Summary
Embryonic stem cells (ESC) maintain high genomic plasticity, essential for their capacity to enter diverse differentiation pathways. Post-transcriptional modifications of chromatin histones play a pivotal role in maintaining this plasticity. We now report that one such modification, monoubiquitylation of histone H2B on lysine 120 (H2Bub1), catalyzed by the E3 ligase RNF20, increases during ESC differentiation and is required for efficient execution of this process. This increase is particularly important for the transcriptional induction of relatively long genes during ESC differentiation. Furthermore, we identify the deubiquitinase USP44 as a negative regulator of H2B ubiquitylation, whose downregulation during ESC differentiation contributes to the increase in H2Bub1. Our findings suggest that optimal ESC differentiation requires dynamic changes in H2B ubiquitylation patterns, which must occur in a timely and well-coordinated manner.
Introduction
Eukaryotic chromatin consists of repeating units of the nucleosome, comprising the core histone proteins (H2A, H2B, H3 and H4) wrapped by 146 base pairs of DNA. Histones undergo posttranslational modifications (PTMs) such as methylation, acetylation, phosphorylation, SUMOylation and ubiquitylation, which occur primarily within their N-terminal and C-terminal tails and play vital roles in regulating chromatin dynamics, gene expression and DNA repair (reviewed in Campos and Reinberg, 2009). Not surprisingly, histone PTMs also impact developmental processes, and their deregulation can instigate a variety of pathologies (Bhaumik et al., 2007; Martin-Subero and Esteller, 2011).
While polyubiquitylation usually tags proteins for degradation via the 26S proteasome, monoubiquitylation mainly modulates the molecular characteristics, and hence function and/or localization, of the substrate protein. Histone H2B is monoubiquitylated on Lys120 in mammals (Thorne et al., 1987). Recently, Lys34 was identified as a second monoubiquitylation site (Wu et al., 2011). In mammals, Lys120-monoubiquitylated histone H2B (hereafter referred to as H2Bub1) is preferentially associated with highly transcribed genes (Minsky et al., 2008). The human RNF20/RNF40 complex is the major H2B E3 ligase (Kim et al., 2005). H2Bub1 can cooperate with FACT and the PAF complex to regulate transcription elongation by RNA Polymerase II (Pavri et al., 2006), and can also facilitate DNA repair (Moyal et al., 2011; Nakamura et al., 2011) and mRNA 3’ end processing (Pirngruber et al., 2009) in human cells. A recent yeast study proposes a role for H2Bub1 also in mRNA export from the nucleus into the cytoplasm (Vitaliano-Prunier et al., 2012). Like other histone PTMs, H2Bub1 has been linked with cancer. USP22, an H2Bub1 deubiquitinase (DUB), is part of a gene signature predictive of a cancer stem cell tumor phenotype of aggressive growth, metastasis and therapy resistance (Zhang et al., 2008). Mammalian RNF20 is a putative tumor suppressor (Shema et al., 2008); its downregulation in mammalian cells promotes migration, anchorage independence and tumorigenesis (Shema et al., 2011; Shema et al., 2008). Recently, reduced H2Bub1 levels were shown in advanced and metastatic breast cancer, parathyroid tumors and seminoma (Chernikova et al., 2012; Hahn et al., 2012; Prenzel et al., 2011).
Several studies implicate H2Bub1 in developmental processes (Buszczak et al., 2009; Schmitz et al., 2009; Zhu et al., 2005). Of note, Drosophila H2B deubiquitylation is essential for stem cell maintenance (Buszczak et al., 2009).
Embryonic stem cells (ESC) are pluripotent cells derived from the inner cell mass of the blastocyst (reviewed in Young, 2011). ESCs maintain high genomic plasticity, essential for the capacity to enter any differentiation pathway. Epigenetic mechanisms, including chromatin structure and histone PTMs, are pivotal in this process. Notably, the chromatin of ESC encompasses bivalent domains, where active chromatin marks (e.g. H3K4me3) exist concomitantly with the repressive mark H3K27me3 (Azuara et al., 2006; Bernstein et al., 2006). Accordingly, ESC differentiation is regulated by the concerted action of chromatin modifying enzymes (reviewd in Ang et al., 2011; Fisher and Fisher, 2011; Meissner, 2010; Melcer and Meshorer, 2010). However, a role for RNF20/40 and histone H2Bub1 in ESC differentiation has not been described.
Here, we report that H2Bub1 increases during induced differentiation of human and mouse ESC, as well as of embryonal carcinoma stem cells (ECSC). This increase is essential for optimal differentiation, and is particularly important for efficient transcriptional induction of long genes during differentiation. Furthermore, we identify the DUB USP44 as a negative regulator of H2B ubiquitylation, whose downregulation during ESC differentiation contributes to the increase in H2Bub1. Overall, our findings demonstrate the importance of properly regulated H2Bub1 turnover for ESC differentiation.
Results
Histone H2B monoubiquitylation increases during embryonic stem cell differentiation
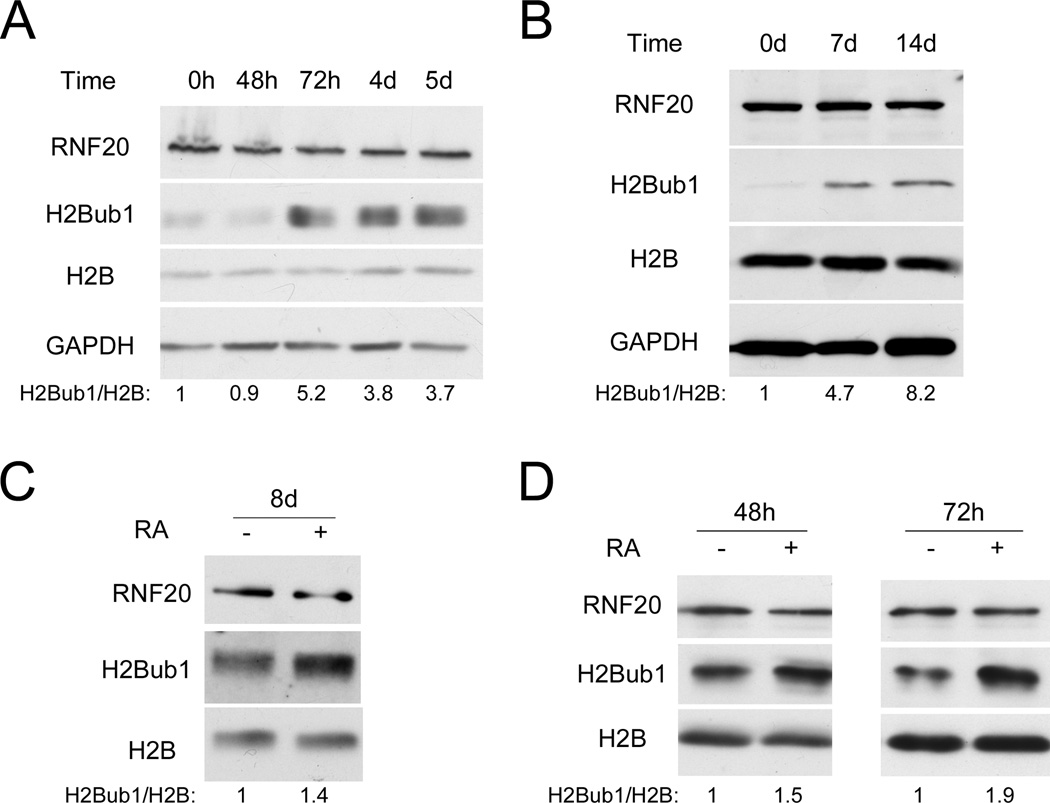
To explore links between H2Bub1 and differentiation, we monitored H2Bub1 levels in mouse embryonic stem cells (mESC) subjected to different differentiation protocols. As seen in Fig. 1A, induction of mESC neuronal differentiation elicited a marked increase in global H2Bub1; differentiation was confirmed by reduced expression of ‘stemness’ genes (Nanog, Oct4) and increased expression of transcripts encoding the pro-neuronal transcription factor Pax6 and the neuronal filament Nestin (Fig. S1A). To determine whether the increase of H2Bub1 is restricted only to neuronal differentiation, a similar analysis was performed on mESC differentiated to Keratinocytes. Keratinocyte differentiation was confirmed by downregulation of Oct4 mRNA and upregulation of cytokeratin K5 mRNA (Fig. S1B). Remarkably, differentiation was accompanied by a gradual increase in H2Bub1, which became very prominent after 2 weeks (Fig. 1B). Likewise, H2Bub1 increased, albeit more modestly, upon retinoic acid (RA)-induced neuronal differentiation of human ESC (hESC, Fig. 1C), confirmed by downregulation of Nanog and Oct4 and upregulation of Pax6 mRNA (Fig. S1C).
