Dying primary liver, NIH 3T3, and HeLa cells can reverse the advanced stage of apoptosis and survive even after incurring DNA damage. Some surviving cells harbor genetic alterations that result in phenotypic diversity, including oncogenic transformation.
Abstract
Apoptosis serves as a protective mechanism by eliminating damaged cells through programmed cell death. After apoptotic cells pass critical checkpoints, including mitochondrial fragmentation, executioner caspase activation, and DNA damage, it is assumed that cell death inevitably follows. However, this assumption has not been tested directly. Here we report an unexpected reversal of late-stage apoptosis in primary liver and heart cells, macrophages, NIH 3T3 fibroblasts, cervical cancer HeLa cells, and brain cells. After exposure to an inducer of apoptosis, cells exhibited multiple morphological and biochemical hallmarks of late-stage apoptosis, including mitochondrial fragmentation, caspase-3 activation, and DNA damage. Surprisingly, the vast majority of dying cells arrested the apoptotic process and recovered when the inducer was washed away. Of importance, some cells acquired permanent genetic changes and underwent oncogenic transformation at a higher frequency than controls. Global gene expression analysis identified a molecular signature of the reversal process. We propose that reversal of apoptosis is an unanticipated mechanism to rescue cells from crisis and propose to name this mechanism “anastasis” (Greek for “rising to life”). Whereas carcinogenesis represents a harmful side effect, potential benefits of anastasis could include preservation of cells that are difficult to replace and stress-induced genetic diversity.
INTRODUCTION
Apoptosis, or programmed cell death, plays essential roles in development and homeostasis by eliminating unwanted, abnormal, injured, or dangerous cells (Kerr et al., 1972; Jacobson et al., 1997; Fuchs and Steller, 2011). In addition, activating apoptotic pathways is an important strategy for treating intractable diseases such as cancer and autoimmunity, whereas limiting apoptosis may be beneficial for treating liver and heart failure and neurodegenerative diseases (Fischer and Schulze-Osthoff, 2005; Narula et al., 2006; Guicciardi and Gores, 2010; Chabaud and Moulin, 2011).
Apoptosis is generally believed to be irreversible after mitochondrial fragmentation and caspase activation (Green and Kroemer, 2004; Riedl and Shi, 2004; Taylor et al., 2008; Chipuk et al., 2010) because mitochondrial dysfunction alone can lead to cell death (Green and Kroemer, 2004; Luthi and Martin, 2007), and mitochondria release cell-executing factors, including apoptosis-inducing factor (AIF), endonuclease G (EndoG) (Li et al., 2001; Susin et al., 1999), and cytochrome c (Riedl and Shi, 2004). Cytochrome c release activates the caspase cascade, and the active proteases then destroy structural and functional components in cells, resulting in the morphological manifestations of apoptosis, such as nuclear condensation, cell shrinkage, and membrane blebbing (Liu et al., 1997; Enari et al., 1998; Coleman et al., 2001; Luthi and Martin, 2007). After activation of executioner caspases such as caspase-3 and -7, apoptosis can follow within minutes (Takemoto et al., 2003; Riedl and Shi, 2004; Taylor et al., 2008; Chipuk et al., 2010), and so this stage of the process is generally considered to be irreversible, although the precise “point of no return” is controversial (Kroemer et al., 2009).
Here we report an unexpected, intrinsic reversibility of execution-stage apoptosis in multiple cell types, including primary liver and heart cells, the embryonic fibroblast NIH 3T3 cell line, and human cervical cancer HeLa cells, which are important models for studies of oncogenesis and general cellular mechanisms, including apoptotic cell death (Masters, 2002; McKillop and Schrum, 2005; Narula et al., 2006; Rubin, 2008). Removal of apoptotic inducers was sufficient to allow the majority of cells to reverse the apoptotic process and recover. Dying cells exhibited multiple morphological and biochemical hallmarks of apoptosis, including cell shrinkage and blebbing, mitochondrial fragmentation, nuclear condensation, caspase-3 activation, and DNA damage. After washing, however, all these signs reversed, and the vast majority of such cells survived and proliferated. Of note, some of the surviving cells exhibited chromosomal abnormalities and oncogenic transformation, suggesting that they harbor mutations from the apoptotic process. Therefore reversal of apoptosis may represent an important, unrecognized mechanism for cell rescue and mutagenesis.
RESULTS
Dying cells recover from execution stage of apoptosis after removal of apoptotic stimuli
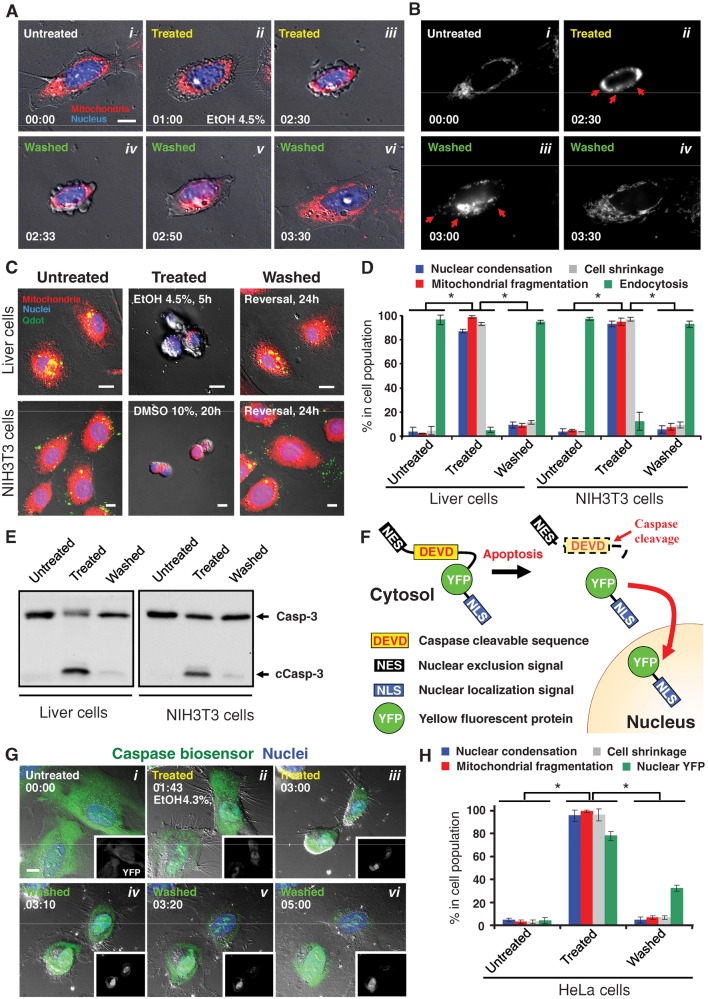
To test directly whether apoptosis is an irreversible process, we exposed cells to an apoptotic inducer until they showed typical hallmarks of apoptosis and then washed the inducer away. Untreated healthy cells spread on the substrate (Figure 1Ai) and contained filamentous mitochondria that extended throughout cytoplasm (Figure 1Bi). After exposure to 4.5% ethanol for 2.5 h, liver cells displayed morphological hallmarks of apoptosis, including cell shrinkage, plasma membrane blebbing, nuclear condensation (Figure 1A, ii and iii), and altered mitochondrial morphology (Figure 1Bii). These are well-recognized signs of effector caspase activation and define the execution stage of apoptosis (Taylor et al., 2008; Kroemer et al., 2009). Of interest, time-lapse live-cell fluorescence microscopy revealed that, upon washing, the same cells that had exhibited the morphological hallmarks of apoptosis recovered normal morphology (Figure 1, A, iv–vi, and B, iii and iv, Supplemental Figure S1, and Supplemental Video S1), ruling out the interpretation that the apparent recovery represented cells that failed to respond to the inducer in the first place.
