Abstract
Purpose.
The perivascular localization of stem cell (SC) niches suggests the presence of a vascular niche. We aimed to determine the angiogenesis potential of limbal niche cells (NCs).
Methods.
Human limbal NCs were isolated and serially passaged on plastic or coated Matrigel in embryonic SC medium containing BFGF and leukemia inhibitory factor before being reseeded in 3D Matrigel. Expression of angiogenesis markers was assessed by RT-qPCR and immunofluorescence staining. Their angiogenesis potential was measured by differentiation into vascular endothelial cells and by supporting vascular tube network formed by human umbilical vein endothelial cells (HUVEC) on Matrigel. Their support of limbal epithelial progenitor cells (LEPC) was examined in sphere growth formed by reunion in 3D Matrigel.
Results.
On plastic, limbal NC could be cultured only up to four passages before turning into myofibroblasts. In contrast, on coated Matrigel, they could be expanded for up to 12 passages with upregulation of markers suggestive of angiogenesis progenitors when reseeded in 3D Matrigel because they could differentiate into vascular endothelial cells and pericytes stabilizing the tube network formed by HUVEC. Although both expanded limbal NCs and HUVEC rejoined with LEPC to form spheres to upregulate expression of ΔNp63α, CK15, and CEBPδ, the former but not the latter abolished expression of CK12 keratin.
Conclusions.
Human limbal NCs continuously expanded on the basement membrane differentiate into angiogenesis progenitors that prevent differentiation of LEPC/SCs. They may partake in formation of the vascular niche and contribute to angiogenesis during wound healing.
Human limbal stromal niche cells have the potential of differentiating into angiogenesis progenitors and preventing corneal epithelial differentiation, implying their partaking in formation of the vascular niche and contributing to angiogenesis during wound healing.
Introduction
Stem cell (SC) niches consist of supporting niche cells (NCs), extracellular matrix, and modulating signals that can maintain SC survival, self-renewal, and fate decision.1 The perivascular localization of SC niches in the bone marrow,2–4 the central nervous system,5 and the testis6 indicates the presence of a vascular niche, which consists of vascular endothelial cells, pericytes, and the basement membrane in between.7 Vascular endothelial cells can support SCs in bone marrow,8,9 the central nervous system,5,10,11 pancreatic islets,12 and muscles.13 There is only one study defining pericytes as stromal cells to support keratinocytes in organotypic cultures.14 Perivascular pericytes have been recognized as a common source of mesenchymal SCs,15–19 which may regulate self-renewal and differentiation of SCs20–23 and, in some in vitro instances, differentiate into vascular endothelial cells.24,25 It remains unclear whether angiogenesis progenitors but not vascular endothelial cells may also serve as NCs.
The corneal epithelial SCs are located at a unique anatomic region termed limbal palisades of Vogt,26,27 where the basement membrane is fenestrated28,29 and undulated with papillae or “pegs” of the limbal stroma extending upward.30 Histological serial sections disclosed that limbal SCs may invade into the limbal stroma, giving rise to a structure called limbal epithelial crypts.28,29 Ultrastructural analyses also disclosed similar limbal crypts surrounded by focal stromal projections (i.e., finger-like projections of stroma containing a central blood vessel).31 These structural features indicate that limbal epithelial SCs lie deeper in the stroma, which is rich in blood vessels; however, it remains unclear whether a vascular niche also exists in the limbal palisades of Vogt.
We have recently reported that collagenase, which cleaves interstitial collagens but not the basement membrane, can isolate a cluster of cells consisting of not only entire limbal epithelial progenitors and SCs but also subjacent vimentin (Vim)+ stromal mesenchymal cells.32 These Vim+ cells are as small as 5 μm in diameter and heterogeneously express embryonic SC (ESC) markers, such as Oct4, Sox2, SSEA4, and Nanog, as well as other SC markers, such as Nestin, N-Cadherin, and CD34.32 Recently, we reported that these small limbal stromal cells could be isolated and expanded on coated Matrigel in the embryonic SC medium (ESCM) supplemented with BFGF and leukemia inhibitory factor (LIF) into Vim+ spindle cells that may reversibly express ESC markers upon being reseeded in 3D Matrigel.33 Herein, we further characterize such expanded spindle cells as angiogenesis progenitors and provide strong evidence that they abolish corneal epithelial differentiation of human limbal epithelial progenitors/SCs more effectively than vascular endothelial cells. The significance of these findings in the role of angiogenesis during wound healing in the human limbal niche is further discussed.
Materials and Methods
Cell Isolation from Human Limbus
Corneolimbal rims from human donors (23 to 70 years old) after corneal transplantation were provided by the Florida Lions Eye Bank (Miami, FL) and handled according to the Declaration of Helsinki. After being rinsed three times with Hank's balanced salt solution, containing 50 mg/mL gentamicin and 1.25 mg/mL amphotericin B, and the removal of excessive sclera, conjunctiva, iris, and corneal endothelium, the rim was cut into 1-clock-hour segments, each including tissue 1 mm within and beyond the anatomic limbus. Limbal segments were digested with 2 mg/mL collagenase A in serum-free ESCM at 37°C for 18 hours under humidified 5% CO2 to generate “collagenase-isolated clusters.''32 In parallel, the limbal segment was digested with 10 mg/mL dispase in ESCM at 4°C for 16 hours to isolate an intact epithelial sheet.34 All materials used for cell isolation and culture are listed in Supplementary Table 1 (see Supplementary Material, http://www.iovs.org/content/53/7/3357/suppl/DC1).
Table 1. .
Serial Passages of the Limbal Stromal NCs on Plastic
|
Passage |
Seeding Density (× 105/cm2) |
Culture Time (d) |
Final Density (× 105/cm2) |
NCDs |
Cumulative NCD |
Population Doubling Time (h) |
| P0 | 0.1 | 16 | 0.13 | 0.38 | 0.38 | 1014 |
| P1 | 0.05 | 16 | 0.25 | 2.32 | 2.70 | 165 |
| P2 | 0.05 | 18 | 0.2 | 2.00 | 4.70 | 216 |
| P3 | 0.05 | 30 | 0.13 | 1.38 | 6.08 | 522 |
| P4 | 0.05 | 30 | 0.06 | 0.26 | 6.34 | 2737 |
The table summarizes the seeding density, culture time, and final density from P0 to P4, from which cumulative NCDs and population doubling time were calculated.
Serial Passages on Plastic or Coated Matrigel
Single cells derived from collagenase-isolated clusters by 0.25% trypsin and 1 mM EDTA (T/E) at 37°C for 15 minutes were seeded at 1 × 105 per cm2 in the 6-well plastic plate with or without coated Matrigel, which was prepared by adding 40 μL of 5% Matrigel per cm2 1 hour before use and cultured in ESCM containing 4 ng/mL BFGF and 10 ng/mL LIF in humidified 5% CO2 with media changed every 3 or 4 days. Cells at 80% to 90% confluence were rendered single cells by T/E and serially expanded at the seeding density of 5 × 103 cells per cm2 for up to 12 passages. The extent of total expansion was measured by the number of cell doubling (NCD) using the following formula: NCD = log10(y/x)/log102, where y is the final density of the cells and x is the initial seeding density of the cells. In parallel, cells were cultured in the 6-well plate without coated Matrigel in the same medium as a control.