Figure 1. Histone H2B is monoubiquitylated during embryonic stem cell differentiation.
(A) R11 mouse embryonic stem cells (mESC) were harvested at the indicated time points after transfer to defined N2B27 differentiation medium, followed by Western blot analysis. Numbers at the bottom were generated by quantification (ImageJ) of the H2Bub1 signal normalized to the H2B signal. (B) CGR8 mESC were differentiated to keratinocytes as described under Experimental Procedures. Cells were harvested at the indicated time points for Western blot analysis. Quantification was as in (A). (C) Human embryonic stem cells (hESC) were transferred to defined N2B27 differentiation medium, supplemented with EGF and FGF, and treated with retinoic acid (RA, +) or DMSO (−). Cells were harvested 8 days later for Western blot analysis. Quantification was as in (A). (D) NT2 cells were treated with retinoic acid (RA, +) or DMSO (−) for 48 or 72 hr. Cell extracts were subjected to Western blot analysis with the indicated antibodies. Quantification was as in (A). See also Fig. S1A,B,C,D for parallel qRT-PCR analysis, Fig. S1E for H2Aub1 and H3K4me3 analysis, and Fig. S1F,G for cell cycle characterization of NT2 cells.
Embryonal carcinoma (EC) cells, the stem cells of teratocarcinomas, resemble pluripotent ESCs of the early embryo (Andrews, 1988). Cells of the human EC cell line NTERA2 clone D1 (NT2) differentiate into mature neurons in presence of RA (Andrews, 1988). NT2 cells are often employed as a surrogate experimental model for studying aspects of hESC biology, since they are easier to maintain and manipulate. Gene expression profiling of NT2 cells confirmed high similarity to hESC; moreover, NT2 cells possess ESC-like chromatin features, including bivalent domains (Schwartz et al., 2005; Shahhoseini et al., 2010). Treatment of NT2 cells with RA also led to elevated H2Bub1 levels (Fig. 1D); neuronal differentiation was confirmed by reduced Nanog and Oct4 and increased Pax6 mRNA (Fig. S1D). Of note, RNF20 protein levels were usually not significantly altered during ESC or NT2 cells differentiation (Fig. 1A,B,C,D), although a mild increase in RNF20 mRNA and protein was occasionally observed in differentiating mESC (e.g Fig. 2C). Interestingly, H3K4me3 and monoubiquitylated H2A (H2Aub1) also increased modestly (Fig. S1E). In sum, in several experimental models, induced differentiation of ESC and ESC-like cells is associated with increased H2B monoubiquitylation.
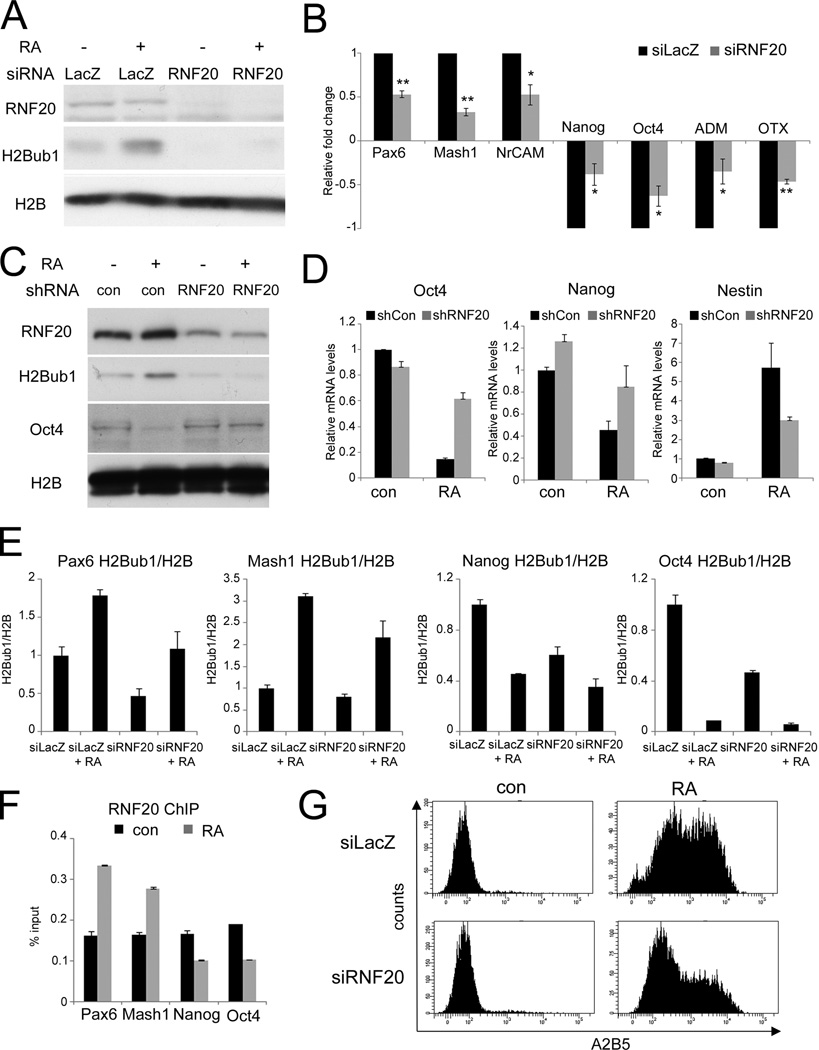
Figure 2. RNF20 is necessary for optimal neuronal differentiation of NT2 and embryonic stem cells.
