TBX3 is a direct target of the retinoic acid (RA) signaling pathway. RA activates endogenous TBX3 expression, which is mediated by a RA–receptor complex directly binding and activating the TBX3 promoter, and this regulation is functionally relevant in mouse embryonic limb development.
Abstract
TBX3, a member of the T-box transcription factor gene family, is a transcriptional repressor that is required for the development of the heart, limbs, and mammary glands. Mutations in TBX3 that result in reduced functional protein lead to ulnar-mammary syndrome, a developmental disorder characterized by limb, mammary gland, tooth, and genital abnormalities. Increased levels of TBX3 have been shown to contribute to the oncogenic process, and TBX3 is overexpressed in several cancers, including breast cancer, liver cancer, and melanoma. Despite its important role in development and postnatal life, little is known about the signaling pathways that modulate TBX3 expression. Here we show, using in vitro and in vivo assays, that retinoic acid (RA) activates endogenous TBX3 expression, which is mediated by an RA–receptor complex directly binding and activating the TBX3 promoter, and we provide evidence that this regulation may be functionally relevant in mouse embryonic limb development. Our data identify TBX3 as a direct target of the RA signaling pathway and extend our understanding of the role and regulation of TBX3 in limb development.
INTRODUCTION
TBX3 is a member of the T-box gene family that encodes DNA-binding transcription factors with well-defined roles in embryonic development. Tbx3 plays critical roles in a variety of developmental processes, including maintenance of stem cells, cell-fate determination, and organogenesis. It has been shown to promote self-renewal of embryonic stem cells through up-regulation of the pluripotency factor Nanog (Ivanova et al., 2006; Niwa et al., 2009), and when coexpressed with Oct4, Sox2, and Klf4, Tbx3 improves the ability of induced pluripotent stem cells to colonize germ tissues and produce viable offspring (Han et al., 2010). Tbx3 has also been shown to control the proliferation and cell-fate determination of multipotent hepatic progenitor cells in the developing liver (Suzuki et al., 2008). During heart development, Tbx3 contributes to the specification of the atrioventricular conduction system, which coordinates contraction of the heart (Bakker et al., 2008), as well as correct development of the outflow tract (Mesbah et al., 2008). In humans, TBX3 is expressed in the fetal lung, kidney, heart, liver, and spleen (Bamshad et al., 1999), and Tbx3 homozygous mutant mice die in utero with abnormalities in the limbs, genitalia, and mammary glands (Davenport et al., 2003; Jerome-Majewska et al., 2005). In the mouse, the hindlimb is truncated, whereas the forelimb is less severely affected with abnormal or missing posterior elements (Davenport et al., 2003).
Of importance, dosage sensitivity to TBX3 has been reported, which is underscored by observations that mutations that lead to haploinsufficiency of the human TBX3 gene result in ulnar-mammary syndrome. This syndrome is characterized by malformations of the apocrine glands, genitalia, hair, teeth, and limbs, including severe reduction of the posterior elements of the forelimb, with rare involvement of the hindlimb (Bamshad et al., 1997), in contrast to the mouse, in which the hindlimb is always more severely affected (Davenport et al., 2003). In addition, several lines of evidence suggest that overexpression of TBX3 may be a critical step in oncogenesis. TBX3 levels are up-regulated in a number of breast cancer cell lines, some small cell lung cancers, rat bladder carcinomas, and liver tumors (Ito et al., 2005; Lomnytska et al., 2006; Renard et al., 2007) and is detected in the plasma of >80% of ovarian cancer and breast cancer patients (Lomnytska et al., 2006). Recent data by Peres et al. (2010) demonstrate that overexpression of TBX3 in melanoma and breast cancer cells is responsible for tumor formation, metastasis, and invasion, potentially through its ability to suppress E-cadherin expression. This is consistent with a study by Rodriguez et al. (2008), which showed that TBX3 directly represses E-cadherin expression and that later stages of melanoma progression correlate with elevated levels of TBX3 and decreased levels of E-cadherin. An understanding of how TBX3 expression is regulated thus has implications for its role in embryonic development, as well as for shedding light on its contribution to oncogenesis. However, only a few signaling pathways governing Tbx3 expression have been described.
A study by Renard et al. (2007) demonstrated that Tbx3 is downstream of the Wnt/β-catenin signaling pathway in liver cancers and in particular that an active, mutant form of β-catenin transcriptionally up-regulated Tbx3. More recently, protein kinase C signaling was shown to activate TBX3 expression via the AP-1 transcription factors c-Jun and JunB, and this was shown to be important in promoting breast cancer cell migration (Mowla et al., 2011). Although the phosphotidylinositol-3-OH kinase-Akt, mitogen-activated protein kinase, and bone morphogenetic protein (BMP) pathways have also been implicated in the regulation of Tbx3 (Behesti et al., 2006; Niwa et al., 2009), the details of the mechanisms involved have yet to be elucidated. The developmentally important retinoic acid (RA) signaling pathway has also been implicated in the regulation of certain T-box family members. For example, chick embryos exposed to ectopic RA showed an overall repression of Tbx1 expression (Roberts et al., 2005) but an expansion of Tbx5 expression in the atrium of the heart (Liberatore et al., 2000). Furthermore, an investigation by Suzuki et al. (2004) revealed that in chick limbs implanted with RA-soaked beads, there was an expansion in expression of both Tbx3 and its closely related family member Tbx2. Consistent with these data, Tümpel et al. (2002) demonstrated a loss of posterior Tbx3 expression in the leg bud of retinoid-deficient quail. Although these studies are suggestive that RA may be able to regulate Tbx3 gene expression, the precise mechanisms of this regulation are not known, and whether these effects are reproducible in a mammalian model system has not been shown.
