Abstract
Genome-wide association studies (GWAS) have identified over a dozen loci associated with colorectal cancer (CRC) risk. Here we examined potential effect-modification between single nucleotide polymorphisms (SNPs) at 10 of these loci and probable or established environmental risk factors for CRC in 7,016 CRC cases and 9,723 controls from nine cohort and case-control studies. We used meta-analysis of an efficient empirical-Bayes estimator to detect potential multiplicative interactions between each of the SNPs [rs16892766 at 8q23.3 (EIF3H/UTP23); rs6983267 at 8q24 (MYC); rs10795668 at 10p14 (FLJ3802842); rs3802842 at11q23 (LOC120376); rs4444235 at 14q22.2 (BMP4); rs4779584 at15q13 (GREM1); rs9929218 at16q22.1 (CDH1); rs4939827 at18q21 (SMAD7); rs10411210 at19q13.1 (RHPN2); and rs961253 at 20p12.3 (BMP2)] and select major CRC risk factors (sex, body mass index, height, smoking status, aspirin/non-steroidal anti-inflammatory drug use, alcohol use, and dietary intake of calcium, folate, red meat, processed meat, vegetables, fruit, and fiber). The strongest statistical evidence for a gene-environment interaction across studies was for vegetable consumption and rs16892766, located on chromosome 8q23.3, near the EIF3H and UTP23 genes (nominal p-interaction =1.3×10–4; adjusted p-value 0.02). The magnitude of the main effect of the SNP increased with increasing levels of vegetable consumption. No other interactions were statistically significant after adjusting for multiple comparisons. Overall, the association of most CRC susceptibility loci identified in initial GWAS appears to be invariant to the other risk factors considered; however, our results suggest potential modification of the rs16892766 effect by vegetable consumption.
Keywords: Colorectal Cancer, Epidemiology, Gene-environment interactions, Genotype phenotype correlations, Polymorphisms in genes that modify dietary exposures
Introduction
Approximately one third of colorectal cancer (CRC), the second leading cancer in the United States (US), is attributable to inherited factors (1). Identification of associated genetic variants may elucidate mechanisms underlying this disease. First results from genome-wide association studies (GWAS) have demonstrated considerable success in identifying genetic variants associated with CRC (2–10). However, these variants currently explain only a small fraction of the genetic heritability (9). Recent work postulates that there may be up to 65 to 70 common loci underlying CRC susceptibility, requiring large sample sizes for detection (11); additional avenues of work are also needed to identify other factors underlying the “missing heritability” (12). Less common genetic variants, and gene-environment interactions (GxE) are postulated to explain an important component (12, 13). In addition, alternative models (e.g., recessive models) have generally not been tested. A full examination of the role of GxE underlying CRC will require genome-wide scans incorporating genetic and environmental factors and interaction terms across the genome. Nonetheless, a logical first step in exploring the GxE contribution is to characterize potential effect modification of genetic risk variants already identified as having marginal effects.
This paper focuses on potential GxE interactions for the first ten CRC GWAS loci identified: 8q24 (MYC); 15q13 (GREM1); 18q21 (SMAD7); 11q23 (LOC120376); 8q23.3 (EIF3H/UTP23); 10p14 (FLJ3802842); 14q22.2 (BMP4); 16q22.1 (CDH1); 19q13.1 (RHPN2); 20p12.3 (BMP2). In the context of this paper, we use the term “environmental risk factors” broadly to include non-SNP risk factors, including sex, which is genetically determined, as well as factors like tobacco use and height, which themselves may be intermediate phenotypes with genetic and environmental determinants. Previous studies have examined gene-environment interactions with selected sets of these known variants for some environmental covariates. However, these studies have either focused only on single variants (14–16) or had relatively small sample sizes and results have been inconsistent (17, 18). Here we perform a more comprehensive examination of these loci and twelve probable or established CRC risk factors [sex, body mass index (BMI), height, smoking status, aspirin/non-steroidal anti-inflammatory drug (NSAID) use, and intake of alcohol, dietary calcium, dietary folate, red meat, processed meat, vegetables, fruit, and fiber] in a combined analysis of nine case-control and nested case-control studies comprising 7,016 CRC cases and 9,723 controls.
Methods
Study participants
The studies used are listed in Table 1 and have been described in detail previously (10). In brief, we used data from five nested case-control studies in prospective US cohorts [Health Professionals Follow-up Study (HPFS); Nurses’ Health Study (NHS); Physician’s Health Study (PHS); Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO); Woman’s Health Initiative (WHI)] and four case-control studies from the US, Canada and Europe [Assessment of Risk in Colorectal Tumors in Canada (ARCTIC); French Association STudy Evaluating RISK for sporadic colorectal cancer (ASTERISK); Darmkrebs: Chancen der Verhuetung durch Screening (DACHS); Diet, Activity and Lifestyle Survey (DALS)]. The ARCTIC study used subjects from the Ontario Colon Cancer Family Registry (19). All cases were defined as invasive colorectal adenocarcinoma (International Classification of Disease Code 153–154) and confirmed by medical record, pathology report, or death certificate. The studies used nested case-control or case-control designs with study-specific eligibility and matching criteria, except for PLCO. For PLCO, controls were drawn from the controls used in previous GWAS studies of prostate cancer and lung cancer available through dbGaP (20, 21). To account for the different eligibility and matching criteria used in those GWAS, sampling fraction weights, based on sex, smoking status, age at entry, and year of entry were used to weight the PLCO case and controls to be representative of eligible subjects in the full PLCO cohort. PHS subjects were matched on smoking status, so that study is excluded from the summary of main effects of smoking-related variables. Due to small numbers, we excluded samples reported as racial/ethnic groups other than “White”; European ancestry was confirmed in GWAS samples using principal components analysis (22).
Table 1.