(A) NT2 cells were transfected with siRNA oligonucleotides (SMARTpool) directed against RNF20 (siRNF20) or against LacZ (siLacZ) as control, and 24 hr later treated with either retinoic acid (RA) or DMSO (−). Following 72 hours of RA treatment, cultures were harvested for Western blot analysis. (B) Cells were treated as in (A). Expression of the indicated genes was quantified by qRT-PCR. All values were normalized to GAPDH mRNA in the same sample. Average relative fold change was calculated from three separate experiments. For each gene, the RA-induced fold change upon RNF20 depletion was normalized to the RA-induced fold change in the control (siLacZ), which was defined as 1.0. * p-value < 0.05. ** p-value < 0.01. See also Fig. S2A for raw values and Fig. S2B for RNF20 depletion with single oligonucleotides. (C) R1 mESC were stably transfected with non-targeting shRNA plasmid (con) or with RNF20 shRNA plasmid (RNF20). Drug-resistant clones were isolated and validated for the expected pattern of RNF20 expression. Cultures were treated with retinoic acid (RA) for 4 days and harvested for Western blot analysis with the indicated antibodies. (D) RNA was extracted from cultures treated as in (C). Expression of the indicated genes was quantified by qRT-PCR. All values were normalized to GAPDH mRNA in the same sample. Bars indicate averages of data from duplicate qPCR reactions; error bars represent standard deviation. Similar data was obtained in three independent experiments. See also Fig. S2C for shRNF20 mESC morphology. (E) Cells were treated as in (A) and subjected to chromatin immunoprecipitation (ChIP) analysis with antibodies specific for H2B and H2Bub1. Immunoprecipitated DNA was quantified by qPCR with primers specific for the 5’ transcribed regions of the indicated genes. Bars indicate H2Bub1 ChIP readings normalized to H2B in the same treatment, and represent averages from duplicate qPCR reactions; error bars represent standard deviation. Similar results were obtained in three independent experiments. (F) Cells were treated as in (A), and subjected to ChIP analysis with RNF20 antibody. Immunoprecipitated DNA and input DNA were quantified by qPCR with primers specific for the 5’ transcribed regions of the indicated genes. Results are average of three separate experiments. (G) NT2 cells were transfected with siRNA oligonucleotides directed against RNF20 (siRNF20) or against LacZ (siLacZ) as control, and 24 hr later were treated either with retinoic acid (RA) or DMSO (con). Following 96 hr of RA treatment, surface levels of the neuronal cell antigen A2B5 were quantified by FACS analysis.
Recently, the Rad6-Bre1 pathway was linked to cell cycle progression in yeast (Zimmermann et al., 2011). To investigate whether the increase in H2Bub1 during differentiation is secondary to cell cycle arrest, NT2 cells were treated with RA for 72 hr and subjected to cell cycle analysis. No significant change in cell cycle distribution was detected at that time point (Fig. S1F); note that full NT2 neuronal differentiation is achieved only after 3 weeks under these conditions. Conversely, treatment with either hydroxyurea or nocodazole, arresting NT2 cells in early S-phase or M phase, respectively (Fig. S1G, right), had no significant effect on H2Bub1 levels (Fig. S1G, left). Hence, the upregulation of H2Bub1 during NT2 differentiation is not due to cell cycle arrest.
Downregulation of RNF20 and H2Bub1 compromises differentiation
Given its correlation with differentiation, we wished to determine whether the increase in H2Bub1 is important for that process. NT2 cells were transfected with pools of RNF20-specific siRNA (siRNF20) or control siRNA oligonucleotides (siLacZ), and exposed to RA for 72 hours. RNF20 knockdown led to substantial reduction of H2Bub1 in both untreated and RA treated cells (Fig. 2A). Remarkably, this compromised significantly the RA-induced downregulation of the stemness genes Nanog and Oct4 and the developmental-related genes ADM and OTX (Fig. 2B, S2A). Likewise, upregulation of mRNA encoding the neuronal adhesion molecule NrCAM and the pro-neural transcription factors Pax6 and Mash1 was also attenuated (Fig. 2B, S2A). Notably, the basal expression of those genes was not significantly influenced by RNF20 depletion (Fig. S2A). To rule out off-target effects, those results were confirmed with single siRNA oligonucleotides directed against RNF20 (Fig. S2B). Thus, downregulation of RNF20 and H2Bub1 compromises the transcriptional reprogramming associated with induced differentiation of NT2 cells.
To determine whether RNF20 and H2Bub1 are important also for the differentiation of authentic ESC, mESC were stably transduced with either a plasmid expressing mouse RNF20 shRNA or a non-targeting shRNA plasmid. As seen in Fig. 2C, stable partial knockdown of RNF20 led to a proportional reduction in H2Bub1 and dampened its RA-induced upregulation. Importantly, this compromised the RA-induced downregulation of Oct4 and Nanog mRNA (Fig. 2D) and Oct4 protein (Fig. 2C), as well as the induction of Nestin mRNA (Fig. 2D). Furthermore, induction of morphological changes characteristic of mESC differentiation was severely impeded (Fig. S2C). Hence, partial depletion of RNF20 and H2Bub1 compromises also mESC differentiation.
Chromatin immunoprecipitation (ChIP) analysis was next performed on several of the above genes. In response to RA treatment of NT2 cells, H2Bub1 and RNF20 levels on those genes were found to change in correlation with the changes in their transcription rates (Fig. 2E,2F). Furthermore, partial depletion of RNF20 decreased both basal and RA-induced levels of H2Bub1 within those genes (Fig. 2E). However, RNF20 depletion did not decrease Oct4 and Nanog mRNA in untreated cells (Fig. 2D, cont), confirming that high H2Bub1 is not required for efficient expression of those pluripotency genes in undifferentiated cells.
To confirm more directly that differentiation was impaired, we monitored the appearance of the neuronal antigen A2B5 on the cell surface of RA-treated NT2 cells. Indeed, RNF20 depletion significantly attenuated surface accumulation of A2B5 (Fig. 2G). Overall, our findings imply that downregulation of RNF20 and H2Bub1 interferes with the differentiation of ESC and ESC-like cells, suggesting a positive involvement of the increased H2Bub1 levels in the differentiation process.