Coculturing with Limbal Epithelial Progenitor Cells in 3D Matrigel
As reported,33,35 cells expanded on coated Matrigel at passage 4 (P4) were reseeded in 3D Matrigel to generate P4/3D aggregates. Single cells obtained from P4/3D aggregates or human umbilical vein endothelial cells (HUVEC) were prelabeled with red fluorescent nanocrystals (Qtracker cell labeling kits; Invitrogen, Carlsbad, CA), mixed with single cells derived from dispase-isolated limbal epithelial sheets at a ratio of 1:4, and seeded at the density of 5 × 104 per cm2 to generate sphere growth. After 10 days of culturing in ESCM, the resultant spheres were collected by digesting Matrigel with 10 mg/mL dispase at 37°C for 2 hours.
Differentiation into Vascular Endothelial Cells
To induce differentiation into vascular endothelial cells, single cells from P4/3D aggregates were seeded at the density of 104 cells per cm2 in 24-well plastic plates for 3 days in the Endothelial Cell Growth Medium 2 (EGM2) supplemented with10 ng/mL VEGF. At 80% to 90% confluence, cells were incubated with 10 μg/mL Dil-Ac-LDL (Invitrogen) for 10 hours at 37°C in the humidified 5% CO2 incubator or fixed with 4% paraformaldehyde for immunofluorescence staining.
Vascular Tube Formation by HUVEC
Single cells obtained from P4/3D aggregates were mixed at a ratio of 1:1 with red fluorescent nanocrystal (Invitrogen) prelabeled HUVEC and seeded at the density of 105 cells per cm2 on the surface of Matrigel, which was prepared by adding 50 μL of 100% Matrigel into 24-well plates for 30 minutes before use, and cultured in EGM2 to elicit vascular tube-like network, as reported.36–38 P4/3D cells or HUVEC alone were also seeded at the same density as the controls.
Immunofluorescence Staining
Cytospin preparation of 4 × 104 single cells per slide was made by Cytofuge (StatSpin, Inc., Westwood, MA) at 1000 rpm for 8 minutes. After being air dried for 5 minutes, cells were fixed with 4% paraformaldehyde for 15 minutes, permeated with 0.2% Triton X-100 in PBS for 20 minutes, and blocked with 2% bovine serum albumin in PBS for 1 hour before addition of primary antibody overnight. The appropriate secondary antibodies were then incubated for 1 hour, and Hoechst 33342 was used to counterstain the nucleus for 5 minutes before image analysis. Isotype-matched nonspecific IgG antibodies were used as the controls. Immunofluorescence micrographs were taken by laser confocal microscope (LSM700; Carl Zeiss, Thornhood, NY). Detailed information about primary and secondary antibodies is listed in Supplementary Table 2 (see Supplementary Material, http://www.iovs.org/content/53/7/3357/suppl/DC1).
Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction
Single cells were subjected to total RNA extraction by RNeasy Mini RNA isolation kit (Qiagen, Valencia, CA), and cDNA was reverse-transcribed from 1 to 2 μg of total RNA by high-capacity reverse transcription kit (Applied Biosystems, Foster City, CA). Quantitative PCR (qPCR) amplification of different genes was carried out in a 20-μL solution containing cDNA, TaqMan Gene Expression Assay Mix, and universal PCR master mix (Applied Biosystems). All assays were performed in triplicate for each condition. The results were normalized by an internal control, glceraldehyde-3-phosphate dehydrogenase. The relative gene expression data were analyzed by the comparative CT method (ΔΔCT). All TaqMan Gene Expression Assays with probe sequences are listed in Supplementary Table 3 (see Supplementary Material, http://www.iovs.org/content/53/7/3357/suppl/DC1).
Western Blot
Proteins were extracted from Day 10 spheres generated by limbal epithelial progenitors alone or mixed with P4/3D cells or HUVEC by radioimmunoprecipitation (RIPA) buffer supplemented with proteinase inhibitors. Equal amounts of proteins measured by the BCA assay (Pierce, Rockford, IL) in total cell extracts were separated by 10% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were then blocked with 5% (wt/vol) fat-free milk in TBST (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.05% [vol/vol] Tween-20), followed by sequential incubation with specific primary antibodies and their respective secondary antibodies using β-actin as the loading control. The immunoreactive bands were visualized by a chemiluminescence reagent (Western Lightning; Pierce). Antibodies used are listed in Supplementary Table 2 (see Supplementary Material, http://www.iovs.org/content/53/7/3357/suppl/DC1).
Statistical Analysis
All assays were performed in triplicate, each with a minimum of three donors. The data are reported as means ± SD. Group means were compared using the appropriate version of Student's unpaired t-test. Test results were reported as two-tailed P values, where P < 0.05 was considered statistically significant.
Results
Serial Passages on Plastic
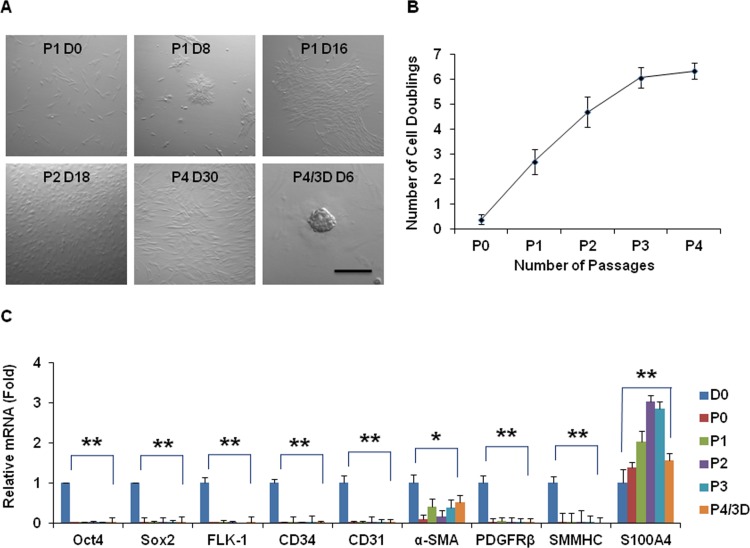
Previously, we successfully expanded limbal NCs on coated Matrigel for up to 4 passages.33 To investigate the significance of Matrigel for such success, we first expanded the limbal NCs on plastic by serial passage in ESCM containing LIF and BFGF using collagenase-isolated cells from a 62-year-old donor. Such culture yielded spindle cells (Fig. 1A) and could only reach P4 with a doubling time of more than 165 hours and NCD of 6 (Fig. 1B). When P3 single cells were reseeded in 3D Matrigel for 6 days, they generated P4/3D aggregates at Day 6 with a smooth contour (Fig. 1A). Compared with D0 cells just isolated, P3 spindle cells did not express Oct4 and Sox2 (i.e., markers of ESC). They also did not express Flk-1, CD34, CD31, and platelet-derived growth factor receptor β (PDGFRβ), markers suggestive of angiogenesis progenitors. Because they expressed transcripts of α-smooth muscle actin (α-SMA) and S100A4, but not smooth muscle myosin heavy chain (SMMHC) (Fig. 1C, n = 3, *P < 0.05, **P < 0.01), limbal NCs expanded on plastic turned into myofibroblasts. Furthermore, the resultant P4/3D cells did not regain expression of ESC and angiogenesis markers even after being reseeded in 3D Matrigel.
Figure 1. .
