Abstract
The CXC chemokine interleukin-8 (IL-8) is an angiogenic growth factor that is overexpressed in various cancers, including non-small cell lung cancer (NSCLC). Previously, IL-8 was shown as a transcriptional target of RAS signaling, raising the possibility of its role in oncogenic KRAS-driven NSCLC. Using microarray analysis, we identified IL-8 as the most downregulated gene by shRNA-mediated KRAS knockdown in NCI-H1792 NSCLC cells where IL-8 is overexpressed. NSCLC cell lines harboring KRAS or EGFR mutations overexpressed IL-8, while IL-8 levels were more prominent in KRAS mutants compared to EGFR mutants. IL-8 expression was downregulated by shRNA-mediated KRAS knockdown in KRAS mutants or by treatment with EGFR tyrosine kinase inhibitors and EGFR siRNAs in EGFR mutants. In our analysis of the relationship of IL-8 expression with clinical parameters and mutation status of KRAS or EGFR in 89 NSCLC surgical specimens, IL-8 expression was shown to be significantly higher in NSCLCs of males, smokers, and elderly patients and those with pleural involvement and KRAS mutated adenocarcinomas. In KRAS mutant cells, the MEK inhibitor markedly decreased IL-8 expression, while the p38 inhibitor increased IL-8 expression. Attenuation of IL-8 function by siRNAs or a neutralizing antibody inhibited cell proliferation and migration of KRAS mutant/IL-8 overexpressing NSCLC cells. These results indicate that activating mutations of KRAS or EGFR upregulate IL-8 expression in NSCLC; IL-8 is highly expressed in NSCLCs from males, smokers, elderly patients, NSCLCs with pleural involvement, and KRAS-mutated adenocarcinomas; and IL-8 plays a role in cell growth and migration in oncogenic KRAS-driven NSCLC.
Keywords: non-small cell lung cancer, KRAS, interleukin-8, molecular target
KRAS encodes a small GTP-binding protein that is involved in many cellular processes including proliferation, differentiation, and apoptosis.1 Wild-type KRAS has intrinsic GTPase activity, which catalyzes the hydrolysis of bound GTP to GDP and thereby inactivates the RAS growth-promoting signal, whereas mutant KRAS is locked into the GTP-bound state, leading to constitutive RAS signaling. Activating KRAS mutations play essential roles in malignant transformation in various human cancers including non-small cell lung cancer (NSCLC).1 KRAS mutations are found in ~ 25% of NSCLC but almost never in small cell lung cancer (SCLC)2,3 and are associated with poor prognosis of NSCLC patients.4 To improve survival for patients with NSCLC, there is an urgent need to develop therapeutic modalities for NSCLC harboring KRAS mutations. Therapeutic approaches targeting oncogenic Ras including farnesyl transferase inhibitors have failed in the treatment of NSCLC5; moreover, KRAS mutations are associated with resistance to EGFR tyrosine kinase inhibitors (EGFR-TKIs) for NSCLC.6,7 Thus, no effective treatment strategies have been established for KRAS mutant NSCLC.
A functional relationship between inflammation and cancer has been suggested for a long time.8 The CXC chemokine interleukin-8 (IL-8), which was originally identified as a neutrophil chemoattractant with inflammatory activity,9 is an important proinflammatory mediator relevant to cancer development.10 Increasing evidence suggests an important role for IL-8 in tumor progression and metastasis by promoting cell proliferation and angiogenesis in NSCLC.11–17 Furthermore, previous studies have reported that elevated IL-8 expression is an unfavorable prognostic factor in NSCLC.16,18,19 In a previous study, IL-8 was shown to be a transcriptional target of RAS signaling,20 raising the possibility of its role in oncogenic KRAS-driven NSCLC.
In a recent study, we performed a microarray analysis to compare gene expression profiling of mutant KRAS-disrupted NSCLC clones to those of the mutant KRAS expressing clones.21 Consequently, we identified IL-8 as the most down-regulated gene (−17.4 fold-change) by mutant KRAS knockdown in NCI-H1792 NSCLC cell line harboring a heterozygous KRAS mutation. In this study, we confirmed that prior to KRAS knockdown, H1792 cells overexpressed IL-8 at both the mRNA and the protein levels and that short hairpin RNA (shRNA)-mediated KRAS knockdown downregulated IL-8 expression. These results led us to examine IL-8 expression in a panel of lung cancer cell lines and clinically annotated surgical resection specimens and to analyze the relationship of IL-8 expression with clinicopathological parameters and KRAS mutation status. We also assessed whether attenuation of IL-8 function inhibited in vitro cell growth and migration of KRAS mutant/IL-8 overexpressing NSCLC cells. Here, we describe the positive association between IL-8 expression, KRAS mutations and certain clinicopathological features and therapeutic significance of IL-8 expression in KRAS mutated NSCLC.
Material and Methods
Cell lines and culture conditions
Twenty-two small cell lung cancer (SCLC) cell lines (NCI-H187, -H209, -H345, -H378, -H524, -H526, -H740, -H865, -H889, -H1045, -H1092, -H1184, -H1238, -H1339, -H1607, -H1618, -H1672, -H1963, -H2141, -H2171, -H2227, and HCC33), 10 NSCLC cell lines harboring KRAS mutations (NCI-H23, -H157, -H358, -H441, -H460, -H1264, -H1792, -H2009, -H2122, and HCC4017), 10 NSCLC cell lines harboring EGFR mutations (NCI-H820, -H1650, -H3255, -H1975, HCC827, HCC2279, HCC2935, HCC4006, HCCC4011, and PC9), 10 NSCLC cell lines with wild-type KRAS/EGFR (NCI-H322, -H520, -H661, -H838, -H1299, -H1395, -H1437, -H2077, -H2126, and HCC95), and immortalized human bronchial epithelial cell lines (HBEC3 and HBEC4, established as described22), were obtained from the Hamon Center collection (University of Texas Southwestern Medical Center). BEAS-2B (ATCC), HBEC3, and HBEC4 cell lines were used as noncancerous controls. Cancer cells were cultured with RPMI 1640 medium supplemented with 5% fetal bovine serum. The immortalized human bronchial epithelial cell lines were cultured with Keratinocyte-SFM (Invitrogen, Carlsbad, CA) medium with 50 μg/ml bovine pituitary extract (Invitrogen) and 5 ng/ml EGF (Invitrogen). All of the cell lines have been DNA fingerprinted for provenance using the PowerPlex 1.2 kit (Promega, Madison, WI) and confirmed to be the same as the DNA fingerprint library maintained either by ATCC or by the Minna/Gazdar lab (which is the primary source of the lines). The lines were also tested to be free of mycoplasma by e-Myco kit (Boca Scientific, Boca Raton, FL).