FIGURE 1:
Reversibility of apoptosis in primary mouse liver, NIH 3T3, and HeLa cells. (A) Time-lapse live-cell fluorescence microscopy of a primary liver cell before, during, and after exposure to ethanol. The same cell before ethanol induction (Untreated, i), induced with 4.5% ethanol in culture medium for 2.5 h (Treated, ii and iii), and then washed and further cultured with fresh medium (Washed, iv–vi). Merged images, mitochondria (red) and nuclei (blue) were visualized by fluorescence and cell morphology by DIC. Time presented as h:min. Scale bar, 10 μm. Also see Supplemental Figure S1 and Supplemental Video S1. (B) Monochrome images of mitochondria from A. Mitochondrial fragmentation is indicated by red arrows. (C) Fluorescence and DIC microscopy of healthy liver and NIH 3T3 cells (Untreated), cells that were exposed to apoptotic inducers (liver cells, 4.5% ethanol for 5 h; NIH 3T3 cells, 10% DMSO for 20 h) (Treated), and treated cells that were washed to remove apoptotic inducers and further cultured for 24 h (Washed). Merged images, mitochondria (red), nuclei (blue), and Quantum Dots (Qdots) taken up by endocytosis (green) were visualized by fluorescence and cell morphology by DIC. Scale bar, 10 μm. (D) Quantification of the apoptotic response and its reversal. Morphological signs of apoptosis included nuclear condensation, mitochondrial fragmentation, and cell shrinkage. Uptake of Quantum Dots by endocytosis is characteristic of healthy cells, whereas the other features are characteristic of apoptotic cells. Apoptosis was induced in liver cells with 4.5% ethanol for 5 h and in NIH 3T3 cells with 10% DMSO for 20 h (Treated). Treated cells were then washed and further cultured for 24 h in standard conditions. *p < 0.01; n = 3 independent experiments. Error bars denote SD. (E) Western blot analysis of the total cell lysate of untreated, treated, and washed liver and NIH 3T3 cells for the protein level of caspase-3 (Casp-3). c, cleaved form. (F) Schematic diagram of the caspase biosensor NES-DEVD-YFP-NLS. (G) Real-time live-cell microscopy of HeLa cells expressing the caspase biosensor before (Untreated, i), during (Treated, ii and iii), and after (Washed, iv-vi) exposure to 4.3% ethanol. Merged images, caspase biosensor (YFP, green) and nuclei (blue) were visualized by confocal microscopy and cell morphology by DIC. Corresponding monochromatic YFP image is shown in each panel. Time presented as h:min. Scale bar, 10 μm. Also see Supplemental Figure S4 and Supplemental Videos S2 and S3. (H) Quantification of the caspase biosensor response in HeLa cells. Treated cells were exposed to 4.3% ethanol for 5 h. After washing, cells were cultured for 2 h in standard conditions. *p< 0.01; n = 3 independent experiments. Error bars denote SD.
The reversal of the signs of apoptosis occurred in the vast majority of both primary liver and NIH 3T3 cells (Figure 1, C and D). After exposure to 4.5% ethanol for 5 h, >80% of liver cells shrank and displayed morphological hallmarks of apoptosis, and >90% of NIH 3T3 cells exhibited multiple hallmarks of apoptosis in response to 10% dimethyl sulfoxide (DMSO) for 20 h (Figure 1D). Strikingly, >90% of both cell types reversed all signs of apoptosis after removal of the inducer for 24 h (Figure 1D). The survival of cells was further assessed by their ability to take up Quantum Dots (Jaiswal et al., 2003) through endocytosis, which was abolished during ethanol induction and recovered following washing (Figure 1, C and D). Moreover, time-lapse microscopy of >30 living cells in a single field showed morphological recovery of the majority of them after removal of the apoptotic stimulus (Supplemental Figure S2). In contrast, virtually all cells died when they were left in the inducer (Supplemental Figure S3).
Cleavage and activation of caspase-3 is a biochemical hallmark of execution-stage apoptosis (Riedl and Shi, 2004; Taylor et al., 2008; Kroemer et al., 2009). Western blotting revealed an increase in cleaved caspase-3 and a decrease in full-length caspase-3 in both liver cells induced with ethanol and NIH 3T3 cells induced with DMSO (Figure 1E). Of importance, after washing and then culturing of the apoptotic cells in fresh medium for 24 h, both full-length and cleaved caspase returned to pretreatment levels (Figure 1E).
Time-lapse live imaging of HeLa cells expressing a caspase biosensor confirmed that dying cells could reverse apoptosis, even after executioner caspase activation (Figure 1, F and G). The biosensor (NES-DEVD-YFP-NLS) is a protein composed of a nuclear exclusion signal (NES), a caspase-cleavable linker peptide (DEVD), which is the preferred cleavage site for caspase-3 (Talanian et al., 1997; Takemoto et al., 2003), yellow fluorescent protein (YFP), and a nuclear localization signal (NLS). In healthy cells, YFP is kept in the cytosol by the NES (Figure 1F). On caspase activation, however the peptide is cleaved and the YFP is free to translocate to nucleus due to the NLS. As expected, the majority of YFP localized in the cytosol at the untreated cells (Figure 1Gi). When exposed to 4.3% ethanol for 3 h, the vast majority of cells exhibited morphological hallmarks of apoptosis, including nuclear condensation, membrane blebbing, and cell shrinkage (Figure 1G, ii and iii, and Supplemental Figure S4). YFP translocated to the nucleus, where it accumulated in many cells, whereas in others it appeared to be degraded. Of interest, after removal of the apoptotic inducer, the same cells regained normal morphology within 2 h (Figure 1G, iv–vi, Supplemental Figure S4, and Supplemental Videos S2 and S3). This indicates that single cells can reverse apoptosis after caspase activation. After removal of the inducer, >90% of cells recovered normal morphology, and 32% of them retained nuclear YFP (Figure 1H). Taken together, our results indicate that cells can reverse apoptosis even after executioner caspase activation.
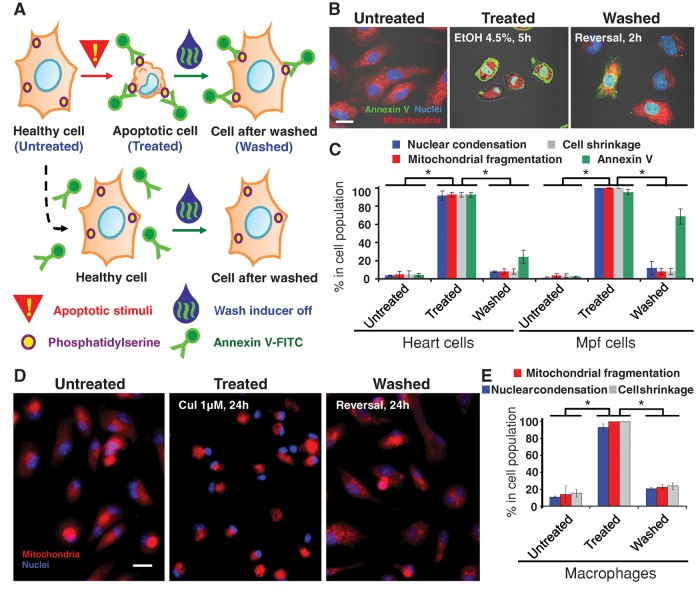
Reversal of apoptosis also occurred in primary rat heart cells exposed to 4.5% ethanol for 5 h, in Mustela putoris furo (Mpf) brain cells (CRL1516) exposed to 2 μM jasplakinolide for 50 h (Figure 2, A–C), and in primary mouse macrophages exposed to 1 μM cucurbitacin I for 24 h (Figure 2, D and E). Greater than 90% of each cell type displayed morphological hallmarks of apoptosis, including nuclear condensation, mitochondrial fragmentation, and cell shrinkage. After removal of the inducer for 24 h, ∼90% of the cells recovered morphology. Fluorescently labeled annexin V was also used to track reversal of apoptosis in heart and brain cells (Figure 2A). Annexin V binds efficiently to phosphatidylserine, which moves from the inner to the outer leaflet of the plasma membrane during apoptosis (Logue et al., 2009). Ten minutes before washing, fluorescein isothiocyanate–conjugated annexin V (annexin V-FITC) was applied to label apoptotic cells (Figure 2B), and >90% of both cell types were labeled (Figure 2, B and C). After removal of the inducer for 2 h to heart and 3 h to brain cells, >20% of heart cells and 60% of brain cells retained the annexin V-FITC label after morphological recovery (Figure 2, B and C). This provides an additional strategy to track cells that undergo apoptosis and survive, without the need of transfection.
FIGURE 2:
Reversibility of apoptosis in primary rat heart cells, ferret brain cells, and primary mouse macrophages. (A) Schematic diagram of approach using annexin V-FITC to track cells that reverse apoptosis. (B) Confocal and DIC microscopy of rat primary heart cells that were exposed to 4.5% ethanol for 5 h (Treated) or not (Untreated). Treated cells were then washed to remove apoptotic inducers and further cultured for 2 h (Washed). Merged images, mitochondria (red), nuclei (blue), and annexin V-FITC (annexin V)–labeled exposed phosphatidylserine (PS) (green) were visualized by fluorescence, and cell morphology was by DIC. Scale bar, 10 μm. (C) Quantification of the apoptotic response and its reversal on primary rat heart cells and Mpf brain cells. Percentage of cells showing morphological signs of apoptosis including mitochondrial fragmentation, nuclear condensation, cell shrinkage, and cell surface phosphatidylserine labeled with annexin V-FITC (Annexin V) for control cells (Untreated), cells treated with apoptotic inducer (heart cells with 4.5% ethanol for 5 h, brain cells with 2 μM jasplakinolide for 50 h) (Treated), and treated cells that were washed and further cultured with fresh medium (heart cells for 2 h, brain cells for 3 h) in standard conditions (Washed). *p < 0.01; n = 3 independent experiments. Error bars denote SD. (D) Fluorescence of healthy, untreated macrophages, those that were exposed to 1 μM cucurbitacin I (CuI) for 24 h (Treated), and treated cells that were washed to remove apoptotic inducers and further cultured for 24 h (Washed). Merged images, mitochondria (red) and nuclei (blue). Scale bar, 30 μm. (E) Percentage of the untreated, treated, and washed macrophages that displayed mitochondrial fragmentation, nuclear condensation, and cell shrinkage. *p < 0.01; n = 3 independent experiments. Error bars denote SD.
