Oxa1 is dynamically redistributed within the inner membrane of mitochondria. Its distribution is influenced by the availability of nuclear-encoded mitochondrial proteins, as well as by mitochondrial protein translation. The findings suggest a spatial compartmentalization of the Oxa1-mediated insertion of nuclear- and mitochondrial-encoded proteins.
Abstract
The Oxa1 protein is a well-conserved integral protein of the inner membrane of mitochondria. It mediates the insertion of both mitochondrial- and nuclear-encoded proteins from the matrix into the inner membrane. We investigated the distribution of budding yeast Oxa1 between the two subdomains of the contiguous inner membrane—the cristae membrane (CM) and the inner boundary membrane (IBM)—under different physiological conditions. We found that under fermentable growth conditions, Oxa1 is enriched in the IBM, whereas under nonfermentable (respiratory) growth conditions, it is predominantly localized in the CM. The enrichment of Oxa1 in the CM requires mitochondrial translation; similarly, deletion of the ribosome-binding domain of Oxa1 prevents an enrichment of Oxa1 in the CM. The predominant localization in the IBM under fermentable growth conditions is prevented by inhibiting mitochondrial protein import. Furthermore, overexpression of the nuclear-encoded Oxa1 substrate Mdl1 shifts the distribution of Oxa1 toward the IBM. Apparently, the availability of nuclear- and mitochondrial-encoded substrates influences the inner-membrane distribution of Oxa1. Our findings show that the distribution of Oxa1 within the inner membrane is dynamic and adapts to different physiological needs.
INTRODUCTION
Compartmentalization is fundamental to many aspects of the function of organelles. Mitochondria feature a complex architecture with two membranes—the outer membrane and the highly convoluted inner membrane, as well as two aqueous compartments—the intermembrane space and the matrix. Electron microscopy images revealing the folding of the inner membrane early on suggested that the inner membrane might be further subdivided into two morphologically and presumably functional domains, namely the inner boundary membrane (IBM), which parallels the outer membrane, and the cristae membrane (CM) (Penniston et al., 1968; Werner and Neupert, 1972). Accumulating evidence demonstrates that both subdomains have different, albeit overlapping protein compositions (Gilkerson et al., 2003; Vogel et al., 2006; Wurm and Jakobs, 2006; Suppanz et al., 2009). Little is known on the dynamics of protein distributions within the inner membrane and the molecular mechanisms that create and maintain the subcompartmentalization.
The proteins of the respiratory chain executing oxidative phosphorylation are enriched in the CM. In the budding yeast Saccharomyces cerevisiae, the respiratory chain complexes III, IV, and V are composed of nuclear- and mitochondrial-encoded subunits. Hence the proper assembly of the inner membrane protein complexes requires a finely tuned coordination of the insertion of imported proteins and of mitochondrial translation products into the inner membrane.
A central component in this process is the well-conserved protein Oxa1. Oxa1 mediates the export of a number of proteins from the matrix into the inner membrane. The Oxa1 pathway is essential for the insertion of the mitochondrial-encoded Cox1, Cox2, and Cox3, all of which are subunits of the cytochrome c oxidase (complex IV). In addition, Oxa1 is also required for the insertion of nuclear-encoded proteins into the inner membrane, including Mdl1 and Oxa1 itself (He and Fox, 1997; Hell et al., 1997, 1998, 2001; Szyrach et al., 2003; Bohnert et al., 2010). Thus Oxa1 is important for the integration of the assembly of nuclear- and mitochondrial-encoded subunits of the respiratory chain complexes and presumably has a role in influencing the compositions of the IBM and the CM.
In this study we show that Oxa1 is dynamically redistributed between the IBM and the CM as a response to different carbon sources. We further show that the availability of either nuclear-encoded or mitochondrial-encoded Oxa1 substrate shifts the distribution of Oxa1 either to the IMB or the CM, respectively.
RESULTS
The distribution of Oxa1 between the IBM and the CM is determined by the available carbon source
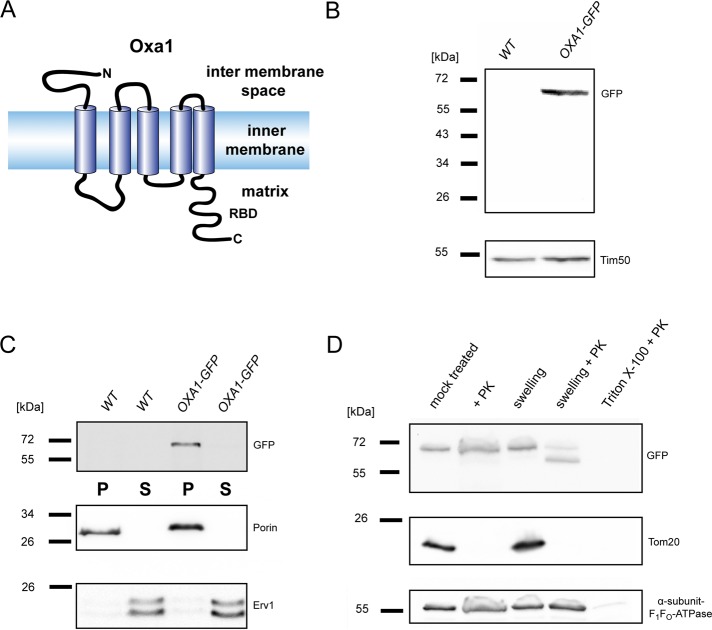
Oxa1 exhibits a well-conserved core region of five transmembrane (TM) domains (Luirink et al., 2001). In S. cerevisiae, the mature protein has an N-terminal domain of ∼80–amino acid residues that protrudes into the intermembrane space and a ∼90–amino acid C-terminal domain that is located on the matrix side of the membrane (Figure 1A).
FIGURE 1:
C-terminally tagged Oxa1 is properly integrated into the inner membrane. (A) Model of the Oxa1 topology in the mitochondrial inner membrane. Adapted from Bonnefoy et al. (2009). (B) Western blot analysis of whole-cell extracts from wild-type and Oxa1-GFP–expressing cells using a GFP-specific antiserum. As a loading control, an antiserum against Tim50 was used. (C) Analysis of the membrane insertion of Oxa1-GFP. Mitochondria were extracted with 100 mM sodium carbonate. The membrane pellet (P) and the soluble supernatant (S) were analyzed by immunoblotting using a GFP-specific antiserum. As controls, antisera against the integral outer membrane protein porin and the soluble intermembrane space protein Erv1 were used. (D) Subfractionation of mitochondria of cells expressing Oxa1-GFP. Isolated mitochondria were incubated with or without 20 μg/ml proteinase K (PK). The outer membrane was selectively opened by incubation in hypotonic buffer (generation of mitoplasts) in the presence or absence of PK. Finally, both membranes were opened by treatment with detergent and PK. The samples were analyzed by Western blotting, using an antiserum against GFP. As a control, antisera against Tom20 (outer membrane) and the α subunit of the F1Fo-ATPase (matrix) were used. All analyses were performed with Δoxa1 cells expressing Oxa1-GFP from a centromeric plasmid under the control of the native promoter.