During development, RA is produced by the conversion of retinaldehyde sourced from maternal retinoids by retinaldehyde dehydrogenases (Raldh1, 2, and 3). RA activates the nuclear RA receptors (RARs) and retinoid X receptors (RXRs; reviewed in Bastien and Rochette-Egly, 2004). These receptors function as a RA-binding heterodimer that regulates target gene transcription via a retinoic acid response element (RARE; Mangelsdorf et al., 1995). The importance of RA signaling in embryogenesis is highlighted by reports describing numerous developmental defects in mouse embryos in which Raldh2 is knocked out or when different isoforms of the RARs are mutated together. These defects include compromised formation of the brain, pancreas, heart, lungs, limbs, and skeleton (Lohnes et al., 1994; Mendelsohn et al., 1994; Niederreither et al., 2001, 2002a; Martín et al., 2005; Ribes et al., 2006). Given that RA and Tbx3 have been implicated in the development of several common organs, we wanted to explore the possibility that Tbx3 may be downstream of the RA signaling pathway. Indeed, here we show, using in vitro and in vivo assays, that the RA–receptor complex directly binds the TBX3 promoter to activate TBX3 expression, and we provide evidence that this regulation is functionally relevant in mouse embryonic limb development.
RESULTS
Retinoic acid transcriptionally activates TBX3 gene expression
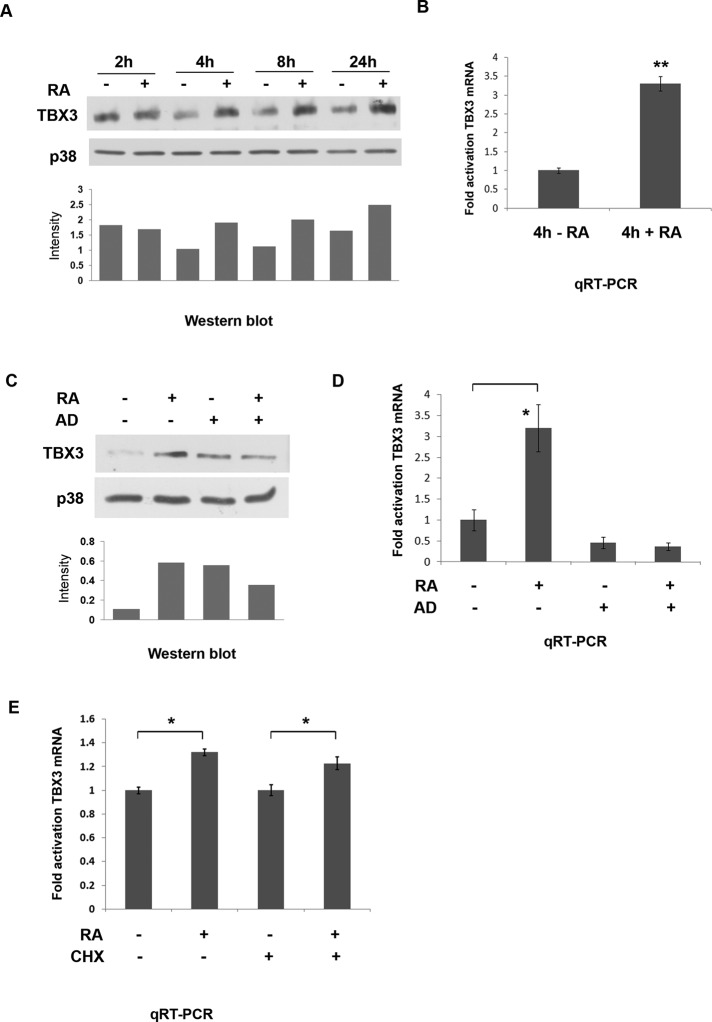
The development of several organs, including the heart, kidney, and limbs require both RA and Tbx3 (Lohnes et al., 1995; Bamshad et al., 1999), raising the interesting possibility that they act in sequence or in parallel during organogenesis. To begin to address this question, we first tested whether Tbx3 expression can be regulated by RA in cultured cells, using the ME1402 human melanoma line, which expresses endogenous TBX3 (Peres et al., 2010). ME1402 cells were treated with 10 μM all-trans retinoic acid or dimethyl sulfoxide (DMSO) vehicle over a time course spanning 2–24 h. Western blot analyses show that TBX3 levels increase with RA at all time points tested (Figure 1A), indicating that RA is able to rapidly up-regulate expression of TBX3 protein. Quantitative real-time PCR (qRT-PCR) experiments carried out on cells treated with RA for 4 h show a corresponding increase in TBX3 mRNA levels (Figure 1B), suggesting that RA may regulate TBX3 transcriptionally. This was confirmed in experiments in which the pretreatment of ME1402 cells with actinomycin D (a transcriptional inhibitor) abolished the 4-h RA-mediated increase of TBX3 protein (Figure 1C) and mRNA (Figure 1D) levels. Moreover, pretreatment of ME1402 cells with cycloheximide (an inhibitor of de novo protein synthesis) did not abolish the activation of TBX3 mRNA levels by RA (Figure 1E), indicating that the RA signaling pathway directly regulates TBX3 transcription.
FIGURE 1:
Retinoic acid activates human TBX3 mRNA and protein levels. ME1402 cells were treated with either vehicle (DMSO) or 10 μM RA. (A) Protein prepared from the indicated times was analyzed by Western blotting with an antibody to TBX3, and p38 was used as a loading control. The bar graph compares the relative intensity of each TBX3 band normalized to the p38 loading control. (B) The cells were treated with RA for 4 h, and total RNA was extracted. Quantitative real-time PCR was performed on reverse-transcribed RNA using primers specific to TBX3, and mRNA levels were normalized to GUSB. (C, D) Cells were pretreated with vehicle (control) or 5 μg/ml actinomycin D (AD) for 1 h and then treated with RA for 4 h, and protein and RNA were harvested for use in (C) Western blot analysis and (D) real-time PCR as described in A and B, respectively. The bar graph in C compares the relative intensity of each TBX3 band normalized to the p38 loading control. (E) Cells were pretreated with vehicle (control) or 30 μg/ml cycloheximide (CHX) for 1 h and then treated with RA for 4 h and RNA harvested for use in real-time PCR as described in B. Bars, SD. *p < 0.05, **p < 0.001.