Cells can reverse apoptosis after DNA damage has occurred
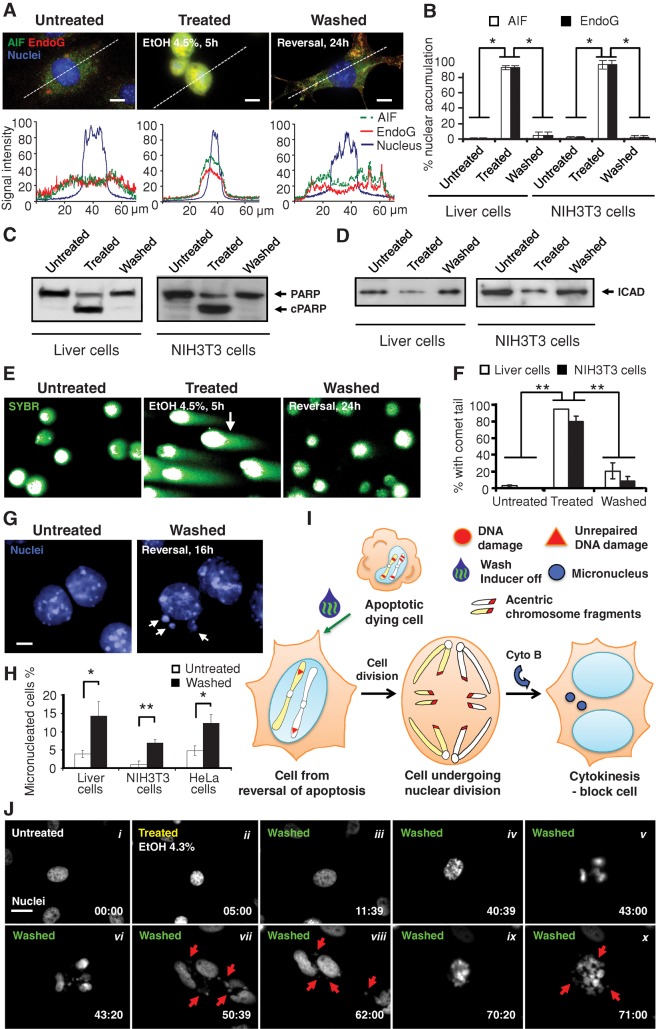
Genomic destruction is a hallmark of apoptosis (Kerr et al., 1972; Taylor et al., 2008), raising the interesting possibility that cells that reverse apoptosis might acquire genetic alterations. During apoptotic induction, nuclear translocation of mitochondrial AIF and EndoG (Figure 3, A and B), which are apoptotic nucleases (Susin et al., 1999; Li et al., 2001), occurred in both liver and NIH 3T3 cells, suggesting that DNA was likely to be damaged.
FIGURE 3:
Damage of DNA in dying cells before reversal of apoptosis. (A) Fluorescence micrographs showing the subcellular localization of AIF (green), EndoG (red), and nuclei (blue) in primary liver cells that were untreated (Untreated) or treated with 4.5% ethanol for 5 h (Treated) and treated cells that were then cultured for 24 h in fresh medium after removal of the ethanol (Washed) from cells. Quantification of the corresponding fluorescence signals of AIF, EndoG, and nucleus along the dotted line as indicated in their respective images. Scale bar, 10 μm. (B) Percentage of liver and NIH 3T3 cells that displayed nuclear accumulation of AIF and EndoG. Treated liver cells were exposed to 4.5% ethanol for 5 h. NIH 3T3 cells were treated with 10% DMSO for 20 h. Treated cells that were then washed to remove apoptotic inducers were further cultured for 24 h (Washed). *p < 0.01; n = 3 independent experiments. Error bars denote SD. (C, D) Western blot analysis of total cell lysates of untreated, treated, and washed liver and NIH 3T3 cells were probed for (C) PARP and (D) ICAD. c, cleaved form. (E) Fluorescence microscopy for the SYBR-stained DNA of untreated, treated, and washed liver cells subjected to the comet assay for DNA damage. Cells embedded in agarose were subjected to electrophoresis. Broken DNA forms comet tails such as the one indicated by an arrow. Intact DNA remains within the nuclear envelop. (F) Percentage of untreated, treated, and washed liver and NIH 3T3 cells that displayed a comet tail. **p < 0.01; n = 3 independent experiments. Error bars denote SD. (G) Fluorescence microscopy on nuclear morphology of untreated as well as treated and washed primary liver cells 16 h after removal of apoptotic inducer. Cytokinesis was blocked with cytochalasin B (Cyto B). Arrows indicate micronuclei in the washed cells. Scale bar, 10 μm. (H) Percentage of untreated, as well as treated and then washed, liver, NIH 3T3, and HeLa cells that displayed micronuclei. Apoptotic inductions for the liver and NIH 3T3 cells were as described in (B) and for HeLa as in (J). See Materials and Methods for detail. *p < 0.05; **p < 0.01; n = 3 independent experiments. Error bars denote SD. (I) Proposed model for the formation of micronuclei in cells that divide after reversal of apoptosis, likely as a result of unrepaired DNA damage. (J) Time-lapse live-cell fluorescence microscopy of HeLa cells before, during, and after exposure to ethanol. The same cell before ethanol induction (Untreated, i), induced with 4.3% ethanol in culture medium for 5 h (Treated, ii), and then washed and further cultured with fresh medium (Washed, iii–x). Monochromatic images of nuclei stained with Hoechst 33342. Arrows indicate some of the micronuclei. Time presented as h:min. Scale bar, 30 μm. See Supplemental Figure S5 and Supplemental Video S4 for images and video with fluorescence microscopy for the corresponding mitochondria and nuclei and phase contrast microscopy for cell morphology of the same cells. Cytochalasin B was not present in this experiment.
We also detected caspase-mediated damage of DNA repair systems. For example, the enzyme poly(ADP)-ribose polymerase-1 (PARP), which is required for genomic stability (Lazebnik et al., 1994; Wang et al., 1997), was cleaved in the induced cell population (Figure 3C). Activated caspases also unlock DNA fragmentation factor/caspase-activated DNase (DFF40/CAD) by cleaving its inhibitor DFF45/ICAD (Figure 3D; Liu et al., 1997; Enari et al., 1998). Thus mitochondria- and caspase-mediated DNA damage mechanisms were activated in the dying cells before washing.
The single-cell gel electrophoresis (comet) assay is a sensitive method for detecting DNA damage, including single- and double-strand breaks (Olive and Banath, 2006). The vast majority of both liver and NIH 3T3 cells treated with apoptotic inducers showed prominent comet tails before washing (Figure 3, E and F). After removal of apoptotic inducers, the comet tails disappeared from most cells, indicating repair of the broken DNA (Figure 3, E and F). In addition, nuclear AIF and EndoG were reduced (Figure 3, A and B), and both full-length and cleaved PARP and ICAD returned to pretreatment levels (Figure 3, C and D). Taken together, these results indicate that cells can reverse the dying process even after DNA damage.
Genetic alterations and transformation occur after reversal of apoptosis
To address how completely the DNA damage could be repaired, we performed cytokinesis-block micronucleus assays. At 16 h after removal of the inducer of apoptosis, although most nuclei appeared normal, there was a significant increase in the number of cells that displayed micronuclei compared with untreated controls in both liver and NIH 3T3 cells (Figure 3, G and H). Micronuclei are biomarkers of DNA damage, including chromosome breakage and/or whole chromosome loss in dividing cells (Fenech, 2007). The formation of micronuclei is evidence of unrepaired DNA damage (Figure 3I), indicating that whereas many breaks were repaired, some remained. Increased in formation of micronuclei also occurred after reversal of ethanol-induced apoptosis in HeLa cells (Figure 3H). Time-lapse live-cell microscopy revealed abnormalities, including formation of micronuclei during the first cell division after reversal of apoptosis (Figure 3J, i–vii, and Supplemental Figure S5). Of note, new micronuclei formed during the division of the daughter cells (Figure 3J, viii–x, Supplemental Figure S5, and Supplemental Video S4), suggesting that some DNA damage persisted.