Tumor Specimens of NSCLC Patients
Tumor specimens were obtained from 89 patients (45 men and 44 women) with primary NSCLC cancer who underwent surgery between July 2003 and May 2008 at the Gunma University School of Medicine Hospital (Gunma, Japan). Of 89 patients, 48 were smokers and 41 were never smokers. Tumors were histologically classified as adenocarcinomas (N = 77) or squamous cell carcinomas (N = 12), according to the criteria of the World Health Organization. We classified the postsurgical pathological stage as stage I in 57 tumors (IA, 38; IB, 19), stage II in 11 tumors (IIA, 6; IIB, 5), and stage III/IV in 21 tumors (IIIA, 16; IIIB, 4; IV, 1), according to the tumor-node-metastasis classification. Noncancerous lung specimens (N = 5) obtained from five patients were used as normal controls for tumor specimens. Mutations in KRAS at codon 12 or EGFR in exons 19 and 21 were analyzed using the Smart Amplification Process version 2 assay (DNAFORM, Kanagawa, Japan) followed by direct sequencing to confirm the presence of these mutations.23 The study protocol was approved by the institutional review board of Gunma University Graduate School of Medicine. The specimens were immediately frozen after collection and stored at −80°C until mRNA extraction was performed.
Use of retroviral shRNA vectors and synthetic small interfering RNA (siRNA)
To achieve mutant-specific KRAS knockdown, the pRS-KRAS-C12 retroviral vector producing shRNA against the KRAS G12C mutation (GGT to TGT) was used.21 Cells were infected with retroviral vectors by previously described methods.24 siRNAs targeting against IL-8 or EGFR were obtained from the siGENOME library (Dharmacon, Lafayette, CO). A siRNA against Tax (the human leukemia virus gene) was used as a nontargeting control.25 One day before transfection, 1 × 105 cells were plated on each well of six well plates. Cells were transfected with 30 nM siRNAs using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer’s protocol. After 72 hr, cells were harvested to verify the silencing effects on the expression of the target genes.
Quantitative real-time RT-PCR
mRNA expressions of IL-8, CXCR1, CXCR2, and EGFR were analyzed by quantitative real-time RT-PCR.26 Briefly, total RNA was extracted using the RNeasy mini kit (Qiagen), and cDNA was synthesized using 2 μg of total RNA with the SuperScript II First-Strand Synthesis using the oligo (dT) primer system (Invitrogen) according to the manufacturer’s protocol. Primers and probes for IL-8, CXCR1, CXCR2, and EGFR were purchased from Applied Biosystems (assay ID: Hs00174103_m1, Hs00174146_m1, Hs00174304_m1, and Hs01076078_m1, respectively). For the quantitative analysis, the TBP gene was used as an internal reference gene to normalize input cDNA. We then standardized the expression levels of the IL-8, CXCR1, CXCR2, and EGFR genes by dividing the value for a tumor cell line by the mean of values obtained in noncancerous bronchial epithelial cell lines (N = 3) for the analysis of cell lines (the mean of the three non-cancerous lines = 1) or by dividing the value for a tumor specimen by the mean of values obtained in normal lung tissues (N = 5) for the analysis of tumor specimens (the mean of the five normal lung tissues = 1). PCR was performed using the Gene Amp 7700 Sequence Detection System and software (Applied Biosystems). The comparative Ct method was used to compute relative expression values.
Western blot analysis
Western blotting was performed using whole cell lysates, separated on SDS/polyacrylamide gel, and electroblotted to nitrocellulose membranes (Schleicher & Schuell, Keene, NH).25 The membranes were incubated with mouse monoclonal anti-KRAS (Santa Cruz, Santa Cruz, CA), mouse monoclonal anti-α-tubulin (Calbiochem, San Diego, CA), rabbit polyclonal anti-extracellular signal-regulated kinase (ERK) 1/2 (Cell Signaling), and rabbit polyclonal anti-phospho-ERK1/2 antibodies (Cell Signaling). The membranes were developed with peroxidase-labeled anti-mouse or anti-rabbit IgG (Amersham Pharmacia, Piscataway, NJ) by Super Signal chemiluminescence substrate (Pierce, Rockford, IL).
Enzyme-linked immunosorbent assay
The concentration of IL-8 in the cultured medium was determined by enzyme-linked immunosorbent assay (ELISA) using the human IL-8 duo set kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s protocol.
MTT and IL-8 neutralization assays
Cell viability was measured using the 3-(4,5 dimethylthiazol-2yl)-2,5-diphenyl-tetrazolium bromide (MTT) Cell Growth Assay Kit (Chemicon International, Temecula, CA).27 Briefly, 48 hr after siRNA transfection, trypan blue-negative viable cells (2,000 cells for H157, H460 and H1792; 4,000 cells for H2122) were re-plated and cultured in 96-well plates in replicates of eight. After 72 hr, the MTT assay was performed as previously described.27 For the IL-8 neutralizing assay, cells were treated with the anti-human IL-8 antibody (R&D Systems) or the IgG1 isotype control antibody (R&D Systems), and the MTT assay was performed 72 hr later.
Colony formation assay
In-vitro growth characteristics were tested by a colony formation assay.25 Briefly, after 48 hr of siRNA transfection, cells were harvested, and 500 trypan blue-negative viable cells were replated in each well of six-well plates. The cells were cultured in RPMI 1640 supplemented with 5% serum, and surviving colonies were counted 10 days later after staining with methylene blue.
Migration assay
Cell migration was measured in a modified Boyden chamber.27 Briefly, polycarbonate filters with 8 μm pores were coated with 100 μg/ml of collagen in 0.5 M acetic acid for 16 hr. The coated filter was then placed on a 12-blind-well chemotaxis chamber, and 105 cells in 100 μl per well were loaded into the upper wells. The cells were incubated for 15 min before loading. After incubation at 37°C in 5% CO2 for 4 hr, the filter was disassembled. The upper side of the filter was then scraped free of cells. The cells on the lower side of the filter were fixed with methanol and stained with a Diff-Quick staining kit (International Reagent, Kobe, Japan). The number of cells that migrated to the lower side of the filter was counted. The number of migrated cells in each experiment was adjusted by the cell viability assay (trypan-blue assay) to correct for anti-proliferation effects of the IL-8 neutralizing antibody (corrected migrated cell number = counted migrated cell number/percentage of viable cells).28
Statistical analysis
Statistical analyses were performed using version 5.0 of GraphPad Prism for Windows (GraphPad Software, San Diego, CA). p < 0.05 was considered significant, and p ≥ 0.05 but p < 0.1 was considered borderline significant.