RA receptors are likely to be direct transcriptional regulators of TBX3
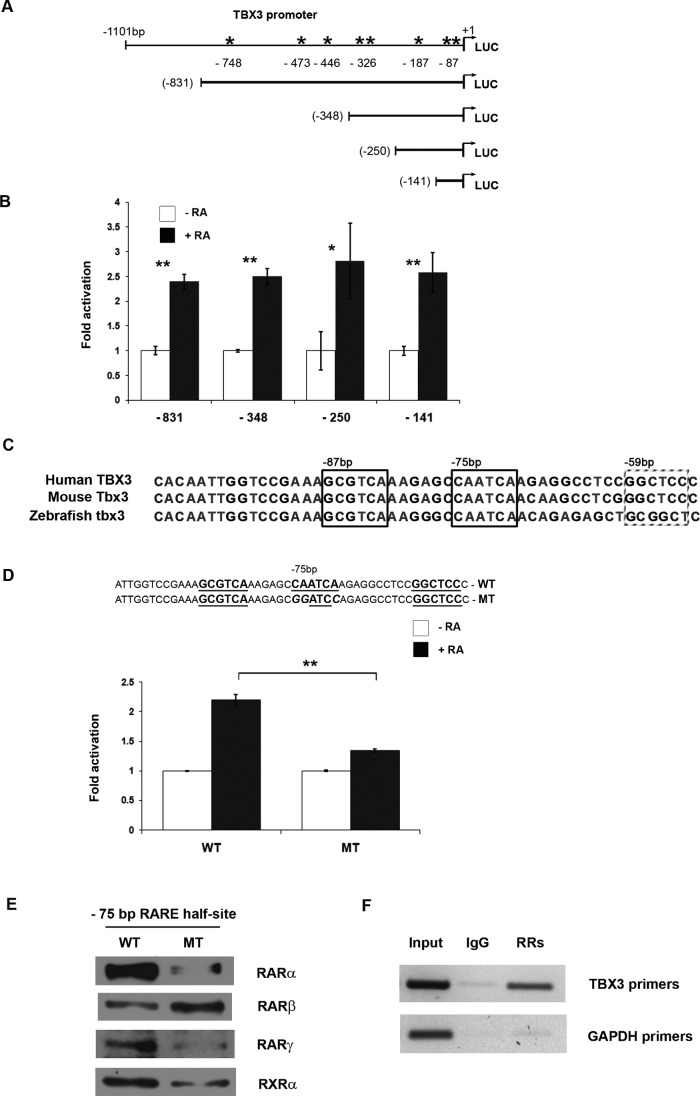
The results shown in Figure 1 suggested that the RA-mediated increase in TBX3 mRNA and protein levels is a direct transcriptional effect. Analysis of the 5′ regulatory region of the TBX3 gene revealed several putative RAREs ranging from −59 to −748 base pairs upstream of the transcription start site (Figure 2A). To investigate whether RA could activate expression via these putative RAREs, we performed transient cotransfection experiments using 501mel cells cotransfected with RAR and RXR expression vectors along with a luciferase vector containing 831 base pairs of TBX3 5′-flanking sequence. In the presence of RA, we observed a significant increase in luciferase reporter activity (Figure 2B), indicating that this region of the TBX3 promoter is likely to harbor a RARE(s). To further localize the putative RARE(s), we tested whether progressively deleting parts of the TBX3 promoter would abolish RA-dependent transactivation. We found that constructs containing 141 base pairs or more of TBX3 5′-flanking sequence could induce RA-dependent reporter expression, indicating that the RARE is likely to reside in the region 141 base pairs upstream from the transcriptional start site (Figure 2B).
FIGURE 2:
Retinoic acid activation of the TBX3 promoter is mediated by a degenerate RARE half site at −75 base pairs. (A) Schematic representation of human TBX3 deletion constructs. The arrow indicates the transcription start site at +1, and the asterisk indicates the putative RAREs. (B) 501mel cells cotransfected with TBX3-Luc deletion constructs and vectors expressing human RARα and RXRα were treated with vehicle (DMSO) or RA (white and black bars, respectively). Luciferase activity was measured 48 h posttransfection. (C) Alignment of the −150–base pair region of human, mouse, and zebrafish Tbx3 promoters. Sequence shown extends from −103 to −53 base pairs in the human TBX3 promoter, with a border outlining each RARE half-site. (D) WT- or MT-TBX3 promoter luciferase constructs shown were transfected into 501mel cells and treated as in B and luciferase activity analyzed. For B and D the fold activation values were calculated by setting promoter activity of untreated cells to 1. Mean values (±) were calculated from three independent experiments. (E) Biotinylated DNA probes of the TBX3 promoter (−137/+36 base pairs) containing the WT or MT RARE were generated by PCR, immobilized on streptavidin beads, and incubated with nuclear extracts from ME1402 cells treated with 10 μM RA for 4 h. The DNA-bound protein complexes were isolated and analyzed by Western blotting using antibodies to RARα, RARβ, RARγ, and RXRα. (F) ME1402 cells were treated with 10 μM RA for 4 h and chromatin immunoprecipitation assays performed with antibodies against RARα, RARβ, RARγ, and RXRα (RRs) or IgG (negative control). Immunoprecipitated DNA was assayed by PCR with primers against the TBX3 promoter or GAPDH (negative control). Bars, SD. *p < 0.05, **p < 0.001.
