Abstract
High activity of aldehyde dehydrogenase (ALDH) is characteristic of normal and cancerous stem cells. Recently, high ALDH expression was shown to be associated with poor prognosis in uterine endometrial adenocarcinoma. The population with high ALDH activity (ALDH-hi) was more invasive, anti-apoptotic, and tumorigenic than that with low activity (ALDH-lo). Here, the transcriptional regulation of ALDH1A1 gene, which is responsible for ALDH activity, was examined in endometrial adenocarcinoma. The promoter region of ALDH1A1 contained CCAAT and octamer binding motifs, and their mutation diminished promoter activity. Among CCAAT-recognizing transcription factors, nuclear factor YA (NFYA) was involved in ALDH1A1 transcription. Two alternatively spliced isoforms of NFYA (NFYA-long and NFYA-short) have been reported. The sorted ALDH-hi population of endometrial adenocarcinoma preferentially expressed NFYA-short, whereas ALDH-lo dominantly expressed NFYA-long. NFYA-short possessed higher transactivation ability than did NFYA-long. In addition, an additive effect of NFYA with Oct-1, which recognizes octamer binding motif, was observed in ALDH1A1 transactivation. These results indicate that the alternatively spliced isoforms of NFYA, in cooperation with Oct-1, play an important role in ALDH1A1 expression in endometrial adenocarcinoma.
Keywords: aldehyde dehydrogenase, nuclear factor Y, alternatively spliced isoform, transcriptional regulation, endometrial adenocarcinoma
Introduction
Tumors consist of heterogeneous cell populations derived from a single clone. Recently, it has been demonstrated that tumor cells with tumorigenic potential are limited to a small population of cells, called cancer-initiating cells (CICs), in several tumors, such as leukemia, breast, brain, and colon cancers.1-8 Aldehyde dehydrogenase (ALDH) oxidizes retinol to retinoic acid in the early stage of stem cell differentiation, and hematopoietic and neural stem cells show high ALDH activity.9,10 Previous studies showed that CICs of human multiple myeloma, acute myeloid leukemia, and cancers of brain, lung, breast, and ovary expressed ALDH at a high level, suggesting that ALDH activity might be a common marker for both normal and malignant stem cell populations.11-21 Very recently, high activity of ALDH was shown to be associated with poor prognosis in uterine endometrial adenocarcinoma.22 The population showing high ALDH activity (ALDH-hi) of endometrial adenocarcinoma was more invasive, anti-apoptotic, and tumorigenic than the population with low activity (ALDH-lo).22
The human ALDH superfamily consists of 19 genes, among which ALDH1A1 is known to be expressed in stem cells.23 Yanagawa et al.24 demonstrated that the CCAAT motif located just upstream from the transcription initiation site (–74 to −70; +1 shows transcription initiation site) was essential for ALDH1A1 gene expression in hepatoma cell line. The gel retardation assay revealed that the transcription factor binding to CCAAT motif was nuclear factor Y (NFY). The additive effect of NFY to Oct-1, which bound the octamer (OCT) sequence adjacent to the CCAAT motif, on ALDH1A1 gene transactivation has not been examined. Elizondo et al25 demonstrated the binding of CCAAT/enhancer binding protein β (C/EBPβ), instead of NFY, to the CCAAT motif, which transactivated the mouse counterpart of human ALDH1A1 gene.
NFY consists of 3 subunits; NFYA, NFYB, and NFYC.23 Two alternatively spliced isoforms of NFYA (NFYA-long and NFYA-short) have been reported.26 NFYA-long contains an exon encoding the majority of a glutamine-rich transactivation domain, whereas NFYA-short lacks this. NFYA-long and NFYA-short show distinct expression patterns; the former is preferentially expressed in epithelial cells and the latter in lymphoid cells.27 The level of NFYA-long increases through mouse and human embryonic stem (ES) cell differentiation, whereas NFYA-short is significantly downregulated.28 Although the functional significance of 2 NFYA isoforms remains to be elucidated, recent data indicate that NFYA-short promotes self-renewal of hematopoietic stem cells.29
Here, we examined the transcriptional regulation of ALDH1A1 gene in human endometrial adenocarcinoma and found that the alternative spliced isoforms of NFYA regulated the expression of ALDH1A1 gene in cooperation with Oct-1.