Overview of the studies included in this analysis
| Study | Case | Control | Total | % Female | % Colon | Median Agea (Range) |
Genotyping Platform |
|---|---|---|---|---|---|---|---|
| ARCTIC | 821 | 883 | 1704 | 50.3 | 66.1 | 62 (27–77) | Affymetrix GWAS platforms |
| ASTERISK | 954 | 1060 | 2014 | 41.6 | 70.8 | 67 (40–99) | BeadXpress |
| DACHS | 1731 | 1742 | 3473 | 40.3 | 60.7 | 69 (33–98) | BeadXpress |
| DALS Ib | 689 | 720 | 1409 | 47.4 | 100 | 66 (30–79) | BeadXpress |
| DALS IIb | 706 | 710 | 1416 | 43.4 | 100 | 66 (30–79) | Illumina GWAS platforms |
| HPFS | 344 | 635 | 979 | 0 | 76.2 | 69 (48–82) | TaqMan OpenArray |
| NHS | 465 | 1009 | 1474 | 100 | 79.8 | 61 (44–69) | TaqMan OpenArray |
| PLCO | 544 | 1976 | 2520 | 26.7 | 94.9 | 64 (55–74) | Illumina GWAS platforms |
| PHS | 288 | 454 | 846 | 0 | 76.3 | 58 (40–84) | TaqMan OpenArray |
| WHI | 474 | 534 | 1008 | 100 | 97 | 68 (50–79) | Illumina GWAS platforms |
Study abbreviations are given in the text.
Age in years
DALS I: Initial GWAS of subjects in the DALS study; DALS II: Follow-up replication for a subset of SNPs for subjects in the DALS study
All participants gave informed consent, and studies were approved by the Institutional Review Board.
Genotype data
We examined 10 SNPs identified through published CRC GWAS prior to September, 2010 (Table 2). For WHI, PLCO, and DALS I, genotype data were generated using Illumina HumanHap300k and 240k (PLCO), 550k (WHI, DALS) and 610k (DALS, PLCO) BeadChip Array Systems on the Infinium platform as previously described (10). ARCTIC samples were genotyped on Affymetrix platforms (3) and imputed with BEAGLE (23), using the phased HapMap release 22 as the reference sample (24). We used imputed SNPs, coded as the best call genotype, for all 10 SNPs in ARCTIC. The imputation quality was moderate for rs4939827 (r2=0.49) and rs10411210 (r2=0.82), and was high for all SNPs with imputation r2 ranging from 0.90–1.00 (see Supplemental Table 1). For DACHS, DALS II, and ASTERISK, samples were genotyped using BeadXpress technology according to the manufacturer’s protocol (25). For DACHS, the 8q24 SNP, rs6983267, was not successfully genotyped on BeadXpress and was replaced, for a subset of the samples (2,849 total), with previous TaqMan genotyping. For ASTERISK, we used TaqMan results for rs10505477 as an LD substitute for rs6983267 (see Supplemental Table 1). The linkage disequilibrium (LD) r2 between rs10505477 and rs6983267 in the HapMap Utah residents with ancestry from northern and western Europe (CEU) population is 0.93. The NHS, HPFS, and PHS samples were genotyped using TaqMan OpenArray technology. All genotyping underwent standard quality control (QC) checks (10), including concordance checks for blinded and unblinded duplicates, examination of sample and SNP call rates, and checking Hardy-Weinberg equilibrium (HWE) in controls. Call rate, HWE p-value, and minor allele frequency for each SNP in each study are included in Supplemental Table 1. One SNP, rs4444235 at 14q22.2, was excluded from the NHS study because of the HWE p-value in controls (p=3×10−5).
Table 2.
Associations between SNPs and CRC risk in previously published reports and the current study
| Chromosomal Location |
Gene/Locus | SNP | Minor Allele |
Other Allele |
MAF | Published OR (95% CI)a |
Published Reference |
Current Study OR (95% CI) a,b |
Current Study p-value a |
Current Study N |
Current Study NAc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 8q23.3 | EIF3H/UTP23 | rs16892766 | C | A | 0.09 | 1.25 (1.19–1.32) | (2) | 1.17 (1.08–1.27) | 1.6 × 10−4 | 16,775 | 68 |
| 8q24 | MYC | rs6983267 | A | C | 0.48 | 0.83 (0.81–0.85) | (3, 7) | 0.88 (0.84–0.93) | 4.1 × 10−7 | 15,657 | 1,186 |
| 10p14 | LOC338591 | rs10795668 | A | G | 0.31 | 0.89 (0.86–0.91) | (2) | 0.97 (0.93–1.02) | 0.25 | 16,782 | 61 |
| 11q23 | LOC120376 | rs3802842 | C | A | 0.29 | 1.11 (1.08–1.15) | (5) | 1.12 (1.06–1.17) | 2.1 × 10−5 | 16,769 | 74 |
| 14q22.2 | BMP4 | rs4444235 | G | A | 0.47 | 1.09 (1.06–1.12) | (8) | 1.05 (1.01–1.11) | 0.03 | 15,322 | 1,521 |
| 15q13 | CRAC1/GREM1 | rs4779584 | A | G | 0.20 | 1.15 (1.10–1.19) | (6) | 1.15 (1.08–1.21) | 2.1 × 10−6 | 16,775 | 68 |
| 16q22.1 | CDH1 | rs9929218 | A | G | 0.30 | 0.91 (0.89–0.94) | (8) | 0.94 (0.89–0.99) | 0.01 | 16,728 | 115 |
| 18q21 | SMAD7 | rs4939827 | G | A | 0.48 | 0.83 (0.81–0.86) | (5) | 0.90 (0.86–0.94) | 2.0 × 10−6 | 16,762 | 81 |
| 19q13.1 | RHPN2 | rs10411210 | A | G | 0.10 | 0.87 (0.83–0.91) | (8) | 0.96 (0.89–1.04) | 0.30 | 16,747 | 96 |
| 20p12.3 | BMP2 | rs961253 | A | C | 0.36 | 1.12 (1.09–1.15) | (8) | 1.12 (1.07–1.18) | 1.3 × 10−6 | 16,783 | 60 |
SNP=single nucleotide polymorphism; MAF=minor allele frequency; OR=odds ratio; CI=confidence interval; N=number; NA=not available.
OR and p-values for log-additive model; odds ratio represents each additional copy of the minor allele.
Presented ORs for current data are based on subjects that overlap with main effects previously published (10).
NA=number missing for each SNP. 8q24 has a higher proportion missing because it was not successfully genotyped using BeadXpress.