Differentiation-associated induction of long genes is selectively sensitive to RNF20 depletion
To further characterize the transcriptional effects of RNF20 depletion during differentiation, expression microarray analysis was performed. Confirming what was observed above for a limited set of differentiation-related genes, the results revealed that the differentiation-associated transcriptional program was significantly impaired by RNF20 knockdown (Fig. 3A). Based on the analysis in Fig. 3A, we identified a subset of genes with RNF20-dependent induction by RA (see Supplemental Methods), whose RA-induced upregulation in the control (siRNA L) was compromised by RNF20 depletion (siRNA R). Analysis of those genes using the DAVID program, revealed significant enrichment for genes involved in developmental processes (Table S1). Interestingly, genes in the “RNF20-dependent” subset were found to be significantly longer than those in the subset of genes whose induction by RA was “RNF20-independent” (Fig. 3B). Moreover, when analyzing all the genes that were induced by RA, a positive correlation was found between gene length and sensitivity to RNF20 depletion (Fig. 3C). Hence the longer a gene is, the more is its induction during differentiation likely to depend on RNF20 and presumably H2Bub1.
Figure 3. Induction of long genes is selectively affected by RNF20 depletion.
(A) NT2 cells were transfected with siRNA oligonucleotides (SMARTpool) directed against RNF20 (R) or LacZ (L) as control, and 24 hr later were treated either with retinoic acid (RA) or DMSO (−). Following 72 hr of RA treatment, RNA was extracted and subjected to expression microarray analysis. The heat-map depicts the expression patterns of the genes whose abundance was significantly upregulated or downregulated (see Supplemental Methods) following addition of RA. For each row (gene) the log-expression values were centered (mean 0) and normalized (STD = 1). Red and blue colors indicate increased and decreased expression, respectively (relative to the mean value of a gene). (B) Empirical probability density functions of the lengths (longest known coding transcripts) of genes in the “RNF20-dependent” group (solid line, median = 86Kb) and “RNF20-independent” group (dashed line, median = 43Kb). The assignment of genes to each group is described in Supplemental Methods P-value was calculated using Rank Sum test, rejecting the null hypothesis that the two groups have equal medians. (C) A Spearman correlation coefficient of 0.27 was found between the length of the longest coding transcript of each gene and the ratio between FC1 and FC2. FC1 was defined as fold change in RA-treated relative to DMSO treated control cells (siLacZ+RA/siLacZ+DMSO), while FC2 denotes fold change in RNF20-depleted cells (siRNF20+RA/siRNF20+DMSO). Each point represents a gene with significant induction by RA in the control cells. (D) Cells were treated as in (A). Mature mRNA and pre-mRNA were analyzed by qRT-PCR employing exon-exon junction primers or intronic primers, respectively. Values were generated as in Fig. 2B, based on three separate experiments. * p-value < 0.05. ** p-value < 0.01. (E) Cells were treated as in (A) and subjected to ChIP analysis as in Fig. 2C. Immunoprecipitated DNA was quantified by qPCR with primers specific for either the 5‘ or 3‘ transcribed regions of the indicated genes. Bars indicate averages of data from duplicate qPCR reactions; error bars represent standard deviation. Similar results were obtained in three independent experiments. See also Fig. S3A for H3K36me3 ChIP, Fig. S3B for RNF20-independent genes H2Bub1 ChIP, Fig. S3C for RNF20 ChIP, Fig. S3D,E for basal expression analysis, and Fig. S4A,B for the effect of WAC depletion on NT2 differentiation.
The effects of RNF20 depletion on the expression of representative genes were further validated by qRT-PCR. Fig. 3D shows the analysis of four RA-induced genes, all longer than 100Kb. In agreement with the microarray data, induction of the RNF20-dependent genes DAAM2, Cdh2 and Sulf1 was significantly attenuated by RNF20 knockdown (upper panel). In contrast, induction of the RNF20-independent gene BMPER was unaffected by RNF20 depletion. Of note, qRT-PCR analysis of heterogeneous nuclear RNA with intron-derived primers, providing an approximation of relative transcription rates (Kuroda et al., 2005; Phelps et al., 2006; Shema et al., 2008), confirmed that the effects of RNF20 depletion were exerted transcriptionally rather than posttranscriptionally (Fig. 3D, lower panel). In further support of transcriptional regulation, RA treatment led to increased H3K36 trimethylation (H3K36me3) within the transcribed regions of those genes (Fig. S3A).
Consistent with their RNF20 dependence, the RA-induced increase in H2Bub1 levels within the DAAM2, Cdh2 and Sulf1 genes was markedly attenuated in RNF20 depleted cells (Fig. 3E), whereas the induction of H2Bub1 within BMPER and additional RNF20-independent genes was affected to a lesser extent or not at all (Fig. S3B). As expected, increased RA-induced recruitment of RNF20 to the transcribed regions of Cdh2 and Sulf1 could be confirmed (Fig. S3C). Surprisingly, this was not the case for DAAM2, raising the possibility that other E3 ligases may mediate the increase in H2Bub1 within the transcribed region of that gene.
While the RNF20-dependent subgroup was enriched in relatively long genes, some other long genes did not exhibit dependence on RNF20 for their induction by RA. Interestingly, the RNF20-independent genes tended to be expressed at higher levels than the RNF20-dependent ones under basal conditions, prior to induction of differentiation (Fig. S3D). When genes were binned according to their size, this differential trend was very significant for the longest genes, but not for the shortest ones (Fig. S3E). These observations suggest that, for relatively long genes, RNF20 is particularly important for turning on genes that are more tightly “off” under basal conditions. In further support of this conjecture, analysis of H3K9me3 ChIP data in undifferentiated NT2, obtained within the ENCODE project, revealed that RNF20-dependent genes tend to have higher basal levels of H3K9me3 within their transcribed regions than the RNF20-independent subgroup (Median 6.9 ± 9 vs. 4.4 ± 6.6, p-value = 1.04E-05; see Supplemental Methods for further details).
The WAC protein was recently identified as a positive regulator of H2B monoubiquitylation (Zhang and Yu, 2011). As expected, WAC depletion resulted in decreased basal H2Bub1 levels in NT2 cells, and compromised their RA-induced increase (Fig. S4A). Notably, this resulted in impaired induction of neuronal differentiation-associated genes such as Pax6 and Mash1, as well as of the RNF20-dependent genes Cdh2, DAAM2 and Sulf1 (Fig. S3E), further arguing that induction of those genes is dependent on efficient H2B monoubiquitylation.
Together, our findings support the conjecture that increased H2B ubiquitylation during differentiation is selectively required for maximal transcriptional induction of relatively long genes.