Serial passages on plastic. Cells isolated from collagenase-isolated clusters from a 62-year-old donor were serially passaged on plastic in ESCM containing LIF and BFGF. They yielded spindle cells (A) and could only reach P4 with a doubling time of more than 165 hours and NCD of 6 (B). When P3 single cells were reseeded in 3D Matrigel for 6 days, they generated P4/3D aggregates at Day 6 with a smooth contour (A). Compared with D0 cells just isolated, P3 spindle cells did not express Oct4, Sox2, Flk-1, CD34, CD31, PDGFRβ, and SMMHC transcripts, but expressed α-SMA and S100A4 transcripts (C, *P < 0.05, **P < 0.01). Furthermore, after being reseeded in 3D Matrigel, the resultant P4/3D cells did not regain expression of the above markers. Scale bar = 200 μm.
Serial Passages on Coated Matrigel
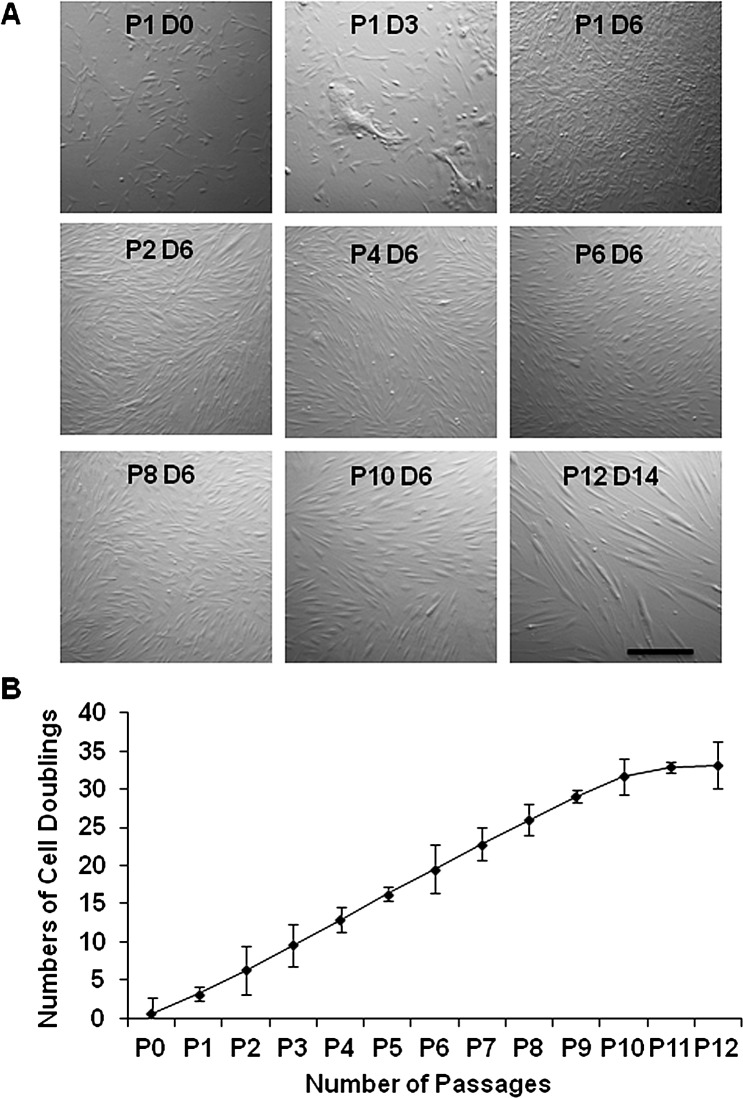
When the above collagenase-isolated cells were serially passaged on coated Matrigel in ESCM with BFGF and LIF, consistent with our recent report,33 spindle-shaped cells could be isolated and expanded by completely eliminating epithelial cells by P2 (Fig. 2A). Unlike the counterpart cultured on plastic, spindle cells could be expanded on coated Matrigel for up to P12, resulting in a total of 33 cell doublings, yielding approximately 1 × 1010 spindle cells from 12 limbal segments. Cells at P1 to P10 exhibited a uniform proliferative rate with a cell doubling time between 43 and 47 hours (Fig. 2B, Table 2).
Figure 2. .
Serial passages on coated Matrigel. Single cells derived from collagenase-isolated limbal clusters from one limbal segment of the same donor as in Figure 1 were serially passaged on coated Matrigel in ESCM with BFGF and LIF. They generated spindle cells from P1 to P12 (A) and had a steady proliferative rate with the doubling time of 43 to 47 hours from P2 to P10 (B). Bar = 100 μm.
Table 2. .
Serial Passages of the Limbal Stromal NCs on Coated Matrigel
|
Passage |
Seeding Density (× 105/cm2) |
Culture Time (d) |
Final density (× 105/cm2) |
NCDs |
Accumulative NCD |
Population Doubling Time (h) |
| P0 | 0.1 | 6 | 0.15 | 0.58 | 0.58 | 246 |
| P1 | 0.05 | 6 | 0.30 | 2.58 | 3.17 | 55 |
| P2 | 0.05 | 6 | 0.45 | 3.17 | 6.34 | 45 |
| P3 | 0.05 | 6 | 0.48 | 3.26 | 9.60 | 44 |
| P4 | 0.05 | 6 | 0.50 | 3.32 | 12.92 | 43 |
| P5 | 0.05 | 6 | 0.51 | 3.35 | 16.28 | 43 |
| P6 | 0.05 | 6 | 0.50 | 3.32 | 19.60 | 43 |
| P7 | 0.05 | 6 | 0.48 | 3.26 | 22.86 | 44 |
| P8 | 0.05 | 6 | 0.45 | 3.17 | 26.03 | 45 |
| P9 | 0.05 | 6 | 0.42 | 3.07 | 29.10 | 47 |
| P10 | 0.05 | 6 | 0.30 | 2.58 | 31.69 | 56 |
| P11 | 0.05 | 10 | 0.12 | 1.26 | 32.95 | 190 |
| P12 | 0.05 | 14 | 0.06 | 0.26 | 33.21 | 1277 |
The table summarizes the seeding density, culture time, and final density from P0 to P12, from which cumulative NCDs and population doubling time were calculated.