Results
IL-8 overexpression is induced by activating mutations of KRAS or EGFR in NSCLC cells
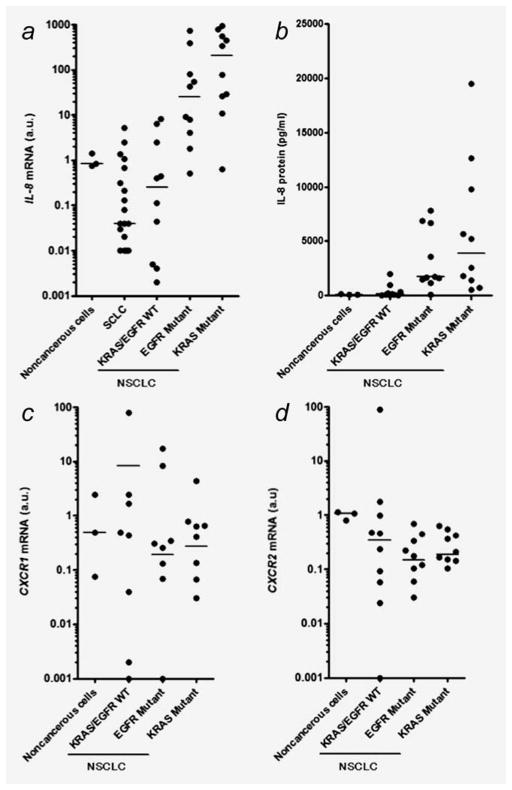
To assess the association between IL-8 expression and KRAS mutations in lung cancer, we first examined IL-8 mRNA expression in a panel of lung cancer cell lines by quantitative RT-PCR analysis. IL-8 mRNA levels were significantly higher in NSCLCs harboring KRAS mutation or EGFR mutations than those with wild-type KRAS/EGFR (Fig. 1a). In contrast, IL-8 expression was low or undetectable in SCLCs, in agreement with a previous study.29 We next examined IL-8 protein levels in cultured NSCLC cell lines. IL-8 protein levels were significantly higher in KRAS mutants or EGFR mutants than those with wild-type KRAS/EGFR (Fig. 1b). Quantitatively, differences in IL-8 levels were more prominent in KRAS mutants than EGFR mutants compared to NSCLCs with wild-type KRAS/EGFR. Notably, we were able to confirm the IL-8 overexpression in NCI-H1792 cells with a baseline protein level of 5,662 pg/ml, and microarray analysis revealed transcriptional downregulation of IL-8 expression (fold-change was −17.4) by shRNA-mediated KRAS knockdown.21 We also examined the expression of IL-8 receptors, CXCR1 and CXCR2, to determine whether these receptors are also differentially expressed in NSCLC cell lines according to the mutation status of EGFR and KRAS. There were no significant differences in the CXCR1 and CXCR2 mRNA expression levels between NSCLCs with or without these mutations (Figs. 1c and 1d).
Figure 1.
(a) Expression of IL-8 mRNA in human bronchial epithelial cell lines (noncancerous cells; N = 3), SCLC cell lines (N = 22), NSCLC cell lines with wild-type KRAS/EGFR (KRAS/EGFR WT; N = 10), NSCLC cell lines harboring EGFR mutations (EGFR mutant; N = 10) or KRAS mutations (KRAS mutant; N = 10). IL-8 levels were significantly higher in KRAS mutant (p < 0.01) or EGFR mutant (p < 0.05) than in KRAS/EGFR WT. (b) IL-8 protein concentration in cultured lung cancer cell lines as determined by ELISA assay. Twenty-four hours after plating 106 cells on 100 mm dishes, culture medium was replaced with 10 ml of RPMI 1640 medium with 5% serum. After cells were cultured for additional 48 hr, medium was collected, and an ELISA assay was performed. IL-8 levels were significantly higher in EGFR Mutant (p < 0.05) or KRAS Mutant (p < 0.01) NSCLCs than in KRAS/EGFR WT. Expressions of (c) CXCR1 and (d) CXCR2 mRNA in the noncancerous and NSCLC cell lines. The arbitrary units (a.u.) of the expressions of IL-8, CXCR1, and CXCR2 were calculated as described in Materials and Methods. Points represent the mean IL-8 levels from three independent experiments. Lines represent the median levels in each group. Differences were analyzed by Kruskal–Wallis test with Dunn’s multiple comparisons.
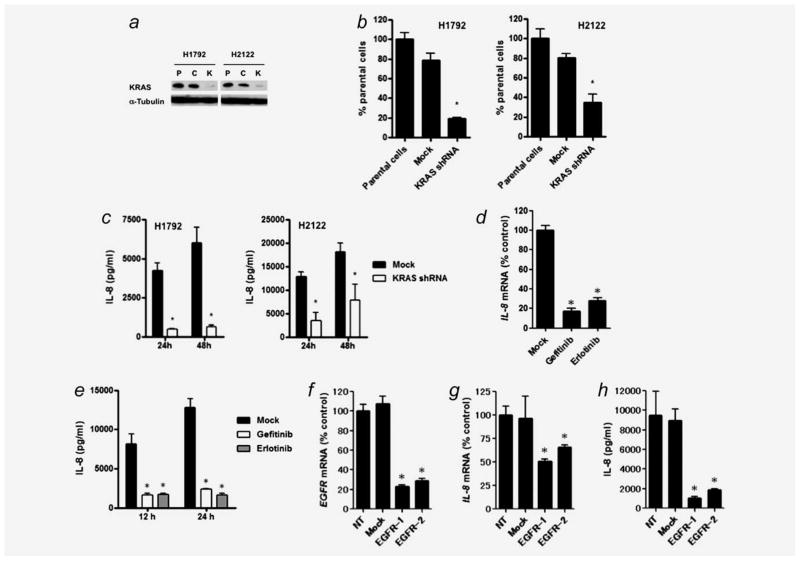
NCI-H2122 NSCLC cells exhibited the highest level of IL-8 protein among all lines (19,477 pg/ml) and had the same type of KRAS mutation at codon 12 (G12C) as H1792 cells. Therefore, we established KRAS-disrupted H2122 clones and KRAS-disrupted H1792 clones using the pRS-KRAS-C12 vector (Fig. 2a) and assessed whether shRNA mediated KRAS knockdown downregulates IL-8 expression in H1792 and H2122 cell lines. In both lines, mutant KRAS knockdown resulted in significant reduction of IL-8 expression at both mRNA and protein levels (Figs. 2b and 2c).
Figure 2.
(a) Stable knockdown of mutant KRAS expression by retroviral-mediated shRNA in H1792 and H2122 cells, both of which harbor the KRAS G12C mutation (GGT to TGT) in codon 12. P: parental cells, C: pRS (non-targeting shRNA) vector-infected cells, K: pRS-KRAS-C12-infected cells. Thirty micrograms of whole cell lysate was loaded per lane, and western blots were performed using α-Tubulin expression levels as a loading control. (b) shRNA-mediated KRAS knockdown reduces IL-8 mRNA expression in H1792 and H2122 cells as determined by quantitative real-time RT-PCR. *p < 0.01 for comparison between KRAS knockdown cells and the parental cells by Kruskal–Wallis test with Dunn’s multiple comparisons. Mean levels of the parental cells are set at 100%. (c) shRNA-mediated KRAS knockdown reduced IL-8 protein levels in cultured medium of H1792 and H2122 cells as determined by ELISA assay. *p < 0.001 for comparisons between KRAS knockdown cells and the parental cells by ANOVA with Bonferroni multiple comparisons. Column represents mean ± SD from four independent experiments. (d) Treatment with gefitinib or erlotinib transcription ally down-regulated IL-8 expression in HCC827 NSCLC cells harboring EGFR mutations. After treatment with gefitinib (1 μM) or erlotinib (1 μM) for 24 hr, cells were harvested for quantitative real-time RT-PCR analysis. Columns represent means ± SD from four independent experiments. *p < 0.001 for comparison of Mock treatment (DMSO alone) by ANOVA with Bonferroni multiple comparisons. (e) Treatment with gefitinib or erlotinib reduced IL-8 protein levels in cultured medium of HCC827 cells. After treatment with gefitinib (1 μM) or erlotinib (1 μM) for 12 or 24 hr, cultured medium was collected, and IL-8 concentration was determined by ELISA assay. Columns represent means ± SD from three independent experiments. *p < 0.001 for comparison of Mock treatment (DMSO alone) by ANOVA with Bonferroni multiple comparisons. (f) siRNA-mediated EGFR knockdown in HCC827 cells as evaluated by quantitative RT-PCR. NT: treatment with medium alone; Mock: treatment with Tax siRNA. Two siRNAs targeting different sites of EGFR mRNA (EGFR-1 and EGFR-2) were used. Columns represent the mean ± SD from four independent experiments. *p < 0.0001 for comparisons between NT and each siRNA treatment by ANOVA with Bonferroni multiple comparisons. shRNA-mediated EGFR knockdown reduced IL-8 expression at (g) mRNA and (h) protein levels in HCC827 cells as evaluated by quantitative real-time RT-PCR and ELISA assays. Twenty-four after siRNA transfection, the medium was replaced by fresh medium. After 48 hr, cells and supernatants were harvested for the assays. *p < 0.0001 for comparisons between NT and each siRNA treatment by ANOVA with Bonferroni multiple comparisons. Mean levels of NT are set at 100% for Fig. 2F and 2G.