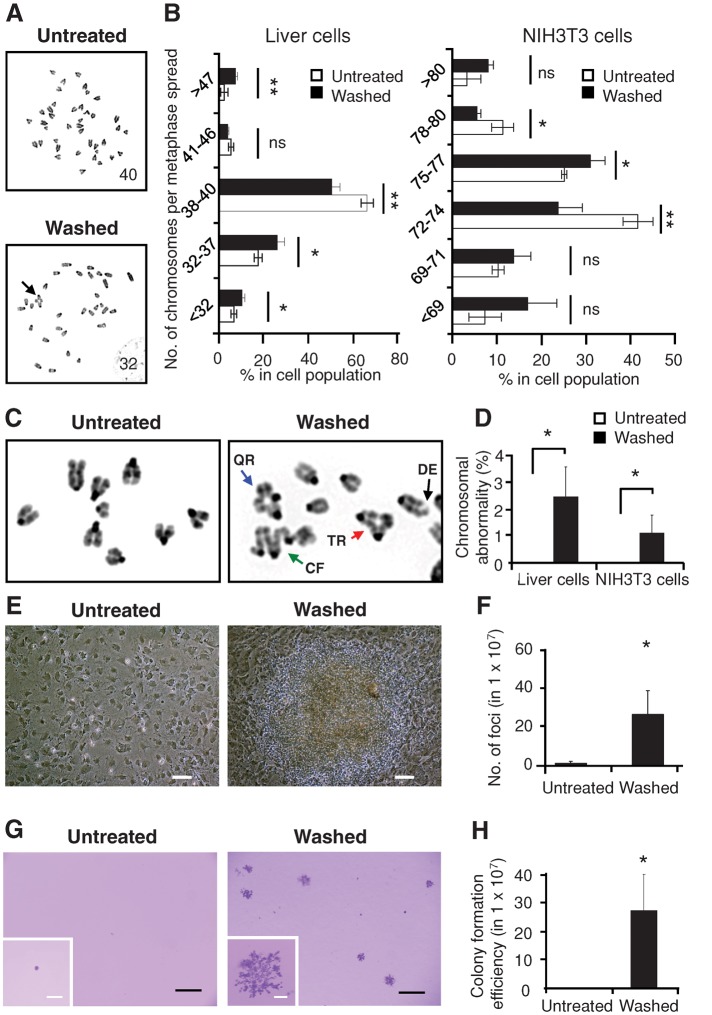
The presence of unrepaired DNA damage in cells after reversal of apoptosis raised the question of whether surviving cells bear chromosomal abnormalities. Therefore we performed karyotyping on colchicine-treated, metaphase-arrested cells 3 d after induction and washing. We found a significant increase, compared with untreated cells, in chromosomal aberrations, including variations in chromosome number (Figure 4, A and B) and radial configurations (Figure 4, C and D), the latter of which result from misjoining of broken chromatids (German, 1964). This indicates the presence of genetic alterations in cells after reversal of apoptosis.
FIGURE 4:
Genetic alterations and transformation after reversal of apoptosis. (A) Inverted DAPI-banding image of metaphase spreads of untreated liver cells (Untreated) compared with cells that were treated with 4.5% ethanol for 5 h, washed, and then cultured for 3 d after removal of apoptotic inducer (Washed). The number of metaphase chromosomes is indicated on the corresponding images. An abnormal chromosomal configuration is indicated by an arrow. (B) Percentage of untreated and washed liver cells (3 d after 5-h exposure to 4.5% ethanol) and NIH 3T3 cells (3 d after 20-h exposure to 10% DMSO) displaying the indicated number of chromosomes in metaphase spreads. ns, p > 0.05; *p < 0.05; **p < 0.01; n = 3 independent experiments. Error bars denote SD. (C) Representative inverted DAPI-banding images of the configuration of metaphase chromosomes of untreated (Untreated) as well as treated and washed (Washed) liver cells. Configurations indicated by arrows: triradial (TR, red); quadriradial (QR, blue); complex figures (CF, green); deletion (DE, black). (D) Percentage of the untreated and the washed liver and NIH 3T3 cells displaying at least one abnormality in chromosomal configuration. *p < 0.05; n = 3 independent experiments. Error bars denote SD. (E) Image of untreated NIH 3T3 cells and the foci from the cells after being treated and washed (Washed) after 3 wk of culture. Scale bars: 50 μm. (F) Number of foci in untreated as well as treated and washed NIH 3T3 cells after 3 wk of culture. *p < 0.05; n = 3 independent experiments. Error bars denote SD. (G) Image of crystal violet–stained colonies in soft agar in untreated as well as treated and washed NIH 3T3 cells from the foci after 5 wk of culture. Inset, enlarged image of a colony. Scale bars, 400 μm (white), 2 mm (black). (H) Number of colonies formed in soft agar in untreated as well as treated and washed NIH 3T3 cells after 5 wk of culture. *p < 0.05; n = 3 independent experiments. Error bars denote SD.
Genetic alterations in individual cells can promote phenotypic diversity (Bloom, 1972; Stratton et al., 2009) and can lead to transformation (Bloom, 1972; Rubin, 2008; Gordon et al., 2012), for which NIH 3T3 cells serve as an important experimental model (Rubin, 2008). A fraction of cells that reversed apoptosis displayed classic transformed phenotypes (Bloom, 1972; Cifone and Fidler, 1980; Rubin, 2008), including focus formation (Figure 4, E and F) and proliferation in soft agar (Figure 4, G and H), indicating loss of contact inhibition of growth and anchorage-independent growth, respectively. In contrast, similar phenotypes were virtually undetectable in untreated control cells. These phenotypes are also hallmarks of cancer cells (Bloom, 1972; Cifone and Fidler, 1980; Rubin, 2008), suggesting that reversal of apoptosis may be carcinogenic.
New transcription is critical to reverse apoptosis
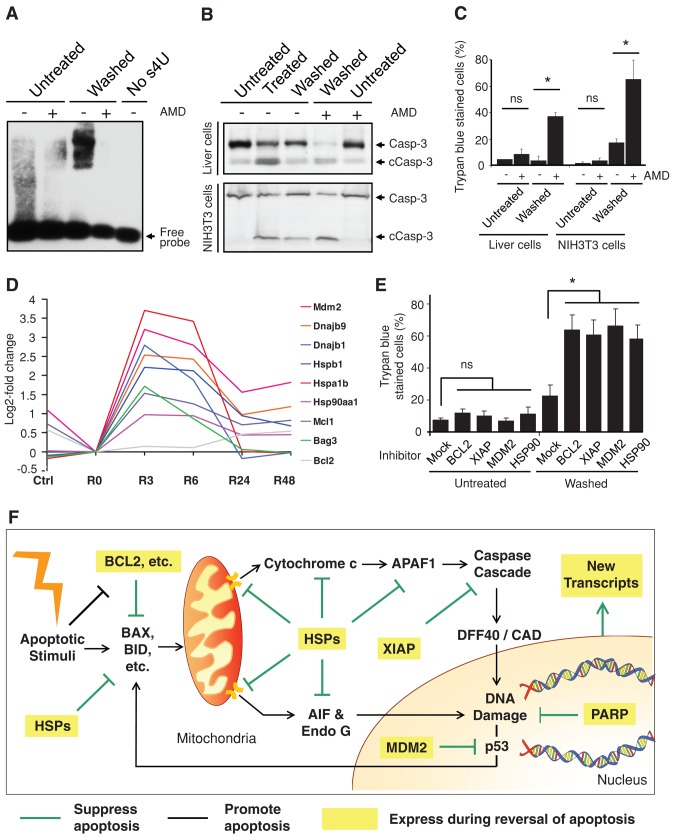
To gain insight into the mechanism by which cells reverse apoptosis and survive, we first tested whether new transcription is required. We detected new RNA synthesis immediately after removal of the apoptotic inducers (Figure 5A), suggesting that new transcription occurs. We then exposed cells transiently to the reversible transcription inhibitor actinomycin D (Sawicki and Godman, 1972). This promoted persistence of cleaved caspase-3 (Figure 5B) and irreversible cell death (Figure 5C) as indicated by trypan blue staining, which labels dead cells due to their plasma membrane permeability (Kroemer et al., 2009). In contrast, the same concentration of actinomycin D did not cause significant cell death in control cells that had not been treated with apoptotic inducers (Figure 5, B and C). These results indicate that new transcription is required for the reversal of apoptosis.
FIGURE 5:
Critical contributing factors in reversal of apoptosis. (A) RNA blot for detecting new RNA synthesis in untreated vs. treated and washed (Washed) liver cells 1 h after a 5-h exposure to 4.5% ethanol and with and without transient exposure (1 h, 1μg/ml) to actinomycin D (AMD). No s4U served as negative control for the probe binding to RNA. The detailed procedure is described in Materials and Methods. (B) Western blot analysis of the cleavage of caspase-3 (Casp-3) on the total lysate of untreated, treated (5-h exposure to 4.5% ethanol for liver cells, 20-h exposure to 10% DMSO for NIH 3T3 cells), and washed liver cells and NIH 3T3 cells with and without transient exposure to actinomycin D (AMD) immediately after removal of apoptotic stimuli. The AMD-exposed cells were cultured in fresh medium for 23 h and then subjected to the analysis. c, cleaved form. (C) Percentage of untreated as well as treated and washed liver cells and NIH 3T3 cells with and without transient AMD exposure that displayed plasma membrane permeability in trypan blue exclusion assay. The AMD exposed-cells were cultured in fresh medium for 23 h and then subjected to the assay. ns, p > 0.05; *p < 0.05; n = 3 independent experiments. Error bars denote SD. (D) A time-course microarray study in liver cells, log2-fold change of gene expression comparison between ethanol-induced apoptotic cells (R0), untreated cells (Ctrl), and induced cells that were then washed and further cultured in fresh medium for 3 (R3), 6 (R6), 24 (R24), and 48 h (R48). The log2 signal values from three biological replicates were averaged (geometric mean) for each time point. (E) Percentage of untreated and washed liver cells with and without 24-h exposure of inhibitors of BCL-2 (ABT 263, 1 μM), XIAP (Embelin, 20 μM), MDM2 (Nutlin-3, 10 μM), and HSP90 (17-allylaminogeldanamycin, 0.5 μM) that displayed full plasma membrane permeability in trypan blue exclusion assay. The corresponding cells were exposed to the inhibitors after apoptotic stimuli had been removed from the treated cells and then further cultured for 24 h. ns, p > 0.05; *P < 0.05; n = 3 independent experiments. Error bars denote SD. (F) Proposed model for reversal of apoptosis. Expression of multiple prosurvival factors and new transcripts during reversal of apoptosis promotes cell survival by suppressing the activated apoptotic pathways and repairing the cells.