Expression of Pericyte Markers by Expanded Spindle Cells
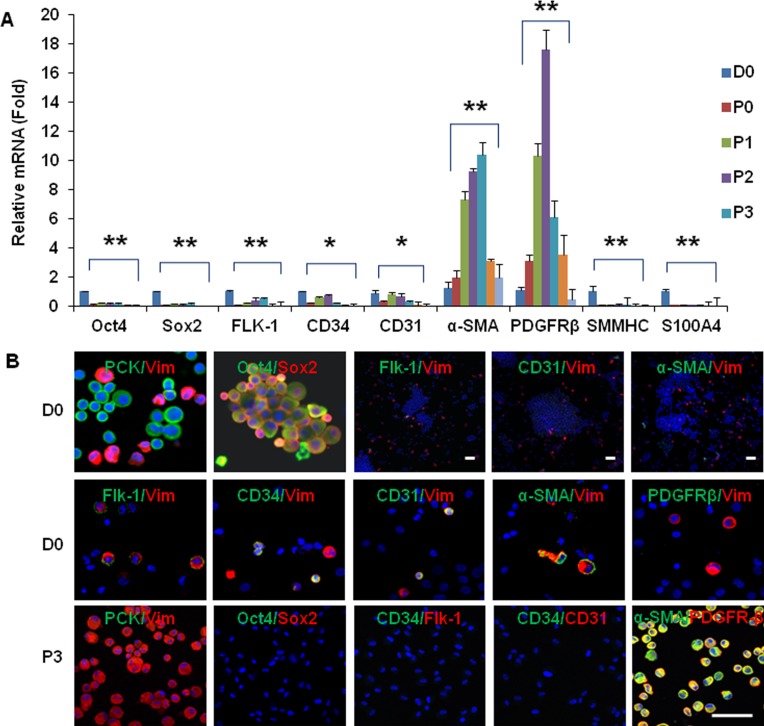
Consistent with our recent reports,32,33 double immunostaining with antibodies against pancytokeratins (PCKs) and Vim showed that collagenase-isolated clusters consisted of approximately 80% PCK+/Vim– epithelial cells and 20% PCK–/Vim+ cells, and that both PCK+ cells and Vim+ cells expressed ESC markers, such as Oct4 and Sox2 (Fig. 3B). Further double immunostaining between the aforementioned markers and Vim showed that Vim+ cells expressed Flk-1, CD34, CD31, and α-SMA, but the overall percentage of colocalization was less than 1% (n = 1000), and none expressed PDGFRβ (Fig. 3B), suggesting that most of these Vim+ NCs did not express markers suggestive of either endothelial progenitor cells or pericytes. As reported,33 PCK+/Vim– epithelial cells were completely eliminated after P2, as confirmed by the disappearance of p63 and CK12 transcripts and negative staining to PCK and p63. In contrast, spindle cells emerged from P3 and uniformly expressed Vim but not PCK (Fig. 3B). RT-qPCR showed that expression of Oct4, Sox2, Flk-1, CD34, CD31, SMMHC, and S100A4 transcripts became undetectable, whereas that of α-SMA and PDGFRβ transcripts were markedly upregulated during serial passage (Fig. 3A, n = 3, P < 0.05). Compared with the expression level at D0 when cells were freshly isolated, expression of the α-SMA transcript was markedly elevated until P12, whereas that of the PDGFRβ transcript was also elevated until P8 (Fig. 3A). This pattern of transcript expression was confirmed by immunofluorescence staining. For example, P3 spindle cells did not express Oct4, Sox2, Flk-1, CD34, and CD31, but strongly expressed α-SMA and PDGFRβ (Fig. 3B). Their lack of expression of SMMHC supported that they were not smooth muscle cells.39 Their positive expression of α-SMA without S100A4 supported that they were not myofibroblasts.40 Collectively, these data indicated that expanded spindle cells upregulated their expression of markers suggestive of pericyte differentiation.
Figure 3. .
Pericyte phenotype promoted by serial passages on coated Matrigel. When compared with freshly isolated cells at D0, RT-qPCR revealed a notable decrease of Oct4, Sox2, Flk-1, CD34, CD31, SMMHC, and S100A4 transcripts but a dramatic increase of α-SMA and PDGFRβ transcripts during serial passages (A, n = 3, *P < 0.05, **P < 0.01). D0 cells consisted of PCK+ and Vim+ cells and expressed Oct4 and Sox2. In addition, Vim+ cells expressed Flk-1, CD34, CD31, or α-SMA (see photos with high magnification), but the overall percentage of colocalization was less than 1% (see photos with low magnification of double labeling of Flk-1/Vim, CD31/Vim and α-SMA/Vim, n = 1000), and none expressed PDGFRβ (B, D0). In contrast, P3 cells were all PCK-/Vim+, α-SMA+, and PDGFRβ+, but negative to Oct4, Sox2, Flk-1, CD34, and CD31 (B, P3). Nuclei were counterstained by Hoechst 33342 (blue). Scale bar = 50 μm.
Angiogenesis Progenitors Promoted by Reseeding in 3D Matrigel
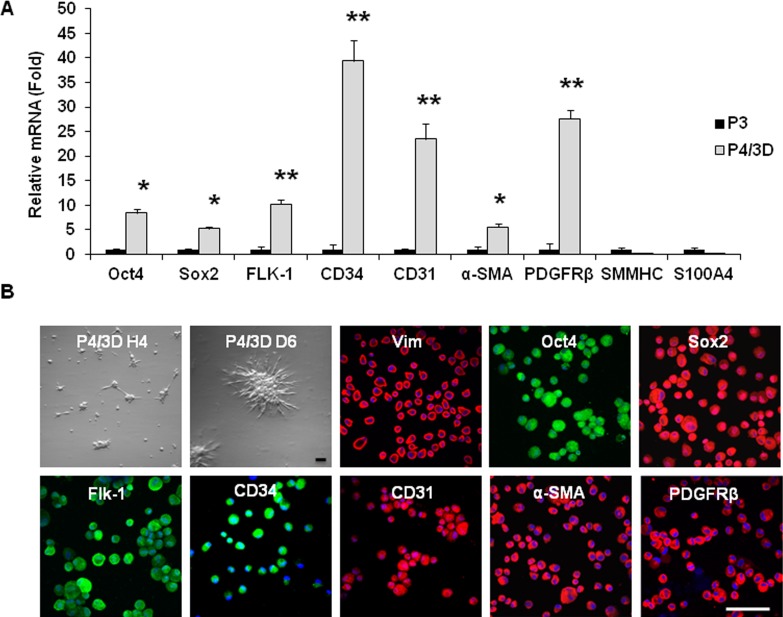
Previously, we discovered that expression of ESC markers could be regained in P3 spindle cells if reseeded in 3D Matrigel.33 We thus wondered whether expression of markers suggestive of angiogenesis progenitors could be influenced by such a maneuver. Single P3 cells formed cell aggregates as early as 4 hours after being reseeded in 3D Matrigel (Fig. 4B). At Day 6, these cell aggregates adopted a stellate contour (Fig. 3B). Consistent with our recent report,33 expression of Oct4 and Sox2 transcripts by P4/3D D6 aggregates was indeed upregulated to 5- and 8-fold when compared with that expressed by P3 spindle cells expanded on coated Matrigel (Fig. 4A). Interestingly, expression of Flk-1, CD34, and CD31 transcripts was markedly upregulated by 10- to 40-fold, whereas that of α-SMA and PDGFRβ transcripts was upregulated by 5- and 27-fold, respectively (Fig. 4A, n = 3, P < 0.05). In contrast, expression of SMMHC and S100A4 transcripts remained undetectable in P4/3D D6 cells. Immunofluroscence staining of single cells released from P4/3D aggregates confirmed positive and uniform expression of Vim and all of the aforementioned angiogenesis markers (Fig. 4B), but negative expression of SMMHC and S100A4 markers (not shown). These results suggested that reseeding back in 3D Matrigel not only restored expression of ESC markers but also promoted expression of markers suggestive of angiogenesis progenitors in the direction of pericytes but not smooth muscle cells or myofibroblasts.
Figure 4. .
Angiogenesis progenitors promoted by reseeding in 3D Matrigel. P3 cells expanded on coated Matrigel were reseeded in 3D Matrigel, they formed P4/3D aggregates; single cells were collected on Day 6. Compared with P3 spindle cells, expression of Oct4, Sox2, Flk-1, CD34, CD31, α-SMA, and PDGFRβ transcripts were markedly upregulated in P4/3D cells (A, n = 3, *P < 0.05, **P < 0.01). In contrast, expression of SMMHC and S100A4 remained lacking. P4/3D aggregates were noted as early as 4 hours and exhibited with a stellate contour at Day 6 (B). Immunostaining of P4/3D cells showed uniform expression of Vim together with the above positive markers (B). Nuclei were counterstained by Hoechst 33342 (blue). Scale bar = 50 μm.