In this study we used two approaches to determine the submitochondrial localization of Oxa1: quantitative immuno–electron microscopy (immuno-EM) on wild-type mitochondria of cryosectioned yeast cells and live-cell microscopy on yeast cells with genetically enlarged mitochondria. To enable the visualization of the distribution of Oxa1, we tagged the protein at its C-terminus with either the green fluorescent protein (GFP) or the 8–amino acid Flag tag. The Oxa1 fusion proteins were not degraded and behaved as integral membrane proteins (Figure 1, B and C, and Supplemental Figure S1). In isolated mitochondria, the C-terminal GFP tag was accessible to added protease only when the inner membrane was opened with detergent (Figure 1D). The proteolytic pattern of Oxa1-GFP after disruption of only the outer membrane was similar to the previously reported proteolytic pattern of Oxa1 (Sato and Mihara, 2009), fully corroborating previous reports showing that Oxa1 tolerates C-terminal tagging (Reif et al., 2005; Jia et al., 2007). We conclude that a C-terminal tag has no influence on the proper integration of Oxa1 into the inner membrane.
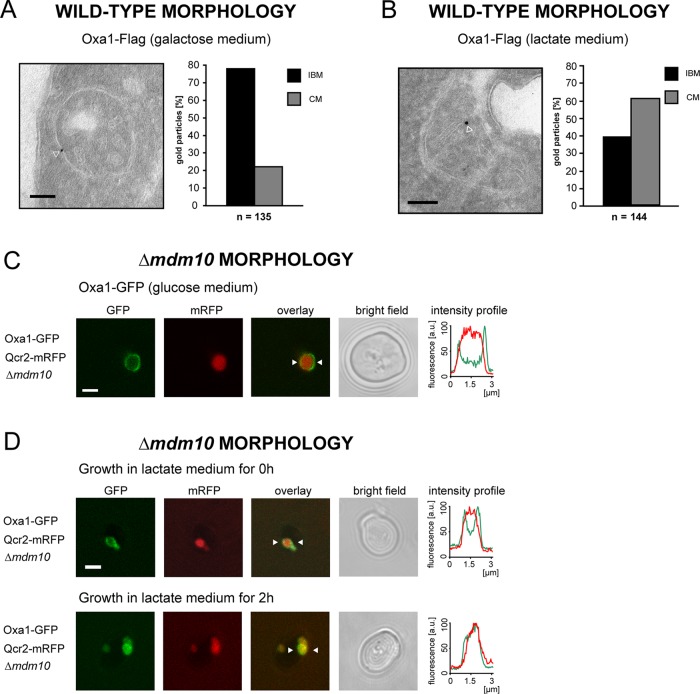
Using immuno-EM, we quantified the distribution of Oxa1 between the IBM and the CM in wild-type mitochondria. To this end, we decorated cryosectioned wild-type yeast cells expressing Oxa1-Flag with an antiserum against the Flag tag. The analyzed cells were devoid of a chromosomal copy of the endogenous OXA1 gene and expressed Oxa1-Flag from a centromeric plasmid; hence the cells expressed close-to-normal levels of Oxa1-Flag (Supplemental Figure S1). The various steps of the labeling procedure were carefully optimized so that nonspecific background labeling was negligible. As a result, the mitochondria of cells expressing Oxa1-Flag were decorated on average with one to two gold particles, and we analyzed at least 100 gold particles to ensure a sufficient statistical basis. We assigned gold particles either to the IBM (distance of the center of the gold particle from the IBM of ≤20 nm) or to the CM. The cells were grown in liquid medium containing fermentable galactose as a carbon source. Under these growth conditions, the length of the CM equals the length of the IBM (Suppanz et al., 2009). In galactose-grown cells expressing Oxa1-Flag 79% of all analyzed gold particles (n = 135) were assigned to the IBM (Figure 2A). This observation of a predominant localization of Oxa1 in the IBM was fully corroborated by analyzing Δoxa1 cells expressing Oxa1-GFP (77% of gold particles [n = 48] in the IBM; Supplemental Figure S2). This finding was in apparent contradiction with a previous study (Vogel et al., 2006), which reported that the majority of Oxa1 is localized in the CM. Because Vogel et al. (2006) propagated the cells in growth media containing lactate, which is a nonfermentable carbon source, we next asked whether the carbon source influences the partitioning of Oxa1 between the IBM and the CM.
FIGURE 2:
The carbon source influences the distribution of Oxa1 between the IBM and the CM. (A) Quantitative immuno-EM analysis of the distribution of Oxa1-Flag in cells grown on the fermentable carbon source galactose. The sections were decorated with a Flag-specific antiserum. Left, representative image. The arrowhead points to a gold particle. Right, quantification of the Oxa1-Flag distribution between the IBM and CM. (B) Quantitative immuno-EM of Oxa1-Flag–expressing cells grown in lactate medium using Flag-specific antiserum. Left, representative image. Right, quantification of the Oxa1-Flag distribution between the CM and the IBM. The quantification was corrected for the absolute lengths of CM and IBM in lactate medium (ratio CM/IBM = 1.5/1). (C) Live-cell fluorescence microscopy of Δmdm10 cells expressing Oxa1-GFP and Qcr2-mRFP grown on a glucose-containing medium. The cells exhibit enlarged mitochondria due to the lack of Mdm10. Qcr2-mRFP labels the CM-containing interior of the mitochondria. Single confocal sections are displayed. The intensity profiles show the fluorescence intensity of Oxa1-GFP (green) and Qcr2-mRFP (red) between the two arrowheads. (D) Fluorescence images of Δmdm10 cells shifted from a glucose-containing medium to a medium containing the nonfermentable lactate. The cells expressed Oxa1-GFP and Qcr2-mRFP. Top, immediately after shifting the cells to lactate medium. Bottom, 2 h after the change of the carbon source. Scale bars, 100 nm (A, B) and 2 μm (C, D).
We indeed found that in Oxa1-Flag–expressing cells grown in lactate, 70% of all gold particles were assigned to the CM (n = 144; Figure 2B). In lactate-grown cells the relative length ratio of CM to IBM is on average 1.5:1 (Vogel et al., 2006). Even if we take this into account, the distribution of Oxa1 in CM and IBM is ∼3:2, demonstrating an enrichment of Oxa1 in the CM under nonfermentable growth conditions. Thus there is a dynamic adaptation of the distribution of Oxa1 to the respective carbon source.
To address the question of how long the adaptation of the Oxa1 distribution to a new carbon source takes, we next resorted to live-cell microscopy of GFP-tagged Oxa1. Owing to the limited optical resolution of conventional fluorescence microscopy, protein distributions within mitochondria are very difficult or even impossible to resolve with optical microscopy (Jakobs, 2006). To investigate the partitioning of Oxa1 between the IBM and the CM in living cells, we used yeast cells in which we enlarged the mitochondria by deleting the gene MDM10, encoding an outer membrane protein (Sogo and Yaffe, 1994). Such Δmdm10 yeast strains were previously established as an in vivo model system to microscopically study protein distributions within the inner membrane (Wurm and Jakobs, 2006; Suppanz et al., 2009). Although enlarged, these mitochondria generally contain cristae membranes (Supplemental Figure S3). In this study, all experiments using Δmdm10 strains were performed at least in triplicate. In each experiment, >100 cells were analyzed. The cells shown represent typical examples, which represent the majority (>90%) of the respective cells with enlarged mitochondria in the particular experiment.