Scanning and sequence alignment of this region revealed a degenerate RARE at −87 base pairs that was highly conserved between human, mouse, and zebrafish Tbx3 promoters (Figure 2C). Furthermore, the half-sites at −87 and −75 base pairs comprising this putative degenerate RARE contain conserved, repeated TCA motifs, which are often observed in RAREs (Chen et al., 2007; Donato et al., 2007; Dhandapani et al., 2011), and a nonconsensus spacer region of 6 base pairs separating the half-sites, which is in the range of previously described spacers of 1, 2, 3, 8, and 11 base pairs (Lucas et al., 1991; Mader et al., 1994; Donato et al., 2007; Chen et al., 2007; Dhandapani et al., 2011). Of interest, the −75–base pair half-site may also form a putative RARE with a half-site at −59 base pairs, although this half-site is not as evolutionarily conserved as the one at −87. To test whether the RARE half-site at −75 base pairs was required for RA-dependent expression of the TBX3-luciferase reporter, we examined the effect of mutating this sequence on promoter activity. We transfected 501mel cells with constructs containing either the wild-type (WT) or mutant (MT) luciferase vectors along with RAR and RXR expression vectors and measured luciferase reporter activity. As expected, the construct containing the WT RARE half-site at −75 base pairs efficiently activated luciferase activity (Figure 2D). In contrast, luciferase activity was much lower in cultures transfected with the mutant TBX3 promoter. It is worth noting that when the RARE half-site at −75 base pairs was mutated, reduction but not loss of basal TBX3 promoter activity was observed, suggesting that this site may also play a role in basal TBX3 promoter activity (unpublished data).
Using a DNA affinity immunoblot (DAI) in vitro assay, we next sought to determine whether RA receptors were able to bind the identified RAREs in the TBX3 promoter. Nuclear extracts from RA-treated ME1402 cells were incubated with biotinylated DNA probes containing either the wild-type or mutant TBX3 RARE. Protein-bound biotinylated DNA was isolated and analyzed by Western blotting using antibodies specific to RARα, RARβ, RARγ, and RXRα to determine which RARs and RXRs could bind to the sequence. The results show that, with the exception of RARβ, all the RA receptors tested bound the probe carrying the WT −75–base pair RARE half-site in the TBX3 promoter and displayed a decreased affinity for this site when it was mutated (Figure 2E). RARβ, however, bound the probe carrying the mutated RARE at −75 base pairs even more strongly than the WT probe.
To establish whether RA receptors are able to bind to the TBX3 promoter in vivo, we performed chromatin immunoprecipitation assays. RA-treated ME1402 cells were fixed with formaldehyde, and the cross-linked chromatin was extracted and sheared. RA-bound chromatin was immunoprecipitated with appropriate antibodies, and after reversing the cross-link, a set of primers spanning the putative RARE half-sites at −87, −75, and −59 base pairs in the TBX3 promoter was used to amplify the immunoprecipitated DNA. The result obtained (Figure 2F) indicated that a specific PCR product was detected when using a mixture of retinoic acid receptor antibodies but not with the nonspecific immunoglobulin G (IgG) control. PCR conducted with primers against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a negative control. Taken together, the data presented here suggest that the RA transcriptional complex both binds to and directly activates TBX3 gene expression.
RA-mediated increase in TBX3 levels correlates with a decrease in cell number
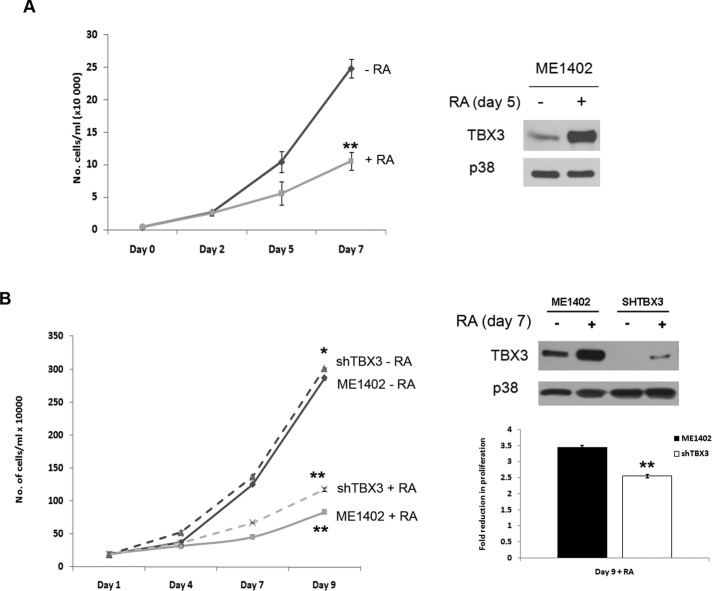
RA has been shown to be an effective chemopreventative agent in clinical trials on breast cancer due to its antiproliferative effects on cell cycle arrest, differentiation, and apoptosis, and we recently showed that TBX3 may be a novel target for anticancer drugs (Peres et al., 2010). Of importance, although knocking down TBX3 in breast cancer and melanoma cells inhibited tumor formation and invasion, it led to an increase in cell proliferation. We were therefore interested to determine the effects of RA on the proliferation of ME1402 melanoma cells over a period of 7 d. As an indirect measure of proliferation, growth curve analyses of cells grown in the presence of RA showed a significant decrease in cell numbers compared with untreated cells (Figure 3A, left), which correlated with an increase in TBX3 protein levels (Figure 3A, right). These results suggested that the RA-mediated decrease in ME1402 cell number might depend on activation of TBX3 expression. To explore this possibility, we generated a stable knockdown TBX3 cell line using a lentiviral system (Figure 3B) and tested its proliferative ability in response to RA by growth curve analyses. The results show that silencing TBX3 reduces the impact of RA on the proliferation of the ME1402 cells, which was shown to be statistically significant when the data were subjected to a two-sample t test (Figure 3B, left). Whereas the ME1402 cells show an almost 3.5-fold reduction in cell number by 9 d of RA treatment, the shTBX3 cells were less responsive under these conditions, showing a 2.5-fold reduction. Western blot analyses show that even in the knockdown TBX3 cell line, RA treatment is still able to induce an increase in TBX3 protein levels, albeit to a much lesser extent than the parental cell line. These results suggest that, in TBX3-associated cancers, the reported antiproliferative effects of RA may be mediated partly through increasing TBX3 expression.