Prosurvival factors contribute to reversal of apoptosis
To determine the transcriptional profiles of liver cells undergoing reversal of apoptosis, we conducted time-course gene expression analysis of 30,774 transcripts using RNA microarrays and quantitative PCR with reverse transcription (qRT-PCR). We observed enhanced expression of multiple prosurvival genes, including antiapoptotic BCL-2 family members (Bag3, Bcl2, and Mcl1), X-linked inhibitor of apoptosis protein (XIAP), the murine double minute (MDM2), and heat shock proteins (Dnajb1, Dnajb9, Hsp90aa1, Hspa1b, and Hspb1), after removal of the apoptotic inducer (Figure 5D and Supplemental Figure S6). Expression of most of these genes peaked at the 3- and 6-h time points after removal of the apoptotic inducer and then dropped again.
The contributions of some of the identified candidates to reversing apoptosis were tested by application of pharmacological inhibitors to liver cells after removal of the apoptotic stimuli. Inhibition of BCL-2, XIAP, MDM2, or HSP90 significantly suppressed reversal of apoptosis, as indicated by the increased percentage of trypan blue–stained cells (Figure 5E) and persistence of cleaved caspase-3 (Supplemental Figure S7), suggesting their roles in reversing apoptosis by suppressing activated proapoptotic pathways (Figure 5F).
DISCUSSION
Apoptosis is critical in multicellular organisms (Kerr et al., 1972; Jacobson et al., 1997; Fuchs and Steller, 2011). It is widely accepted that there are both proapoptotic and antiapoptotic signals in cells, and the balance between the two determines whether a cell lives or dies. Although the precise point of no return may vary in different cell types and conditions (Kroemer et al., 2009), it has generally been assumed that apoptosis is irreversible after caspase activation and DNA damage (Riedl and Shi, 2004; Taylor et al., 2008; Chipuk et al., 2010). Strikingly, the time-lapse live imaging and biochemical studies reported here reveal that apoptotic cells can survive and reverse the process of dying even after they have reached an advanced stage of apoptosis, including caspase-3 activation and even DNA damage. This phenomenon was not limited to one cell type or apoptotic inducer but rather was observed for different types of cells and inducers. We propose to name this process anastasis, 1 which is a Greek word that means “rising to life” and thus represents an apt contrast to the word apoptosis (Kerr et al., 1972), which is a Greek word meaning “falling to death.”
Although it was surprising to observe cells recovering normal cell morphology after reaching such an advanced stage of apoptosis, in a few other biological contexts it is known that substantial caspase activation does not obligatorily lead to cell death. One example is sperm maturation in Drosophila melanogaster (Arama et al., 2003). Late in sperm development, most of the spermatocyte cytoplasmic contents are destroyed in a process that requires apoptosome components and caspase activity. In this case, apoptosis proceeds partway and then stops but does not actually reverse. Presumably sperm DNA is protected from damage, although this has not been examined directly.
In another example, we previously reported reversible apoptosis in human cancer cells after various inductions (Tang et al., 2009). Here we show that removal of apoptotic inducers was sufficient to promote anastasis in primary cells, indicating that it is an endogenous mechanism and suggesting that it is a normal physiological process rather than a function restricted to abnormal cells. The present study also suggests that anastasis may be a general phenomenon in mammalian cells.
Of interest, chromosomal abnormalities and oncogenic transformation of primary liver cells and NIH 3T3 fibroblasts occurred after reversal of apoptosis. These observations suggest that anastasis could represent a previously unrecognized cause of genetic alterations. It also represents a form of stress-induced mutagenesis, possibly analogous to that described in prokaryotic and eukaryotic organisms (McClintock, 1984; Capy et al., 2000; Rosenberg, 2001; Jiang et al., 2003). In bacteria and plants, stress-induced mutagenesis has been proposed to accelerate evolution by natural selection by increasing the mutation frequency precisely when organisms find themselves maladapted to their environments.
The observation that anastasis can occur in normal cells has multiple intriguing implications. For example, anastasis could represent one mechanism underlying the observation that repeated injury increases the risk of cancer in a variety of tissues (Boffetta and Hashibe, 2006). Alcohol abusers are more prone to liver cancer (McKillop and Schrum, 2005), and massive apoptosis has been observed in mouse liver after exposure to ethanol (Goldin et al., 1993). Here we show that transient exposure of primary liver cells to high concentrations of ethanol caused them to initiate and then reverse apoptosis, increasing the frequency of oncogenic transformation. Therefore reversal of apoptosis could be a mechanism that allows cells to survive a transient crisis but then results in genomic rearrangements similar to those recently reported in cancer genome sequencing studies (Liu et al., 2011; Stephens et al., 2011).
Reversal of apoptosis could also be a mechanism by which cancer cells initially survive chemotherapy and radiation treatments and later evolve resistance. As we show here, cells that reverse apoptosis may acquire new mutations from the dying process, in addition to the direct mutagenic effects of radiation and chemotherapy drugs (Ross, 1999; Johnstone et al., 2002; Fu et al., 2012). Therefore cancer cells that undergo reversal of apoptosis after anticancer treatment could acquire new mutations and thus transform into more aggressive and metastatic cancers. Acquisition of mutations in normal cells as a consequence of reversal of apoptosis might also cause new tumors after cycles of anticancer therapy.
In addition to its pathophysiological implications such as oncogenesis, a possible beneficial effect of anastasis could be to preserve injured cells that are difficult to replace, such as mature neurons and heart cells. Neuronal cell death occurs, for example, in the retinas of flies, rabbits, and rats after exposure to excess light (McKechnie and Foulds, 1980; Milligan et al., 1997; Gordon et al., 2002). Of interest, the features of apoptosis vanish quickly in the corresponding tissues when the environment improves (McKechnie and Foulds, 1980; Milligan et al., 1997; Gordon et al., 2002). During heart failure, significant numbers of cells release cytochrome c and activate caspase-3 but manage to maintain normal nuclear morphology (Narula et al., 1999; Reed and Paternostro, 1999), suggesting arrest of the apoptotic process in heart cells (Narula et al., 2006). The results shown here suggest that heart cells can even reverse the apoptotic process.
Environmental stresses also trigger apoptosis in germ cells in mammals and other organisms (Aitken et al., 2011). For example, in Drosophila melanogaster and Caenorhabditis elegans, germ cells undergo apoptosis in response to starvation (Drummond-Barbosa and Spradling, 2001; Salinas et al., 2006). In this setting, reversal of apoptosis, if it occurs, could in principle promote genetic diversity and thereby accelerate evolution by natural selection, specifically in stressful environmental conditions. Further investigations of this newly discovered process will be necessary to test these hypotheses, with significant implications for our understanding of development, homeostasis, diseases, and evolution.
MATERIALS AND METHODS
Cell culture
Primary liver cells were isolated from BALB/c mice by collagenase B according to the manufacturer's instructions (Worthington Biochemical, Lakewood, NJ) and cultured as described (Zurlo and Arterburn, 1996). Primary cultures of heart cells were dissociated from ventricles of 1- or 2-d-old neonatal Sprague Dawley rats (Harlan, Indianapolis, IN) by enzymatic digestion of 0.1% trypsin overnight (US Biochemicals, Cleveland, OH) and then 0.1% collagenase (Worthington Biochemical) for dissociation as previously described (Iravanian et al., 2003). Peritoneal macrophages were isolated from BALB/c mice as previously described (Hu et al., 2009). NIH 3T3 cells, human cervical cancer HeLa cells, and Mpf brain cells (CRL 1656) were purchased from the American Type Culture Collection (Manassas, VA). The cells were cultured in DMEM/F-12 (DMEM:nutrient mixture F-12) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies, Carlsbad, CA) at 37°C under an atmosphere of 5% CO2/95% air. Cells were seeded onto tissue culture plates for 2 d with 70% confluence before being subjected to each experiment. The medium was changed every 24–36 h.
