Abstract
In this issue of Cell Stem Cell, Curtis et. al. (2010) reveal that the identities of lung cancer stem cell populations differ depending on the specific tumor oncogenotype in three murine lung adenocarcinoma models. These findings highlight the importance of determining the cancer stem cell oncogenotype for genotypically diverse malignancies.
According to the cancer stem cell model, only self-renewing, stem-like tumor cells possess the capacity to initiate tumor formation and distant metastases. In this regard a stem cell hierarchy, possibly inherited from a transformed adult stem cell, is thought to exist in many cancers, and that long-term clinical remission can only be achieved in these cancers by eliminating or incapacitating the cancer stem cell population. Indeed, the prospect for stem cells serving as the driving force for tumor initiation and progression has lead to a concerted effort to identify and characterize stem cells and cancer stem cells in many tissue types. Although cancer stem cells have been identified and described in numerous human malignancies, the biology of lung cancer stem cells remains less well studied. This difference is due in part to the lack of validated human lung stem cell markers as well as the genotypic and histological diversity found in lung cancer (Sullivan et al., 2010).
The lung is a complex organ, comprising regionally and functionally distinct cell phenotypes, and driving the development and turnover of these populations are a diverse class of lung stem cells. In the distal region of the murine lung, stem cell turnover has been previously reported in the bronchioalveolar duct junction. Termed bronchioalveolar stem cells or BASCs, these cells self-renew and differentiate to repopulate damaged epithelium and, during Kras driven oncogenesis, initiate the formation of lung adenocarcinomas (Dovey et al., 2008; Kim et al., 2005). Like their Kras-transformed counterparts, BASCs express the alveolar type 1 cell marker SP-C, the Clara cell marker CCA (aka CC10 or CCSP) and the panmurine stem cell marker Sca-1. In the case of murine adenocarcinomas, oncogenically manipulated BASCs represent a likely reservoir of lung cancer stem cells. However in this issue of Cell Stem Cell, Curtis et al. (2010) discover that the identity of the lung cancer stem cell population that promotes adenocarcinogenesis is likely dependent on the oncogenotype driving the malignancy.
It has been shown in lung cancer, as well as in other epithelial tumors, that oncogenes not only drive tumor formation but also confer cell lineage and tumor histotype specificity (Bass et al., 2009; Kwei et al., 2008). This oncogene phenotype specificity is also manifest in the tumor stem cell population where, for example in mouse models of breast cancer, tumor stem cells from MMTV-Wnt-1 and MMTV-Neu murine tumors were identified by the expression of different stem cell markers (Cho et al., 2008; Liu et al., 2007).
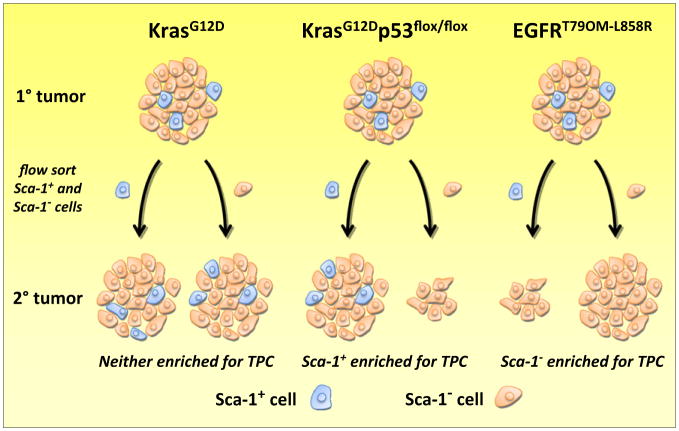
Seizing on the notion that cancer stem cells may be heterogeneous, Curtis et al. investigated the tumor propagating capacity of cells that expressed BASC markers in three oncogenetic models of murine lung adenocarcinoma: mutant Kras, mutant Kras with a p53 deficiency (Kras; p53-flox), and mutant EGFR-driven lung adenocarcinomas. The authors used an orthotopic transplantation assay with limiting cell dilutions to determine the “stemness” of each tumor genotype. The authors found that the abundance of tumor stem cells, operationally defined as tumor propagating cells (TPCs) on the basis of the functional assay used in their studies, were remarkably similar in Kras and Kras; p53-flox lung adenocarcinomas. In addition, tumors from each genotype possessed similar proportions of CD45−/CD31−/Sca-1+ cells, a putative BASC-like population was previously thought to be the tumor stem cell pool. For determining whether this putative cancer stem cell population is enriched in TPCs, limiting dilutions of Sca-1+ and Sca-1− cells isolated by FACS were transplanted into recipient mice. Remarkably, the ability of the BASC marker Sca-1 to enrich for TPCs varied greatly between tumor genotypes. In adenocarcinomas driven by Kras alone, Sca-1 expression was not associated with TPC activity, whereas in Kras; p53-flox adenocarcinomas, Sca-1 expression was associated with a greater than 5-fold enrichment in TPCs. Importantly, Kras; p53-flox Sca-1+ tumor cells fit the profile of a population enriched in lung cancer stem cell as they were found to form robust tumors in secondary and tertiary mice from as few as 100 cells. Conversely, the few secondary tumors that formed from Kras; p53-flox Sca-1− cells were relatively small, had no detectable Sca-1+ cells, and could not generate tertiary tumors. In striking contrast, in the case of EGFR-driven lung adenocarcinomas, the selection of Sca-1− tumor cells was found to greatly enrich for TPC activity (Figure 1).