We also assessed whether attenuation of oncogenic EGFR activity by siRNA-mediated EGFR knockdown or by treatment with EGFR-TKIs (gefitinib and erlotinib) downregulates IL-8 expression in HCC827 cells, which exhibit the highest basal IL-8 protein levels (7,791 pg/ml) among all EGFR mutant NSCLCs. The treatments with EGFR-TKIs or EGFR siR-NAs resulted in significant reduction of IL-8 expression at both mRNA and protein levels (Figs. 2d–2h), suggesting that activating EGFR mutations transcriptionally upregulate IL-8 expression.
Association of IL-8 expression with clinicopathological parameters and mutation status of KRAS or EGFR in NSCLC
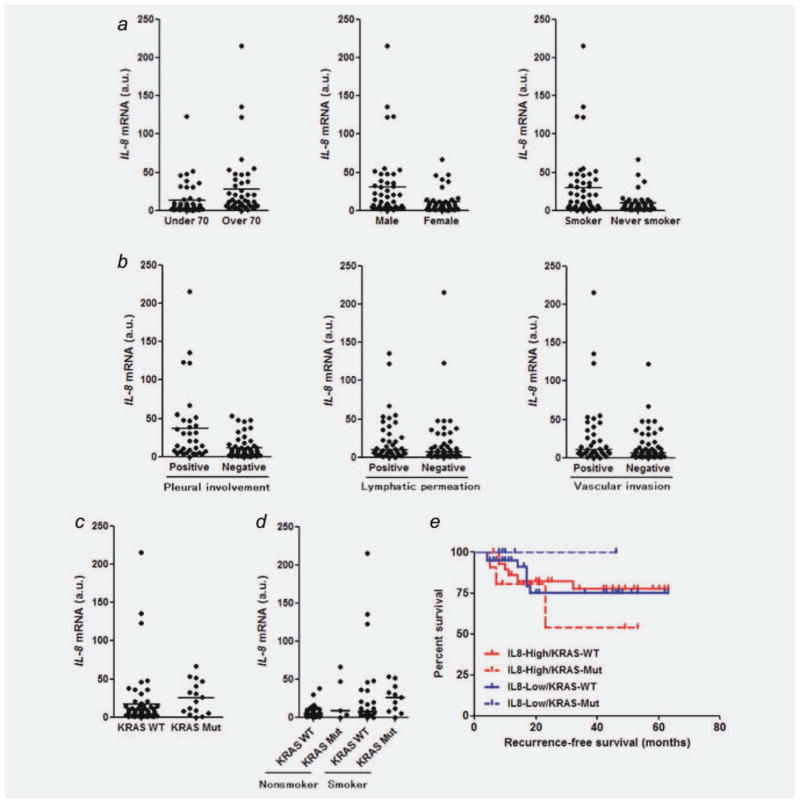
To evaluate the association between IL-8 expression and clinicopathological parameters of NSCLC, IL-8 mRNA expression was examined in surgical specimens of 89 NSCLC patients by quantitative RT-PCR. IL-8 expression was significantly higher in the specimens from males, smokers and elderly patients (Fig. 3a), although IL-8 expression was elevated in subsets of patients from each clinicopathological group. In contrast, there were no significant differences in IL-8 expression levels according to histological subtypes and pathological stages (data not shown). IL-8 expression was significantly higher in NSCLC specimens with pleural involvement and was higher in those with lymphatic permeation or vascular involvement, both with borderline significance (Fig. 3b). We also evaluated the association between IL-8 expression and mutation status of KRAS or EGFR in a subgroup of lung adenocarcinomas to avoid histological bias since mutations of KRAS or EGFR are observed in adenocarcinomas but rarely in squamous cell lung carcinomas.30,31 In fact, all KRAS and EGFR mutations were found in adenocarcinomas but not in squamous cell carcinomas in our study. IL-8 expression was significantly higher in lung adenocarcinomas with KRAS mutations than those with wild-type KRAS (Fig. 3c), whereas we did not observe higher IL-8 expression in adenocarcinomas with EGFR mutations than those with wild-type EGFR (data not shown). Furthermore, we divided the adenocarcinoma population into four groups according to KRAS mutation status and smoking history and then compared the IL-8 expression levels to avoid smoking-related bias since the possible association between KRAS mutations and smoking status in lung adenocarcinoma has been indicated.32 In the adenocarcinoma population, there was a significant difference in the IL-8 expression levels among these four groups (Fig. 3d). Thus, IL-8 appears to be preferentially overexpressed in lung adenocarcinomas with KRAS mutations compared to those with EGFR mutations. In addition, several lung adenocarcinomas with wild-type KRAS/EGFR had elevated levels of IL-8 expression, possibly through other mechanisms.
Figure 3.
(a) Comparisons of IL-8 mRNA expression levels between patients with NSCLC under the age of 70 and those over 70 years old (p = 0.0016), males and females (p = 0.0056), smokers and nonsmokers (p = 0.007); (b) NSCLC specimens with or without pleural involvement (p = 0.0003), NSCLC specimens with or without lymphatic permeation (p = 0.0871), NSCLC specimens with or without vascular invasion (p = 0.054); (c) adenocarcinoma specimens with or without KRAS mutations (p = 0.0355). IL-8 expression levels in NSCLC surgical specimens were normalized to the mean (= 1 a.u.) of values obtained from five different noncancerous lung tissues. Points represent the mean IL-8 levels obtained from three independent experiments. Lines represent median IL-8 levels in each group. Differences were analyzed by Mann–Whitney test. (d) Comparison of IL-8 mRNA expression among the four groups of KRAS wild-type/nonsmoker, KRAS mutant/nonsmoker, KRAS wild-type/smoker and KRAS mutant/smoker (the median levels of these groups were 5.9, 9.2, 8.7, and 26.3, respectively; p = 0.0279 by Kruskal–Wallis test). (e) Kaplan–Meier analysis of disease-free survival (month) for NSCLC patients who were divided according to the IL-8 expression levels and KRAS mutation status. KRAS-WT: KRAS wild-type; KRAS-Mut: KRAS mutation; IL8-High: ≤7.5 a.u.; IL8-Low: >7.5 a.u. Note that the median IL-8 level of all NSCLC tumor specimens was 7.5 a.u.
To evaluate the prognostic significance of IL-8 expression and KRAS mutation status in NSCLC, we divided patients with NSCLC into four groups according to the IL-8 expression level and KRAS mutation status and compared disease-free survival among these groups. There was a tendency of poor survival in patients with NSCLC with IL-8 high/KRAS mutant versus IL-8 low/KRAS mutant (Fig. 3e), although the patient numbers are too small to draw any firm conclusions. Overall, IL-8 overexpression appears to be associated with unfavorable outcome for patients with NSCLC harboring KRAS mutations.