Results
Promoter analysis of ALDH1A1 gene
To identify the cis-acting elements enhancing promoter activity of ALDH1A1, promoter region starting from nt −291 (Fig. 1A) was cloned into luciferase reporter plasmid. When the reporter plasmid was transfected to HEC-1 cells abundantly transcribing ALDH1A1 mRNA, strong luciferase activity was detected (Fig. 1A). When CCAAT and OCT motifs, previously reported to mediate ALDH1A1 transactivation, were mutated, the luciferase activity decreased significantly (Fig. 1B).
Figure 1.
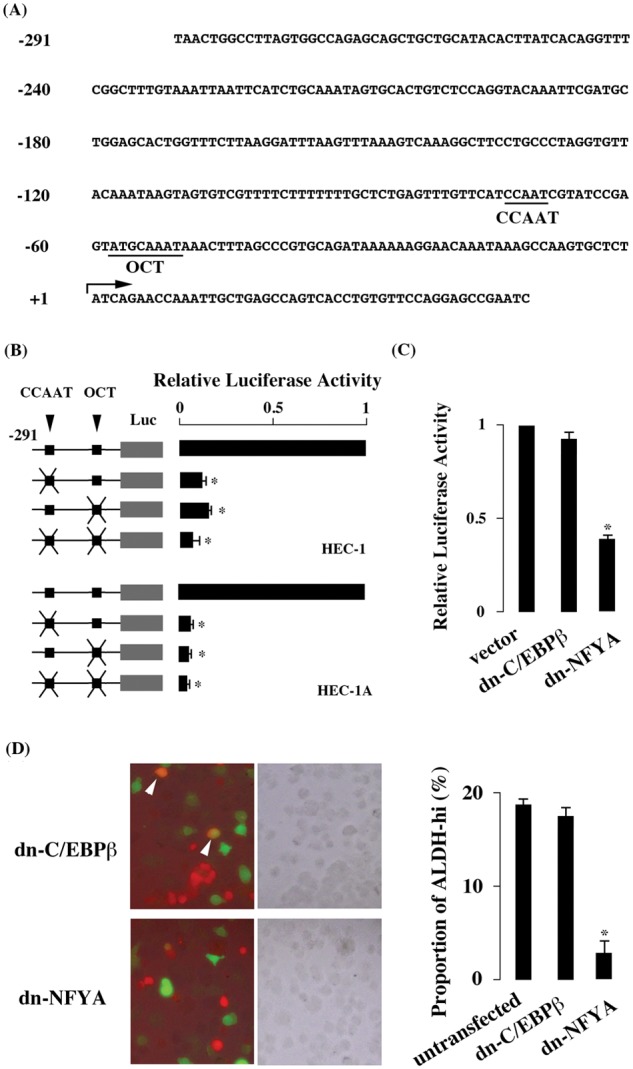
Promoter analysis of ALDH1A1 gene. (A) Promoter region of ALDH1A1 gene. The CCAAT and OCT motifs are underlined. The transcription initiation site (+1) is shown with an arrow. (B) Luciferase activity in HEC-1 and HEC-1A cells. The luciferase activity obtained in the reporter plasmid mutated at CCAAT or OCT motif was divided by that of the reporter plasmid containing intact motifs, and the result is shown as relative luciferase activity. (C) Effect of dominant negative C/EBPβ and NFYA. (D) ALDH activity in HEC-1 cells overexpressing dominant negative forms of C/EBPβ and NFYA. ALDH-hi cells were stained in green, and cells overexpressing dominant negative forms were stained in red. The merged (left) and phase-contrast (middle) images and the proportion of ALDH-hi cells (right) are shown. ALDH-hi cells overexpressing dominant negative forms are indicated with arrowheads. The values are mean ± SE of 3 experiments. *P ≤ 0.05.