Harmonization of environmental data
Information on basic demographics and environmental risk factors was collected by self-report using in-person interviews and/or structured questionnaires, as detailed previously (19, 26–34). We carried out a multi-step data harmonization procedure, reconciling each study’s unique protocols and data-collection instruments. First, we defined common data elements (CDEs). We examined the questionnaires and data dictionaries for each study to identify study specific data elements that could be mapped to the CDEs. Through an iterative process, we communicated with each data contributor to obtain relevant data and coding information. The data elements were written to a common data platform, transformed via a SQL programming script, and combined into a single dataset with common definitions, standardized permissible values, and standardized coding. The mapping and resulting data were reviewed for quality assurance, and range and logic checks were performed to assess data and data distributions within and between studies. Outlying samples were truncated to the minimum or maximum value of established range for each variable. The reference time for cohort studies was time of enrollment (WHI and PLCO) or blood draw (HPFS, NHS and PHS). The data elements considered were analyzed as continuous variables (BMI and height); dichotomous variables [sex (male/female), smoking (ever/never at reference time), aspirin/NSAID use (yes/no for regular use at reference time; see exact definitions in Supplemental Table 2)]; ordered categorical variables [alcohol consumption (three categories defined by g/day)]; study-specific quartiles for smoking pack years (using never smokers as reference, other quartiles coded 1–4); and sex- and study-specific quartiles, where the quartile groups were coded with the median value of the quartile within each study and sex and scaled to a unit scale reflective of the distribution for that variable [dietary calcium (units of 500 mg/day), dietary folate (units of 500 mcg/day), red meat (units of servings/day), processed meat (units of servings/day), fruit (units of 5 servings/day), vegetables (units of 5 servings/day), and dietary fiber (units of 10 g/day)]. We use scales such as 500 mg/day for calcium, to provide more meaningful and easier to interpret effect sizes. All quartile variables had 4 categories for each sex within each study. Because some studies collected dietary information in categories that could not be converted to study-specific quartiles, we also examined red meat, processed meat, vegetables, and fruit as dichotomous variables, cut at sex- and study-specific medians. We accounted for the multiple testing burden and potential correlation between these additional variables using permutation testing, as described in the statistical methods section. For all variables, the lowest category of exposure (or no use) was used as the reference.
Statistical methods
Unless otherwise indicated, we adjusted all regression analyses described below for age, center, and sex, as appropriate. We used fixed-effects meta-analysis methods to obtain summary odds ratios (ORs) and 95% confidence intervals (CIs) across studies. The p-values from the meta-analysis, unadjusted for multiple comparisons, are termed nominal p-values. We report the p-value for heterogeneity, and examine forest plots for results showing evidence for heterogeneity. For PLCO, we used inverse sampling fractions as weights in all analyses to account for study design; for all other studies, we used equal weights.
Inadequate modeling of the marginal association can bias interaction testing (35). Therefore, for each SNP and environmental factor, we employed a screening method, based on logistic regression main-effect associations, to find a reasonable form to use for GxE testing. Nested models were compared using likelihood ratio tests, with a p-value <0.05 indicating significantly better performance. For SNPs, we considered assumptions of log-additive (SNPs coded 0/1/2, representing counts of the minor allele) and recessive (SNPs coded 0/1 where 0 represents homozygous for common allele or heterozygous and 1 represents homozygous for the minor allele) modes of inheritance in comparison to an unrestricted model with indicator variables for heterozygote and homozygote minor alleles. We did not consider a dominant mode of inheritance, because the log-additive model usually does not lose power if the true model is dominant. If the unrestricted model did not significantly outperform the log-additive model, we used the log-additive model. If the unrestricted model performed significantly better than the log-additive model, but not the recessive model, we used the recessive model. If the unrestricted model performed significantly better than the log-additive and the recessive, we used the unrestricted model. Under this procedure, we selected the recessive model for rs6983267, and the log-additive model for the other nine SNPs. Dichotomous environmental variables were coded 0/1 and did not require model selection. For the continuous variables, BMI and height, we compared main-effects models with and without a quadratic term. In both cases, the model with the quadratic term did not perform significantly better, so we modeled these variables using only a linear term. For the categorical variables (alcohol, pack years, and the quartile version of the dietary variables), we compared a model using a group-linear variable to a saturated model with indicator variables for each non-reference category. For alcohol, the saturated model performed significantly better, so we modeled alcohol with indicator variables. In contrast, for the other variables, the saturated model was not significantly better than a model with a single group-linear term. Thus, we modeled these variables with their sex- and study-specific medians, as described above in the section on data harmonization.
To test for interactions between SNPs and environmental risk factors, we used an efficient empirical-Bayes (EB) shrinkage method (36). This method creates a weighted average of the standard case-only and case-control estimators, which is weighted towards the unbiased case-control estimator when the assumption of gene-environment independence in the population is suspect and towards the more efficient case-only estimator when the assumption is supported by the data. We modeled both the main effect and interaction based on the model selected from the main effects, as described above. Subjects missing data for a particular SNP or environmental factor were dropped from the analysis for that SNP × factor interaction test.
Because we performed 180 tests (10 SNPs × 18 versions of the environmental risk factors), with correlation among some tests, we used permutations to account for multiple testing. We ran the analysis 1000 times using a permuted case-control status in each run. Then we used the Westfall & Young step-down procedure (37) to derive the adjusted p-value for each GxE interaction based on the permuted p-values. We term these the adjusted p-values, and used them to evaluate the statistical significance of a given interaction at the 0.05 level.
For situations where the EB interaction-term adjusted p-value was <0.05, we also examined the results from the traditional logistic regression case-control estimate and examined results adjusting for additional covariates (smoking history, BMI, alcohol consumption and red meat consumption). As follow-up analysis, we examined the main effect for the SNP in strata defined by the environmental risk factor. We also pooled the data across studies and examined a) the main effect of the environmental factor in strata defined by the SNP; and b) the combined effect in strata defined by both the SNP and the environmental factor. As a supplemental analysis, we examined all 180 SNP × environmental factor GxE interactions in substratum analyses restricted to colon only and rectal only cases.
Data harmonization was performed using SAS and T-SQL. All other analyses were conducted using the R programming language.