Effects of the MEK and p38 inhibitors on IL-8 expression in NSCLC cells with mutations of KRAS or EGFR
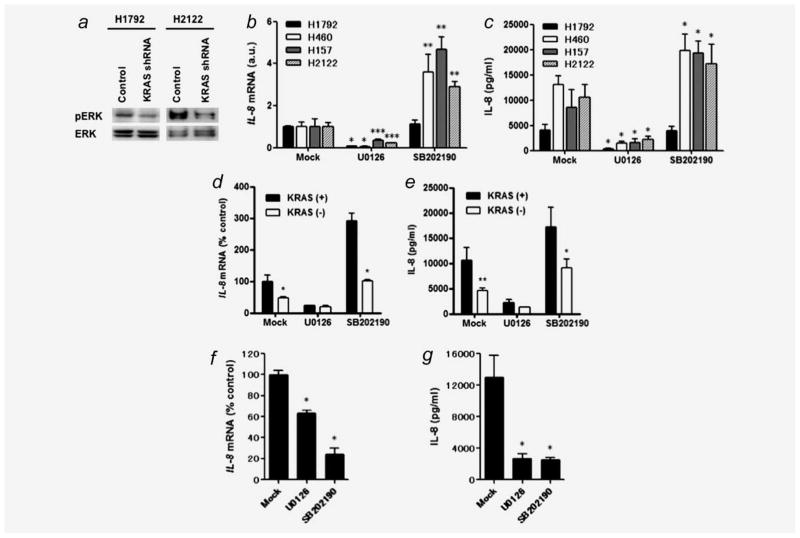
A previous study demonstrated that IL-8 is transcriptionally upregulated through the ERK-MAPK pathway activation.20 In the current study, we found that KRAS knockdown was accompanied with ERK dephosphorylation in H1792 and H2122 cells (Fig. 4a), suggesting that oncogenic KRAS induces ERK activation in KRAS mutant NSCLC cells. To further assess whether IL-8 upregulation is involved in ERK-MAPK pathway activation, we examined the effect of the MEK inhibitor U0126 on IL-8 expression in KRAS mutant NSCLC cell lines H1792, H157, H460, and H2122, in which IL-8 was most highly expressed at basal levels of 5,662, 9,772, 12,663, and 19,477 pg/ml, respectively. In all cases, U0126 markedly downregulated IL-8 expression at both mRNA and protein levels (Figs. 4b and 4c). These lines were also treated with the p38 inhibitor SB202190 because p38 was shown as an upstream mediator of IL-8 transcription.33–35 Unexpectedly, the p38 inhibitor upregulated IL-8 expression in three NSCLC lines (H460, H157, and H2122) while having no effect on H1792 cells (Figs. 4b and 4c). Furthermore, in studies of H2122 cells with shRNA-mediated KRAS knockdown, we found that U0126 still was able to downregulate IL-8 mRNA and protein expression, while KRAS knockdown blocked SB202190-mediated IL-8 upregulation (Figs. 4d and 4e). We also examined the effect of U0126 and SB202190 on IL-8 expression in the EGFR mutant HCC827 cells. Unlike the case of KRAS mutant NSCLC cells, both inhibitors significantly reduced IL-8 expression at mRNA and protein levels in HCC827 cells (Figs. 4f and 4g).
Figure 4.
(a) shRNA-mediated KRAS knockdown reduces phosphorylated ERK levels in H1792 and H2122 cells. Thirty microgram of whole cell lysate was loaded per lane, and western blots were performed using total ERK expression levels as a loading control. (b) Effects of U0126 and SB202190 on IL-8 mRNA expression in KRAS mutant/IL-8 overexpressing NSCLC cell lines. Twenty-four hour after 5 × 105 cells were plated in each well of 6-well plates, cultured medium was replaced with 2 ml of the growth medium with U0126 (10 μM) or SB202190 (10 μM). After additional culture for 24 hr, cells were harvested for real-time RT-PCR analysis. Columns represent means ± SD from four independent experiments. *p < 0.01, **p < 0.001, ***p < 0.05 for comparisons of the treatment with DMSO alone (Mock) to the treatment with U0126 or SB202190 on each cell line by ANOVA with multiple comparisons. (c) Effects of U0126 or SB202190 on IL-8 protein expression from KRAS mutant/IL-8 overexpressing NSCLC cells. After treatments with U0126 or SB202190 for 24 h, cultured medium was collected, and IL-8 concentration was determined by ELISA. Columns represent means ± SD from four independent experiments. *p < 0.001 for comparisons of the treatment with DMSO alone (Mock) to the treatment with U0126 or SB202190 on each cell line by ANOVA with Bonferroni multiple comparisons. Effects of KRAS knockdown on IL-8 expression at (d) mRNA and (e) protein levels in H2122 cells treated with U0126 or SB202190. IL-8 levels were determined at 24 hr posttreatment. Columns represent means ± SD from four independent experiments. *p < 0.001, **p < 0.01 for comparison between mutant KRAS-disrupted cells (white square) and mutant KRAS-expressing (pRS vector-infected) cells (black square) on each treatment by ANOVA with Bonferroni multiple comparisons. Effects of U0126 and SB202190 on IL-8 expression at (f) mRNA and (g) protein levels in HCC827 cells. The experimental procedures and conditions were the same as Figures 4b and 4c. *p < 0.001 for comparisons of the treatment with DMSO alone (Mock) to the treatment with U0126 or SB202190 by ANOVA with Bonferroni multiple comparisons.
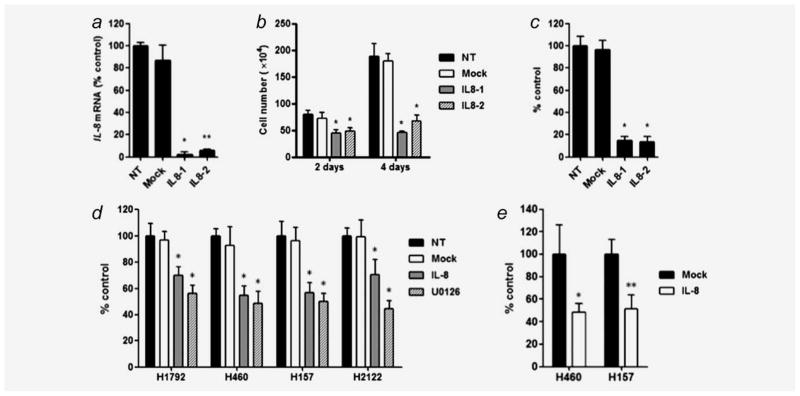
IL-8 siRNAs or an IL-8 neutralizing antibody abolishes in vitro cell growth and migration of KRAS mutant NSCLC cells
To assess whether attenuation of IL-8 function leads to growth inhibition of KRAS mutant NSCLC cells, we examined the effect of siRNA mediated IL-8 knockdown on in vitro cell growth of H1792 cells. IL-8 siRNAs silenced IL-8 expression compared to the untreated H1792 cells (Fig. 5a). Under this condition, IL-8 siRNAs (but not mock siRNA) significantly inhibited cell proliferation (Fig. 5b) and colony formation (Fig. 5c) in H1792 cells, suggesting that siRNA-mediated IL-8 knockdown abolishes in vitro cell growth of KRAS mutant NSCLC cells.