Apoptotic inductions
Unless specifically mentioned, apoptosis in the primary mouse liver cells and primary rat heart cells was induced by 5-h exposure to 4.5% ethanol (Scharlau, Barcelona, Spain), HeLa cells by 5-h exposure to 4.3% ethanol, and NIH 3T3 cells by 20-h exposure in 10% DMSO (Sigma-Aldrich, St. Louis, MO) in culture medium (vol/vol). Mpf cells were induced by 50-h incubation in 2 μM jasplakinolide (Invitrogen, Carlsbad, CA) and primary mouse macrophages by 24-h incubation in 1 μM cucurbitacin I (ChromaDex, Irvine, CA) in culture medium. These conditions were chosen because they represented the lowest concentration of inducer that caused >90% of cells to undergo apoptosis. For removal of apoptotic inducers, cells were washed three times with culture medium and then cultured for the period of time indicated in the individual experiments.
Immunocytochemistry and fluorescence and confocal microscopy
Mitochondria and nuclei were stained in living cells with 50 nM Mitotracker Red CMXRos and 250 ng/ml Hoechst 33342 (Invitrogen), respectively, for 20 min in culture medium. Cells with active endocytosis were labeled by green fluorescence–emitting Quantum Dots from Qtracker 525 Cell Labeling Kit (Invitrogen) as described (Jaiswal et al., 2003). ApoAlert pCaspase3-Sensor Vector (BD Clontech, San Jose, CA) was transfected to HeLa cells using X-tremeGENE 9 DNA Transfection Reagent (Roche, Indianapolis, IN). Cell surface membrane phosphatidylserine (PS) was detected by using Annexin V Apoptosis Detection Kit according to manufacturer's instructions (BioVision, Milpitas, CA). The cells were fixed with 3.7% (wt/vol) paraformaldehyde in phosphate-buffer saline (PBS) solution for 20 min at room temperature, and incubated with 0.1% Triton X-100 (vol/vol) (Sigma-Aldrich) for 10 min before immunostaining. Endogenous AIF and EndoG were stained with anti-AIF and anti-EndoG primary antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) and conjugated with green fluorescent Alexa Fluor 488 and red fluorescent Alexa 594 anti–rabbit immunoglobulin G secondary antibodies with Zenon Tricolor Labeling kit (Invitrogen), respectively. Cell images were captured with a monochromatic CoolSNAP FX camera (Roper Scientific, Tucson, AZ) on an inverted fluorescence microscope Cell Observer Z1 using a 63×, numerical aperture (NA) 1.4 plan-Apochromat objective or LSM 710 on an upright microscope Axio Examiner using a 40×, NA 1.1W Corr LD C-Apochromat objective (Carl Zeiss, Jena, Germany). Images were analyzed by using AxioVision 4.2 software (Carl Zeiss).
Real-time live-cell microscopy
Cells were cultured in CO2-independent medium (Invitrogen) on a glass-bottom culture dish (MatTek Corporation, Ashland, MA) or a thermo-cell culture FCS2 chamber (Bioptechs, Butler, PA), which was mounted onto the adapter in the stage of an inverted fluorescence microscope. Ethanol (4.5% for liver, 4.3% for HeLa cells [vol/vol]) in culture medium was introduced to the cells through perfusion tubes (Bioptechs), which were connected to the cell chamber. The ethanol was removed, and fresh medium was then introduced to the chamber through these tubes after apoptotic induction. Fluorescence signals of mitochondria and nuclei were visualized by fluorescence with excitation at 561 and 405 nm, respectively, and cell morphology by differential interference contrast (DIC) or phase contrast microscopy. Time-lapse images were captured by Cell Observer Z1 (Carl Zeiss) with a monochromatic CoolSNAP FX camera (Roper) or Evolve 128 EMCCD (Photometrics, Tucson, AZ) using a 63×, NA 1.4 Plan-Apochromat objective or a 40×, NA 0.95 Corr Plan-Apochromat objective (Carl Zeiss), a BioStation IMQ time-lapse imaging system (CELL-S2) using a 40×, NA 0.8 Plan-Apochromat objective (Nikon, Melville, NY), or a C2 Confocal on a TiE Invert microscope using a 60×, NA 1.4 Plan-Apochromat objective (Nikon). Images and heat map were analyzed by using AxioVision 4.2 software.
Western blot analysis
Approximately 3 μg of protein from total cell lysate per lane was separated on a 12% SDS–PAGE gel and transferred onto a Hybond ECL membrane (Amersham Biosciences, Chalfont St Giles, United Kingdom). After blocking, the membrane was incubated overnight at 4°C with primary antibody detecting targeted protein as stated in the text with 1:1000 dilution, followed by another hour of incubation with the corresponding horseradish peroxidase–conjugated secondary antibody (Bio-Rad, Hercules, CA) at room temperature with 1:5000 dilution. Primary antibodies used were as follows: anti-caspase-3, anti-PARP (Cell Signaling Technology, Danvers, MA), and anti-ICAD (BD PharMingen, BD Biosciences, Le Pont de Clax, France). The signal from the secondary antibody was detected with the ECL Western blotting detection system (Amersham-Pharmacia Biotech, GE Healthcare Bio-Sciences, Piscataway, NJ).
Single-cell gel electrophoresis (comet) assay
Comet assay was performed by using the Trevigen Comet Assay kit (Trevigen, Gaithersburg, MD) according to manufacturer's instructions. Alkaline electrophoresis of gelled slides was performed using Ready Sub-Cell GT Cells (Bio-Rad) on ice with 20 V and 200 mA for 30 min. The current was adjusted by the volume of the buffer in the gel tank. The DNA was visualized by SYBR Green staining (Trevigen), followed by fluorescence microscopy.
Cytokinesis-block micronucleus assay
Cells were grown on glass coverslips (Marienfeld, Lauda-Künigahofen, Germany) with 70% confluence in six-well cell culture plates (Nunc, Roskilde, Denmark) and were induced to apoptosis as described in the section Apoptotic inductions. To study genomic damage in apoptotic cells that reversed apoptosis and proliferated, the apoptosis-induced cells were washed and cultured for 16 h in fresh medium that contained cytokinesis-blocking cytochalasin B (3 μg/ml; Sigma-Aldrich). Cells without apoptotic induction served as control. The cells were then fixed by incubating in methanol/acetic acid (5:1, vol/vol) twice for 15 min, followed by overnight fixation at 4°C. After washing three times with PBS, the fixed cells were stained for nuclei by incubation with 250 ng/ml Hoechst 33342 in PBS for 20 min at room temperature. Slides were then prepared as described for fluorescence microscopy and observed under a 63× objective to image micronucleus. The micronuclei in the cytokinesis-blocked cells were scored as described (Fenech, 2007). Only binucleated cells were scored, so as to include only cells that divided once after addition of cytochalasin B. The two main nuclei should have a clear boundary from each other, and micronuclei were counted only when clear boundary from the main nuclei was observed. The diameter of a micronucleus should lie between 1/16 and 1/3 that of the main nuclear diameter. Triplicates were performed, with >100 cells for each condition per set.
Karyotyping
Metaphase chromosome spreads were prepared as described (MacLeod et al., 2007) with modifications. Briefly, cells were arrested at metaphase by adding colchicine (Sigma-Aldrich) at a final concentration of 1 μg/ml into growing culture for 6 h. The arrested cells were then collected by trypsinization with immediate neutralization with cell culture medium and then followed by 5 min of centrifugation at 400 × g. After discarding the supernatant, we loosened the cell pellet by gentle flicking in residual medium. To swell the mitotic cells, we incubated the cell suspension in hypotonic buffer, potassium chloride (5.59 g/l in double-distilled water), and sodium citrate (9.0 g/l in double-distilled water) in 1:1 (vol/vol), for 8 and 15 min for NIH 3T3 and primary liver cells, respectively, at 37ºC. The cells were then pelleted at 400 × g for 5 min to remove the hypotonic buffer. The cells were then fixed by gently adding freshly prepared ice-cold fixative (methanol/acetic acid, 3:1, vol/vol) to the pellet while agitating the centrifuge tube for the whole time so as to prevent cell clumps formation and ensure thorough mixing. The fixative was changed once, and then the cells were fixed overnight at 4ºC. Next, the cells were concentrated in fixative of a volume such that the suspension became slightly cloudy for optimal cell concentration. To spread the metaphase of the fixed cells onto slides, we dropped the cell suspension from height onto a chilly, precleaned SuperFrost Plus microscopic slide (Gerhard Menzel, Braunschweig, Germany) slightly sloped on a freezer block. Then the slides were breathed on to enhance spreading and were mounted with 4′,6-diamidino-2-phenylindole (DAPI)/Antifade kit (MetaSystems, Altlussheim, Germany) after drying. The metaphase chromosomes of metaphase-arrested cells were identified and captured by an automated cytogenetic scanner workstation (MetaSystems) for analysis. Only metaphases of distinctly separated chromosomes and of chromosome spreading patterns from one nucleus were counted in order to avoid overlapped metaphases. Three replicates of >100 metaphases each were counted for the presence of radial configurations in each corresponding metaphase spread for chromosomal abnormality.