FIGURE 3:
RA-mediated reduction in ME1402 cell numbers correlates with increased TBX3 levels. (A) Growth curve assay of ME1402 cells treated with vehicle or RA. Right, Western blot analysis of protein harvested from the growth curve assay. (B) Growth curve assay of ME1402 cells and shTBX3-ME1402 cells treated with vehicle or RA. Right, Western blot analysis of protein harvested from the growth curve assay. The bar graph compares the fold reduction in ME1402 and shTBX3 cell numbers at day 9 of the growth curve assay. Bars, SD. *p < 0.05, **p < 0.001.
Retinoic acid activates Tbx3 expression during mouse embryonic development
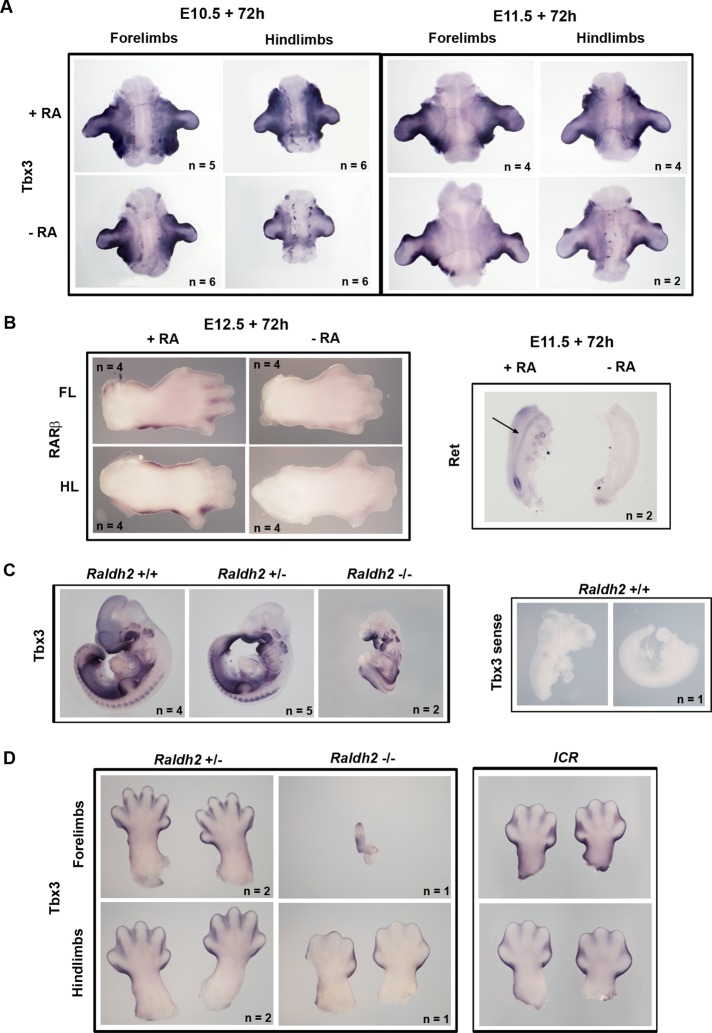
As mentioned earlier, the RA signaling pathway plays an important role in the development of the heart, kidney, and limbs, and Tbx3 has also been implicated in the development of these organs. We therefore speculated that the regulation of TBX3 by RA in cancer might reflect a similar relationship during embryonic development. To explore this hypothesis, we used a limb organ culture assay to test whether RA could activate Tbx3 in tissue. Limb buds were dissected from embryonic day 10.5 (E10.5) and E11.5 ICR mouse embryos and cultured in vitro for 72 h in serum-free medium with serum extender (which contains defined growth factors but no retinoids) with or without added RA. When analyzed for Tbx3 expression by whole-mount in situ hybridization, the results show that in the presence of RA, Tbx3 was appropriately expressed in the margins of the limb buds and in the flank (Figure 4A). However, in the absence of RA, forelimb and hindlimb bud explants from both E10.5 and E11.5 embryos showed a modest but consistent decrease in Tbx3 expression (Figure 4A). Of interest, compared with E10.5 explants, Tbx3 levels were lower in E11.5 forelimb and hindlimb cultures and appeared to be less RA responsive at this stage. It is worth noting that in the same culture system, when RA was depleted from E10.5 embryos by omitting fetal bovine serum (FBS) without serum extender a similar decrease in Tbx3 expression was observed (unpublished data). As a positive control for this in vitro culture system, forelimb and hindlimb buds from E12.5 embryos were cultured in serum-free medium with or without RA and analyzed for expression of the RA-responsive gene RARβ, which is expressed in the handplate and footplate from E12.5 (Mendelsohn et al., 1991). The results in Figure 4B indeed show a decrease in RARβ expression in the absence of RA. As an additional control, genital tracts from E11.5 embryos cultured as described for the limb buds showed decreased expression of the RA-responsive gene Ret (Chia et al., 2011) in the Wolffian duct (Figure 4B). Taken together, these results suggest that RA is able to regulate Tbx3 expression during mouse embryonic development.
FIGURE 4:
RA activates Tbx3 expression during mouse embryonic development. (A) Whole-mount in situ hybridization with Tbx3 probe of forelimb and hindlimb explants from E10.5 and E11.5 ICR mouse embryos cultured in serum-free medium with serum extender, with or without 200 nM all-trans and 9-cis RA for 72 h. (B) Left, whole-mount in situ hybridization with RARβ probe of handplates and footplates from E12.5 embryo forelimbs (FL) and hindlimbs (HL) cultured with or without RA. Right, whole-mount in situ hybridization with c-Ret probe of genital tracts from E11.5 embryos cultured with or without RA. Arrow indicates the Wolffian duct. (C) Left, whole-mount in situ hybridization of Raldh2 wild-type and mutant E10.5 embryos using antisense Tbx3 probe. Right, whole-mount in situ hybridisation of a wild-type E10.5 embryo with a sense Tbx3 probe. Note that this embryo was damaged in dissection, and therefore two halves are shown. Genotypes are as indicated. (D) Whole-mount in situ hybridization showing Tbx3 expression in handplates and footplates from E13.5 Raldh2+/− and Raldh−/− embryos. Right, handplates and footplates from a stage-matched E13.5 ICR embryo for comparison.