Figure 5.
(a) siRNA mediated knockdown of IL-8 expression in H1792 cells as evaluated by quantitative RT-PCR. NT: treatment with medium alone; Mock: treatment with Tax siRNA as a negative control. Two siRNAs targeting different sites of IL-8 mRNA (IL8-1 and IL8-2) were used to knockdown IL-8. Columns represent the mean ± SD from four independent experiments. *p < 0.001, **p < 0.01 for comparisons between NT and each siRNA treatment by Kruskal–Wallis test with Dunn’s multiple comparisons. siRNA-mediated IL-8 knockdown inhibited cell proliferation and colony formation as evaluated by (b) cell growth analysis using trypan-blue staining and (c) colony formation assay in H1792 cells. Columns represent the mean ± SD from six independent experiments for cell growth analysis and three independent experiments for colony formation assay. *p < 0.001 for comparison between NT and each siRNA treatment by ANOVA with Bonferroni multiple comparisons. (d) IL-8 neutralization mediated by an IL-8 antibody inhibits cell proliferation of KRAS mutant/IL-8 overexpressing NSCLC cell lines as evaluated by MTT assay. Cells were treated with medium alone (NT), IgG1 control antibody (Mock; 10 μg/ml), IL-8 antibody (IL-8; 10 μg/ml) and U0126 (10 μM) for 72 hr. *p < 0.001 for comparison between NT and each treatment by ANOVA with Bonferroni multiple comparisons. Columns represent the mean ± SD from replicates of eight from two independent experiments (e) IL-8 neutralization inhibits cell migration of H460 and H157 cells. *p < 0.01; **p < 0.05 for comparison between Mock and IL-8 antibody treatments by unpaired t-test. Columns represent means ± SD from three independent experiments.
To confirm that the growth-inhibitory effect of IL-8 attenuation is not specific for H1792 but is commonly observed in other KRAS mutant/IL-8 overexpressing NSCLC cells, we examined the effect of an IL-8 neutralizing antibody on in vitro cell growth in four NSCLC lines with KRAS mutations and IL-8 overexpression (H1792, H157, H460, and H2122). In all cases, IL-8 neutralization resulted in a modest but significant inhibition of cell growth (Fig. 5d). In addition, we confirmed that the growth-inhibitory effects of IL-8 neutralization were similar to that of U0126 (Fig. 5d), which markedly downregulated IL-8 expression in these lines (Figs. 4b and 4c). We also assessed the effect of IL-8 neutralization on cell migration of H460 and H157 cell lines, which were assessable by the migration assay. IL-8 neutralization resulted in a significant reduction of migrated cells compared to those with the control antibody (Fig. 5e). Overall, these results suggest that attenuation of IL-8 function abolishes in vitro cell growth and migration of KRAS mutant NSCLC cells.
Discussion
KRAS and EGFR mutations are currently recognized as important molecular abnormalities in NSCLC because of their clinical implications in EGFR-TKI therapy. EGFR mutations are associated with a favorable response to EGFR-TKIs,36–39 while KRAS mutations are associated with resistance to EGFR-TKIs.6,40 Although several studies have reported IL-8 overexpression in NSCLC,12,16,17,19,29 the association between IL-8 expression and mutations of KRAS or EGFR remains unknown. In this study, using a panel of lung cancer cell lines and NSCLC specimens, we found that IL-8 was abundantly expressed in NSCLCs with KRAS mutations and a subset of NSCLCs with EGFR mutations. IL-8 overexpression was downregulated by shRNA-mediated KRAS knockdown in KRAS mutant NSCLC cells, consistent with the previous findings that ectopic expression of mutant KRAS resulted in IL-8 upregulation in human cells.20,41 Our findings demonstrate that activating KRAS mutations induce IL-8 overexpression in NSCLC, highlighting important roles of IL-8 in the development of NSCLC with KRAS mutations. Furthermore, we found for the first time that a subset of EGFR mutant NSCLC cell lines and tumor specimens (which all occurred in never smokers) showed elevated IL-8 expression and EGFR-TKIs or EGFR siRNAs downregulated IL-8 expression in EGFR mutant cells, suggesting that activating EGFR mutation also induces IL-8 overexpression in NSCLC. In contrast, we could not observe significantly higher IL-8 expression in EGFR mutant tumor specimens compared to those with wild-type EGFR. Thus, activating EGFR mutations are unlikely to be common mechanisms of IL-8 overexpression in NSCLC.
IL-8 expression analysis in NSCLC tumor specimens revealed that some NSCLCs with wild-type KRAS/EGFR also overexpressed IL-8, suggesting that there are other mechanisms of IL-8 upregulation. Previous studies have reported that cigarette smoking leads to IL-8 upregulation by inducing IL-8 release from bronchial epithelial cells42–44 and that IL-8 is differentially expressed in bronchial epithelial cells in smokers having lung cancer compared to smokers without lung cancer.45 In our study, IL-8 expression was significantly higher in NSCLC tumors from smokers than in those from nonsmokers. Of note, in a subgroup of NSCLCs with wild-type KRAS/EGFR, IL-8 expression was significantly higher in the tumors derived from smokers (p = 0.0143), whereas in a subgroup of KRAS mutant NSCLCs, no significant difference was observed between smokers and nonsmokers. Therefore, it is possible that IL-8 plays a role in tobacco-related carcinogenesis of NSCLC, which may partially explain the mechanisms of KRAS/EGFR independent IL-8 overexpression.
The ERK-MAPK pathway plays a central role in oncogenic KRAS-driven malignant phenotypes of NSCLC.2,21 We found that shRNA-mediated KRAS knockdown was accompanied by ERK dephosphorylation and that the MEK inhibitor completely blocked IL-8 expression in KRAS mutant NSCLC cells. These results are in agreement with a previous study showing that IL-8 is transcriptionally upregulated through the ERK-MAPK pathway activation.20 Our findings that MEK inhibitor-mediated IL-8 downregulation was unaffected by KRAS knockdown suggest that oncogenic KRAS-induced IL-8 overexpression is highly responsible for ERK-MAPK pathway activation in NSCLC. Interestingly, treatment with the p38 inhibitor resulted in IL-8 upregulation, which was inhibited by KRAS knockdown. This finding suggests that the negative feedback from p38 to ERK is present in NSCLC as observed in other types of cancers46 and that oncogenic KRAS upregulates this negative feedback.
We found that both of the U0126 and p38 inhibitors downregulated IL-8 expression in EGFR mutant HCC827 cells. A recent study indicated that EGFR related IL-8 production from NSCLC cells is stimulated through the ERK activation.47 Since HCC827 cells highly express phosphorylated-ERK and EGFR-TKIs are able to inhibit the ERK phosphorylation in this cell line,48,49 it is likely that oncogenic EGFR mutations upregulate IL-8 expression in NSCLC cells, at least in part, through the ERK-MAPK pathway activation. In addition, unlike the cases of KRAS mutant NSCLC cells, the p38-MAPK pathway activation may also be one of the mechanisms of IL-8 upregulation in EGFR mutant NSCLC cells.