Differentiation into Vascular Endothelial Cells
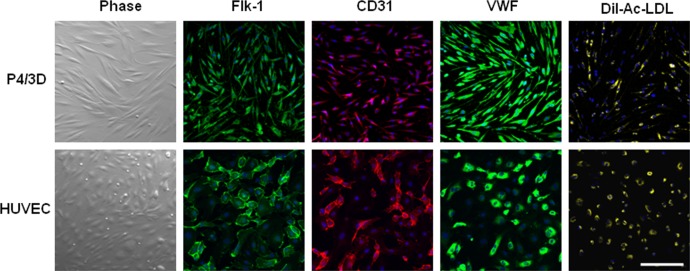
To confirm that the aforementioned P4/3D cells were indeed angiogenesis progenitors, cells were released from 3D Matrigel by dispase digestion, rendered into single cells by T/E, and seeded on plastic in EGM2 supplemented with10 ng/mL VEGF-A according to a reported method.41 After 3 days of culturing, the resultant cells exhibited spindle cells similar to HUVEC (Fig. 5). They also expressed positive immunofluorescence staining to Flk-1, CD31, and von Willebrand factor and took up Dil-Ac-LDL (Fig. 5, top) in a similar fashion to the positive control of HUVEC (Fig. 5, bottom). These data indicated that P4/3D cells indeed could differentiate into vascular endothelial cells.
Figure 5. .
Differentiation into vascular endothelial cells. Single cells of P4/3D aggregates were cultured on plastic in EGM2 with VEGF for 3 days, yielding spindle cells similar to HUVEC. They uniformly expressed Flk-1, CD31, and von Willebrand factor, and took up Dil-Ac-LDL (top) in a similar fashion to the positive control of HUVEC cultured in the same condition (bottom). Nuclei were counterstained by Hoechst 33342 (blue). Scale bar = 100 μm.
Support of HUVEC-Formed Vascular Tube Network
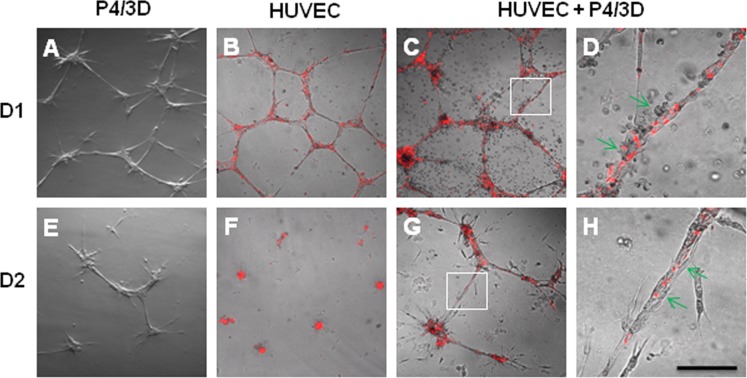
One important step in the process of angiogenesis is to stabilize the vascular network formed by vascular endothelial cells by pericytes.42 To confirm that P4/3D cells were indeed angiogenesis progenitors, we examined whether they also possessed the phenotype of pericytes. To recapitulate such a function of pericytes, we seeded single HUVEC, single P4/3D cells, and a combination of both on the surface of 100% Matrigel in EGM2 as previously reported.36–38 Both single P4/3D cells and prelabeled (red) HUVEC formed networks at Day 1 (Figs. 6A, 6B); however, such networks were largely disintegrated by Day 2 (Figs. 6E, 6F). In contrast, the network formed by cocultured P4/3D cells and HUVEC (Fig. 6C) was maintained at Day 2 (Fig. 6G) and Day 5 (not shown). Higher magnification of such network confirmed tight adherence of P4/3D cells onto HUVEC (red) (Fig. 6D, 6H). These results confirmed that P4/3D cells indeed possessed the pericyte phenotype to stabilize the vascular tube-like network formed by HUVEC.
Figure 6. .
Support of HUVEC vascular tube network. Fluorescence pre-labeled (red) HUVEC and P4/3D cells were seeded alone or together on the surface of 100% Matrigel in EGM2. Although vascular tube-like network was noted in all three conditions at Day 1 (A–C), such network in P4/3D cells (A) or HUVEC (B) alone was disintegrated by Day 2 (E, F). In contrast, the network formed by cocultured P4/3D cells and HUVEC (C) was maintained at Day 2 (G) and Day 5 (not shown). High magnification of insets (C, G) revealed close association between P4/3D cells and HUVEC (red) in the network at Day 1 (D) and Day 2 (H). Scale bar = 200 μm for A–C and E–G, and 50 μm for D and H.
Prevention of Differentiation of Limbal Epithelial Progenitors
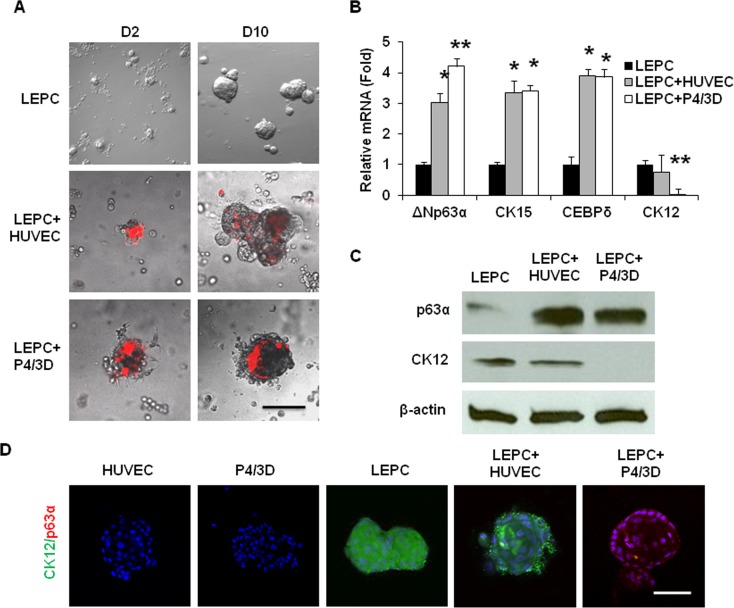
Compared with PCK+ cells in collagenase-isolated clusters, those in dispase-isolated sheets expressed less p63α and CK15, but more CK12.32 Thus, dispase isolated more differentiated limbal epithelial progenitor cells (LEPC) than collagenase based on the findings that p63α signifies limbal basal progenitors including SC,43,44 CK15 is expressed by limbal-basal epithelial cells,45–47 and CK12 is a marker of corneal epithelial differentiation.48,49 Single PCK+ epithelial cells and Vim+ stromal cells from collagenase-isolated clusters could reunite to generate sphere growth in 3D Matrigel and such reunion helps to maintain epithelial clonal growth and prevent corneal epithelial differentiation.35 We thus examined whether LEPC obtained from dispase-isolated epithelial sheets could also form reunion with prelabeled (red) P4/3D cells or HUVEC in 3D Matrigel. As shown in Figure 6, reunion indeed occurred at Day 2 and gradually developed into a larger sphere by Day 10, similar to those formed by LEPC alone (Fig. 7A). Compared with spheres formed by LEPC alone, spheres formed by LEPC+HUVEC and LEPC+P4/3D had significantly higher transcript expression of ΔNp63α, CK15, and CEBPδ, of which the latter plays a role in maintaining quiescence of limbal epithelial SCs50 (Fig. 7B, n = 3, all P < 0.05). Expression of the CK12 transcript by LEPC+HUVEC was not different from LEPC alone (Fig. 7B, n = 3, P > 0.05), but that by LEPC+P4/3D was significantly reduced to an undetectable level (Fig. 7B, n = 3, P < 0.01). Western blot analysis confirmed that the protein level of p63α was elevated to 6.5- and 6.1-fold in LEPC+HUVEC and LEPC+P4/3D respectively, when compared with LEPC alone (Fig. 7C, n = 3, P < 0.05). The protein level of CK12 was not changed in LEPC+HUVEC (Fig. 7C, n = 3, P > 0.05) but was reduced to an undetectable level in LEPC+P4/3D (Fig. 7C, n = 3, P < 0.01). Double staining with p63α and CK12 also confirmed that HUVEC or P4/3D cells alone did not express p63α or CK12, and that CK12 was expressed by LEPC alone and LEPC+HUVEC, but not LEPC+P4/3D (Fig. 7D). Collectively, these findings indicated that although both the P4/3D cells and HUVEC could join with LEPC to generate sphere growth in 3D Matrigel to promote expression of epithelial progenitor/SC markers, the former but not the latter could prevent differentiation of LEPC.