USP44 promotes H2Bub1 deubiquitylation
Unlike the increase in H2Bub1 during differentiation, RNF20 protein levels did not increase significantly in most cases (Fig. 1), nor were there observable changes in the subcellular localization of RNF20 (Fig. S5A), its association with chromatin (Fig. S5B), or its interaction with RNF40 (Fig. S5C), whose binding to RNF20 is required for efficient H2B monoubiquitylation (Kim et al., 2009). Hence, the increase in H2Bub1 during differentiation may be due to mechanisms distinct from changes in RNF20. One such alternative could be downregulation of one or more DUBs that target H2Bub1. Both USP3 and USP22 have been implicated as H2Bub1-specific DUBs (Nicassio et al., 2007; Zhang et al., 2008). Although the mRNA levels of those DUBs did not decrease markedly in response to RA (Fig. S5E), it remains possible that they are more active in untreated NT2 cells, thereby keeping H2Bub1 low. However, transient knockdown of those USPs, or of the H2Aub1 DUB USP16 (Joo et al., 2007), affected neither basal H2Bub1 levels nor its induction by RA (Fig. S5D, E), suggesting a role for other DUBs. Notably, the microarray data indicated a pronounced drop in the mRNA encoding the DUB USP44 in RA-treated NT2 cells, validated by qRT-PCR (Fig. 4A). A similar trend was observed during neuronal differentiation of hESC and keratinocytic differentiation of mESC (Fig. 4A). USP44, a direct target of Oct4 (Boyer et al., 2005), was indeed already reported to be downregulated during ESC differentiation (Jung et al., 2010). Interestingly, USP44 shares substantial homology with the histone DUBs ubp8, USP22, USP16 and USP3 (Henry et al., 2003; Joo et al., 2007; Nicassio et al., 2007; Zhang et al., 2008). Moreover, USP44 is localized to the nucleus (Stegmeier et al., 2007; Suresh et al., 2010) and associates with chromatin (Fig. 4B). Together, these findings evoked a possible role for USP44 in the turnover of H2Bub1 during ESC differentiation.
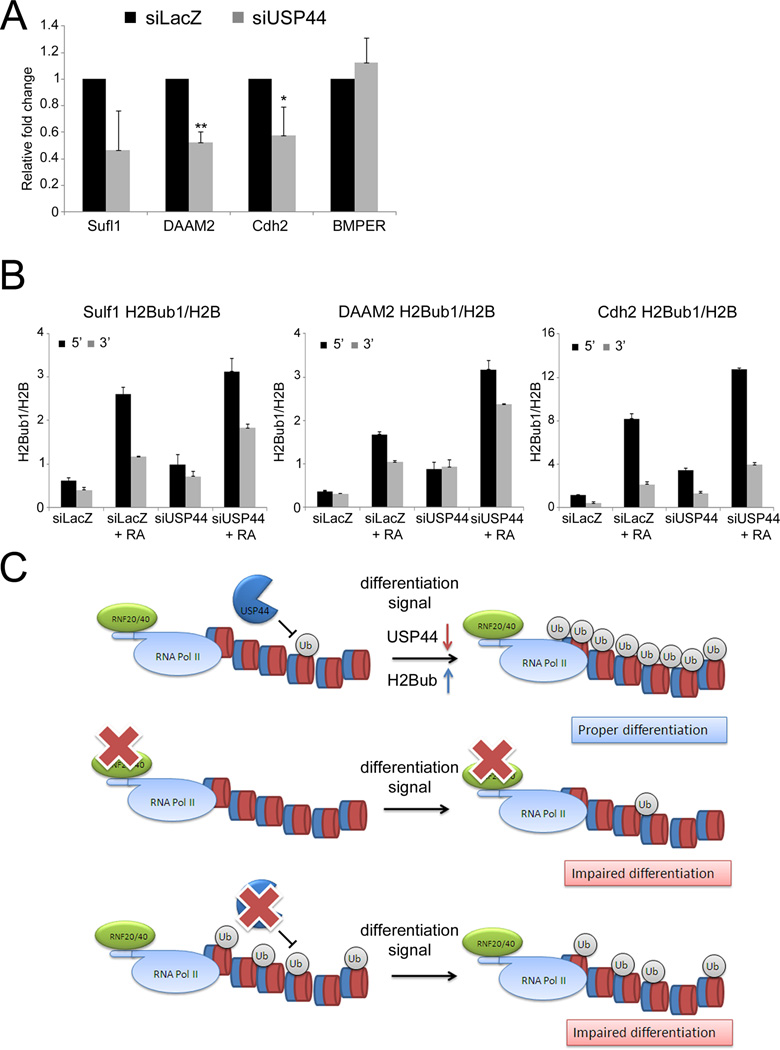
Figure 4. USP44 is downregulated during differentiation to promote H2B ubiquitylation.
(A) qRT-PCR analysis of USP44 mRNA during differentiation. Left: NT2 treated with RA for 72 hr. Middle: hESC treated with RA for 8 days. Right: mESC differentiated to keratinocytes for 7 or 14 days. Levels in undifferentiated cells were set as 1. For NT2 and hESC, values were normalized to β-ACTIN mRNA in the same sample; for mESC, normalization was for 36B4 mRNA. Bars indicate averages from duplicate qPCR reactions; error bars represent standard deviation. (B) HeLa cells were transfected with control empty vector (Vec) or Myc-USP44 expression plasmid. Cells were harvested 48 hr later. Chromatin-bound and unbound proteins were separated and subjected to Western blot analysis. (C) NT2 cells were transfected with siRNA oligonucleotides (SMARTpool) directed against USP44 (siUSP44) or siLacZ as control and harvested 72 hr later for Western blot analysis. Right panel: quantification (ImageJ) of the H2Bub1 signal normalized to the H2B signal, averaged from three independent experiments. The H2Bub1/H2B signal in the siLacZ sample of each experiment was set as 1. * p-value < 0.05. (D) HeLa cells were transfected with either control empty vector (Vec), Myc-tagged USP44 (Myc-USP44 WT) or catalytically inactive Myc-tagged USP44 (Myc-USP44 CI). Cells were harvested 72 hr later and subjected to Western blot analysis. Right panel: quantification as in (C), averaged from three independent experiments; the H2Bub1/H2B value in the empty vector control (Vec) of each experiment was set as 1. ** p-value < 0.01. See also Fig. S5A,B,C for subcellular localization of RNF20 and interaction with RNF40 during differentiation, and Fig. S5D,E,F,G for the effect of siUSP3, siUSP16, siUSP22 and siUSP44 on H2Bub1.
To investigate whether USP44 might be responsible for the differences in H2Bub1 between non-differentiated and differentiated cells, we knocked down USP44 in NT2 cells, using a pool of siRNA oligonucleotides (Fig. S5F). USP44 depletion elicited a significant increase in global H2Bub1 (Fig. 4C), comparable to that seen upon treatment of NT2 cells with RA (Fig. 1D). This was confirmed with single siRNA oligos (Fig. S5G), ruling out off-target effects. Furthermore, overexpression of USP44 in HeLa cells, possessing low endogenous USP44, significantly reduced H2Bub1 levels: in contrast, catalytically inactive USP44 failed to exert such an effect (Fig. 4D). However, pure recombinant USP44 protein was unable to directly deubiquitylate H2Bub1 in vitro (data not shown). Hence, while this might imply that USP44 needs additional protein partners in order to act as an H2Bub1 DUB, we can presently not formally exclude the possibility that USP44 acts indirectly, e.g through deubiquitylating another DUB. Nevertheless, given the similarity of USP44 to validated histone DUBs, it is most likely that USP44 is indeed an H2Bub1-specific DUB, whose downregulation during ESC differentiation contributes to increased H2Bub1 levels.