Several transcription factors, such as NFYA and C/EBPβ, recognize the CCAAT motif. Yanagawa et al24 reported that NFYA recognizes the CCAAT motif of the ALDH1A1 promoter, whereas Elizondo et al25 reported that C/EBPβ, instead of NFYA, recognizes it. In HEC-1 cells, the dominant negative form of NFYA but not of C/EBPβ reduced luciferase activity (Fig. 1C), indicating that NFYA but not C/EBPβ was involved in ALDH1A1 promoter activity. Moreover, the dominant negative form of NFYA but not of C/EBPβ reduced the proportion of cells with high ALDH activity (Fig. 1D).
The luciferase assay was also done with HEC-1A, which abundantly expresses ALDH1A1, to examine whether CCAAT and OCT motifs were effective in cell lines other than HEC-1. Comparable results were obtained in HEC-1A cells (Fig. 1B), indicating that CCAAT and OCT motifs mediated transactivation of ALDH1A1 gene in both HEC-1 and HEC-1A cells.
Co-expression of ALDH1A1 with NFYA and Oct-1
Co-expression of ALDH1A1 with NFYA and Oct-1 was immunohistochemically examined. ALDH1A1-expressing endometrial adenocarcinoma cells expressed NFYA abundantly (Fig. 2A and 2B, arrows). ALDH1A1 expression negative adenocarcinoma cells also expressed NFYA (Fig. 2A and 2B, arrowheads). The Oct-1 expression pattern was similar to NFYA; both ALDH1A1 expression positive and negative cells expressed Oct-1 (Fig. 2C and 2D, arrows and arrowheads, respectively).
Figure 2.
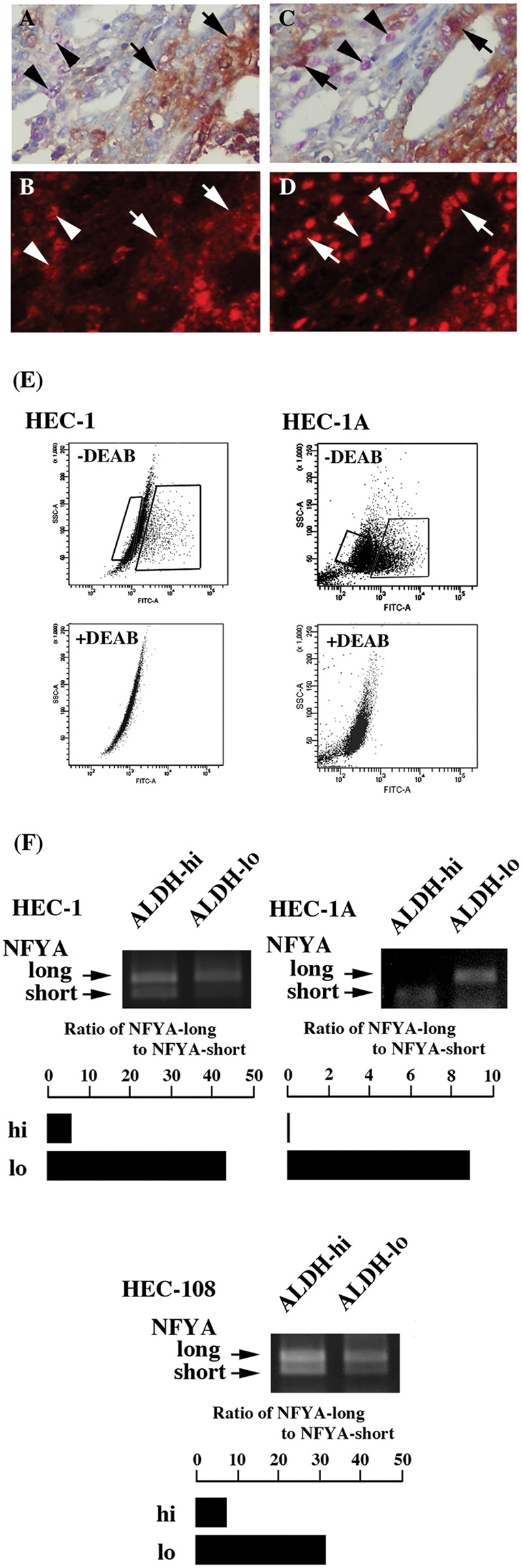
Co-expression of ALDH1A1 with NFYA and Oct-1 in endometrial adenocarcinoma and the preferential expression of alternatively spliced isoforms of NFYA. Double staining of ALDH1A1 with NFYA (A and B) and with Oct-1 (C and D). ALDH1A1 expression is shown in brown (A) and NFYA in red (B). Tumor cells expressing both ALDH1A1 and NFYA are shown with arrows and those expressing NFYA alone with arrowheads. ALDH1A1 expression is shown in brown (C) and Oct-1 in red (D). Tumor cells expressing both ALDH1A1 and Oct-1 are shown with arrows and those expressing Oct-1 alone with arrowheads. Magnification is 400×. (E) Dot blot analysis of ALDH-hi and ALDH-lo populations of HEC-1 and HEC-1A. These 2 populations are boxed. (F) RT-PCR analysis of NFYA-long and NFYA-short in the sorted ALDH-hi and ALDH-lo populations. The intensity of both NFYA-long and NFYA-short is quantified, and its ratio is shown. Experiments were repeated 3 times, and the representative result is shown.