Figure 7. .
Epithelial sphere growth in 3D Matrigel. LEPCs derived from dispase-isolated epithelial sheets alone or mixed with fluorescence prelabeled (red) HUVEC or P4/3D cells to generate sphere growth from Day 2 to Day 10 in 3D Matrigel (A). Compared with those formed by LEPC alone, spheres formed by LEPC+HUVEC and by LEPC+P4/3D expressed significantly more ΔNp63α, CK15, and CEBPδ transcripts (B, n = 3, *P < 0.05, **P < 0.01). In addition, expression of CK12 transcripts by LEPC+HUVEC was not different from that of LEPC alone (B, n = 3, P > 0.05), whereas that by LEPC+P4/3D was not detectable (B, n = 3, P < 0.01). Compared with LEPC alone, expression of p63α protein was elevated in both LEPC+HUVEC and LEPC+P4/3D (C, n = 3, P < 0.05). In contrast, expression of CK12 protein was not reduced in LEPC+HUVEC (C, n = 3, P > 0.05) but was reduced to a nondetectable level in LEPC+P4/3D using β-actin as a loading control (C, n = 3, P < 0.05). Double staining with p63α and CK12 showed that HUVEC or P4/3D cells alone did not expresss p63α or CK12, whereas CK12 was expressed by LEPC alone and LEPC+HUVEC but not LEPC+P4/3D (D). Scale bar = 200 μm for A and 100 μm for D.
Discussion
Embarking on what we have established in identification, isolation, and expansion of limbal NCs,32,33,35 the present study was undertaken to provide evidence that there might exist a vascular niche in limbal palisades of Vogt by showing that these limbal NCs had the capacity of differentiating into angiogenesis progenitors during serial passages on coated Matrigel. Furthermore, like pericytes, they could support networks formed by vascular endothelial cells. Although like vascular endothelial cells these angiogenesis progenitors could serve as NCs to maintain expression of epithelial SC markers, but unlike vascular endothelial cells, they prevented corneal epithelial differentiation.
It is believed that angiogenesis progenitors could be isolated and expanded from the perivascular location of a variety of tissues.18,51–53 Herein, our findings suggested that another source of such angiogenesis progenitors could come from a subset of Vim+ cells that lie immediately subjacent to the limbal-basal epithelial cells. Imminently isolated by collagenase digestion from the in vivo environment, these small Vim+ cells are tightly associated with PCK+/p63α+ epithelial cells in the midst of basement membrane remnants and heterogeneously express several ESC markers.32 We subsequently found that expression of ESC markers is crucial for limbal NCs to support epithelial SC function during sphere growth.33 Although the expression of ESC markers can be preserved if limbal NCs are immediately seeded in 3D Matrigel on collagenase isolation, they do not proliferate.33,35 To circumvent this limitation, we discovered that it was possible to expand limbal NCs by serial passage on coated Matrigel.33 Herein, we extended what was reported earlier (up to P3) to a total of 12 passages and 33 doublings to expand these limbal NCs (Fig. 2, Table 2). Although expression of ESC markers was transiently lost during serial passages on coated Matrigel (Fig. 3), it could be regained when cells were reseeded in 3D Matrigel (Fig. 4) to support epithelial clonal growth in sphere cultures.33 Previously, we noted that cells cultured on coated Matrigel in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, but not ESCM containing BFGF and LIF, resulted in an irreversible loss of expression of ESC markers and a lack of support of limbal epithelial progenitors/SCs in sphere growth.33 Herein, we further demonstrated that cells cultured on plastic (without coated Matrigel) in ESCM containing BFGF and LIF also irreversibly lost the expression of ESC markers and turned into myofibroblasts expressing α-SMA and S-100A4 but not PDGFRβ and SMMHC (Fig. 1). Collectively, these findings suggest that continuous contact with the basement membrane substrate, such as Matrigel, and culturing in the serum-free ESCM containing BFGF and LIF are both crucial to ensure the preservation of expression of ESC markers bestowing expanded cells with the NC status. The lack of such continuous contact results in myofibroblast differentiation (Fig. 1), suggesting that matrix environment plays a key role in maintaining NC phenotype. Future studies are needed to investigate how matrix rigidity/stiffness of Matrigel might promote proliferation without irreversible loss of the NC phenotype.
Notwithstanding the exact modulating mechanism, the maneuver by switching from coated Matrigel to 3D Matrigel also gave rise to angiogenesis progenitors. During serial passages on coated Matrigel, Vim+ spindle cells lost expression of ESC markers but upregulated expression of α-SMA and PDGFRβ without SMMHC and S100A4 (Fig. 3). On being reseeded in 3D Matrigel, these spindle cells regained ESC markers while upregulating expression of additional angiogenesis markers such as Flk-1, CD34, and CD31 (Fig. 4). Expression of Flk-1, CD34, and CD31 is used to denote endothelial progenitor cells isolated from the human tissues54–58 or derived from ESCs.41,59–61 Expression of both α-SMA and PDGFRβ has also been used to mark pericytes.62–66 SMMHC is specific for smooth muscle cells,39 whereas S100A4 predicates differentiation of myofibroblast in addition to the expression of α-SMA.40 We thus speculated that Vim+ cells in P4/3D stellate aggregates were angiogenesis progenitors in the direction of pericytes but not smooth muscle cells or myofibroblasts because they not only expressed the above angiogenesis markers, but they also differentiated into vascular endothelial cells (Fig. 5) and pericytes to support vascular network formed by HUVEC (Fig. 6). Hence, continuous contact with the basement membrane and culturing in ESCM containing BFGF and LIF preserved expression of not only ESC markers but also angiogenesis markers. The loss of the phenotype of angiogenesis progenitors by serial passages on plastic without coated Matrigel (Fig. 1) supported their dependence on the basement membrane and bode well with the in vivo state of angiogenesis progenitor cells, which are normally embedded in the basement membrane.7,38 Thus, our finding shed new light on how destruction of the basement membrane might lead to limbal niche dysfunction leading to fibrosis in limbal stem cell deficiency.