Transformation assays
For the focus formation assay, cells were seeded in 10-cm2 culture dishes (Nunc) to reach 70% confluence. They were induced and washed as described. Then the culture medium was changed every 3 d. After 3 wk of culture, morphologically transformed foci whose diameter exceeded 0.5 mm were counted. The assay was performed three times. From each replicate, at least five transformed foci were isolated by picking with sterile pipette tip and were then cultured for soft agar assay. Anchorage-independent colony formation of NIH 3T3 cells undergoing anastasis was determined as described previously (Cifone and Fidler, 1980) with some modifications. Briefly, the cells were harvested by trypsinization. A total of 3 × 103 cells were resuspended in 1.5 ml of complete cell culture medium containing 0.3% agarose. The suspensions were cultured in single wells of six-well cell culture plates (Nunc) above a layer of solidified 0.5% agarose in the medium. After incubation at 37°C under an atmosphere of 5% CO2/95% air for 5 wk, plates were stained with 0.5 ml of 0.005% crystal violet solution (Sigma-Aldrich) for 1 h before being subjected to microscopy.
New RNA detection
For newly synthesized RNA detection, cells were incubated in the presence of 50 μM 4-thiouridine (s4U; Sigma-Aldrich) for 1 h and total RNA was extracted by TRIzol (Invitrogen). The RNAs were then subjected to biotinylation as described (Zeiner et al., 2008). The biotin-labeled RNAs were agarose electrophoresed, transferred to nylon membrane (Bio-Rad) with Trans-Blot SD DNA/RNA Blotting Kit (Bio-Rad), and then detected by chemiluminescence using LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, IL).
Microarray and gene expression data analysis
Mouse primary liver cells were treated with 4.5% ethanol for 5 h (R0) and then washed and cultured in fresh medium for 3 (R3), 6 (R6), 24 (R24), and 48 (R48) h. The untreated cells were used as control (Ctrl). Total RNA was isolated and purified by RNeasy Mini Kit (Qiagen, Cologne, Germany). As a first level of quality control, to detect possible batch effects or sample outliers, we performed principal component analysis with Partek Genomics Suit 6.5 (Partek, St. Louis, MO) on log2-normalized signal values for all samples; biological replicate samples were observed to cluster together. Furthermore, an analysis of variation was run on all data, including variables for cell sample, batch, and error, in which analysis also showed an excellent ratio of signal (biological variation) to noise (batch and error). The RNA was subjected to reverse transcription using SABiosciences C-03 RT2 First Strand Kit (SABiosciences-Qiagen, Frederick, MD). These cDNA samples were analyzed on the Illumina MouseWG-6 v2.0 Expression BeadChip (Illumina, San Diego, CA).
Processed Illumina signal value data were imported into the Partek and Spotfire DecisionSite 9.1 (TIBCO, Palo Alto, CA) platforms for evaluation for expression fold change at the gene level between time points and for fold change across time when compared with a common starting point. First, all signal values were converted into log2 space and quality control tests run to ensure data integrity. Signals for the three biological replicates at each time point were taken together for comparison to other time points; Student's t test used to determine statistical significance as p values, and fold change was based on averaged values. For our time-course analysis, all time points were compared with time point R0, the time at which ethanol treatment ended. Functional Gene Ontology (The Gene Ontology Consortium, www.geneontology.org) and pathway analyses were run with Spotfire's Gene Ontology Browser and Ingenuity Pathway Analysis (Ingenuity Systems, www.ingenuity.com) software, respectively.
qRT-PCR
Total RNA was isolated and purified by RNeasy Mini Kit (Qiagen, Cologne, Germany), and 1.5 μg of the total RNA was reverse transcribed into cDNA via the M-MLV reverse transcriptase (Invitrogen) with oligo-dT as primer. RT-PCR was performed on an IQ5 machine (Bio-Rad) using SYBR GreenER qPCR SuperMix (Invitrogen) in 25 μl of reaction with the following PCR cycle parameters: 10 min at 95°C (pre-denaturation and hot start); 40 cycles of 35 s at 95°C; 35 s at 58°C; 30 s at 72°C (denaturation/annealing/amplification). The following primers were used for detection of their corresponding mRNA. Bcl2 forward primer sequence, 5′-CCT GTG GAT GAC TGA GTA CC-3′; reverse primer sequence, 5′-GAG ACA GCC AGG AGA AAT CA-3′ (Sigma-Aldrich). Xiap forward primer sequence, 5′-CTG AAA AAA CAC CAC CGC TAA C-3′; reverse primer sequence, 5′-CTA AAT CCC ATT CGT ATA GCT TCT TG-3′. Mdm2 forward primer sequence, 5′-CGG CCT AAA AAT GGC TGC AT-3′; reverse primer sequence, 5′-TTT GCA CAC GTG AAA CAT GAC A-3′. Hsp90aa forward primer sequence, 5′CTC CAA TTC ATC GGA CGC TCT G 3′; reverse primer sequence, 5′ TCA AGT CGG CCT TGG TCA TTC C 3′. Gapdh forward primer sequence, 5′-TGC CTC CTG CAC CAC CAA CT-3′; reverse primer sequence, 5′-CGC CTG CTT CAC CAC CTT C-3′. All RT-PCR assays were completed in triplicate, and the threshold cycle of each reaction was converted to DNA equivalent by reading against its corresponding standard curve generated by amplifying dilutions of cDNA containing the relevant target sequences. The relative mRNA expression levels of the target genes were normalized to the mean of Gapdh, which served as the internal control.
Inhibitor treatment
Immediately after removal of apoptotic inducers, specific inhibitor targeting BCL-2 (ABT 263, 1 μM; Selleck Chemicals, Houston, TX), XIAP (Embelin, 20 μM; Sigma-Aldrich), MDM2 (MDM2 inhibitor, 20 μM; Sigma-Aldrich), or HSP90 (17-allylaminogeldanamycin, 0.5 μM; Sigma-Aldrich) was applied to the cells together with fresh culture medium. A mock experiment was also performed in the untreated cells.
Statistics
Statistical comparison was performed using two-tailed (unless stated otherwise) Student's t test (except in microarray and gene expression data analysis, as described in the corresponding section). Differences were considered to be significant when the p value was <0.05.
Supplementary Material
Acknowledgments
We thank Hoi Yan Law and Tony Woo (Carl Zeiss Far East Co., Hong Kong Special Administrative Region, China) for their support with regard to live-cell microscopy; Danny K. Y. Poon, Kevin Neoh, and Bryan Yuen (MetaSystems Asia Co., Hong Kong Special Administrative Region, China) for the use of the automated cytogenetic scanner workstation; Jeffry Reidler and Jason Hill (Nikon Instruments, Melville, NY) for access to the Confocal Laser Point Scanning Microscope and BioStation IMQ time-lapse imaging system; Wing Yin Chiu, Pui Ying Choi, Hoi I Ting, and Hill Tone Leung (Chinese University of Hong Kong, Hong Kong, China) for assistance in cytogenetic studies; David Yue and Worawan Limpitikul (Johns Hopkins University School of Medicine, Baltimore, MD) for isolation of primary heart cells; and Clinton Jones (University of Nebraska, Lincoln, NE) and Heiko J. Luhmann (Johannes Gutenberg-University, Aminz, Germany) for providing us pCaspase3-Sensor plasmid. Statistical and functional analyses of microarray expression data were performed at the John Hopkins Medical Institute Deep Sequencing and Microarray Core, Johns Hopkins University. We thank Ralph Bohlmann and James Voelz for their ideas and suggestions for the word anastasis. This work was supported by a Fulbright Grant (007-2009), Sir Edward Youde Memorial Fellowship 2009/10 and Walter Szeto Memorial Scholarship 2009/10 to H.L.T., a National Institutes of Health Grant (R01GM46425) to D.J.M., and a grant from the University Grants Committee of the Hong Kong Special Administrative Region, China (Project No. AoE/B-07/99), the Lee Hysan Foundation, the Lo Kwee-Seong Biomedical Research Endowment Fund, and a donation from Saskia van der Stap to M.C.F.
Abbreviations used:
- AIF
apoptosis-inducing factor
- APAF1
apoptotic protease activating factor 1
- BAX
BCL2-associated X protein
- BCL2
B cell CLL/lymphoma-2
- BID
BH3 interacting-domain death agonist
- Casp-3
caspase-3
- cCasp-3
cleaved caspase-3
- cPARP
cleaved poly(ADP)-ribose polymerase-1
- DEVD
caspase cleavable sequence
- DFF40/CAD
DNA fragmentation factor/caspase-activated DNase
- DIC
differential interference contrast microscopy
- EndoG
endonuclease G
- Gapdh
glyceraldehyde 3-phosphate dehydrogenase
- HSPs
heat shock proteins
- IAP
inhibitor of apoptosis protein
- ICAD
inhibitor of caspase-activated DNase
- MDM2
murine double minute
- NES
nuclear exclusion signal
- NLS
nuclear localization signal
- PARP
poly(ADP)-ribose polymerase-1
- XIAP
X chromosome-encoded IAP
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-11-0926) on April 25, 2012.