USP44 depletion impairs differentiation and induction of RNF20-dependent genes
To investigate the impact of USP44 depletion on differentiation, NT2 cells were transfected with USP44 siRNA or LacZ siRNA and exposed to RA. As expected, basal H2Bub1 in untreated cells increased upon USP44 knockdown (Fig. 5A, compare lanes 1 and 3), although it did not reach the levels of RA-treated cells (lanes 2 and 3). Of note, USP44 depletion compromised the RA-driven H2Bub1 upregulation (lanes 2 and 4 and bar graph on the right).
Figure 5. USP44 depletion impairs differentiation.
(A) NT2 cells were transfected with siRNA oligos directed against US44 (siUSP44) or siLacZ as control. 24 hr later, retinoic acid (RA) or DMSO (−) was added for an additional 48 hr, followed by extraction and Western blot analysis. Right panel: quantification of relative H2Bub signal, done as in Fig. 4C. * p-value < 0.05. (B) NT2 cells were transfected and treated as in (A). RNA was extracted and analyzed by qRT-PCR with the indicated primers. Values were calculated as in Fig. 2B, averaging from three separate experiments. * p-value < 0.05. ** p-value < 0.01. See also Fig. S6 for raw values. (C) Cells transfected as in (A) were treated with RA or DMSO for 72 hr and subjected to ChIP analysis as in Fig. 2C. Similar results were obtained in three independent experiments. (D) NT2 cells were transfected with siRNA oligonucleotides directed against USP44 (siUSP44) or against LacZ (siLacZ) as control. 24 hr later, cultures were treated with either retinoic acid (RA) or DMSO (con) for an additional 96 hr, and subjected to FACS analysis of surface A2B5 as in Fig. 2E.
Remarkably, USP44 knockdown compromised the repression of Nanog and Oct4 and the induction of Pax6 (Fig. 5B, S6A), while increasing the association of H2Bub1 with those genes at both basal and differentiated states (Fig. 5C). The ability of USP44 depletion to dampen the transcriptional effects of RA suggested a differentiation defect. Indeed, just like RNF20 depletion, USP44 knockdown significantly attenuated the accumulation of A2B5 on the surface of RA-treated cells (Fig. 5D). Together these observations argue that, similar to RNF20, proper expression of USP44 is necessary for optimal execution of the differentiation program. While at first glance counterintuitive, this may actually imply that dynamic H2B ubiquitylation-deubiquitylation cycles are required for the onset of the differentiation process. In the absence of either ubiquitylation (siRNF20) or deubiquitylation (siUSP44), this process may abort.
To further compare the effects of RNF20 depletion and USP44 depletion on differentiation, we monitored Cdh2, DAAM2 and Sulf1 mRNA levels. As seen in Fig. 6A, RA-driven induction of those RNF20-dependent genes was attenuated in USP44-depleted NT2 cells. In contrast, induction of the RNF20-independent gene BMPER was unaffected by USP44 knockdown (Fig. 6A), as was the case also for other RNF20-indenependt transcripts (Fig. S6B). As seen in Fig. 6B, USP44 depletion resulted in excessive H2Bub1 levels at both 5’ and 3’ ends of the transcribed regions of the Cdh2, DAAM2 and Sufl1 genes. Hence, USP44 depletion and abnormally elevated H2Bub1 levels result in impaired induction of RNF20-dependent genes. These findings support the notion that transcriptional activation of these genes in response to differentiation signals requires optimal H2Bub1 turnover. In conclusion, initiation of ESC differentiation may entail dynamic changes in H2B monoubiquitylation patterns, which must occur in a timely and well-coordinated manner (Fig. 6C).
Figure 6. USP44 depletion compromises the induction of RNF20-dependent genes.
(A) NT2 cells were transfected with siRNA oligos directed against USP44 (siUSP44) or siLacZ as control and 24 hr later were treated for 72 hr with either retinoic acid (RA) or DMSO. RNA was extracted and analyzed by qRT-PCR with the indicated primers, and relative fold change upon RA treatment was calculated as in Fig. 2B, averaging from three separate experiments. * p-value < 0.05. ** p-value < 0.01. (B) Cells treated as in (A) were subjected to ChIP analysis as in Fig. 3E. Similar results were obtained in three independent experiments. (C) Proposed model for regulation of H2Bub1 during ESC differentiation. Upper panel: in the undifferentiated state, USP44 is highly expressed and maintains low levels of H2Bub. Differentiation signals decrease USP44 levels, increasing H2Bub1 to enable efficient differentiation. Middle: RNF20 depletion abolishes the increase in H2Bub1, impairing differentiation. Lower: USP44 depletion prior to the differentiation signal increases H2Bub1 levels; however, the dynamic turnover of H2Bub1 is impaired, disabling proper execution of the differentiation program.
Discussion
Chromatin modifications play a pivotal role in cell differentiation. We report here that the onset of differentiation in ESC and ESC-like cells is accompanied by a global increase in H2Bub1, which is required for optimal execution of the differentiation program. In conjunction with the findings of Karpiuk et al., these facts position H2B ubiquitylation as an important regulator of stem cell differentiation. Yet, the link between H2Bub1 and differentiation may also depend on cellular context; thus, a recent report suggests that at least in some progenitor cells there may actually be a negative correlation between H2Bub1 and differentiation (Vethantham et al., 2012).
Our data imply that H2Bub1 is particularly important for the efficient induction of relatively long genes in response to differentiation signals. Yeast data also demonstrate a role for H2Bub1 in transcription of long genes (Gaillard et al., 2009). It is thus conceivable that the expression of differentiation-induced longer genes is preferentially subject to negative regulation in ESC, which presumably occurs also at the elongation step and is relieved by H2Bub1. In this regard, we have observed that relatively long genes whose induction during differentiation is RNF20-dependnet possess high H3K9me3 levels within their transcribed region. H2Bub1 was reported to be required for the methylation of histone H3K4 at gene promoters (Kim et al., 2009; Lee et al., 2007; Sun and Allis, 2002). However, since H2Bub1 is found mainly within the transcribed region of genes, it is possible that it has an additional distinct role in recruiting various histone demethylases in order to remove H3K9me3 from lineage-specific genes. Such possibility would be consistent with the facts that H2Bub1 abounds in the transcribed region and that H3K9me3 can be observed as extended “blocks” within transcribed regions (Hawkins et al., 2010). Given that RNF20-dependent longer genes tend to display lower basal expression levels and higher H3K9me3 in the undifferentiated state, we propose that H2Bub1 may be selectively required for switching such genes into a more open chromatin state. In support of a role for H2Bub1 in switching the chromatin into a more open state, a recent study employing chemically defined nucleosome arrays found that H2Bub monoubiquitylation interferes with chromatin compaction and leads to an open and biochemically more accessible fiber conformation (Fierz et al., 2011). Such open conformation may be needed in order to increase the overall transcription elongation rate rather than to overcome a localized transcriptional block. It is conceivable that failure of such “switch”, as elicited experimentally by RNF20 knockdown, may selectively affect longer genes whose optimal transcriptional elongation is more critical for successful production of full-length transcripts.