Δmdm10 cells exhibiting enlarged mitochondria and expressing Oxa1-GFP were initially grown on agar plates containing glucose and were subsequently transferred into a liquid medium with the nonfermentable lactate as the sole carbon source. Fully corroborating the EM data, we found that in the mitochondria of Δmdm10 cells grown on media containing glucose, Oxa1-GFP was enriched in the IBM (Figure 2C). As expected, Qcr2 (Cor2), a subunit of the respiratory chain complex III, tagged with the monomeric red fluorescent protein (mRFP), was localized in the cristae-containing interior of enlarged mitochondria, in full agreement with previous reports showing an enrichment of Qcr2 in the CM (Gilkerson et al., 2003). However, already after 2 h in lactate medium, a shift in the distribution of Oxa1-GFP toward the CM was observed (Figure 2D).
Taken together, Oxa1 is dynamically redistributed within the inner membrane of mitochondria, depending on the available carbon source. In mitochondria of cells grown on a fermentable carbon source like glucose or galactose, Oxa1 is predominantly localized in the IBM, whereas on a nonfermentable carbon source like lactate it is enriched in the CM. On a shift of the carbon source from glucose to lactate, the redistribution occurs in a time frame of <2 h.
Protein synthesis at mitochondrial ribosomes is required for the predominant localization of Oxa1 in the CM under respiratory growth conditions
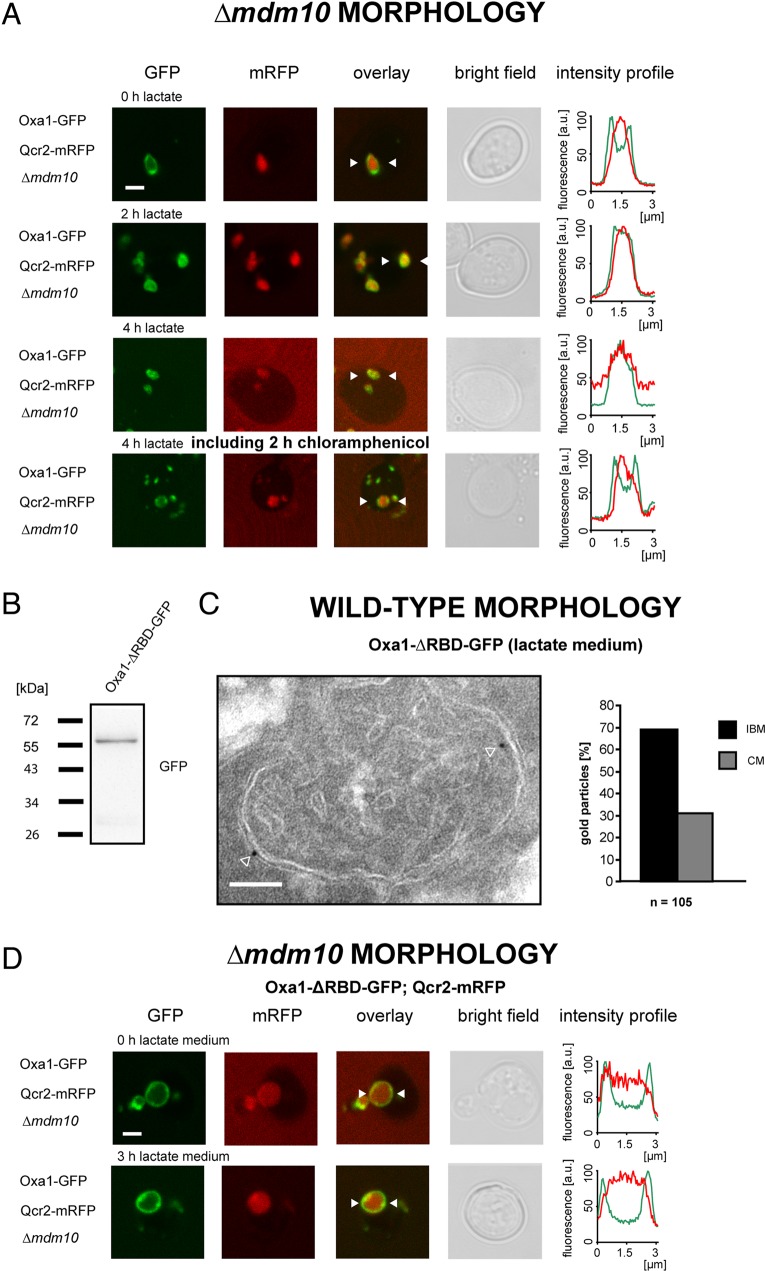
Growth on a nonfermentable carbon source requires the respiratory chain, of which several proteins are encoded by the mitochondrial genome and are inserted by the Oxa1 machinery. To investigate whether active mitochondrial translation affects the enrichment of Oxa1 in the CM, we analyzed the influence of chloramphenicol, a specific inhibitor of mitochondrial translation (Denslow and O'Brien, 1978; Tenson and Mankin, 2006), on the distribution of Oxa1. To this end, we grew Oxa1-GFP–expressing cells harboring enlarged mitochondria on glucose-containing agar plates into small colonies to ensure that the cells were dividing and not in a stationary growth phase. Subsequently a single colony was picked, and the cells were transferred for 2 h in liquid growth medium containing lactate. As already shown (Figure 2D), after 2 h on lactate, Oxa1-GFP was found in the cristae-containing interior of the enlarged mitochondria (Figure 3A). Next, after 2 h in lactate, the cell suspension was divided into two parts. One half was treated for 2 h with chloramphenicol, whereas the other half was mock treated. We found that the treatment with chloramphenicol reversed the distribution from enrichment in the CM back to an enrichment in the IBM. This finding points to participation of mitochondrial translation in determining the localization of Oxa1 in the CM when the cells are propagated on a nonfermentable carbon source.