The likelihood that limbal NCs might adopt the fate of angiogenesis progenitors also unravels not only their supporting role of epithelial SCs but also their possible involvement in regeneration during wound healing. In organotypic cultures of keratinocytes, pericytes promote keratinocyte renewal by synthesizing and secreting Laminin 5.14 The present study showed for the first time that both P4/3D cells and HUVEC could support the expression of such limbal basal epithelial progenitors, including SC markers as ΔNp63α,43,44 CK15, a marker for limbal epithelial basal cells,45–47 and CEBPδ, a marker for limbal SC quiescence,50 during sphere growth (Fig. 7). This result suggested that vascular endothelial cells in the limbal niche might serve as NCs to maintain “stemness” of the limbal epithelial progenitor/SCs. However, expression of CK12, a marker for corneal epithelial differentiation,48,49 was abolished only in spheres containing P4/3D cells, but not HUVEC (Fig. 7). Hence, angiogenesis progenitors derived from limbal NCs might play a crucial role in preventing corneal epithelial differentiation. Future studies of how aforementioned angiogenesis progenitors may regulate function of limbal epithelial progenitors/SCs should help us better understand the role of vascularization in the limbal niche and look into the issue of whether a vascular niche in the limbal niche might be generated by limbal NCs.
In summary, human limbal stromal NCs have the potential to differentiate into angiogenesis progenitors and to prevent corneal epithelial progenitor/SC differentiation, implying that the cells partake in formation of the vascular niche and contribute to angiogenesis during wound healing.
Supplementary Material
Acknowledgments
The authors thank Angela Y. Tseng, Hua He, Suzhen Zhang, Lorraine Chua, and Ek Kia Tan for technical assistance on article preparation.
Footnotes
Supported by Grant RO1 EY06819 from the National Eye Institute, National Institutes of Health, Bethesda, Maryland (SCGT); partially by the Nature Science Foundation of Hubei Province (2010CDB09802); and Wuhan Chen-Guang Plan Grant 201150431124, Wuhan, People's Republic of China (GGL).
Disclosure: G.-G. Li, None; S.-Y. Chen, None; H.-T. Xie, None; Y.-T. Zhu, None; S.C.G. Tseng, None
References
- 1. Greco V, Guo S. Compartmentalized organization: a common and required feature of stem cell niches? Development. 2010;137:1586–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Avecilla ST, Hattori K, Heissig B, et al. Chemokine-mediated interaction of hematopoietic progenitors with the bone marrow vascular niche is required for thrombopoiesis. Nat Med. 2004;10:64–71 [DOI] [PubMed] [Google Scholar]
- 3. Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121 [DOI] [PubMed] [Google Scholar]
- 4. Kopp HG, Avecilla ST, Hooper AT, Rafii S. The bone marrow vascular niche: home of HSC differentiation and mobilization. Physiology (Bethesda). 2005;20:349–356 [DOI] [PubMed] [Google Scholar]
- 5. Tavazoie M, van der Veken L, Silva-Vargas V, et al. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoshida S, Sukeno M, Nabeshima Y. A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science. 2007;317:1722–1726 [DOI] [PubMed] [Google Scholar]
- 7. Nikolova G, Strilic B, Lammert E. The vascular niche and its basement membrane. Trends Cell Biol. 2007;17:19–25 [DOI] [PubMed] [Google Scholar]
- 8. Garrett RW, Emerson SG. Bone and blood vessels: the hard and the soft of hematopoietic stem cell niches. Cell Stem Cell. 2009;4:503–506 [DOI] [PubMed] [Google Scholar]
- 9. Butler JM, Nolan DJ, Vertes EL, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen Q, Goderie SK, Jin L, et al. Endothelial cells stimulate self-renewal and expand neurogenesis of neural stem cells. Science. 2004;304:1338–1340 [DOI] [PubMed] [Google Scholar]
- 11. Whitman MC, Greer CA. Adult neurogenesis and the olfactory system. Prog Neurobiol. 2009;89:162–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567 [DOI] [PubMed] [Google Scholar]
- 13. Christov C, Chretien F, Abou-Khalil R, et al. Muscle satellite cells and endothelial cells: close neighbors and privileged partners. Mol Biol Cell. 2007;18:1397–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paquet-Fifield S, Schluter H, Li A, et al. A role for pericytes as microenvironmental regulators of human skin tissue regeneration. J Clin Invest. 2009;119:2795–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi S, Gronthos S. Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res. 2003;18:696–704 [DOI] [PubMed] [Google Scholar]
- 16. Caplan AI. All MSCs are pericytes? Cell Stem Cell. 2008;3:229–230 [DOI] [PubMed] [Google Scholar]
- 17. Crisan M, Yap S, Casteilla L, et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313 [DOI] [PubMed] [Google Scholar]
- 18. He W, Nieponice A, Soletti L, et al. Pericyte-based human tissue engineered vascular grafts. Biomaterials. 2010;31:8235–8244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bianco P, Robey PG, Saggio I, Riminucci M. “Mesenchymal” stem cells in human bone marrow (skeletal stem cells): a critical discussion of their nature, identity, and significance in incurable skeletal disease. Hum Gene Ther. 2010;21:1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caplan AI, Dennis JE. Mesenchymal stem cells as trophic mediators. J Cell Biochem. 2006;98:1076–1084 [DOI] [PubMed] [Google Scholar]
- 21. Hardy SA, Maltman DJ, Przyborski SA. Mesenchymal stem cells as mediators of neural differentiation. Curr Stem Cell Res Ther. 2008;3:43–52 [DOI] [PubMed] [Google Scholar]
- 22. Yoo SW, Kim SS, Lee SY, et al. Mesenchymal stem cells promote proliferation of endogenous neural stem cells and survival of newborn cells in a rat stroke model. Exp Mol Med. 2008;40:387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Walter MN, Wright KT, Fuller HR, MacNeil S, Johnson WE. Mesenchymal stem cell-conditioned medium accelerates skin wound healing: an in vitro study of fibroblast and keratinocyte scratch assays. Exp Cell Res. 2010;316:1271–1281 [DOI] [PubMed] [Google Scholar]
- 24. Haack-Sorensen M, Friis T, Bindslev L, Mortensen S, Johnsen HE, Kastrup J. Comparison of different culture conditions for human mesenchymal stromal cells for clinical stem cell therapy. Scand J Clin Lab Invest. 2008;68:192–203 [DOI] [PubMed] [Google Scholar]
- 25. Friis T, Haack-Sorensen M, Mathiasen AB, et al. Mesenchymal stromal cell derived endothelial progenitor treatment in patients with refractory angina. Scand Cardiovasc J. 2011;45:161–168 [DOI] [PubMed] [Google Scholar]
- 26. Schermer A, Galvin S, Sun T-T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lavker RM, Tseng SC, Sun TT. Corneal epithelial stem cells at the limbus: looking at some old problems from a new angle. Exp Eye Res. 2004;78:433–446 [DOI] [PubMed] [Google Scholar]
- 28. Shanmuganathan VA, Foster T, Kulkarni BB, et al. Morphological characteristics of the limbal epithelial crypt. Br J Ophthalmol. 2007;91:514–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dua HS, Shanmuganathan VA, Powell-Richards A, Tiqhe PJ, Joseph A. Limbal epithelial crypts: a novel anatomical structure and a putative limbal stem cell niche. Br J Ophthalmol. 2005;89:529–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gipson IK. The epithelial basement membrane zone of the limbus. Eye. 1989;3; 132–140 [DOI] [PubMed] [Google Scholar]