Distinct expression of NFYA-long and NFYA-short in ALDH-hi and ALDH-lo populations
The amount of NFYA-long and -short in the sorted ALDH-hi and ALDH-lo populations of HEC-1 cells (Fig. 2E) was examined by RT-PCR. To check the purity of sorted cells, ALDH activity was examined just after sorting. Although a limited number of ALDH-lo cells were detected, most of cells showed high ALDH activity in the sorted ALDH-hi population (Suppl. Fig. S1). The intensity of NFYA-long was comparable to that of NFYA-short in the ALDH-hi population (Fig. 2F). In contrast, the intensity of NFYA-long was significantly stronger than that of NFYA-short in the ALDH-lo population. The ratio of NFYA-long to -short was significantly lower in the ALDH-hi population, indicating that ALDH-hi and ALDH-lo populations of HEC-1 preferentially expressed NFYA-short and NFYA-long, respectively. The distinct expression pattern of NFYA-long and NFYA-short was also detected in HEC-1A (Fig. 2F). In HEC-108 cells, another endometrial adenocarcinoma cell line, NFYA-long and NFYA-short showed a distinct expression pattern between ALDH-hi and ALDH-lo populations (Fig. 2F).
Additive effect of NFYA with Oct-1 on ALDH1A1 promoter activity
When transfected with both NFYA-long and Oct-1, the luciferase activity increased compared with that obtained by transfection with NFYA-long alone (Fig. 3). The co-transfection of NFYA-short with Oct-1 increased luciferase activity compared with that obtained by NFYA-short alone (Fig. 3). These findings indicated that NFYA and Oct-1 additively transactivated ALDH1A1 promoter. NFYA-short showed higher luciferase activity than NFYA-long (Fig. 3), suggesting that the transactivation effect on ALDH1A1 gene was stronger in NFYA-short than that in NFYA-long.
Figure 3.
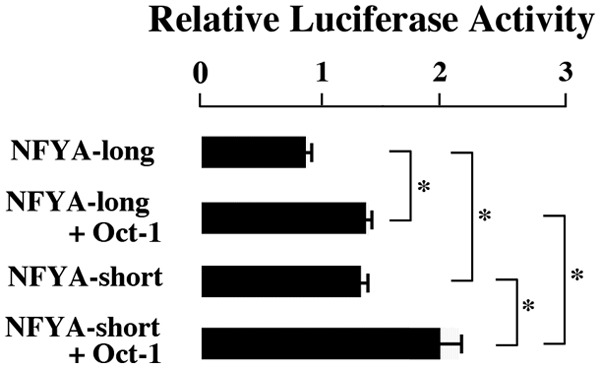
Synergistic effect of NFYA and Oct-1 and the comparison of NFYA-long and NFYA-short transactivation abilities. The values are mean ± SE of 3 experiments. *P ≤ 0.05.