FIGURE 3:
The predominant localization of Oxa1 in the CM under respiratory growth conditions depends on mitochondrial translation and the interaction of Oxa1 with ribosomes. (A) Live-cell fluorescence microscopy of Δmdm10 cells expressing Oxa1-GFP and Qcr2-mRFP. The cells were initially grown on glucose-containing agar plates and then transferred into lactate-containing liquid growth medium. From top to bottom, immediately and 2 h after shifting the cells to lactate medium. After 2 h on lactate-containing medium, the cells were mock treated or incubated with chloramphenicol and grown for a further 2 h. Chloramphenicol inhibits translation at mitochondrial ribosomes. Single confocal sections are displayed. (B) Whole-cell extract from cells expressing Oxa1-∆RBD-GFP analyzed by immunoblotting using a GFP-specific antiserum. (C) Distribution of Oxa1-∆RBD-GFP in cells grown in lactate-containing medium analyzed by quantitative immuno-EM using a GFP-specific antiserum. Left, representative image of a cell expressing Oxa1-∆RBD-GFP in addition to the authentic Oxa1. The arrowheads point to gold particles. Right, quantification of its distribution between the IBM and the CM. The quantified distributions were corrected for the absolute lengths of CM and IBM (ratio CM/IBM in lactate medium, 1.5/1). (D) Fluorescent micrographs of Δmdm10 cells expressing Qcr2-mRFP and Oxa1-∆RBD-GFP. The cells expressed in addition the authentic Oxa1 from its native genomic locus to maintain the respiratory activity of the mitochondria. The cells were initially grown on glucose-containing agar plates and then transferred into lactate-containing medium. Images were taken immediately after the transfer (top) or 3 h after the transfer (bottom). The intensity profiles show the fluorescence intensity of Oxa1-ΔRBD-GFP (green) and Qcr2-mRFP (red) between the two arrowheads in the respective images. Scale bars, 2 μm (A, D) and 100 nm (C).
To further investigate the role of mitochondrial translation on the distribution of Oxa1, we next analyzed mitochondria in wild-type cells. The C-terminal tail of Oxa1 is matrix localized and contains a ribosome-binding domain (RBD), which directly interacts with mitochondrial ribosomes (Jia et al., 2003; Szyrach et al., 2003). We asked whether this interaction is required for the enrichment of Oxa1 in the CM under respiratory growth conditions. Because the RBD is essential for respiratory growth, we expressed GFP-tagged Oxa1 lacking the RBD in addition to the endogenous Oxa1. Western blotting showed that the deletion of the RBD did not induce degradation of the protein (Figure 3B). Surprisingly, we found, using immunogold-EM, that in cells grown on lactate, Oxa1-ΔRBD-GFP was enriched in the IBM (Figure 3C), whereas under the same growth conditions, Oxa1-Flag (Figure 2B) was enriched in the CM. This finding demonstrates that the RBD is required for the enrichment of Oxa1 in the CM under respiratory conditions. Consistently, also in the enlarged mitochondria of Δmdm10 cells, the RBD-deficient mutant of Oxa1 no longer shifted its predominant localization upon a change of the carbon source from the IBM to the CM (Figure 3D).
Together these findings strongly suggest that a direct interaction of Oxa1 with the actively translating ribosome is required for the enrichment of Oxa1 in the CM under respiratory conditions. The results also suggest that the predominant localization of Oxa1 in the CM under respiratory conditions is an active mechanism driven by the interaction with the ribosome rather than a passive consequence of different protein compositions in the inner membrane.
The accumulation of Oxa1 in the IBM depends on active mitochondrial protein import
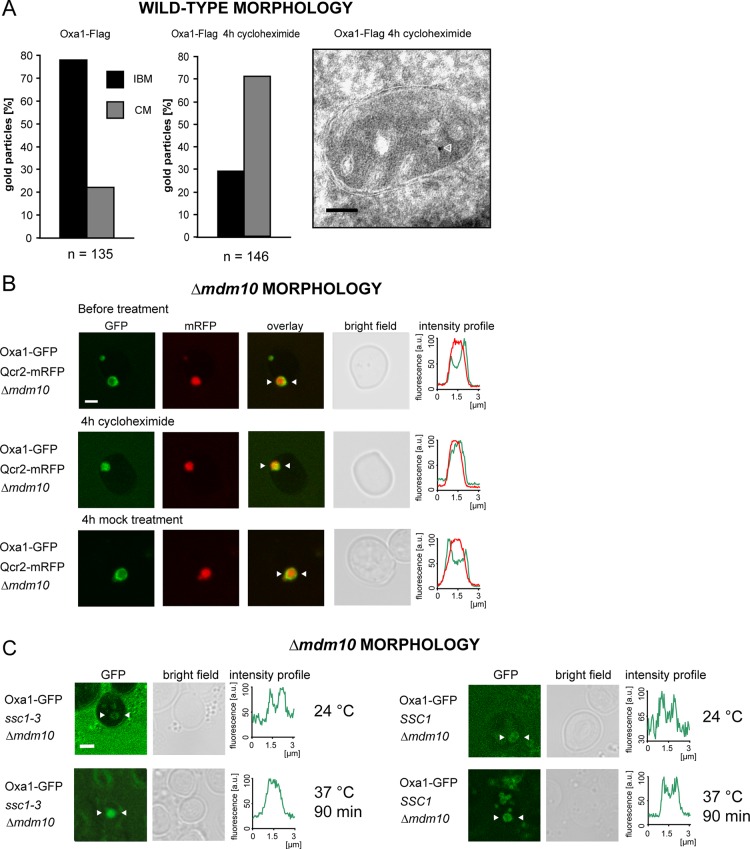
Oxa1 is not only required for the insertion of mitochondrial-encoded proteins in the inner membrane, but it also mediates the insertion of some nuclear-encoded proteins. So we next analyzed whether the import of nuclear-encoded proteins into mitochondria is required for the enrichment of Oxa1 in the IBM under fermentable growth conditions. To address this question, we incubated yeast cells expressing either Oxa1-Flag or Oxa1-GFP with cycloheximide, a potent inhibitor of cytoplasmic translation, and monitored the distribution of the tagged Oxa1 by quantitative immuno-EM (Figure 4A) in wild-type mitochondria, as well as by live-cell fluorescence microscopy in cells with enlarged mitochondria (Figure 4B). Four hours after application of cycloheximide, both approaches revealed a redistribution of Oxa1 toward the CM. Immuno-EM demonstrated that after this time period Oxa1 is no longer enriched in the IBM but is predominantly located in the CM (71% of all gold particles in the CM, n = 146). On mock treatment with the solvent dimethyl sulfoxide (DMSO), no redistribution was observed, demonstrating that the change in the submitochondrial distribution of Oxa1 was due to an inhibition of cytoplasmic translation.
FIGURE 4:
The predominant localization of Oxa1 in the IBM under fermentable growth conditions depends on the import of nuclear-encoded proteins. (A) Quantitative immuno-EM of the distribution of Oxa1-Flag in cells treated for 4 h with (middle) or without (left) cycloheximide, an inhibitor of cytoplasmic translation. The cells were grown in galactose-containing medium. Right, representative image. The arrowhead points to a gold particle. (B) Live-cell fluorescence microscopy of Δmdm10 cells expressing Oxa1-GFP and Qcr2-mRFP. The cells were grown on glucose-containing agar plates and transferred into fresh liquid growth medium containing glucose. They were imaged before (top) and after 4 h treatment with cycloheximide. As a control, the cells were incubated for 4 h with the solvent DMSO. The intensity profiles show the distribution of the fluorescence signals between the arrowheads. (C) Distribution of Oxa1-GFP in ssc1-3 cells exhibiting enlarged mitochondria due to a lack of MDM10. In ssc1-3 cells, mitochondrial import is largely normal at the permissive temperature (24°C), whereas at the nonpermissive temperature (37°C) import is inhibited. Left, distribution of Oxa1-GFP in ssc1-3 cells grown in glucose-containing medium at both temperatures. Right, control experiments with SSC1 cells. Scale bars, 100 nm (A) and 2 μm (B, C).