Results
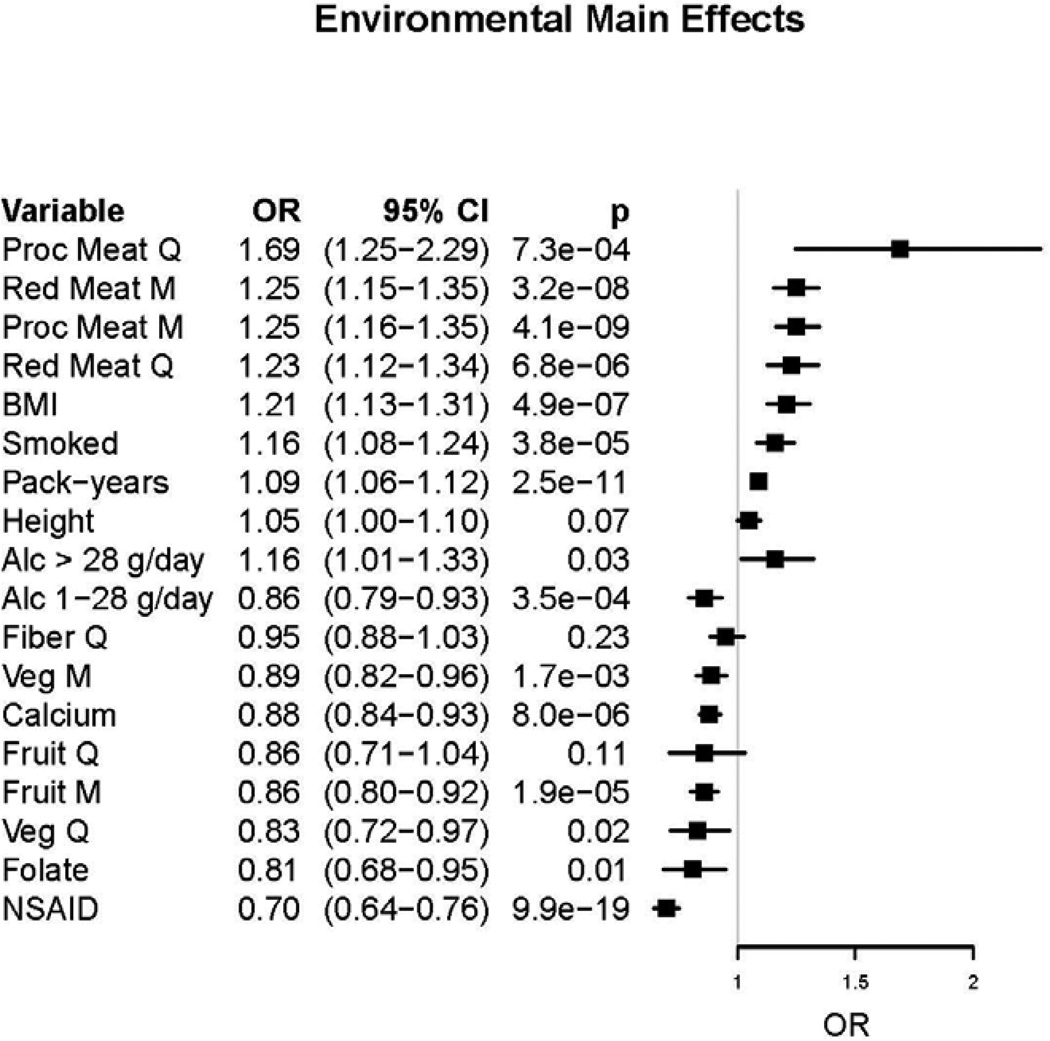
Study characteristics are described in Table 1 and Supplemental Table 3. Table 2 shows the marginal results for each SNP. As we have previously reported using an overlapping set of subjects (10), 8 of the 10 loci show statistical evidence for association with CRC with nominal p-values ranging from 0.03 to 4.1×10−7. One SNP (rs16892766) had a heterogeneity p-value of 0.03; the heterogeneity p-value for all other SNPs ranged from 0.18–0.96, indicating little evidence for heterogeneity in the main effects of SNPs across studies. The two established SNPs not showing statistical evidence for association are rs10795668 at 10p14 and rs10411210 at 19q13; however, both showed a statistically non-significant odds ratio in the same direction of association as previous reports (Table 2). Our model selection procedure indicated a recessive model for rs6983267 (8q24): the OR for the AA genotype (homozygous for the minor allele) compared to the AC+CC genotype was 0.82 (0.78–0.89, p = 1.55 × 10−6). Focusing on marginal effects for the environmental risk factors (Figure 1), we observed statistical support for an increased risk of CRC with increased processed meat and red meat consumption (both derived as quartiles and as median cut points), increasing BMI, ever smoking, and increasing number of pack-years of smoking. We observed statistical evidence for a decreased risk for CRC with increased vegetable consumption (both quartiles and median cut), high fruit consumption, increased dietary folate, and any aspirin/NSAID use. Alcohol consumption showed a reduced risk for light drinkers (1–28 g/day) and increased risk for heavy drinkers (>28 g/day) compared to those who consumed less than 1 gram of alcohol per day. The main effects for quartiles of fiber and fruit intake were not statistically significant, but showed expected trends towards inverse associations. We did not investigate sex as a main effect, because most of the studies either matched on sex, or were restricted to one sex.
Figure 1.
The results for the 180 gene-environment interactions tested are presented in Supplemental Table 4. Six SNP/environmental factor interactions showed nominal p<0.01 (Table 3; forest plots for individual study results are in Supplemental Figure 1). The lowest nominal p-value was for rs16892766, with vegetables as quartiles (interaction OR=1.88, 95% CI: 1.36–2.59; nominal p-interaction =1.3×10−4). rs16892766 has a minor allele frequency (MAF) of 0.1 in the CEU population and is located on chromosome 8q23.3. This was the only finding with an adjusted p<0.05 (adjusted p-value=0.02). Because of potential correlations between the environmental factors tested, we used permutations methods to adjust for multiple comparisons. A Bonferroni correction assumes the tests are independent. For the permutations, the cut-off that corresponds to a family wise error rate of 0.05 can be calculated by taking the 5th percentile of the minimum of p-values of all tests across all permutation runs. For our data, it was 3.75 × 10−4, slightly less conservative than the Bonferroni cut off 0.05/180 = 2.78 × 10−4. The rs16892766/vegetable consumption interaction was statistically significant with either correction. This same SNP had a nominal p-value for interaction <0.01 for processed fiber as quartiles (nominal p-interaction=6.0×10−4; adjusted p-value p=0.09) and for vegetables dichotomized at sex- and study-specific medians (nominal p-interaction=3.5×10−3; adjusted p-value=0.40). The correlation between vegetable quartiles and fiber quartiles in this data set was 0.65. Table 4 shows the association with colorectal cancer risk in strata defined by quartiles of vegetable consumption. The magnitude of the main effect of the minor (C) allele for this SNP increased with increasing levels of vegetable consumption, ranging from no evidence for association (OR= 0.94; 95% CI: 0.77–1.15; nominal p=0.54) in the lowest quartile to a relatively strong association for a common genetic factor (OR=1.40; 95% CI: 1.13–1.74; nominal p=0.002) in the highest quartile. Results of the pooled analysis showing associations for vegetables in strata defined by levels of the SNP, and the combined association in strata defined by rs16892766 genotype and vegetable consumption are shown in supplemental materials (Supplemental Tables 5 and 6).