Discussion
In the current study, ALDH1A1 transcriptional regulation was examined in endometrial adenocarcinoma. As previously observed in hepatoma,24 the CCAAT motif was important for ALDH1A1 transcription in endometrial adenocarcinoma, because the promoter activity was completely abolished by the mutation at CCAAT motif. The current study revealed that the mutation at OCT motif abolished ALDH1A1 promoter activity in endometrial adenocarcinoma, which was consistent with the previous study that Oct-1, a transcription factor recognizing OCT motif located adjacent to CCAAT, affects NFYA binding in hepatoma.24 Both NFYA and Oct-1 appeared to additively transactivate ALDH1A1 promoter in endometrial adenocarcinoma.
Several transcription factors are known to recognize CCAAT motif. Elizondo et al25 reported that C/EBPβ, instead of NFYA, recognizes the CCAAT motif in the mouse counterpart of the human ALDH1A1 gene. To examine which transcription factor was involved in ALDH1A1 transactivation, the dominant negative C/EBPβ or NFYA was overexpressed in endometrial adenocarcinoma. No effect was observed on ALDH1A1 promoter activity when the dominant negative C/EBPβ was expressed In contrast, the dominant negative NFYA reduced the promoter activity, indicating that the transcription factor involved in ALDH1A1 transcription was NFYA in human endometrial adenocarcinoma.
The simplest model for transcriptional regulation is that the presence of a transcription factor that enhances promoter activity is correlated with the expression of target gene; that is, the transcription factor expresses only when the target gene is activated. When expression of NFYA or Oct-1 is positively correlated with ALDH1A1 transcription, the transcriptional regulation of ALDH1A1 gene could be explained by this simplest model. The co-expression of ALDH1A1 with NFYA and Oct-1 was immunohistochemically examined. Unexpectedly, NFYA and Oct-1 were present not only in ALDH1A1 expression positive cells but also in negative cells.
Another explanation for transcriptional regulation was that the character of transcription factors was different between target gene-on and target gene-off status. Two alternatively spliced isoforms of NFYA were reported: NFYA-long and NFYA-short.27 Mouse and human ES cells in immature status dominantly express NFYA-short. During the differentiation process of ES cells, the expression level of NFYA-short reduces and that of NFYA-long increases.28 These findings suggest that NFYA-short may play an important role for maintenance of stemness.28,29 The ALDH-hi population in HEC-1, HEC-1A, and HEC-108 preferentially expressed NFYA-short, whereas the ALDH-lo population dominantly expressed NFYA-long. The transactivation ability of NFYA-short on ALDH1A1 gene was stronger than that of NFYA-long. These findings suggested that the high expression of ALDH1A1 appeared to be due to a predominantly expressed isoform of NFYA, NFYA-short. Since ALDH1A1 was a marker for CICs of endometrial adenocarcinoma, the current results are consistent with the notion that NFYA-short was involved in the maintenance of stemness.28,29
Taken together, the transcription of ALDH1A1 gene was additively regulated by NFY and Oct-1. In particular, the alternatively spliced isoforms of NFYA played an important role for ALDH1A1 expression in endometrial adenocarcinoma.
Materials and Methods
Cells
Endometrial adenocarcinoma cell lines HEC-1, HEC-1A, and HEC-108 were obtained from the Health Science Research Resources Bank of Osaka, Japan. Cells were cultured in DMEM (Wako, Tokyo, Japan) supplemented with 10% fetal calf serum (FCS, Nippon Bio-Supply Center, Tokyo, Japan).
Promoter analysis by luciferase assay
The promoter region of ALDH1A1 gene starting from nt −291 was amplified with PrimeStar Max DNA polymerase (Takara, Kyoto, Japan) and cloned into pSP-Luc reporter vector containing luciferase gene as described previously.30 The mutation at CCAAT motif (CCAAT to CTGAT) and OCT motif (ATGCAAAT to CTGAACAT) was introduced by PCR and verified by sequencing. The coding region of NFYA (both NFYA-long and NFYA-short) and Oct-1 was also amplified with PrimeStar Max DNA polymerase and cloned into p3xFlag-CMV7.1 expression vector (Sigma, St Louis, Missouri). The constructed reporter plasmid (1 µg) was transiently transfected into HEC-1 or HEC-1A, together with 0.2 µg of the phRL-TK containing Renilla luciferase gene (Promega, Madison, Wisconsin) using Transfast transfection reagent (Promega). Twenty-four hours after transfection, the luciferase activity was measured by Dual Luciferase Reporter Assay Kit (Promega). In some experiments, the dominant-negative form of C/EBPβ31 tagged with Myc and NFYA32 tagged with HA, or the expression plasmid containing NFYA or Oct-1 was co-transfected.