We next addressed the question of whether the redistribution of Oxa1 is caused by reduced overall protein import rates (due to the lack of available import substrates in cycloheximide-treated cells) or whether it is a rather indirect consequence of the inhibition of cytoplasmic protein translation. To this end, we used yeast cells harboring the temperature-sensitive (ts) ssc1-3 allele of SSC1 (ENS1; mtHsp70) (Gambill et al., 1993), coding for a constituent of the mitochondrial import motor, which is required for the transport of preproteins from the cytosol into the matrix but is dispensable for mitochondrial protein synthesis (Herrmann et al., 1994). At the nonpermissive temperature of 37°C, translocation of preproteins across the inner membrane is inhibited in ssc1-3 mitochondria, whereas at the permissive temperature (24°C) protein import is largely normal (Gambill et al., 1993). We found that in ssc1-3 cells with enlarged mitochondria, Oxa1-GFP was at the permissive temperature predominantly localized in the IBM when the cells were grown on glucose-containing media (Figure 4C). At the nonpermissive temperature, when mitochondrial import was blocked, enrichment in the IBM was no longer detectable, suggesting that active protein import is essential to maintain the predominant localization of Oxa1 in the IBM. In control cells with intact SSC1 the temperature shift did not influence the distribution of Oxa1 (Figure 4C).
We conclude that the import of nuclear-encoded mitochondrial proteins is necessary for the maintenance of the predominant localization of Oxa1 in the IBM in cells grown under fermentable conditions.
Overexpression of the Oxa1 substrate Mdl1 shifts the location of Oxa1 to the IBM
To further investigate the effect of mitochondrial import on the distribution of Oxa1, we next asked whether overexpression of a nuclear-encoded Oxa1 substrate influences the inner-mitochondrial distribution of Oxa1. The nuclear-encoded, multidrug-resistance–like protein Mdl1 exhibits six transmembrane segments (Young et al., 2001; Zutz et al., 2009). Oxa1 cooperates with the Tim23 complex in the stepwise integration of this polytopic protein (Bohnert et al., 2010). Hence Mdl1 is a genuine substrate of the Oxa1 machinery. We found that neither overexpression nor deletion of Mdl1 has any noticeable effect on the growth phenotype of cells propagated on fermentable or nonfermentable growth media at temperatures between 25°C and 37°C (Supplemental Figure S4), corroborating other reports (Young et al., 2001; Chloupkova et al., 2003).
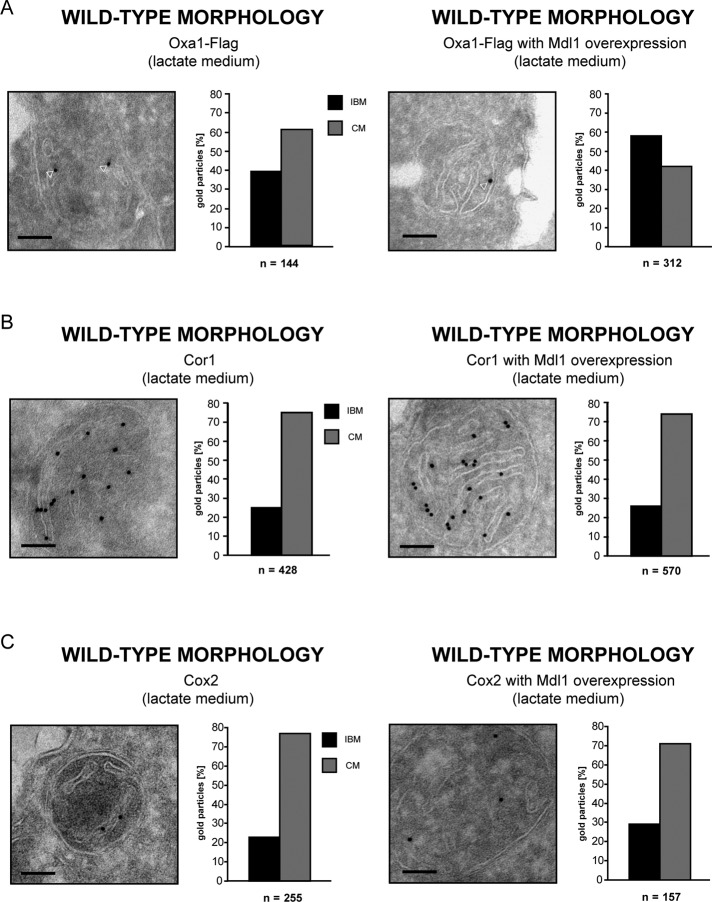
As shown earlier (Figure 2B), in mitochondria of wild-type cells, Oxa1 is enriched in the CM when the cells are propagated on the nonfermentable lactate. When HA-tagged Mdl1 was overexpressed, the Oxa1 distribution was shifted so that Oxa1 was enriched in the IBM (Figure 5A). Hence the overexpression of a nuclear-encoded substrate of Oxa1 influences the distribution of Oxa1.
FIGURE 5:
Overexpression of the nuclear-encoded Oxa1 substrate Mdl1 specifically shifts the distribution of Oxa1 toward the IBM. (A–C) Quantitative immuno-EM analysis of the influence of the overexpression of Mdl1 on the inner-mitochondrial distribution of Oxa1-Flag (A), Cor1 (B), and Cox2 (C). The samples were decorated with antisera specific for the Flag tag, the endogenous Cor1, or Cox2, respectively. In each case: left, cells carrying only an empty vector; right, cells overexpressing Mdl1 from a high-copy-number plasmid. The cells were grown under nonfermentable growth conditions (lactate). Shown are representative images and the corresponding quantification based on a large set of images. The quantified distributions were corrected for the absolute lengths of CM and IBM (ratio CM/IBM in lactate medium, 1.5/1). The arrowheads point to gold particles. Scale bars, 100 nm.
Next we asked whether the overexpression of Mdl1 specifically affects the distribution of Oxa1. We found that the enrichment of Cor1 (Qcr1), a core subunit of complex III of the respiratory chain, in the CM was not affected by overexpression of Mdl1 (75 or 74%, respectively, were assigned to the CM) (Figure 5B). Of interest, comparison of the distribution of Cox2 in wild-type cells with the distribution in cells overexpressing Mdl1 indicated a slight (∼6%) redistribution of Cox2 toward the IBM upon overexpression of Mdl1 (Figure 5C). Cox2 is integrated into the inner membrane by Oxa1 (He and Fox, 1997; Hell et al., 1997; Fiumera et al., 2009), and hence the substantial shift (∼19%) of the distribution of Oxa1 upon overexpression of Mdl1 might account for the small shift in the distribution of Cox2. These findings indicate that the overexpression of Mdl1, a substrate of Oxa1, specifically shifts the distribution of Oxa1 from the CM to the IBM, but that the overexpression of Mdl1 does not affect the distribution of inner membrane proteins unrelated to Oxa1.
