotoxicities from deep-sea fungus, Aspergillus sydowi YH11-2. Arch Pharm Res. 2007;30(9):1051–4. doi: 10.1007/BF02980236. [DOI] [PubMed] [Google Scholar]
- 32.Fribley AM, et al. Large-Scale Analysis of UPR-Mediated Apoptosis in Human Cells. Methods Enzymol. 491:57–71. doi: 10.1016/B978-0-12-385928-0.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 34.Poupko R, Luz Z, Destro R. Carbon-13 NMR of Citrinin in the Solid State and in Solutions. The Journal of Physical Chemistry A. 1997;101(28):5097–5102. [Google Scholar]
- 35.Bernhoft A, et al. Combined effects of selected Penicillium mycotoxins on in vitro proliferation of porcine lymphocytes. Mycopathologia. 2004;158(4):441–50. doi: 10.1007/s11046-004-2843-z. [DOI] [PubMed] [Google Scholar]
- 36.Liu BH, et al. Evaluation of genotoxic risk and oxidative DNA damage in mammalian cells exposed to mycotoxins, patulin and citrinin. Toxicol Appl Pharmacol. 2003;191(3):255–63. doi: 10.1016/s0041-008x(03)00254-0. [DOI] [PubMed] [Google Scholar]
- 37.Liu BH, et al. Induction of oxidative stress response by the mycotoxin patulin in mammalian cells. Toxicol Sci. 2007;95(2):340–7. doi: 10.1093/toxsci/kfl156. [DOI] [PubMed] [Google Scholar]
- 38.Bennett JW, Klich M. Mycotoxins. Clin Microbiol Rev. 2003;16(3):497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Council MR. Report of Patulin Clinical Trials Committee. The Lancet. 1944;244(ii):373–5. [Google Scholar]
- 40.Fliege R, Metzler M. Electrophilic properties of patulin. N-acetylcysteine and glutathione adducts. Chem Res Toxicol. 2000;13(5):373–81. doi: 10.1021/tx9901480. [DOI] [PubMed] [Google Scholar]
- 41.Pfeiffer E, Diwald TT, Metzler M. Patulin reduces glutathione level and enzyme activities in rat liver slices. Mol Nutr Food Res. 2005;49(4):329–36. doi: 10.1002/mnfr.200400089. [DOI] [PubMed] [Google Scholar]
- 42.Malhotra JD, et al. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci U S A. 2008;105(47):18525–30. doi: 10.1073/pnas.0809677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reverberi M, et al. Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl Microbiol Biotechnol. 2010;87(3):899–911. doi: 10.1007/s00253-010-2657-5. [DOI] [PubMed] [Google Scholar]
- 44.Seigle-Murandi F, et al. Antitumor activity of patulin and structural analogs. Pharmazie. 1992;47(4):288–91. [PubMed] [Google Scholar]
- 45.Saxena N, et al. Patulin causes DNA damage leading to cell cycle arrest and apoptosis through modulation of Bax, p(53) and p(21/WAF1) proteins in skin of mice. Toxicol Appl Pharmacol. 2009;234(2):192–201. doi: 10.1016/j.taap.2008.09.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.