H.L.T., H.M.T., and K.H.M. developed protocols and optimized all experiments and contributed to live-cell microscopy and Western blot analysis and worked with S.H., S.S.W., C.S.W., and H.T.L. on cytogenetic studies. S.H. and H.Y.W. worked on transformation assays. S.H., K.M.W., C.S.W., and K.L. worked on qRT-PCR studies. H.T.L. prepared schematic diagrams. C.C.T. worked on microarray data analysis. W.K.L. provided critical reagents. H.L.T., D.J.M., and M.C.F. designed experiments and wrote the article together with H.M.T. All authors discussed the results and commented on the manuscript.
1We thank Ralph Bohlmann and James Voelz for their ideas and suggestions for the word anastasis. The word apoptosis is a compound made up of a Greek prefix (apo) attached to a Greek stem (ptosis) and refers to the dropping or falling of petals from a flower or leaves from a tree (Kerr et al., 1972). Anastasis is a compound consisting of a prefix (ana) and stem (stasis) and is commonly used to denote “standing again” or “rising to life.” In Luke 2:34, the Greek phrase for “falling and rising” is ptosin kai anastasin (in the accusative case, ending in “n” because of the preposition “unto”). Because the key concept in ptosis is “falling to death” and in anastasis is “rising to life,” anastasis seems a good antonym to the term apoptosis. We therefore propose the word anastasis to describe the process of reversal of apoptosis.
REFERENCES
- Aitken RJ, Findlay JK, Hutt KJ, Kerr JB. Apoptosis in the germ line. Reproduction. 2011;141:139–150. doi: 10.1530/REP-10-0232. [DOI] [PubMed] [Google Scholar]
- Arama E, Agapite J, Steller H. Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev Cell. 2003;4:687–697. doi: 10.1016/s1534-5807(03)00120-5. [DOI] [PubMed] [Google Scholar]
- Bloom AD. Induced chromosomal aberrations: biological and clinical significance. J Pediatr. 1972;81:1–8. doi: 10.1016/s0022-3476(72)80365-2. [DOI] [PubMed] [Google Scholar]
- Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncol. 2006;7:149–156. doi: 10.1016/S1470-2045(06)70577-0. [DOI] [PubMed] [Google Scholar]
- Capy P, Gasperi G, Biemont C, Bazin C. Stress and transposable elements: co-evolution or useful parasites? Heredity. 2000;85:101–106. doi: 10.1046/j.1365-2540.2000.00751.x. [DOI] [PubMed] [Google Scholar]
- Chabaud S, Moulin VJ. Apoptosis modulation as a promising target for treatment of systemic sclerosis. Int J Rheumatol. 2011;2011:495792. doi: 10.1155/2011/495792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifone MA, Fidler IJ. Correlation of patterns of anchorage-independent growth with in vivo behavior of cells from a murine fibrosarcoma. Proc Natl Acad Sci USA. 1980;77:1039–1043. doi: 10.1073/pnas.77.2.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3:339–345. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- Fischer U, Schulze-Osthoff K. Apoptosis-based therapies and drug targets. Cell Death Differ. 2005;12(Suppl 1):942–961. doi: 10.1038/sj.cdd.4401556. [DOI] [PubMed] [Google Scholar]
- Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Cytological evidence for crossing-over in vitro in human lymphoid cells. Science. 1964;144:298–301. doi: 10.1126/science.144.3616.298. [DOI] [PubMed] [Google Scholar]
- Goldin RD, Hunt NC, Clark J, Wickramasinghe SN. Apoptotic bodies in a murine model of alcoholic liver disease: reversibility of ethanol-induced changes. J Pathol. 1993;171:73–76. doi: 10.1002/path.1711710115. [DOI] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Gordon WC, Casey DM, Lukiw WJ, Bazan NG. DNA damage and repair in light-induced photoreceptor degeneration. Invest Ophthalmol Visual Sci. 2002;43:3511–3521. [PubMed] [Google Scholar]
- Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- Guicciardi ME, Gores GJ. Apoptosis as a mechanism for liver disease progression. Semin Liver Dis. 2010;30:402–410. doi: 10.1055/s-0030-1267540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wu Z, Yang L, Fung MC. Molecular cloning and expression of a functional anti-inflammatory protein, Sj16, of Schistosoma japonicum. Int J Parasitol. 2009;39:191–200. doi: 10.1016/j.ijpara.2008.06.017. [DOI] [PubMed] [Google Scholar]
- Iravanian S, Nabutovsky Y, Kong CR, Saha S, Bursac N, Tung L. Functional reentry in cultured monolayers of neonatal rat cardiac cells. Am J Physiol Heart Circ Physiol. 2003;285:H449–H456. doi: 10.1152/ajpheart.00896.2002. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Jaiswal JK, Mattoussi H, Mauro JM, Simon SM. Long-term multiple color imaging of live cells using Quantum Dot bioconjugates. Nat Biotechnol. 2003;21:47–51. doi: 10.1038/nbt767. [DOI] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: a link between cancer genetics and chemotherapy. Cell. 2002;108:153–164. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371:346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Liu P, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zou H, Slaughter C, Wang X. DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Logue SE, Elgendy M, Martin SJ. Expression, purification and use of recombinant annexin V for the detection of apoptotic cells. Nat Protoc. 2009;4:1383–1395. doi: 10.1038/nprot.2009.143. [DOI] [PubMed] [Google Scholar]
- Luthi AU, Martin SJ. The CASBAH: a searchable database of caspase substrates. Cell Death Differ. 2007;14:641–650. doi: 10.1038/sj.cdd.4402103. [DOI] [PubMed] [Google Scholar]
- MacLeod RA, Kaufmann M, Drexler HG. Cytogenetic harvesting of commonly used tumor cell lines. Nat Protoc. 2007;2:372–382. doi: 10.1038/nprot.2007.29. [DOI] [PubMed] [Google Scholar]
- Masters JR. HeLa cells 50 years on: the good, the bad and the ugly. Nat Rev Cancer. 2002;2:315–319. doi: 10.1038/nrc775. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- McKechnie NM, Foulds WS. Recovery of the rabbit retina after light damage (preliminary observations) Albrecht Von Graefes Arch Klin Exp Ophthalmol. 1980;212:271–283. doi: 10.1007/BF00410521. [DOI] [PubMed] [Google Scholar]
- McKillop IH, Schrum LW. Alcohol and liver cancer. Alcohol. 2005;35:195–203. doi: 10.1016/j.alcohol.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Milligan SC, Alb JG, Jr, Elagina RB, Bankaitis VA, Hyde DR. The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J Cell Biol. 1997;139:351–363. doi: 10.1083/jcb.139.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narula J, Haider N, Arbustini E, Chandrashekhar Y. Mechanisms of disease: apoptosis in heart failure—seeing hope in death. Nat Clin Pract Cardiovasc Med. 2006;3:681–688. doi: 10.1038/ncpcardio0710. [DOI] [PubMed] [Google Scholar]
- Narula J, et al. Apoptosis in heart failure: release of cytochrome c from mitochondria and activation of caspase-3 in human cardiomyopathy. Proc Natl Acad Sci USA. 1999;96:8144–8149. doi: 10.1073/pnas.96.14.8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- Reed JC, Paternostro G. Postmitochondrial regulation of apoptosis during heart failure. Proc Natl Acad Sci USA. 1999;96:7614–7616. doi: 10.1073/pnas.96.14.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM. Evolving responsively: adaptive mutation. Nat Rev Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- Ross GM. Induction of cell death by radiotherapy. Endocr Related Cancer. 1999;6:41–44. doi: 10.1677/erc.0.0060041. [DOI] [PubMed] [Google Scholar]
- Rubin H. Cell-cell contact interactions conditionally determine suppression and selection of the neoplastic phenotype. Proc Natl Acad Sci USA. 2008;105:6215–6221. doi: 10.1073/pnas.0800747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas LS, Maldonado E, Navarro RE. Stress-induced germ cell apoptosis by a p53 independent pathway in Caenorhabditis elegans. Cell Death Differ. 2006;13:2129–2139. doi: 10.1038/sj.cdd.4401976. [DOI] [PubMed] [Google Scholar]
- Sawicki SG, Godman GC. On the recovery of transcription after inhibition by actinomycin D. J Cell Biol. 1972;55:299–309. doi: 10.1083/jcb.55.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- Takemoto K, Nagai T, Miyawaki A, Miura M. Spatio-temporal activation of caspase revealed by indicator that is insensitive to environmental effects. J Cell Biol. 2003;160:235–243. doi: 10.1083/jcb.200207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, Ghayur T, Brady KD, Wong WW. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- Tang HL, Yuen KL, Tang HM, Fung MC. Reversibility of apoptosis in cancer cells. Br J Cancer. 2009;100:118–122. doi: 10.1038/sj.bjc.6604802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiner GM, Cleary MD, Fouts AE, Meiring CD, Mocarski ES, Boothroyd JC. RNA analysis by biosynthetic tagging using 4-thiouracil and uracil phosphoribosyltransferase. Methods Mol Biol. 2008;419:135–146. doi: 10.1007/978-1-59745-033-1_9. [DOI] [PubMed] [Google Scholar]
- Zurlo J, Arterburn LM. Characterization of a primary hepatocyte culture system for toxicological studies. In Vitro Cell Dev Biol Anim. 1996;32:211–220. doi: 10.1007/BF02722948. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.