Taken together, our results show that Oxa1 is dynamically redistributed between the IBM and the CM depending on the available carbon source. The distribution of Oxa1 between IBM and CM is influenced by the availability of its substrates, that is, of nuclear-encoded or mitochondrial-encoded proteins targeted to the inner membrane. The observation that the RBD of Oxa1, which facilitates the binding to mitochondrial ribosomes, is required for an enrichment of Oxa1 in the CM, whereas overexpression of a nuclear-encoded substrate favors enrichment in the IBM, further indicates that the distribution of Oxa1 is at least in part actively determined and not the consequence of a passive process.
DISCUSSION
For the analysis of the submitochondrial distribution of Oxa1, we used two approaches: quantitative immunogold electron microscopy on cryosectioned wild-type yeast cells and live-cell fluorescence microscopy on living yeast cells harboring enlarged mitochondria and expressing GFP-tagged Oxa1. The former approach provides quantitative localization information of proteins in unchanged wild-type mitochondria, and thus this method may be seen as the most suitable. Still, immuno-EM is time consuming and inevitably restricted to fixed cells. The use of live-cell fluorescence microscopy allowed us to follow the dynamical adaptation of the Oxa1-GFP distribution in living cells as a response to a change of the carbon source or the addition of inhibitors. Although the latter approach is convenient for screening and the analysis of inner-mitochondrial dynamics in living cells, it cannot replace quantitative immuno-EM, because the deletion of MDM10 may influence the distribution of some proteins (Wurm and Jakobs, 2006; Suppanz et al., 2009).
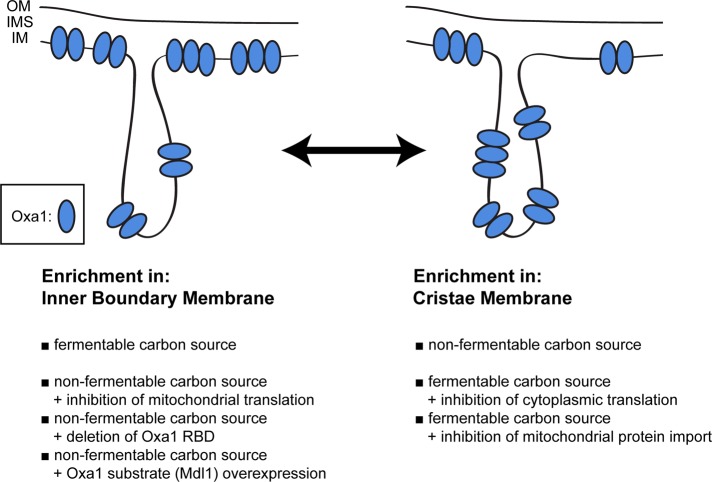
This study demonstrates that the distribution of Oxa1 between the CM and the IBM is dynamic. It is influenced by the growth conditions and by the availability of substrates for membrane insertion by the Oxa1 machinery (Figure 6 and Supplemental Figure S5).
FIGURE 6:
Schematic representation of the factors that influence the distribution of Oxa1 between the IMB and the CM. Oxa1 is enriched in the IBM when grown on a fermentable carbon source and redistributed to the CM upon a change to a nonfermentable carbon source. These distributions can be influenced by various means, as listed. IM, inner membrane; IMS, intermembrane space; OM, outer membrane.
But what determines the distribution of Oxa1? Given the fact that the inner membrane of mitochondria is exceptionally protein rich, with a protein/lipid mass ratio of ∼3:1 (Ardail et al., 1990; Simbeni et al., 1991), the distribution of Oxa1 could be a passive consequence of the presence of other proteins competing for space with Oxa1 and resulting in an extrusion of Oxa1 from the respective subcompartment. Several lines of evidence presented in this article, however, suggest that the distribution of Oxa1 is actively regulated. The finding that (under respiratory conditions) both the inhibition of mitochondrial translation as well as the deletion of the ribosome-binding site of Oxa1 prevent the enrichment of Oxa1 in the CM suggests that interactions of Oxa1 with actively translating mitochondrial ribosomes are important for the CM localization. On the other hand (under fermentable growth conditions), inhibition of either cytoplasmic translation or of protein import prevents enrichment in the IBM. Moreover, the overexpression of the nuclear-encoded Oxa1 substrate Mdl1 shifts the distribution of Oxa1 but not the distribution of Cor1 toward the IBM, indicating that the interactions of Oxa1 with the newly imported nuclear-encoded proteins (or the active import machinery) favor a localization of Oxa1 in the IBM. Together these data indicate that the respective distribution of Oxa1 is at least partially determined by the localization of the available substrates.
Certainly, other factors coinfluencing the distribution of Oxa1 in the inner membrane are possible. These may include further transient low-affinity interactions. Also, the finding of a synthetic growth defect in Δoxa1Δphb1 cells might point to a role of membrane lipids for the distribution of Oxa1 in the inner membrane (Osman et al., 2009). Structural components that may be involved in the regulation of the composition of the IBM and the CM are the so-called cristae junctions (Mannella et al., 1994; Perkins et al., 1997; Perkins and Frey, 2000). Cristae junctions are relatively uniform, small tubular structures that connect the CM with the IBM and may hold sieve-like functions for proteins moving between the two subdomains of the inner membrane. Supporting the concept of a diffusion barrier between CM and IBM, it has been shown that the precursor form of Ccp1 (pCcp1), which has no further reported binding partners, is enriched in the IBM in mutants deficient in the processing of Ccp1 (Suppanz et al., 2009). This observation would be in agreement with the concept of the cristae junctions being not absolute barriers but kinetic or diffusion barriers for specific proteins. It is possible that the distribution of Oxa1 is also influenced by the cristae junctions, although our data do not provide further evidence for this assumption.
On inoculation of S. cerevisiae cells into a medium rich in glucose, the sugar is catabolized mainly by fermentation, with the production of ethanol. As glucose becomes exhausted, the cells turn to alternative carbon sources, for example, ethanol, glycerol, or lactate. This switch from anaerobic growth to aerobic respiration, called diauxic shift, has been studied intensely on the mRNA level (DeRisi et al., 1997; Boy-Marcotte et al., 1998; Haurie et al., 2001; Galdieri et al., 2010). A study using two-dimensional protein gels demonstrated that apart from the general increase of the mitochondrial proteome, the changes in the relative amounts of individual mitochondrial proteins were surprisingly small compared with the transcriptional changes upon diauxic shift (Ohlmeier et al., 2004). The apparent discrepancy between the transcriptome and the proteome data points to additional regulatory mechanism on the posttranscriptional level, including differing protein turnover rates after the diauxic shift. Our observation that Oxa1 changes its distribution upon diauxic shift suggests that the availability of the Oxa1 substrates is altered after changing the carbon source. The observed Oxa1 distributions indicate that under fermentable growth conditions relatively more nuclear-encoded substrates are available, whereas on a nonfermentable carbon source the proportion is shifted to mitochondrial-encoded substrates.