Table 3.
Gene-environment interactions with interaction p-value < 0.01
| Fixed Effects Meta-Analysis | ||||||
|---|---|---|---|---|---|---|
| SNP/Chromosomal Location Environmental Variable |
ORINT | 95% CI | nom.p | adj.p | het.p | Studies |
| rs16892766/8q23.3 Vegetable quartile medians | 1.88 | 1.36–2.59 | 1.3 × 10−4 | 0.02 | 0.68 | ARCTIC/DALS/PLCO/WHI/HPFS/NHS/ PHS |
| rs16892766/8q23.3 Fiber quartile medians | 1.33 | 1.13–1.56 | 6.0 × 10−4 | 0.09 | 0.87 | DALS/PLCO/WHI/HPFS/NHS |
| rs4939827/18q21 Red meat above/below median | 1.14 | 1.05–1.24 | 2.9 × 10−3 | 0.36 | 0.59 | ARCTIC/DALS/PLCO/WHI/DACHS/AST ERISK/HPFS/NHS/PHS |
| rs16892766/8q23.3 Vegetables above/below median | 1.29 | 1.09–1.53 | 3.5 × 10−3 | 0.40 | 0.31 | ARCTIC/DALS/PLCO/WHI/DACHS/AST ERISK/HPFS/NHS/PHS |
| rs3802842/11q23 Folate quartile medians | 1.34 | 1.08–1.67 | 8.2 × 10−3 | 0.71 | 0.65 | DALS/PLCO/WHI/HPFS/NHS |
| rs3802842/11q23 Red meat quartile medians | 1.17 | 1.04–1.32 | 8.6 × 10−3 | 0.73 | 0.58 | ARCTIC/DALS/PLCO/WHI/HPFS/NHS/PHS |
OR=odds ratio; CI=confidence interval; ORINT=multiplicative interaction odds ratio from empirical-Bayes; 95% CI=95% confidence interval for interaction OR; nom.p = nominal p-value; adj.p = adjusted p-value based on permutations; het.p = heterogeneity p-value; Studes=studies included in estimating the interaction term. Permutation-based significance threshold: 3.75×10−4; Bonferroni-based significance threshold: 2.78×10−4.
Table 4.
Main effect of rs16892766 overall and by quartiles of vegetable consumption
| Group | ORa | 95% CI | p-valuea | |
|---|---|---|---|---|
| Overall | 1.17 | (1.08 – 1.27) | 1.6 × 10−4 | |
| By Vegetable Quartiles | ||||
| Quartile 1 | 0.94 | (0.77 – 1.15) | 0.541 | |
| Quartile 2 | 1.19 | (0.96 – 1.47) | 0.114 | |
| Quartile 3 | 1.26 | (1.02 – 1.55) | 0.029 | |
| Quartile 4 | 1.40 | (1.13 – 1.74) | 2.2 × 10−3 | |
OR=odds ratio; CI=confidence interval;
OR and p-values for log-additive model; odds ratios represent each additional copy of minor (C) allele for rs16892766.
The rs16892766/vegetable-consumption results were not altered when we adjusted for additional covariates; interaction OR (adjusted for ever-smoked, BMI, alcohol use, red meat, and processed meat consumption) =1.90 (95% CI: 1.35–2.67; nominal p-interaction=2.48×10−4). A similar magnitude of interaction was seen using traditional case-control logistic analysis (interaction OR=1.79; 95% CI: 1.23–2.59; nominal p-interaction=2.3x10−3).
In supplemental analyses of all GxE interactions stratified by cancer site (colon vs. rectum) (Supplemental Table 7), the strongest statistical evidence for gene-environment interaction among colon cancer cases were for the same rs16892766/vegetable-consumption (interaction OR=1.79; 95% CI: 1.28–2.51; nominal p-interaction=6.5×10−4) and rs16892766/fiber (interaction OR=1.31; 95% CI: 1.12–1.53; nominal p-interaction=9.8×10−4) interactions observed for the combined CRC. For rectal cases, with a smaller sample size, the rs16892766/vegetable-consumption interaction was not statistically significant (interaction OR for rectal cancer=1.51; 95% CI: 0.57–4.03; p-interaction=0.41), and the only interaction with nominal p<0.01 was rs4779584 and dietary calcium (nominal p=6.7×10−3).
Discussion
We performed an evaluation of GxE interactions for 10 SNPs identified through CRC GWAS with probable and established environmental risk factors. Our analysis of over 7,000 CRC cases and 9,700 controls from nine well-characterized cohort and case-control studies showed evidence of an interaction between the rs16892766 SNP and quartiles of vegetable consumption (nominal p-interaction =1.3×10−4; adjusted p-value 0.02). None of the other gene-environment interactions examined was statistically significant after accounting for multiple testing.
The rs16892766 SNP is in an LD region on chromosome 8q23.3. Two studies have fine-mapped this region in relation to CRC risk (38, 39). Both found the strongest signals for a cluster of five SNPs, including rs16892766, that are in high LD; Pittman et al. also identified a sixth SNP in the cluster that is not in the public databases (39). Neither study found evidence for secondary independent signals in this region. The eukaryotic translation initiation factor 3 subunit H (EIF3H) gene is the closest gene to this cluster, with the identified SNPs ~140kb downstream from the gene transcript. Initial functional studies indicated that the rs16892766 region interacts with the EIF3H promoter and represses gene expression (39); however, a subsequent examination of ENCODE data and eQTLs suggests that the variants in this region may be influencing expression levels of the neighboring UTP23, small subunit (SSU) processome component, homolog (yeast) (UTP23) gene, rather than EIF3H itself. The variants may also impact expression of both genes (38). Additional work is needed to elucidate the functional relationship between EIF3H or UTP23, or both, and CRC etiology. As the functional role of this SNP and other variants in the region is unknown, we cannot currently make informed speculations on how it might relate to vegetable consumption.