Despite many studies indicating an essential role for, and therapeutic significance of, oncogenic KRAS in NSCLC, no effective strategies for the treatment of NSCLC harboring KRAS mutations have been established. Recently, the VEGF monoclonal antibody bevacizumab has been approved and several other antiangiogenic agents are being tested for treatment of NSCLC patients50; thus, tumor-related angiogenesis has become an attractive therapeutic target for NSCLC. Given the fact that IL-8 functions as an angiogenic factor and IL-8 neutralization suppresses in vivo tumor growth and angiogenesis of NSCLC,14,15 our findings of oncogenic KRAS-induced IL-8 overexpression raise the possibility of anti-IL-8 therapy for KRAS mutant NSCLC. Also, we found that IL-8 siRNAs or the IL-8 neutralizing antibody inhibited cell proliferation and migration of KRAS mutant NSCLC cells, consistent with previous studies demonstrating that IL-8 neutralization inhibited cell proliferation and migration in NSCLC cell lines including H460 cells.12,14 These findings strongly suggest that IL-8 could be a therapeutic target for KRAS mutant NSCLC.
In previous studies, IL-8 expression was associated with angiogenesis, lymph node metastasis, and unfavorable outcome in patients with NSCLC.16–19 We found that IL-8 was overexpressed in NSCLC tumors with pleural involvement, lymphatic permeation and vascular invasion, suggesting that IL-8 overexpression potentially contributes to the aggressive phenotypes of NSCLC. Furthermore, in KRAS mutant NSCLC patients, disease-free survival of patients with higher IL-8 expression tended to be shorter compared to those with lower IL-8 expression. Because patient numbers were small in this study, further investigation with a larger number of patients with NSCLC will likely elucidate the prognostic significance of IL-8 expression in KRAS mutant NSCLC.
In conclusion, the present study demonstrates the positive association between KRAS mutations, IL-8 overexpression, and certain clinicopathological features in NSCLC. Further in vivo studies will be needed to evaluate the effectiveness of an anti-IL-8 treatment strategy for KRAS mutant NSCLC. The findings of IL-8 overexpression in a subset of NSCLCs with EGFR mutations or wild-type KRAS/EGFR also suggest that IL-8 could be upregulated in some of these lung cancers as well; thus, mechanisms of IL-8 upregulation irrelevant to KRAS mutations should be elucidated.
Acknowledgments
Grant sponsor: SPORE; Grant number: P50CA70907; Grant sponsor: DOD PROSPECT, Texas Higher Education Coordinating Board Advanced Technology Program; Grant number: 01001901392003; Grant sponsor: NASA Specialized Center of Research; Grant number: NNJ05HD36G; Grant sponsor: Ministry of Education, Culture, Sports, Science and Technology, Japan; Grant number: 18790532; Grant sponsor: Gillson Longenbaugh Foundation
DSS was supported by IASLC post-doctoral fellowship.
References
- 1.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 2.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–343. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 3.Buttitta F, Barassi F, Fresu G, Felicioni L, Chella A, Paolizzi D, Lattanzio G, Salvatore S, Camplese PP, Rosini S, Iarussi T, Mucilli F, et al. Mutational analysis of the HER2 gene in lung tumors from Caucasian patients: mutations are mainly present in adenocarcinomas with bronchioloalveolar features. Int J Cancer. 2006;119:2586–2591. doi: 10.1002/ijc.22143. [DOI] [PubMed] [Google Scholar]
- 4.Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert AP, Noel S, Lafitte JJ, Sculier JP. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isobe T, Herbst RS, Onn A. Current management of advanced non-small cell lung cancer: targeted therapy. Semin Oncol. 2005;32:315–328. doi: 10.1053/j.seminoncol.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 8.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 9.Baggiolini M, Walz A, Kunkel SL. Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J Clin Invest. 1989;84:1045–1049. doi: 10.1172/JCI114265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14:6735–6741. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 11.Luppi F, Longo AM, de Boer WI, Rabe KF, Hiemstra PS. Interleukin-8 stimulates cell proliferation in non-small cell lung cancer through epidermal growth factor receptor transactivation. Lung Cancer. 2007;56:25–33. doi: 10.1016/j.lungcan.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 12.Zhu YM, Webster SJ, Flower D, Woll PJ. Interleukin-8/CXCL8 is a growth factor for human lung cancer cells. Br J Cancer. 2004;91:1970–1976. doi: 10.1038/sj.bjc.6602227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao PL, Lin YC, Wang CH, Huang YC, Liao WY, Wang SS, Chen JJ, Yang PC. Autocrine and paracrine regulation of interleukin-8 expression in lung cancer cells. Am J Respir Cell Mol Biol. 2005;32:540–547. doi: 10.1165/rcmb.2004-0223OC. [DOI] [PubMed] [Google Scholar]
- 14.Smith DR, Polverini PJ, Kunkel SL, Orringer MB, Whyte RI, Burdick MD, Wilke CA, Strieter RM. Inhibition of interleukin 8 attenuates angiogenesis in bronchogenic carcinoma. J Exp Med. 1994;179:1409–1415. doi: 10.1084/jem.179.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arenberg DA, Kunkel SL, Polverini PJ, Glass M, Burdick MD, Strieter RM. Inhibition of interleukin-8 reduces tumorigenesis of human non-small cell lung cancer in SCID mice. J Clin Invest. 1996;97:2792–2802. doi: 10.1172/JCI118734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan A, Yang PC, Yu CJ, Chen WJ, Lin FY, Kuo SH, Luh KT. Interleukin-8 messenger ribonucleic acid expression correlates with tumor progression, tumor angiogenesis, patient survival, and timing of relapse in non-small-cell lung cancer. Am J Respir Crit Care Med. 2000;162:1957–1963. doi: 10.1164/ajrccm.162.5.2002108. [DOI] [PubMed] [Google Scholar]
- 17.Masuya D, Huang C, Liu D, Kameyama K, Hayashi E, Yamauchi A, Kobayashi S, Haba R, Yokomise H. The intratumoral expression of vascular endothelial growth factor and interleukin-8 associated with angiogenesis in nonsmall cell lung carcinoma patients. Cancer. 2001;92:2628–2638. doi: 10.1002/1097-0142(20011115)92:10<2628::aid-cncr1616>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 18.Seike M, Yanaihara N, Bowman ED, Zanetti KA, Budhu A, Kumamoto K, Mechanic LE, Matsumoto S, Yokota J, Shibata T, Sugimura H, Gemma A, et al. Use of a cytokine gene expression signature in lung adenocarcinoma and the surrounding tissue as a prognostic classifier. J Natl Cancer Inst. 2007;99:1257–1269. doi: 10.1093/jnci/djm083. [DOI] [PubMed] [Google Scholar]
- 19.Chen JJ, Yao PL, Yuan A, Hong TM, Shun CT, Kuo ML, Lee YC, Yang PC. Up-regulation of tumor interleukin-8 expression by infiltrating macrophages: its correlation with tumor angiogenesis and patient survival in non-small cell lung cancer. Clin Cancer Res. 2003;9:729–737. [PubMed] [Google Scholar]