The Oxa1 distributions also suggest that mitochondrial-encoded Oxa1 substrates are preferentially inserted in the CM, whereas nuclear-encoded substrates are preferentially inserted in the IBM. In the future, it will be of utmost relevance to clarify how the distribution and activity of the Oxa1 machinery are coordinated with the assembly and quality control of the mitochondrial supercomplexes.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth media
All S. cerevisiae strains used were isogenic to BY4741 or BY4743 (Brachmann et al., 1998). Genomic tagging and gene disruption was performed as described previously (Wurm and Jakobs, 2006; Suppanz et al., 2009). For the generation of Δoxa1 strains expressing an OXA1 construct from a plasmid, we transformed diploid Δoxa1/OXA1 cells with a plasmid encoding the respective construct. After sporulation the appropriate spores were selected and verified by PCR. All strains were tested for their ability to grow on nonfermentable carbon sources.
For the expression of GFP or Flag fusion genes we modified the pUG36 (Niedenthal et al., 1996) yeast expression vector. All fusion proteins used in this study have the GFP or the Flag tag at the C-terminus. For the construction of the required vectors, the coding sequence for GFP (without stop codon) in pUG36 was replaced by either a sequence coding for GFP-S65T or by a sequence coding for the Flag tag, allowing C-terminal fusions. These vectors, referred to hereafter as pUG-ST-C-GFP or pUG-ST-C-Flag were used as a basis for the construction of the various OXA1 fusion constructs. All OXA1 fusion constructs used in this study were under the control of the OXA1 promoter.
For the overexpression of the Oxa1 substrate Mdl1, we cloned the corresponding open reading frame into the plasmid pAG425GPD-ccdB-HA (Addgene, Cambridge, MA).
Yeast cells were cultivated according to standard protocols (Sherman, 2002) at 30°C (except stated otherwise) in liquid media or on agar plates containing 2% (wt/vol) glucose, 2% (wt/vol) galactose, or 2% (wt/vol) lactate.
Preparation of whole-cell extracts, isolation of mitochondria, carbonate extraction, and subfractionation
For the preparation of whole-cell extracts, logarithmical growing cells were harvested by centrifugation and washed with buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], 150 mM potassium acetate, 2.5 mM EDTA, 1% [wt/vol] Triton-X-100, protease inhibitors [Complete Protease Inhibitor Cocktail; Roche, Basel, Switzerland], 18 mM phenylmethylsulfonyl fluoride [PMSF], Pepstatin A [140 μg/ml], pH 7.2) at 4°C. Subsequently, the cells were lysed by vortexing in the presence of glass beads at 4°C. The debris was separated from the soluble components by centrifugation (16,000 × g). Carbonate extraction, isolation of mitochondria, and subfractionation were essentially performed as described (Diekert et al., 2001).
Confocal light microscopy
Light microscopic analysis of cells with enlarged mitochondria was performed as described (Wurm and Jakobs, 2006). For the analysis, heat-shock-competent Δmdm10 cells expressing Qcr2-mRFP from the genomic locus were prepared and stored at −80°C. Alternatively, MDM10 was deleted immediately before an imaging experiment. For each experiment, the cells were freshly transformed with a plasmid encoding the respective OXA1 construct. Cells from single colonies were analyzed using confocal light microscopy 5–9 d after the transformation. Only enlarged mitochondria with diameters ranging from 0.8 to 2 μm were analyzed. Imaging was performed with a beam scanning confocal microscope (TCS SP1, or TCS SP5; Leica Microsystems, Wetzlar, Germany) equipped with 1.4 numerical aperture oil immersion lenses (100×, PlanApo, or 63×, PlanApo; Leica). Dual-color images were obtained by sequential scanning. All images were averaged twice. Except for contrast stretching and smoothing, no image processing was applied.
Quantitative immuno–electron microscopy
The yeast cells used for immuno–EM were cultivated at 30°C in liquid media containing 2% (wt/vol) galactose or 2% (wt/vol) lactate as the sole carbon source and were harvested during the logarithmic growth phase. Ultrathin cryosections of the yeast cells were prepared as described previously (Tokuyasu, 1973; Suppanz et al., 2009).
The ultrathin sections (80 nm) were incubated with a polyclonal rabbit antibody against GFP (Abcam, Cambridge, United Kingdom) or a monoclonal mouse antibody against the Flag tag (Sigma-Aldrich, Munich, Germany) for 20 min, followed by an incubation with protein A–gold (10 nm; G. Posthuma, Utrecht University, Utrecht, Netherlands) for 20 min to detect the rabbit primary antibody or a gold-coupled goat secondary antibody against mouse (10 nm, immunoglobulin G; Aurion, Wageningen, Netherlands), respectively. The cryosections were thoroughly washed (five times for 3 min in Tris-buffered saline [TBS]/0.5% [wt/vol] bovine serum albumin; five times for 3 min in TBS), contrasted with uranyl acetate/methyl cellulose for 10 min on ice, embedded in the same solution, and finally examined with an electron microscope (CM120; Philips, Eindhoven, Netherlands).
For quantification, the submitochondrial locations of at least 100 individual gold particles were analyzed. To this end, the distance between the gold particle and the inner boundary membrane was measured. Gold particles within a distance of ≤20 nm to the inner boundary membrane were assigned to the mitochondrial inner boundary membrane. Gold particles within the organelle and exhibiting a distance >20 nm were assigned to the cristae membranes.
To determine the relative surface areas of the IBM and the CM for the respective conditions, representative images showing excellent structural preservation were chosen, and the membrane lengths were measured using ImageJ (National Institutes of Health, Bethesda, MD). The determined ratios of IBM to CM fully corroborated results of previous studies, that is, 1:1 for cells grown on galactose as the sole carbon source and 1:1.5 for cells grown on lactate (Vogel et al., 2006; Suppanz et al., 2009).
Supplementary Material
Acknowledgments
We are grateful to K. Hell (University of Munich, Munich Germany) and D. Rapaport (University of Tübingen, Tübingen, Germany) for providing antisera and thank P. Rehling (University of Göttingen, Göttingen, Germany) for providing the ssc1-3 strain. We thank Stefan W. Hell for continuous support. We acknowledge R. Schmitz-Salue for excellent technical assistance and J. Jethwa for carefully reading the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant JA 1129/3 and the DFG Research Center for Molecular Physiology of the Brain (both to S.J.) and Deutsche Forschungsgemeinschaft Grant FOR967 (to J.M.H.).
Abbreviations used:
- CM
cristae membrane
- DMSO
dimethyl sulfoxide
- EM
electron microscopy
- GFP
green fluorescent protein
- HA
human influenza hemagglutinin
- IBM
inner boundary membrane
- IM
inner membrane
- IMS
intermembrane space
- OM
outer membrane
- PK
proteinase K
- RBD
ribosome-binding domain
- RFP
red fluorescent protein
- TM
transmembrane
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-06-0538) on April 18, 2012.