In addition to its well documented role in transcription control, H2Bub1 was reported to be required for chromatin boundary integrity (Ma et al., 2011). It is tempting to speculate that H2Bub1 contributes to differentiation not only by facilitating the transcription of individual genes, but also by affecting more broadly specific chromatin boundary regions, thereby perhaps setting a barrier to restrict differentiated cells from reverting back to a pluripotent state.
We also implicate USP44 as a regulator of H2Bub1 deubiquitylation. USP44 expression is markedly downregulated in response to differentiation signals. While additional mechanisms may also contribute to the differentiation-associated increase in H2Bub1, our data argue strongly that the turn-off of USP44 expression plays an important role in this process. Like the yeast H2B DUB Ubp8 and the mammalian H2Bub1 DUBs USP22 and USP3, USP44 possesses a zinc-finger ubiquitin-specific protease (ZnF-UBP) domain, proposed to play a crucial role in regulating USPs toward ubiquitylated histones (Bonnet et al., 2008). Moreover, in a screen for interactors of USP44, histones H2A and H2B, and the subunit of the RNA polymerase II – POLR2G were significantly enriched (Sowa et al., 2009). Notably, in differentiated cells USP44 was identified as a critical regulator of the mitotic spindle checkpoint, through deubiquitylation of the Cdc20 protein (Stegmeier et al., 2007). It is plausible that Cdc20 is a high affinity USP44 target, rendering it a substrate for that DUB even in differentiated cells that express rather little USP44. In contrast, efficient deubiquitylation of the much more abundant H2B may require greater amounts of USP44, normally found only in ESC. This would render USP44 practically an ESC-restricted H2Bub1 DUB. Additional members of the ZnF-UBP DUB family, such as USP45 and USP49, might play similar unique roles in other cell types in which they are highly expressed. Lastly, our data do not rule out additional, H2Bub1-independent roles of USP44 in ESC regulation, e.g via deubiquitylation of transcription factors or other regulatory proteins.
Given the antagonistic biochemical activities of USP44 and RNF20 and the facts that RNF20 is a candidate tumor suppressor (Shema et al., 2008) and that H2Bub1 levels are strongly reduced in advanced human breast cancer, parathyroid tumors and seminoma (Chernikova et al., 2012; Hahn et al., 2012; Prenzel et al., 2011), one might expect to observe excess USP44 in cancer. Indeed, high USP44 expression was recently reported in human T-cell leukemia (Zhang et al., 2011). It will be of interest to find out whether, in addition to deregulating the spindle checkpoint, USP44 contributes to the pathology of this and perhaps other malignancies by quenching H2Bub1 and enforcing a less differentiated phenotype.
Experimental Procedures
Cell culture and differentiation
NTera2/D1 cells were grown as described (Gutekunst et al., 2011) with 1mM sodium pyruvate and 0.1mM nonessential amino acids. Neuronal differentiation was induced with 10 µM RA. hESC were grown with feeder layer in hESC medium, and transferred into defined N2B27 differentiation medium containing 20ng/ml EGF, 12 ng/ml FGF and 0.1 µM RA for 8 days. CGR8 embryonic stem cells were used for keratinocytic differentiation. Culture conditions and protocols for epidermal differentiation have been described previously (Medawar et al., 2008). R11 and R1 mESC were maintained with feeder layer in ES medium. Neuronal differentiation of R11 cells was induced under monolayer culture conditions as described (Ying et al., 2003), except that B27 without vitamin A was used. Neuronal differentiation of R1 cells was induced with 1 µM RA. See Supplemental Experimental Procedures for further details.
Microarray hybridization and analysis
For oligonucleotide microarray hybridization, RNA was extracted with RNeasy Mini Kit (Qiagen). 10µg RNA was labeled and hybridized to Affymetrix GeneChip Human Exon 1.0 ST arrays. For analysis, the Affymetrix Expression Console (parameters: Annotation confidence—full, Summarization method—iterPLIER include DABG, Background—PM-GCBG, Normalization method—none) was used, followed by normalization of all arrays together using a Lowess multi-array algorithm (Ballman et al., 2004). Intensity-dependent estimation of noise (Zeisel et al., 2010) was used for statistical analysis of differential expression.
Supplementary Material
Highlights.
Differentiation is associated with an increase in histone H2B monoubiquitylation
RNF20 is necessary for optimal differentiation
RNF20 is required for induction of long genes during differentiation
USP44 negatively regulates H2B ubiquitylation
Acknowledgements
We thank Steve Elledge and Frank Stegmeier for the USP44 plasmids, Yael Aylon, Debora-Rosa Bublik, Tingting Yao and Robert E. Cohen for insightful discussions, Elena Ainbinder and Gilad Beck for generating shRNF20 mESC, Gilgi Friedlander for initial microarray analysis and Tamar Unger and Dikla Hiya for recombinant USP44. This work was supported in part by grant R37 CA40099 from the National Cancer Institute, Grant 293438 (RUBICAN) from the European Research Council, the Dr. Miriam and Sheldon Adelson Medical Research Foundation, The Lower Saxony-Israeli Association (to MO), the Leir Charitable Foundation (to ED) and the Edmond J. Safra Foundation (to AB). MO is incumbent of the Andre Lwoff chair in Molecular Biology, ED is incumbent of the Henry J. Leir Professorial Chair. ES is supported by the Adams Fellowship Program of the Israel Academy of Sciences and Humanities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The GEO accession number for the data in this paper is GSE33214.