Figure 1. Tumor Propagating Capacity of Sca-1+ Lung Cancer Cells.
Transgenic mice harboring mutant Kras (left), mutant Kras with a p53 deficiency (center) or mutant EGFR (right) all develop lung adenocarcinomas that harbor a similar proportion of cells expressing the mouse stem cell (and BASC) marker Sca-1 (blue cells). The tumor propagating capacity (TPC) of Sca-1+ and Sca-1− cells from each primary tumor genotype was tested by implanting small numbers of sorted cells into the lungs of recipient mice. When isolated from primary Kras tumors, both Sca-1+ and Sca-1− cells generated secondary tumors that recapitulated the Sca-1 cell heterogeneity found in the primary tumor (left). However, in tumors with mutant Kras and p53 deficiency, Sca-1+ cells possessed a greater capacity for secondary tumor formation (represented as a larger tumor) than Sca-1− cells (center). In addition, secondary tumors derived from Sca-1+ cells possessed a similar proportion of Sca-1+ cells observed in the primary tumor, whereas the few small tumors derived from Sca-1− cells did not have a detectable Sca-1+ population. These data would suggest Sca-1+ cells in this tumor genotype are enriched in lung cancer stem cells. The opposite appears to be true in mutant EGFR adenocarcinomas, in which Sca-1− cells exhibited a greater capacity for generating secondary tumors (right). The distribution of Sca-1+ cells in secondary mutant EGFR tumors was not determined.
These findings will be of great interest to clinicians trying to use “personalized medicine” to cure lung cancer with combined therapy strategies that in part target specific oncogenotypes and also take aim against cancer stem cells. Kras, p53, and EGFR are among the most commonly mutated genes in lung cancer and their oncogenotypes are known to influence response to targeted therapy (Ding et al., 2008). For example, patients with EGFR mutant lung tumors stand to benefit from EGFR tyrosine kinase inhibitor (TKI) therapy, whereas patients with mutant Kras and/or wild-type EGFR tumors do not (Sun et al., 2007). In fact, testing of non-small cell lung cancers (NSCLCs) for the presence of Kras mutations is being used to select against therapy with EGFR TKIs. If the results of Curtis et al. are shown to be consistent in human NSCLCs it would appear different lung cancer stem cell populations will needed to be therapeutically targeted. Thus, it will be of great interest to see whether the different murine lung cancer stem cells are sensitive to the same or different stem cell-targeted therapies, and of course how do these therapies impact cancer stem cell function in human tumors. There are important caveats to these studies. The authors used an “orthotopic” model, in that the cells to be tested for TPC were seeded in the tracheas of mice. Of course, lung adenocarcinomas normally develop in the lung parenchyma, not the trachea. In addition, the authors used “nude” mice as recipients, which are devoid of T lymphocytes, but other more immune-deprived mouse models are available. Thus, it will be important to further test for TPC after implantation at the sites where these tumors actually arise, and also to test TPC in other immune deprived mouse models, such as NOD/SCID mice.
The findings presented by Curtis et al. suggest that tumor oncogenotype may also be an arbiter of cancer stem cell (or TPC) properties. This study suggests caution in the use of one marker to define putative cancer stem cells in genotypically diverse malignancies and raises interesting questions for future studies. Because the BASC cell identity does not define TPCs in each adenocarcinoma genotype, it raises the question: are these adenocarcinomas initiated and/or driven by another lung stem cell population (i.e., non-BASC-like) or, for that matter, derived from a non-stem cell population? Also do EGFR mutant tumors contain a cancer stem cell? This point is also interesting considering that the vast majority of EGFR mutant NSCLCs arise in patients who have never smoked, whereas NSCLCs with wild-type EGFR nearly always arise in a person with a smoking history. Although we see dramatic clinical responses to EGFR TKI therapy in EGFR mutant NSCLCs, these patients invariably relapse, often with widely metastatic disease. Although mechanisms of EGFR TKI resistance have been well documented, are these resistant cells also enriched in cancer stem cells? Further investigation of the characteristics and potential vulnerabilities of cancer stem cells in lung cancers with different oncogenotypes is clearly of high priority.
Acknowledgments
This work was supported by grant P50CA70907, NASA NNJ05HD36GD, IASLC Fellowship (J.P.S.).
References
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, et al. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RW, Wang X, Diehn M, Shedden K, Chen GY, Sherlock G, Gurney A, Lewicki J, Clarke MF. Stem Cells. 2008;26:364–371. doi: 10.1634/stemcells.2007-0440. [DOI] [PubMed] [Google Scholar]
- Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, Woolfenden AE, Kirsch DG, Wong K-K, Kim CF. Cell Stem Cell. 2010;7(this issue):127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey JS, Zacharek SJ, Kim CF, Lees JA. Proc Natl Acad Sci USA. 2008;105:11857–11862. doi: 10.1073/pnas.0803574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, Lee J, Choi YL, Sato M, Wang P, et al. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JC, Deng T, Lehal RS, Kim J, Zacksenhaus E. Cancer Res. 2007;67:8671–8681. doi: 10.1158/0008-5472.CAN-07-1486. [DOI] [PubMed] [Google Scholar]
- Sullivan JP, Minna JD, Shay JW. Cancer Metastasis Rev. 2010;29:61–72. doi: 10.1007/s10555-010-9216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Schiller JH, Gazdar AF. Nat Rev Cancer. 2007;7:778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]