Vegetable consumption has long been hypothesized to be protective against CRC (40), although epidemiologic studies are not fully consistent (see review in (41)). A recent meta-analysis of 1,694,236 participants including 16,057 colorectal cases with data on vegetable consumption from prospective cohort studies found a statistically significant nonlinear inverse association between both fruit and vegetable intake with CRC risk and the summary relative risk for the highest vs. lowest intake for vegetables was 0.91 (95% CI: 0.86–0.96) (42). The postulated mechanisms have primarily focused on vegetables as a source of fiber and micronutrients, including folate (43). We also observed some evidence for interaction between the rs16892766 SNP and quartiles of both fiber intake (interaction OR=1.33; 95% CI (1.13–1.56); p-interaction= 6.0×10−4; adjusted p-value=0.09), and dietary folate intake (interaction OR=1.34; 95% CI: 1.08–1.67; p-interaction=8.2×10−3; adjusted p-value=0.71). As with vegetable consumption, the pattern was for an increased risk associated with the minor (C) allele at higher levels of consumption. Vegetable consumption shows a positive correlation with both fiber (correlation=0.65) and folate (correlation=0.49) in these studies and it is difficult to disentangle the different measures using reported dietary-intake measures. Future follow-up of this interaction could focus more specifically on biomarkers for different dietary components.
Although we did not observe statistically significant evidence for heterogeneity in the rs16892766/vegetable-consumption interaction, we did observe minor evidence for heterogeneity for the main effect of the rs16892766 SNP in the full sample (heterogeneity p=0.030). We considered the possibility that the underlying GxE interaction may have been contributing to the observed heterogeneity. However, we observed similar evidence of heterogeneity for the main effect of rs16892766 in strata defined by levels of vegetable consumption (heterogeneity p-values ranging from 0.02 to 0.20). These results indicate that the minor level of observed heterogeneity for this SNP did not result from the rs16892766/vegetable-consumption interaction. Additional avenues would need to be explored for the source of this potential heterogeneity in association.
Previous studies of CRC risk have reported potential interactions with the 10 known loci in relation to age, family history, and sex (2, 5, 7, 17, 44, 45); however, the results have been inconsistent. Additional studies have looked at GxE for a broader range of environmental factors (14–18), but ours is the first to report a statistically significant interaction between rs16892766 and vegetable consumption. Using the DALS study, Slattery et al. observed an interaction between rs4939827, on 18q21 near the SMAD7 gene, and recent aspirin/NSAID use (16). We observed evidence for that interaction in the DALS study alone (p-interaction=0.03). However, we did not observe evidence for this association across the other studies, including analysis restricted to colon cancer only. This may reflect differences in how aspirin/NSAID use was collected across studies (Supplementary Table 2): for example, the time frame was 2 years prior to diagnosis for DALS and the other case-control studies, whereas for the cohort studies, baseline data describe a variable number of years prior to diagnosis. It might also reflect other underlying differences among the studies, a false positive in the initial report, or a false negative in the present study. Using a discovery set not included in this report, Figueiredo et al. examined GxE interactions for these same 10 loci with over 10 environmental factors in a sample of 1,191 and 999 unrelated population-based controls (18). They observed several suggestive gene-environment interactions, although none were replicated in an independent sample that overlaps with the ARCTIC sample used in this paper. Further, that analysis was restricted to MSS/MSI-L CRC cases, which have different environmental risk factors (46) and, therefore, perhaps different underlying gene-environment interactions than the more broadly defined CRC cases used in this study.
Strengths of this study include the large sample size and standardized harmonization. We adopted a flexible approach to retrospective harmonization, using methods similar to those proposed by other projects (47, 48). Not every study was included for some of the environmental factors considered, either because they did not collect that particular variable or because they did not collect information in a way that was considered inferentially equivalent. We limited our study to variables that could be combined across at least 50% of the studies and we used yes/no and study-specific quartiles as forms of variables. These forms are most easily comparable across studies. As in many epidemiologic studies, measurement error may be leading to false negatives. We may be missing interactions that would have been found through inclusion of other environmental factors, through different assessments of the environmental variables or through different models, including fully saturated models (35).
The lack of evidence for other GxE interactions for most loci identified through initial GWAS is similar to what has been observed in prostate and breast cancer (49–52). This is not surprising given that the loci were identified through large-scale discovery and replication. SNPs with a strong GxE might show more heterogeneity across studies and may be less likely to appear as the strongest marginal signals. A full examination of the role of gene-environment interactions in CRC will require large, well-powered, genome-wide investigations with well measured and harmonized environmental risk factor data.
Supplementary Material
Acknowledgments
DACHS: We thank all participants and cooperating clinicians, and Ute Handte-Daub, Muhabbet Celik and Ursula Eilber for excellent technical assistance.
HPFS, NHS, PHS: We would like to acknowledge Patrice Soule and Hardeep Ranu of the Dana Farber Harvard Cancer Center Highthroughput Genotyping Core who assisted in the genotyping for NHS, HPFS, and PHS under the supervision of David J. Hunter. Carolyn Guo who assisted in programming for NHS and HPFS, and Haiyan Zhang who assisted in programming for the PHS.
We would like to thank the participants and staff of the Nurses' Health Study and the Health Professionals Follow-Up Study, for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
PLCO: The authors thank Drs. Christine Berg and Philip Prorok, Division of Cancer Prevention, at the National Cancer Institute, the screening center investigators and staff of the PLCO Cancer Screening Trial, Mr. Thomas Riley and staff at Information Management Services, Inc., and Ms. Barbara O’Brien and staff at Westat, Inc. for their contributions to the PLCO Cancer Screening Trial. Most importantly, we acknowledge the study participants for their contributions to making this study possible.
WHI: The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found on the WHI website (53).