- 20.Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell. 2004;6:447–458. doi: 10.1016/j.ccr.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 21.Sunaga N, Shames DS, Girard L, Peyton M, Larsen JE, Imai H, Soh J, Sato M, Yanagitani N, Kaira K, Xie Y, Gazdar AF, et al. Knockdown of oncogenic KRAS in non-small cell lung cancers suppresses tumor growth and sensitizes tumor cells to targeted therapy. Mol Cancer Ther. 2011;10:336–346. doi: 10.1158/1535-7163.MCT-10-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 23.Mitani Y, Lezhava A, Kawai Y, Kikuchi T, Oguchi-Katayama A, Kogo Y, Itoh M, Miyagi T, Takakura H, Hoshi K, Kato C, Arakawa T, et al. Rapid SNP diagnostics using asymmetric isothermal amplification and a new mismatch-suppression technology. Nat Methods. 2007;4:257–262. doi: 10.1038/nmeth1007. [DOI] [PubMed] [Google Scholar]
- 24.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW, Minna JD. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 25.Sunaga N, Miyajima K, Suzuki M, Sato M, White MA, Ramirez RD, Shay JW, Gazdar AF, Minna JD. Different roles for caveolin-1 in the development of non-small cell lung cancer versus small cell lung cancer. Cancer Res. 2004;64:4277–4285. doi: 10.1158/0008-5472.CAN-03-3941. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki M, Sunaga N, Shames DS, Toyooka S, Gazdar AF, Minna JD. RNA interference-mediated knockdown of DNA methyltransferase 1 leads to promoter demethylation and gene re-expression in human lung and breast cancer cells. Cancer Res. 2004;64:3137–3143. doi: 10.1158/0008-5472.can-03-3046. [DOI] [PubMed] [Google Scholar]
- 27.Imai H, Sunaga N, Shimizu Y, Kakegawa S, Shimizu K, Sano T, Ishizuka T, Oyama T, Saito R, Minna JD, Mori M. Clinicopathological and therapeutic significance of CXCL12 expression in lung cancer. Int J Immunopathol Pharmacol. 2010;23:153–164. doi: 10.1177/039463201002300114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu CY, Cha ST, Chang CC, Hsiao CH, Tan CT, Lu YC, Jee SH, Kuo ML. Involvement of matrix metalloproteinase-13 in stromal-cell-derived factor 1 alpha-directed invasion of human basal cell carcinoma cells. Oncogene. 2007;26:2491–2501. doi: 10.1038/sj.onc.1210040. [DOI] [PubMed] [Google Scholar]
- 29.Yatsunami J, Tsuruta N, Ogata K, Wakamatsu K, Takayama K, Kawasaki M, Nakanishi Y, Hara N, Hayashi S. Interleukin-8 participates in angiogenesis in non-small cell, but not small cell carcinoma of the lung. Cancer Lett. 1997;120:101–108. doi: 10.1016/s0304-3835(97)00296-6. [DOI] [PubMed] [Google Scholar]
- 30.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, Roth JA, Albelda SM, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 31.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118:257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 32.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Na YJ, Jeon YJ, Suh JH, Kang JS, Yang KH, Kim HM. Suppression of IL-8 gene expression by radicicol is mediated through the inhibition of ERK1/2 and p38 signaling and negative regulation of NF-kappaB and AP-1. Int Immunopharmacol. 2001;1:1877–1887. doi: 10.1016/s1567-5769(01)00113-8. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Kartha S, Iasvovskaia S, Tan A, Bhat RK, Manaligod JM, Page K, Brasier AR, Hershenson MB. Regulation of human airway epithelial cell IL-8 expression by MAP kinases. Am J Physiol Lung Cell Mol Physiol. 2002;283:L690–L699. doi: 10.1152/ajplung.00060.2002. [DOI] [PubMed] [Google Scholar]
- 35.Estrada Y, Dong J, Ossowski L. Positive crosstalk between ERK and p38 in melanoma stimulates migration and in vivo proliferation. Pigment Cell Melanoma Res. 2009;22:66–76. doi: 10.1111/j.1755-148X.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- 36.Morita S, Okamoto I, Kobayashi K, Yamazaki K, Asahina H, Inoue A, Hagiwara K, Sunaga N, Yanagitani N, Hida T, Yoshida K, Hirashima T, et al. Combined survival analysis of prospective clinical trials of gefitinib for non-small cell lung cancer with EGFR mutations. Clin Cancer Res. 2009;15:4493–4498. doi: 10.1158/1078-0432.CCR-09-0391. [DOI] [PubMed] [Google Scholar]
- 37.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 38.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 39.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 40.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, Bekele BN, Herbst RS, Wistuba II. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 41.Wislez M, Fujimoto N, Izzo JG, Hanna AE, Cody DD, Langley RR, Tang H, Burdick MD, Sato M, Minna JD, Mao L, Wistuba I, et al. High expression of ligands for chemokine receptor CXCR2 in alveolar epithelial neoplasia induced by oncogenic kras. Cancer Res. 2006;66:4198–4207. doi: 10.1158/0008-5472.CAN-05-3842. [DOI] [PubMed] [Google Scholar]
- 42.Chalmers GW, MacLeod KJ, Thomson L, Little SA, McSharry C, Thomson NC. Smoking and airway inflammation in patients with mild asthma. Chest. 2001;120:1917–1922. doi: 10.1378/chest.120.6.1917. [DOI] [PubMed] [Google Scholar]
- 43.Kuschner WG, D’Alessandro A, Wong H, Blanc PD. Dose-dependent cigarette smoking-related inflammatory responses in healthy adults. Eur Respir J. 1996;9:1989–1994. doi: 10.1183/09031936.96.09101989. [DOI] [PubMed] [Google Scholar]
- 44.Mio T, Romberger DJ, Thompson AB, Robbins RA, Heires A, Rennard SI. Cigarette smoke induces interleukin-8 release from human bronchial epithelial cells. Am J Respir Crit Care Med. 1997;155:1770–1776. doi: 10.1164/ajrccm.155.5.9154890. [DOI] [PubMed] [Google Scholar]
- 45.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, Sridhar S, Beamis J, et al. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med. 2007;13:361–366. doi: 10.1038/nm1556. [DOI] [PubMed] [Google Scholar]
- 46.Aguirre-Ghiso JA, Estrada Y, Liu D, Ossowski L. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK) Cancer Res. 2003;63:1684–1695. [PubMed] [Google Scholar]
- 47.Zhang Y, Wang L, Zhang M, Jin M, Wang X. Potential mechanism of interleukin-8 production from lung cancer cells: an involvement of EGF-EGFR-PI3K-Atk-Erk pathway. J Cell Physiol. doi: 10.1002/jcp.22722. in press. [DOI] [PubMed] [Google Scholar]
- 48.Amann J, Kalyankrishna S, Massion PP, Ohm JE, Girard L, Shigematsu H, Peyton M, Juroske D, Huang Y, Stuart Salmon J, Kim YH, Pollack JR, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65:226–235. [PubMed] [Google Scholar]
- 49.Zhang Z, Kobayashi S, Borczuk AC, Leidner RS, Laframboise T, Levine AD, Halmos B. Dual specificity phosphatase 6 (DUSP6) is an ETS-regulated negative feedback mediator of oncogenic ERK signaling in lung cancer cells. Carcinogenesis. 2010;31:577–586. doi: 10.1093/carcin/bgq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horn L, Sandler AB. Angiogenesis in the treatment of non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:206–217. doi: 10.1513/pats.200807-066LC. [DOI] [PMC free article] [PubMed] [Google Scholar]