ALDH activity in cells overexpressing dominant negative forms of C/EBPβ and NFYA
Plasmids containing the dominant-negative form of C/EBPβ tagged with Myc and NFYA tagged with HA were transiently transfected to HEC-1 cells. To examine ALDH enzymatic activity in transfected cells, the Aldefluor kit (Stem Cell Technologies, Vancouver, Canada) was used according to the manufacturer’s instruction. Briefly, cells were suspended in Aldefluor assay buffer containing ALDH-substrate BODIPY-aminoacetaldehyde (BAAA). BAAA was taken up by living cells and converted by intracellular ALDH into BODIPY-aminoacetate, which causes the cells to become brightly fluorescent. As a negative control, cells were stained under the identical conditions with the specific ALDH inhibitor, diethylaminobenzaldehyde (DEAB, Sigma). After staining with BAAA, cells were incubated with anti-Myc or anti-HA (Clontech, Palo Alto, California), followed by incubation with Alexa546-conjugated secondary antibodies. The brightly green fluorescent ALDH-expressing cells and the red fluorescent dominant negative C/EBPβ and NFYA overexpressing cells were detected with fluorescence microscope (Biozero, Keyence, Osaka, Japan).
Double staining of ALDH1A1 with NFYA and Oct-1
The expression of ALDH1A1, NFYA, and Oct-1 was examined with anti-ALDH1 (BD Biosciences, Franklin Lakes, New Jersey), NFYA (Abcam Ltd, Cambridge, UK), and Oct-1 (Thermo Scientific, Rockford, Illinois), respectively. A histological specimen of a patient with endometrial adenocarcinoma was fixed in 10% formalin and routinely processed for paraffin embedding. Paraffin-embedded specimens were stored in a dark room in the Department of Pathology of Osaka University Hospital at room temperature, sectioned at 4 μm thickness at the time of staining, and stained with immunoperoxidase procedure. Double staining of ALDH1A1 with NFYA and Oct-1 was done with EnVision G/2 doublestain system (DAKO Denmark, Glostrup) according to the manufacturer’s protocol: ALDH1A1 expression was colored with DAB and NFYA or Oct-1 with Permanent Red. Since the red fluorescence is released from Permanent Red, the signal of NFYA and Oct-1 was detected with fluorescence microscope (Biozero, Keyence, Osaka, Japan). The study was approved by the ethical review board of Graduate School of Medicine, Osaka University.
Sorting of ALDH-hi and ALDH-lo populations
To isolate populations with high ALDH enzymatic activity, the Aldefluor kit (Stem Cell Technologies) was used. The brightly fluorescent ALDH-expressing cells were detected with FACS Aria (BD Biosciences). Data were analyzed by using Cell Quest software (BD Biosciences). Cells with bright fluorescence are judged as ALDH-hi and those with no or faint fluorescence as ALDH-lo. ALDH-hi and ALDH-lo populations were sorted separately with FACS Aria.
RT-PCR
RNA was extracted from the sorted ALDH-hi and ALDH-lo cells using an RNeasy kit (Qiagen, Valencia, California) with DNase I treatment. Total RNA was subjected to reverse transcription by Superscript III (Invitrogen, Carlsbad, California), and the single strand cDNA was obtained. The alternatively spliced isoforms of NFYA were amplified with primers (5′-ATGGAGCAGTATACAG CAAACAGCAATAGT and 5′-TGATGGTTTGACCTT GTCCA) and electrophoresed in agarose gel. The density of NFYA-short and NFYA-long fragments was quantified with ImageJ software (http://rsbweb.nih.gov/ij/).
Statistical analysis
Statistical analysis for experimental studies was carried out using Student t test. The values are shown as the mean ± standard error of at least 3 experiments. P ≤ 0.05 was considered to indicate statistical significance.