Grant Support
ARCTIC : This work was supported by a GL2 grant from the Ontario Research Fund, the Canadian Institutes of Health Research, and the Cancer Risk Evaluation (CaRE) Program grant from the Canadian Cancer Society Research Institute. TJH and BWZ are recipients of Senior Investigator Awards from the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research.
ASTERISK: This work is funded by a regional Hospital Clinical Research Program (PHRC) and supported by the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la LUtte contre le Cancer (GEFLUC), the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer (LRCC).
CCFR: This work is supported by the National Cancer Institute, National Institutes of Health under RFA # CA-95-011 and through cooperative agreements with members of the Colon Cancer Family Registry and P.I.s. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating institutions or investigators in the Colon CFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the Colon CFR.
The Colon CFR Center, Ontario Registry for Studies of Familial Colorectal Cancer, contributed data to this manuscript and was supported by (U01 CA074783).
DACHS: This work is supported by grants from the German Research Council (Deutsche Forschungsgemeinschaft, BR 1704/6-1, BR 1704/6-3, BR 1704/6-4 and CH 117/1-1), and the German Federal Ministry of Education and Research (01KH0404 and 01ER0814).
DALS: This work is supported by the National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (R01 CA48998 to M.L.S.).
DALS, PLCO, WHI GWAS and GECCO: Funding for the genome-wide scan of DALS, PLCO, and WHI was provided by the National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (R01 CA059045 to U.P.). C.M.H. was supported by a training grant from the National Cancer Institute, Institutes of Health, U.S. Department of Health and Human Services (R25 CA094880). Funding for GECCO infrastructure is supported by National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088 to U.P.).
HPFS, NHS and PHS: HPFS was supported by the National Institutes of Health (P01 CA 055075 to C.S.F., R01 137178 to A.T.C, and P50 CA 127003 to C.S.F.), NHS by the National Institutes of Health (R01 137178 to A.T.C., P50 CA 127003 to C.S.F., and P01 CA 087969 to E.L.G.) and PHS by the National Institutes of Health (CA41281).
PLCO: This research was supported in part by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health, US Department of Health and Human Services.
Control samples were genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS) prostate cancer scan and were supported by the Intramural Research Program of the National Cancer Institute. The datasets used in this analysis were accessed with appropriate approval through the dbGaP online resource (54) through dbGaP accession number 000207v.1p1.c1(20). Control samples were also genotyped as part of the GWAS of Lung Cancer and Smoking. Funding for this work was provided through the National Institutes of Health, Genes, Environment and Health Initiative [NIH GEI] (Z01 CP 010200). The human subjects participating in the GWAS are derived from the Prostate, Lung, Colon and Ovarian Screening Trial and the study is supported by intramural resources of the National Cancer Institute. Assistance with genotype cleaning, as well as with general study coordination, was provided by the Gene Environment Association Studies, GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NHI GEI (U01 HG 004438). The datasets used for the analyses described in this manuscript were obtained from dbGaP through NCBI (55), through dbGaP accession number ph000093.v2.p2.c1.
WHI: The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100001C-4C, HHSN268201100046C and HHSN271201100004C and NO1WH4421.
Footnotes
Conflicts of Interest:
Andrew T. Chan declares a minor conflict of interest in his role as a consultant/advisory board member of Bayer HealthCare, Pfizer Inc. and Millenium Pharmaceuticals.
Andrea Z. LaCroix declares a minor conflict of interest in her role as consultant/advisory board member of Amgen and the University of Massachusetts.
References
- 1.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 3.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 4.Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 5.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 8.Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nature Genetics. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, et al. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–977. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2011 doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JH, Wacholder S, Gail MH, Peters U, Jacobs KB, Chanock SJ, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–575. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berndt SI, Potter JD, Hazra A, Yeager M, Thomas G, Makar KW, et al. Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Hum Mol Genet. 2008;17:2665–2672. doi: 10.1093/hmg/ddn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutter CM, Slattery ML, Duggan DJ, Muehling J, Curtin K, Hsu L, et al. Characterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysis. BMC Cancer. 2010;10:670. doi: 10.1186/1471-2407-10-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slattery ML, Herrick J, Curtin K, Samowitz W, Wolff RK, Caan BJ, et al. Increased risk of colon cancer associated with a genetic polymorphism of SMAD7. Cancer Res. 2010;70:1479–1485. doi: 10.1158/0008-5472.CAN-08-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J, Wilkens LR, Stram DO, Kolonel LN, Henderson BE, Wu AH, et al. Generalizability and epidemiologic characterization of eleven colorectal cancer GWAS hits in multiple populations. Cancer Epidemiol Biomarkers Prev. 2011;20:70–81. doi: 10.1158/1055-9965.EPI-10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueiredo JC, Lewinger JP, Song C, Campbell PT, Conti DV, Edlund CK, et al. Genotype-environment interactions in microsatellite stable/microsatellite instability-low colorectal cancer: results from a genome-wide association study. Cancer Epidemiol Biomarkers Prev. 2011;20:758–766. doi: 10.1158/1055-9965.EPI-10-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb PA, Baron J, Cotterchio M, Gallinger S, Grove J, Haile R, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 20.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 21.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.International HapMap Project [internet] [updated 2010 July7; cited 2012 Feb 22] Phased HapMap release 22 data; Available from: http://ftp.hapmap.org/phasing/2007-08_rel22/ [Google Scholar]
- 25.Illumina.com [internet] San Diego: Illumina, Inc; c 2012. [cited 2012 Feb 22] Data Sheet on BeadXpress system. Available from: http://www.illumina.com/documents/products/datasheets/datasheet_beadxpress_reader.pdf. [Google Scholar]