We next investigated whether the regulation of Tbx3 by RA could be demonstrated in vivo, using the retinaldehyde dehydrogenase 2–knockout (Raldh2−/−) mouse model, in which Raldh2, an enzyme required for most RA synthesis in the embryo, is inactivated. In these experiments a sense Tbx3 probe was used as a negative control to confirm the specificity of the staining. At E10.5 the mutant embryos were much smaller than controls (Figure 4C) and exhibited a pattern of malformations including craniofacial, heart, somite, and limb defects (Niederreither et al., 1999). Of interest, the intensity of Tbx3 staining in the Raldh2−/− embryos was close to that seen for the Raldh2+/− and Raldh2+/+ embryos (Figure 4C). It is worth noting that in order to extend the life of the mutant embryos to E10.5, RA was administered in the food of the pregnant dam between E7.5 and E9.5 to circumvent lethal heart abnormalities. On the basis of previous studies, the half-life of RA under conditions used for dietary supplementation is 24 h or less (Mic et al., 2002), and hence by E10.5, it would not be surprising if residual RA was able to maintain Tbx3 expression or delay its down-regulation in mutants. To address this possibility, we analyzed Tbx3 expression in the mutants at later stages after cessation of supplementation at E9.5. At E13.5, forelimbs of the single mutant recovered were barely formed; however, hindlimbs were present and showed a decrease in Tbx3 expression in the interdigital tissue compared with the control embryos (Figure 4D). Because the mutant hindlimbs show a slight developmental delay compared with littermate controls, we compared Tbx3 staining in the hind limbs of a stage-matched wild-type E13.5 ICR embryo, confirming that interdigital Tbx3 expression is diminished in the Raldh2−/− hindlimb compared with stage-matched controls. Although we were only able to recover a single Raldh2 mutant, together with the in vitro results these data suggest that RA is able to regulate Tbx3 expression during mouse embryonic development and that this may be a physiologically relevant relationship in limb morphogenesis.
DISCUSSION
Alterations in TBX3 levels have catastrophic effects, as evidenced by haploinsufficiency of the human TBX3 gene resulting in ulnar-mammary syndrome (Bamshad et al., 1997) and the overexpression of TBX3 being associated with at least eight different types of cancers (Mahlamäki et al., 2002; Fan et al., 2004; Rowley et al., 2004; Ito et al., 2005; Lomnytska et al., 2006; Lyng et al., 2006; Renard et al., 2007; Rodriguez et al., 2008).The identification and understanding of biochemical pathways that regulate TBX3 function are thus of critical importance. Here we demonstrate for the first time that the RA signaling pathway up-regulates TBX3 mRNA and protein levels and we provide evidence that this up-regulation is a direct transcriptional effect mediated by binding of RA receptors to the TBX3 promoter. Using both wild-type in vitro limb bud cultures and Raldh2-knockout mice lacking endogenous RA, we confirm that this regulation is relevant in mammalian systems.
Although RA signaling has long been known to play an essential role in embryogenesis and numerous RA-responsive genes have been identified, relatively few of these have been unequivocally shown to be direct targets of the RA–receptor complex (Balmer and Blomhoff, 2002). Among the well-known direct targets of RA signaling are the RARβ and CYP26 genes, as well as CRABP-II and several members of the Hox gene family (Hoxa1, Hoxa4, Hoxb1, Hoxb4), all of which are activated by RA–receptor complexes binding to RAREs in their promoters (Sucov et al., 1990; Aström et al., 1994; Frasch et al., 1995; Ogura and Evans, 1995; Gould et al., 1998; Packer et al., 1998; Loudig et al., 2000). Our data reveal that the RA receptors bind a conserved RARE half-site at −75 base pairs in the TBX3 promoter through which they activate TBX3 expression and identify TBX3 as a new direct target of RA signaling. It is worth noting that this half-site is common to two putative RAREs, involving half-sites at −87 and −59 base pairs. Although it is tempting to speculate that the RARE involving the evolutionarily highly conserved −87 half-site is more likely to be responsible for mediating RA-induced TBX3 expression, we cannot rule out the possibility that the site at −59 base pairs may also play a role. Furthermore, we show this positive regulation may be relevant during mouse embryonic development, since RA-depleted limb bud cultures of wild-type E10.5 and E11.5 mice have reduced Tbx3 expression, and there is a loss of interdigital Tbx3 expression in the hindlimbs of E13.5 Raldh2 mutant embryos. The latter result in particular fits well with the role of RA signaling in the interdigital soft tissue recession required for digit separation. RARβ;RARγ double-mutant embryos show interdigital webbing up to E15.5 (Lussier et al., 1993; Ghyselinck et al., 1997) and decreased interdigital apoptosis at E13.5 (Dupé et al., 1999), which is also seen for E13.5 Raldh2 mutants (Zhao et al., 2010). Of interest, we demonstrate that Tbx3 expression is lost in the interdigital mesenchyme of E13.5 Raldh2 mutants, which is reminiscent of data reported by Zhao et al., (2010) for RARβ, BMP-7, and MMP11 in Raldh2 mutants. It is therefore possible that Tbx3, along with BMP-7 and MMP11, may mediate RA-induced apoptosis and tissue remodeling of the interdigital mesenchyme during digit separation. This suggestion is of particular interest in light of data from this study showing that the increase in TBX3 levels in RA-treated ME1402 cells correlated with decreased cell numbers.