REFERENCES
- Ardail D, Privat JP, Egret-Charlier M, Levrat C, Lerme F, Louisot P. Mitochondrial contact sites. Lipid composition and dynamics. J Biol Chem. 1990;265:18797–18802. [PubMed] [Google Scholar]
- Bohnert M, Rehling P, Guiard B, Herrmann JM, Pfanner N, van der Laan M. Cooperation of stop-transfer and conservative sorting mechanisms in mitochondrial protein transport. Curr Biol. 2010;20:1227–1232. doi: 10.1016/j.cub.2010.05.058. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Fiumera HL, Dujardin G, Fox TD. Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim Biophys Acta. 2009;1793:60–70. doi: 10.1016/j.bbamcr.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J Bacteriol. 1998;180:1044–1052. doi: 10.1128/jb.180.5.1044-1052.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li JC, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288c—a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chloupkova M, LeBard LS, Koeller DM. MDL1 is a high copy suppressor of ATM1: evidence for a role in resistance to oxidative stress. J Mol Biol. 2003;331:155–165. doi: 10.1016/s0022-2836(03)00666-1. [DOI] [PubMed] [Google Scholar]
- Denslow ND, O'Brien TW. Antibiotic susceptibility of the peptidyl transferase locus of bovine mitochondrial ribosomes. Eur J Biochem. 1978;91:441–448. doi: 10.1111/j.1432-1033.1978.tb12696.x. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Diekert K, de Kroon AI, Kispal G, Lill R. Isolation and subfractionation of mitochondria from the yeastSaccharomyces cerevisiae. Methods Cell Biol. 2001;65:37–51. doi: 10.1016/s0091-679x(01)65003-9. [DOI] [PubMed] [Google Scholar]
- Fiumera HL, Dunham MJ, Saracco SA, Butler CA, Kelly JA, Fox TD. Translocation and assembly of mitochondrially coded Saccharomyces cerevisiae cytochrome c oxidase subunit Cox2 by Oxa1 and Yme1 in the absence of Cox18. Genetics. 2009;182:519–528. doi: 10.1534/genetics.109.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdieri L, Mehrotra S, Yu S, Vancura A. Transcriptional regulation in yeast during diauxic shift and stationary phase. OMICS. 2010;14:629–638. doi: 10.1089/omi.2010.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson RW, Selker JML, Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003;546:355–358. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- Haurie V, Perrot M, Mini T, Jeno P, Sagliocco F, Boucherie H. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J Biol Chem. 2001;276:76–85. doi: 10.1074/jbc.M008752200. [DOI] [PubMed] [Google Scholar]
- He S, Fox TD. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Herrmann J, Pratje E, Neupert W, Stuart RA. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- Hell K, Herrmann JM, Pratje E, Neupert W, Stuart RA. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Neupert W, Stuart RA. Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J. 2001;20:1281–1288. doi: 10.1093/emboj/20.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Stuart RA, Craig EA, Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs S. High resolution imaging of live mitochondria. Biochim Biophys. Acta. 2006;1763:561–575. doi: 10.1016/j.bbamcr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Dienhart MK, Stuart RA. Oxa1 directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex. Mol Biol Cell. 2007;18:1897–1908. doi: 10.1091/mbc.E06-10-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J, Samuelsson T, de Gier JW. YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 2001;501:1–5. doi: 10.1016/s0014-5793(01)02616-3. [DOI] [PubMed] [Google Scholar]
- Mannella CA, Marko M, Penczek P, Barnard D, Frank J. The internal compartmentation of rat-liver mitochondria: tomographic study using the high-voltage transmission electron microscope. Microsc Res Tech. 1994;27:278–283. doi: 10.1002/jemt.1070270403. [DOI] [PubMed] [Google Scholar]
- Niedenthal RK, Riles L, Johnston M, Hegemann JH. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ohlmeier S, Kastaniotis AJ, Hiltunen JK, Bergmann U. The yeast mitochondrial proteome, a study of fermentative and respiratory growth. J Biol Chem. 2004;279:3956–3979. doi: 10.1074/jbc.M310160200. [DOI] [PubMed] [Google Scholar]
- Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brugger B, Westermann B, Langer T. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J Cell Biol. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penniston JT, Harris RA, Asai J, Green DE. The conformational basis of energy transformations in membrane systems. I. Conformational changes in mitochondria. Proc Natl Acad Sci USA. 1968;59:624–631. doi: 10.1073/pnas.59.2.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G, Renken C, Martone ME, Young SJ, Ellisman M, Frey T. Electron tomography of neuronal mitochondria—three-dimensional structure and organization of cristae and membrane contacts. J Struct Biol. 1997;119:260–272. doi: 10.1006/jsbi.1997.3885. [DOI] [PubMed] [Google Scholar]
- Perkins GA, Frey TG. Recent structural insight into mitochondria gained by microscopy. Micron. 2000;31:97–111. doi: 10.1016/s0968-4328(99)00065-7. [DOI] [PubMed] [Google Scholar]
- Reif S, Randelj O, Domanska G, Dian EA, Krimmer T, Motz C, Rassow J. Conserved mechanism of Oxa1 insertion into the mitochondrial inner membrane. J Mol Biol. 2005;354:520–528. doi: 10.1016/j.jmb.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Sato T, Mihara K. Topogenesis of mammalian Oxa1, a component of the mitochondrial inner membrane protein export machinery. J Biol Chem. 2009;284:14819–14827. doi: 10.1074/jbc.M809520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. In: Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular and Cell Biology, Part B. London: Academic Press; 2002. pp. 3–41. [Google Scholar]
- Simbeni R, Pon L, Zinser E, Paltauf F, Daum G. Mitochondrial membrane contact sites of yeast. Characterization of lipid components and possible involvement in intramitochondrial translocation of phospholipids. J Biol Chem. 1991;266:10047–10049. [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppanz IE, Wurm CA, Wenzel D, Jakobs S. The m-AAA protease processes cytochrome c peroxidase preferentially at the inner boundary membrane of mitochondria. Mol Biol Cell. 2009;20:572–580. doi: 10.1091/mbc.E07-11-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, Mankin A. Antibiotics and the ribosome. Mol Microbiol. 2006;59:1664–1677. doi: 10.1111/j.1365-2958.2006.05063.x. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT. A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F, Bornhovd C, Neupert W, Reichert AS. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Neupert W. Functional and biogenetical heterogeneity of the inner membrane of rat-liver mitochondria. Eur J Biochem. 1972;25:379–396. doi: 10.1111/j.1432-1033.1972.tb01707.x. [DOI] [PubMed] [Google Scholar]
- Wurm CA, Jakobs S. Differential protein distributions define two subcompartments of the mitochondrial inner membrane in yeast. FEBS Lett. 2006;580:5628–5634. doi: 10.1016/j.febslet.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- Zutz A, Gompf S, Schagger H, Tampe R. Mitochondrial ABC proteins in health and disease. Biochim Biophys Acta. 2009;1787:681–690. doi: 10.1016/j.bbabio.2009.02.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.