- 26.Slattery ML, Potter J, Caan B, Edwards S, Coates A, Ma KN, et al. Energy balance and colon cancer--beyond physical activity. Cancer Res. 1997;57:75–80. [PubMed] [Google Scholar]
- 27.Christen WG, Gaziano JM, Hennekens CH. Design of Physicians' Health Study II--a randomized trial of beta-carotene, vitamins E and C, multivitamins, in prevention of cancer, cardiovascular disease, and eye disease, and review of results of completed trials. Ann Epidemiol. 2000;10:125–134. doi: 10.1016/s1047-2797(99)00042-3. [DOI] [PubMed] [Google Scholar]
- 28.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, Crawford ED, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 29.The Women's Health Initiative Study Group. Design of the Women's Health Initiative clinical trial and observational study. Control Clin Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 30.Hoffmeister M, Raum E, Krtschil A, Chang-Claude J, Brenner H. No evidence for variation in colorectal cancer risk associated with different types of postmenopausal hormone therapy. Clin Pharmacol Ther. 2009;86:416–424. doi: 10.1038/clpt.2009.134. [DOI] [PubMed] [Google Scholar]
- 31.Brenner H, Chang-Claude J, Seiler CM, Rickert A, Hoffmeister M. Protection from colorectal cancer after colonoscopy: population-based case-control study. Ann Intern Med. 2010 doi: 10.7326/0003-4819-154-1-201101040-00004. [DOI] [PubMed] [Google Scholar]
- 32.Kury S, Buecher B, Robiou-du-Pont S, Scoul C, Sebille V, Colman H, et al. Combinations of cytochrome P450 gene polymorphisms enhancing the risk for sporadic colorectal cancer related to red meat consumption. Cancer Epidemiol Biomarkers Prev. 2007;16:1460–1467. doi: 10.1158/1055-9965.EPI-07-0236. [DOI] [PubMed] [Google Scholar]
- 33.Colditz GA, Hankinson SE. The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 34.Giovannucci E, Rimm EB, Stampfer MJ, Colditz GA, Ascherio A, Willett WC. Aspirin use and the risk for colorectal cancer and adenoma in male health professionals. Ann Intern Med. 1994;121:241–246. doi: 10.7326/0003-4819-121-4-199408150-00001. [DOI] [PubMed] [Google Scholar]
- 35.Prentice RL. Empirical evaluation of gene and environment interactions: methods and potential. J Natl Cancer Inst. 2011;103:1209–1210. doi: 10.1093/jnci/djr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukherjee B, Chatterjee N. Exploiting gene-environment independence for analysis of case-control studies: an empirical Bayes-type shrinkage estimator to trade-off between bias and efficiency. biometrics. 2008;64:685–694. doi: 10.1111/j.1541-0420.2007.00953.x. [DOI] [PubMed] [Google Scholar]
- 37.Westfall PH, Young SS. Probability and Mathematical Statistics. Wiley; 1993. Resampling-based multiple testing: examples and methods for p-value adjustment. [Google Scholar]
- 38.Carvajal-Carmona LG, Cazier JB, Jones AM, Howarth K, Broderick P, Pittman A, et al. Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pittman AM, Naranjo S, Jalava SE, Twiss P, Ma Y, Olver B, et al. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001126. e1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinmetz KA, Potter JD. Vegetables, fruit, and cancer. I. Epidemiology. Cancer Causes Control. 1991;2:325–357. doi: 10.1007/BF00051672. [DOI] [PubMed] [Google Scholar]
- 41.World Cancer Research Fund and American Institute for Cancer Research. Food, Nutrition, Physical Activity, and the Prevention of Cancer: a Global Perspective. Washington, DC: AICR; 2007. [Google Scholar]
- 42.Aune D, Lau R, Chan DS, Vieira R, Greenwood DC, Kampman E, et al. Nonlinear reduction in risk for colorectal cancer by fruit and vegetable intake based on meta-analysis of prospective studies. Gastroenterology. 2011;141:106–118. doi: 10.1053/j.gastro.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Lee JE, Chan AT. Fruit, vegetables, and folate: cultivating the evidence for cancer prevention. Gastroenterology. 2011;141:16–20. doi: 10.1053/j.gastro.2011.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Holst S, Picelli S, Edler D, Lenander C, Dalen J, Hjern F, et al. Association studies on 11 published colorectal cancer risk loci. Br J Cancer. 2010;103:575–580. doi: 10.1038/sj.bjc.6605774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho JW, Choi SC, Lee YF, Hui TC, Cherny SS, Garcia-Barcelo MM, et al. Replication study of SNP associations for colorectal cancer in Hong Kong Chinese. Br J Cancer. 2011;104:369–375. doi: 10.1038/sj.bjc.6605977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slattery ML, Curtin K, Anderson K, Ma KN, Ballard L, Edwards S, et al. Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. J Natl Cancer Inst. 2000;92:1831–1836. doi: 10.1093/jnci/92.22.1831. [DOI] [PubMed] [Google Scholar]
- 47.Fortier I, Doiron D, Little J, Ferretti V, L'Heureux F, Stolk RP, et al. Is rigorous retrospective harmonization possible? Application of the DataSHaPER approach across 53 large studies. Int J Epidemiol. 2011;40:1314–1328. doi: 10.1093/ije/dyr106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett SN, Caporaso N, Fitzpatrick AL, Agrawal A, Barnes K, Boyd HA, et al. Phenotype harmonization and cross-study collaboration in GWAS consortia: the GENEVA experience. Genet Epidemiol. 2011;35:159–173. doi: 10.1002/gepi.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campa D, Kaaks R, Le ML, Haiman CA, Travis RC, Berg CD, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103:1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindstrom S, Schumacher F, Siddiq A, Travis RC, Campa D, Berndt SI, et al. Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers--results from BPC3. PLoS One. 2011;6:e17142. doi: 10.1371/journal.pone.0017142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Travis RC, Reeves GK, Green J, Bull D, Tipper SJ, Baker K, et al. Gene-environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. Lancet. 2010;375:2143–2151. doi: 10.1016/S0140-6736(10)60636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milne RL, Gaudet MM, Spurdle AB, Fasching PA, Couch FJ, Benitez J, et al. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast Cancer Res. 2010;12:R110. doi: 10.1186/bcr2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Women’s Health Initiative Scientific Resources Website [internet] [cited 2012 Feb 22] WHI investigators shortlist. Available from: https://cleo.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Short%20List.pdf. [Google Scholar]
- 54.CGEMS. Bethesda: National Cancer Institute; 2009. cancer.gov [internet] [cited 2012 Feb 22] Genetic Markers of Susceptibility (CGEMS) data website. Available from: http://www.cgems cancer gov/data/ [Google Scholar]
- 55.Bethesda: National Center for Biotechnology Information; NCBI [internet] [cited 2012 Feb 22] The database of Genotypes and Phenotypes. Available from: http://www.ncbi.nlm.nih.gov/gap. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.