- 31. Shortt AJ, Secker GA, Munro PM, Khaw PT, Tuft SJ, Daniels JT. Characterization of the limbal epithelial stem cell niche: novel imaging techniques permit in vivo observation and targeted biopsy of limbal epithelial stem cells. Stem Cells. 2007;25:1402–1409 [DOI] [PubMed] [Google Scholar]
- 32. Chen SY, Hayashida Y, Chen MY, Xie HT, Tseng SC. New isolation method of human limbal progenitor cells by maintaining close association with their niche cells. Tissue Eng Part C Methods. 2011;17:537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie HT, Chen SY, Li GG, Tseng SC. Isolation and expansion of human limbal stromal niche cells. Invest Ophthalmol Vis Sci. 2012;53:279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Espana EM, Romano AC, Kawakita T, Di Pascuale M, Smiddy R, Tseng SC. Novel enzymatic isolation of an entire viable human limbal epithelial sheet. Invest Ophthalmol Vis Sci. 2003;44:4275–4281 [DOI] [PubMed] [Google Scholar]
- 35. Xie HT, Chen SY, Li GG, Tseng SC. Limbal epithelial stem/progenitor cells attract stromal niche cells by SDF-1/CXCR4 signaling to prevent differentiation. Stem Cells. 2011;29:1874–1975 [DOI] [PubMed] [Google Scholar]
- 36. Song S, Ewald AJ, Stallcup W, Werb Z, Bergers G. PDGFRbeta+ perivascular progenitor cells in tumours regulate pericyte differentiation and vascular survival. Nat Cell Biol. 2005;7:870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Traktuev DO, Merfeld-Clauss S, Li J, et al. A population of multipotent CD34-positive adipose stromal cells share pericyte and mesenchymal surface markers, reside in a periendothelial location, and stabilize endothelial networks. Circ Res. 2008;102:77–85 [DOI] [PubMed] [Google Scholar]
- 38. Stratman AN, Malotte KM, Mahan RD, Davis MJ, Davis GE. Pericyte recruitment during vasculogenic tube assembly stimulates endothelial basement membrane matrix formation. Blood. 2009;114:5091–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumar MS, Owens GK. Combinatorial control of smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol. 2003;23:737–747 [DOI] [PubMed] [Google Scholar]
- 40. Quante M, Tu SP, Tomita H, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park SW, Jun KY, Jeon J, et al. Efficient differentiation of human pluripotent stem cells into functional CD34+ progenitor cells by combined modulation of the MEK/ERK and BMP4 signaling pathways. Blood. 2010;116:5762–5772 [DOI] [PubMed] [Google Scholar]
- 42. Dore-Duffy P. Pericytes: pluripotent cells of the blood brain barrier. Curr Pharm Des. 2008;14:1581–1593 [DOI] [PubMed] [Google Scholar]
- 43. Pellegrini G, Dellambra E, Golisano O, et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci U S A. 2001;98:3156–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McKeon F. p63 and the epithelial stem cell: more than status quo? Genes Dev. 2004;18:465–469 [DOI] [PubMed] [Google Scholar]
- 45. Yoshida S, Shimmura S, Kawakita T, et al. Cytokeratin 15 can be used to identify the limbal phenotype in normal and diseased ocular surfaces. Invest Ophthalmol Vis Sci. 2006;47:4780–4786 [DOI] [PubMed] [Google Scholar]
- 46. Adachi W, Ulanovsky H, Li Y, Norman B, Davis J, Piatigorsky J. Serial analysis of gene expression (SAGE) in the rat limbal and central corneal epithelium. Invest Ophthalmol Vis Sci. 2006;47:3801–3810 [DOI] [PubMed] [Google Scholar]
- 47. Figueira EC, Di GN, Coroneo MT, Wakefield D. The phenotype of limbal epithelial stem cells. Invest Ophthalmol Vis Sci. 2007;48:144–156 [DOI] [PubMed] [Google Scholar]
- 48. Chen WY, Mui MM, Kao WW, Liu CY, Tseng SC. Conjunctival epithelial cells do not transdifferentiate in organotypic cultures: expression of K12 keratin is restricted to corneal epithelium. Curr Eye Res. 1994;13:765–778 [DOI] [PubMed] [Google Scholar]
- 49. Liu C-Y, Zhu G, Converse R, et al. Characterization and chromosomal localization of the cornea-specific murine keratin gene Krt1.12. J Biol Chem. 1994;269:24627–24636 [PubMed] [Google Scholar]
- 50. Barbaro V, Testa A, Di IE, Mavilio F, Pellegrini G, De LM. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J Cell Biol. 2007;177:1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bryan BA, D'Amore PA. Pericyte isolation and use in endothelial/pericyte coculture models. Methods Enzymol. 2008;443:315–331 [DOI] [PubMed] [Google Scholar]
- 52. Psaltis PJ, Harbuzariu A, Delacroix S, Holroyd EW, Simari RD. Resident vascular progenitor cells—diverse origins, phenotype, and function. J Cardiovasc Transl Res. 2011;4:161–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Torsney E, Xu Q. Resident vascular progenitor cells. J Mol Cell Cardiol. 2011;50:304–311 [DOI] [PubMed] [Google Scholar]
- 54. Peichev M, Naiyer AJ, Pereira D, et al. Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood. 2000;95:952–958 [PubMed] [Google Scholar]
- 55. Zengin E, Chalajour F, Gehling UM, et al. Vascular wall resident progenitor cells: a source for postnatal vasculogenesis. Development. 2006;133:1543–1551 [DOI] [PubMed] [Google Scholar]
- 56. Liu C, Wang S, Metharom P, Caplice NM. Myeloid lineage of human endothelial outgrowth cells circulating in blood and vasculogenic endothelial-like cells in the diseased vessel wall. J Vasc Res. 2009;46:581–591 [DOI] [PubMed] [Google Scholar]
- 57. Timmermans F, Plum J, Yoder MC, Ingram DA, Vandekerckhove B, Case J. Endothelial progenitor cells: identity defined? J Cell Mol Med. 2009;13:87–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sen S, McDonald SP, Coates PT, Bonder CS. Endothelial progenitor cells: novel biomarker and promising cell therapy for cardiovascular disease. Clin Sci (Lond). 2011;120:263–283 [DOI] [PubMed] [Google Scholar]
- 59. Ferreira LS, Gerecht S, Shieh HF, et al. Vascular progenitor cells isolated from human embryonic stem cells give rise to endothelial and smooth muscle like cells and form vascular networks in vivo. Circ Res. 2007;101:286–294 [DOI] [PubMed] [Google Scholar]
- 60. Goldman O, Feraud O, Boyer-Di PJ, et al. A boost of BMP4 accelerates the commitment of human embryonic stem cells to the endothelial lineage. Stem Cells. 2009;27:1750–1759 [DOI] [PubMed] [Google Scholar]
- 61. Noghero A, Bussolino F, Gualandris A. Role of the microenvironment in the specification of endothelial progenitors derived from embryonic stem cells. Microvasc Res. 2010;79:178–183 [DOI] [PubMed] [Google Scholar]
- 62. Hellstrom M, Kalen M, Lindahl P, Abramsson A, Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126:3047–3055 [DOI] [PubMed] [Google Scholar]
- 63. Isolation Dore-Duffy P. and characterization of cerebral microvascular pericytes. Methods Mol Med. 2003;89:375–382 [DOI] [PubMed] [Google Scholar]
- 64. Armulik A, Genove G, Mae M, et al. Pericytes regulate the blood-brain barrier. Nature. 2010;468:557–561 [DOI] [PubMed] [Google Scholar]
- 65. Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010;468:562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bell RD, Winkler EA, Sagare AP, et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron. 2010;68:409–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.