It is worth noting that although Tbx3 expression was lost in E13.5 Raldh2 mutant embryos, it was detected at E10.5. This may be explained by maternal RA administration from E7.5 to E9.5 to prevent lethal heart defects in Raldh2 mutants. Although supplemented RA is believed to be cleared by 24 h, as judged by an artificial promoter driving a LacZ reporter (Mic et al., 2002), it is also possible that its effects on RA-responsive endogenous gene expression may be sustained for a longer period. Indeed, defining the role of RA signaling in forelimb development, which is initiated at E9.5, has been difficult due to these technical problems and has led to some controversy (Tickle et al., 1982; Riddle et al., 1993; Mercader et al., 2000; Niederreither et al., 2002b; Yashiro et al., 2004; Zhao et al., 2009; Cooper et al., 2011; Roselló-Díez et al., 2011). Nevertheless, we cannot discount the possibility that at this stage of embryogenesis, Tbx3 is not responsive to RA. However, our data showing loss of interdigital Tbx3 expression in an E13.5 Raldh2 mutant does suggest that Tbx3 may be mediating RA signaling, at the very least, during handplate formation.
In addition to its role in embryogenesis, RA also holds potential as a chemotherapeutic agent through its ability to promote cell cycle arrest, differentiation, and apoptosis (reviewed in Tang and Gudas, 2011). Elucidating downstream targets of RA signaling and their contributions to these antitumorigenic processes is therefore valuable for our understanding of how retinoid-based chemotherapies work. This has implications for identifying mechanisms to improve their efficacy, as well as sensitizing cancers known to be RA resistant. Taken together, our data showing that a robust increase in TBX3 expression corresponds with a decrease in the number of RA-treated melanoma cells and our observations that silencing TBX3 in these cells reduces this effect suggest that TBX3 may be one of the mediators of RA's chemotherapeutic activity. It is also in keeping with our previously published data implicating TBX3 as an antiproliferative factor in other cancer cell lines (Peres et al., 2010). This capacity of TBX3 to limit proliferation is in contrast to much of the literature, which describes TBX3 as a proproliferative factor that functions by repressing the cyclin-dependent kinase inhibitor p14ARF (p19ARF in mouse) to bypass senescence (Carlson et al., 2001; Brummelkamp et al., 2002; Lingbeek et al., 2002). Although these apparently opposing roles may seem contradictory, a similar duality is also seen in development. In the heart, Tbx3 is expressed in the nonchamber myocardium of the atrioventricular canal, which has less proliferative ability than chamber myocardium (Hoogaars et al., 2004; Greulich et al., 2011). Ectopic expression of Tbx3 results in suppression of chamber myocardium and a decrease in the number of dividing cells, whereas silencing Tbx3 promotes proliferation (Ribeiro et al., 2007; Bakker et al., 2008). However, during liver development, formation of the liver is much smaller in Tbx3-null mice, and hepatoblasts isolated from these mice showed decreased proliferation and increased p19ARF expression, suggesting that Tbx3 induces proliferation in hepatoblasts through repression of p19ARF (Suzuki et al., 2008). We suggest that the evidence supports dual roles for TBX3 as a regulator of proliferation in both development and cancer, with its pro proliferative and antiproliferative functions dependent on cellular context, such as the regulation by RA signaling described in this study.
In conclusion, we present data that identify Tbx3 as a novel direct target of RA signaling in mouse embryonic development and elucidate a mechanism by which this gene, the deregulated levels of which have dire consequences, is regulated. Furthermore, our observations that TBX3 is involved in mediating the antiproliferative effects of RA in melanoma cell lines suggest that it is potentially a novel target for anticancer drugs.
MATERIALS AND METHODS
Cell lines and culture conditions
The human melanoma cell lines ME1402 and 501mel were maintained in RPMI 1640 medium (Highveld Biological, Lyndhurst, United Kingdom) supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin and maintained at 37°C in an atmosphere of 5% CO2. For RA treatment, we used 10 μM all-trans RA (Sigma-Aldrich, St. Louis, MO). For transcription inhibitor experiments, cells were pretreated with 5 μg/ml actinomycin D (Sigma-Aldrich) for 1 h and then treated with RA for 4 h. For inhibition of de novo protein synthesis experiments, cells were pretreated with 30 μg/ml cycloheximide (Sigma-Aldrich) for 1 h and then treated with RA for 4 h.
Western blot analysis
Cells were harvested and solubilized at 4°C in radioimmunoprecipitation assay buffer (150 mM NaCl, 1% Triton X-100, 0.1% SDS, 20 mM Tris, pH 7.5, 1% deoxycholate, and a cocktail of protease inhibitors), centrifuged at 12,000 × g for 20 min at 4°C, and the supernatants recovered. Protein concentrations were determined using the bicinchoninic acid assay (Pierce, Rockford, IL), according to the manufacturer's instructions, with bovine serum albumin (BSA) as the standard. Equal amounts of protein were loaded and separated on 8–10% SDS-polyacrylamide gels and transferred to Hybond C (GE Healthcare Life Sciences, Amersham, United Kingdom). Membranes were blocked for 1 h at room temperature with phosphate-buffered saline (PBS) containing 5% nonfat dry milk, probed with appropriate primary antibodies, followed by peroxidase-conjugated anti-rabbit antibody (1:5000), and visualized by enhanced chemiluminescence (Pierce). The primary antibodies used were rabbit polyclonal anti-TBX3 (42-4800; Zymed, San Francisco, CA) and rabbit polyclonal anti-p38 (Sigma-Aldrich).
qRT-PCR
Total RNA was extracted from cells using the RNeasy Plus Mini Kit (Qiagen, Valencia, CA). Reverse transcription of RNA (1 μg) was performed according to the manufacturer's instructions using the InProm-II reverse transcription system (A3800; Promega, Madison, WI). Using 2 μl of cDNA, we conducted PCR with the SensiMix Lite Kit (QT 405-05; Quantace, Taunton, MA) according to the manufacturer's protocol. Real-time PCR was performed on a LightCycler Version 3 (Roche, Basle, Switzerland) using the following parameters: denaturation (15 min at 95°C), annealing and amplification at 35 cycles (15 s at 94ºC; 20 s at 55ºC; 20 s at 72°C), melting temperature (15 s at 65°C), and a cooling step (30 s at 40°C). Each DNA sample was quantified in duplicate, and a negative control without cDNA template was run with every assay to assess the overall specificity. Melting curve analyses were carried out to ensure product specificity, and data were analyzed using the 2−ΔΔCt method. Relative mRNA expression levels were normalized to glucuronidase beta (GUSB) for each reaction with PCR efficiency correction calculated using the formula Ratio = EtargetCPtarget(control – sample)/ErefCPref(control – sample), where E is the real-time PCR efficiency and CP is the crossing point. Primers used to amplify the human TBX3 (QT00022484) and GUSB (QT00046046) were purchased from Qiagen.