Acknowledgments
The authors thank Ms. Megumi Nihei-Sugano, Ms. Etsuko Maeno, Ms. Takako Sawamura, and Mr. Masaharu Kohara for their technical assistance.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Japan (#23590425) and from the Ministry of Health, Labour and Welfare, Japan (#KH22Q151b).
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
References
- 1. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730-7 [DOI] [PubMed] [Google Scholar]
- 2. Lessard J, Sauvageau G. Bmi-1 determined the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255-60 [DOI] [PubMed] [Google Scholar]
- 3. Al-Hajj M, Wicha MS, Benito-Hernandez A, Moriison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;423:396-401 [DOI] [PubMed] [Google Scholar]
- 5. Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-11 [DOI] [PubMed] [Google Scholar]
- 6. O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumor growth in immunodeficient mice. Nature. 2007;445:106-10 [DOI] [PubMed] [Google Scholar]
- 7. Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111-5 [DOI] [PubMed] [Google Scholar]
- 8. Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101:781-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006;103:11707-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corti S, Locatelli F, Papadimitriou D, et al. Identification of a primitive brain-derived normal stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006;24:975-85 [DOI] [PubMed] [Google Scholar]
- 11. Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jiang F, Qiu Q, Khanna A, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsui W, Wang Q, Barber JP, et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68:190-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pearce DJ, Taussig D, Simpson C, et al. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23:752-60 [DOI] [PubMed] [Google Scholar]
- 15. Rasheed ZA, Yang J, Wang Q, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Su Y, Qiu Q, Zhang X, et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:327-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li T, Su Y, Mei Y, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab Invest. 2010;90:234-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cell. Blood. 1990;75:1947-50 [PubMed] [Google Scholar]
- 19. Silva IA, Bai S, McLean K, et al. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991-4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Penumatsa K, Edassery SL, Barua A, Bradaric MJ, Luborsky JL. Differential expression of aldehyde dehydrogenase 1a1 (ALDH1) in normal ovary and serous ovarian tumors. J Ovarian Res. 2010;3:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deng S, Yang X, Lassus H, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rahadiani N, Ikeda J-I, Mamat S, et al. Expression of aldehyde dehydrogenase 1 in endometrioid adenocarcinoma and its clinical implications. Cancer Sci. 2011;102:903-8 [DOI] [PubMed] [Google Scholar]
- 23. Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011;7:292-306 [DOI] [PubMed] [Google Scholar]
- 24. Yanagawa Y, Chen JC, Hsu LC, Yoshida A. The transcriptional regulation of human aldehyde dehydrogenase 1 gene. J Biol Chem. 1995;270:17521-7 [DOI] [PubMed] [Google Scholar]
- 25. Elizondo G, Medina-Diaz IM, Cruz R, Gonzaiez FJ, Vega L. Retinoic acid modulates retinaldehyde dehydrogenase 1 gene expression through the induction of GADD153-C/EBPβ interaction. Biochem Pharmacol. 2009;77:248-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maity S, Sinha S, Ruteshouser E, deCrombrugghe B. Three different polypeptides are necessary for DNA binding of the mammalian heteromeric CCAAT binding factor. J Biol Chem. 1992;267:16574-80 [PubMed] [Google Scholar]
- 27. Ishimaru F, Mari B, Shipp MA. The type 2 CD10/neutral endopeptidase 24.11 promoter: functional characterization and tissue-specific regulation by CBF/NF-Y isoforms. Blood. 1997;89:4136-45 [PubMed] [Google Scholar]
- 28. Grskovic M, Chaivorapol C, Gaspar-Maia A, Ramalho-Santos M. Systematic identification of cic-regulatory sequences active in mouse and human embryonic stem cells. PLos Genet. 2007;3:e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG. NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc Natl Acad Sci U S A. 2005;102:11728-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morii E, Oboki K. MITF is necessary for generation of prostaglandin D2 in mouse mast cells. J Biol Chem. 2004;279:48923-9 [DOI] [PubMed] [Google Scholar]
- 31. Zhang J-W, Tang Q-Q, Vinson C, Lane MD. Dominant-negative C/EBP disrupts mitotic clonal expansion and differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci U S A. 2004;101:43-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mantovani R, Li X-Y, Pessara U, et al. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340-6 [PubMed] [Google Scholar]