References
- Andrews PW. Human teratocarcinomas. Biochim Biophys Acta. 1988;948:17–36. doi: 10.1016/0304-419x(88)90003-0. [DOI] [PubMed] [Google Scholar]
- Ang YS, Gaspar-Maia A, Lemischka IR, Bernstein E. Stem cells and reprogramming: breaking the epigenetic barrier? Trends Pharmacol Sci. 2011;32:394–401. doi: 10.1016/j.tips.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Ballman KV, Grill DE, Oberg AL, Therneau TM. Faster cyclic loess: normalizing RNA arrays via linear models. Bioinformatics. 2004;20:2778–2786. doi: 10.1093/bioinformatics/bth327. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Romier C, Tora L, Devys D. Zinc-finger UBPs: regulators of deubiquitylation. Trends Biochem Sci. 2008;33:369–375. doi: 10.1016/j.tibs.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Spradling AC. Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science. 2009;323:248–251. doi: 10.1126/science.1165678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA, Chernikova DA, Kwok S, Brooks JD, Bailey SM, et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CL, Fisher AG. Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr Opin Genet Dev. 2011;21:140–146. doi: 10.1016/j.gde.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Gaillard H, Tous C, Botet J, Gonzalez-Aguilera C, Quintero MJ, Viladevall L, Garcia-Rubio ML, Rodriguez-Gil A, Marin A, Arino J, et al. Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-not in transcription-coupled repair. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000364. e1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst M, Oren M, Weilbacher A, Dengler MA, Markwardt C, Thomale J, Aulitzky WE, van der Kuip H. p53 hypersensitivity is the predominant mechanism of the unique responsiveness of testicular germ cell tumor (TGCT) cells to cisplatin. PLoS One. 2011;6:e19198. doi: 10.1371/journal.pone.0019198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Dickson KA, Jackson S, Clarkson A, Gill AJ, Marsh DJ. The tumor suppressor CDC73 interacts with the ring finger proteins RNF20 and RNF40 and is required for the maintenance of histone 2B monoubiquitination. Hum Mol Genet. 2012;21:559–568. doi: 10.1093/hmg/ddr490. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- Jung M, Peterson H, Chavez L, Kahlem P, Lehrach H, Vilo J, Adjaye J. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoS One. 2010;5:e10709. doi: 10.1371/journal.pone.0010709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Oikawa K, Yoshida K, Takeuchi A, Takeuchi M, Usui M, Umezawa A, Mukai K. Effects of 3-methylcholanthrene on the transcriptional activity and mRNA accumulation of the oncogene hWAPL. Cancer Lett. 2005;221:21–28. doi: 10.1016/j.canlet.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Ma MK, Heath C, Hair A, West AG. Histone crosstalk directed by H2B ubiquitination is required for chromatin boundary integrity. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002175. e1002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Subero JI, Esteller M. Profiling epigenetic alterations in disease. Adv Exp Med Biol. 2011;711:162–177. doi: 10.1007/978-1-4419-8216-2_12. [DOI] [PubMed] [Google Scholar]
- Medawar A, Virolle T, Rostagno P, de la Forest-Divonne S, Gambaro K, Rouleau M, Aberdam D. DeltaNp63 is essential for epidermal commitment of embryonic stem cells. PLoS One. 2008;3:e3441. doi: 10.1371/journal.pone.0003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- Melcer S, Meshorer E. Chromatin plasticity in pluripotent cells. Essays Biochem. 2010;48:245–262. doi: 10.1042/bse0480245. [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Nicassio F, Corrado N, Vissers JH, Areces LB, Bergink S, Marteijn JA, Geverts B, Houtsmuller AB, Vermeulen W, Di Fiore PP, Citterio E. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr Biol. 2007;17:1972–1977. doi: 10.1016/j.cub.2007.10.034. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Phelps ED, Updike DL, Bullen EC, Grammas P, Howard EW. Transcriptional and posttranscriptional regulation of angiopoietin-2 expression mediated by IGF and PDGF in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2006;290:C352–C361. doi: 10.1152/ajpcell.00050.2005. [DOI] [PubMed] [Google Scholar]
- Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3'-end processing. EMBO Rep. 2009;10:894–900. doi: 10.1038/embor.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, Hintermair C, Eick D, Kremmer E, Simons M, et al. Estrogen-Dependent Gene Transcription in Human Breast Cancer Cells Relies upon Proteasome-Dependent Monoubiquitination of Histone H2B. Cancer Res. 2011;71:5739–5753. doi: 10.1158/0008-5472.CAN-11-1896. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Tamada Y, Doyle MR, Zhang X, Amasino RM. Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol. 2009;149:1196–1204. doi: 10.1104/pp.108.131508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CM, Spivak CE, Baker SC, McDaniel TK, Loring JF, Nguyen C, Chrest FJ, Wersto R, Arenas E, Zeng X, et al. NTera2: a model system to study dopaminergic differentiation of human embryonic stem cells. Stem Cells Dev. 2005;14:517–534. doi: 10.1089/scd.2005.14.517. [DOI] [PubMed] [Google Scholar]
- Shahhoseini M, Taei A, Mehrjardi NZ, Salekdeh GH, Baharvand H. Epigenetic analysis of human embryonic carcinoma cells during retinoic acid-induced neural differentiation. Biochem Cell Biol. 2010;88:527–538. doi: 10.1139/o09-181. [DOI] [PubMed] [Google Scholar]
- Shema E, Kim J, Roeder RG, Oren M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol Cell. 2011;42:477–488. doi: 10.1016/j.molcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER, 3rd, Li MZ, Hannon GJ, Sorger PK, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Suresh B, Ramakrishna S, Lee HJ, Choi JH, Kim JY, Ahn WS, Baek KH. K48- and K63-linked polyubiquitination of deubiquitinating enzyme USP44. Cell Biol Int. 2010;34:799–808. doi: 10.1042/CBI20090144. [DOI] [PubMed] [Google Scholar]
- Thorne AW, Sautiere P, Briand G, Crane-Robinson C. The structure of ubiquitinated histone H2B. Embo J. 1987;6:1005–1010. doi: 10.1002/j.1460-2075.1987.tb04852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Yang Y, Bowman C, Asp P, Lee JH, Skalnik DG, Dynlacht BD. Dynamic Loss of H2B Ubiquitylation without Corresponding Changes in H3K4 Trimethylation during Myogenic Differentiation. Mol Cell Biol. 2012;32:1044–1055. doi: 10.1128/MCB.06026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Babour A, Herissant L, Apponi L, Margaritis T, Holstege FC, Corbett AH, Gwizdek C, Dargemont C. H2B ubiquitylation controls the formation of export-competent mRNP. Mol Cell. 2012;45:132–139. doi: 10.1016/j.molcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zee BM, Wang Y, Garcia BA, Dou Y. The RING finger protein MSL2 in the MOF complex is an E3 ubiquitin ligase for H2B K34 and is involved in crosstalk with H3 K4 and K79 methylation. Mol Cell. 2011;43:132–144. doi: 10.1016/j.molcel.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Amir A, Kostler WJ, Domany E. Intensity dependent estimation of noise in microarrays improves detection of differentially expressed genes. BMC Bioinformatics. 2010;11:400. doi: 10.1186/1471-2105-11-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell. 2011;41:384–397. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, van Deursen J, Galardy PJ. Overexpression of ubiquitin specific protease 44 (USP44) induces chromosomal instability and is frequently observed in human T-cell leukemia. PLoS One. 2011;6:e23389. doi: 10.1371/journal.pone.0023389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Zimmermann C, Chymkowitch P, Eldholm V, Putnam CD, Lindvall JM, Omerzu M, Bjoras M, Kolodner RD, Enserink JM. A chemical-genetic screen to unravel the genetic network of CDC28/CDK1 links ubiquitin and Rad6-Bre1 to cell cycle progression. Proc Natl Acad Sci U S A. 2011;108:18748–18753. doi: 10.1073/pnas.1115885108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.