Plasmid constructs
The human TBX3 promoter luciferase reporter constructs have been described previously (Mowla et al., 2011). The human pSG5-RARα and pSVL-RXRα expression plasmids were kindly provided by Pierre Chambon (Institut de Genetique et de Biologie Moleculaire et Cellulaire, Strasbourg, France). The pTIG-shTBX3 lentiviral plasmid was a kind gift from Marco Weinberg (University of the Witwatersrand, Johannesburg, South Africa), who also designed and cloned the knockdown sequence into the pHIV7-TetRIRESeGFP vector (pTIG; Aagaard et al., 2007).
Transfection and luciferase assays
Transient transfections were performed using FuGENE HD (Roche, Grenzach-Wyhlen, Germany) according to the manufacturer's instructions. We used 501 melanoma cells in these assays in place of ME1402 cells, as the latter have very low transfection efficiency. Cells were plated at 1.5 × 105 cells/ml in 35-mm cell culture dishes 16 h before transfection. We cotransfected 501mel cells with 400 ng each of TBX3-luciferase vector and RARα and RXRα expression plasmids overnight. The next day, the medium was replaced with fresh medium containing RA and incubated at 37°C for a further 24 h. Cell extracts were assayed for firefly luciferase activity using the dual luciferase assay system (Promega, Madison, WI). Luciferase activities were measured using the Luminoskan Ascent Luminometer (Thermo Labsystems, Franklin, MA). All transfections were performed in duplicate, and at least three independent experiments were done to confirm reproducibility.
Site-directed mutagenesis of the TBX3 promoter
The −141–base pair TBX3-luciferase plasmid was modified by site-directed mutagenesis using an adapted protocol based on the Stratagene QuikChange system using Pfu DNA polymerase (Promega). Mutations were introduced as follows. The right half-site of the RARE at –87 was mutated from CAATCA to GGATCC using the primer pair 5′-AAAGCGTCAAAGAGCGGATCCAGAGGCCTCCGGCT-3′ and 5′-AGCCGGAGGCCTCTGGACCGCTC TTTGACGCTTT-3′. Successful introduction of the mutation was confirmed by automated BigDye Terminator sequencing using the AB3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
DNA affinity immunoblot assay
Biotinylated DNA probes spanning from −137 to +36 base pairs relative to the TBX3 transcriptional start site were generated by PCR using primer pairs with the reverse primer containing a 5′-biotin modification (forward, 5′-CACTCGACCTGTGAAAACCAC-3′; reverse, 5′-Bio-AGAGCTCCTCGCCACCCT-3′). The −87 RARE wild-type and mutant TBX3-luciferase plasmids were used as templates. The probes were purified and immobilized on streptavidin coupled beads (Dynabeads M-280 streptavidin; Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Subconfluent ME1402 cells were treated with 10 μM RA for 4 h and nuclear extracts isolated as described previously (Lee and Green, 1990). For each DNA-binding reaction, 40 μg of nuclear extract was incubated with 1 μg of biotinylated DNA probe in 200 μl of binding buffer (20 mM Tris-HCl pH 7.6, 50 mM NaCl, 1 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol, and 10 ng/μl poly(dI-dC)). The beads were extensively washed with binding buffer and boiled in 25 μl of 2× protein loading buffer (125 mM Tris-HCl, pH 6.5, 0.4% SDS, 10% β-mercaptoethanol, and 20% glycerol). Proteins bound to the biotinylated probes were analyzed by SDS–PAGE, followed by immunoblotting using rabbit polyclonal anti-RARα (C-20), anti-RARβ (C-19), anti-RARγ (C-19), and anti-RXRα (D-20) antibodies (Santa Cruz Biotechnology, Santa Cruz, CA).
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays were carried out as previously described (Prince et al., 2004). Briefly, ME1402 cells plated in 15-cm dishes at a total of (10–20) × 106 cells were treated with RA for 4 h and then fixed in 1% formaldehyde at room temperature for 10 min. The reaction was quenched with 125 mM glycine, and cells were collected by scraping in PBS and lysed in lysis buffer (10 mM EDTA, 50 mM Tris-Cl, pH 8.1, 0.5% Nonidet P-40, 1% SDS) plus protease inhibitors. The chromatin was then sonicated to obtain fragments of 300–500 base pairs, precleared with protein A/G agarose beads (sc-2003; Santa Cruz Biotechnology), and incubated with 2 μg each of specific antibody (rabbit polyclonal anti-RARα [C-20], anti-RARβ [C-19], anti-RARγ [C-19], and anti-RXRα [D-20]) or 8 μg of nonspecific IgG (rabbit polyclonal IgG) (all Santa Cruz Biotechnology) at 4°C overnight. Five percent of supernatant volume was reserved as input. Samples were immunoprecipitated with protein A/G agarose beads, and washed and bound DNA was extracted and purified using a PCR purification kit (Qiagen) according to the manufacturer's instructions. PCR was conducted using GoTaq Green PCR Master Mix (Promega) with 4 μl of DNA in a final volume of 20 μl, using the following parameters: 5 min at 94°C for one cycle; 30 s at 94°C, 30 s at 54°C, 1 min at 60°C for 30 cycles; 7 min at 72°C for one cycle. PCR products were visualized after electrophoresis on a 2% agarose gel. The primer pairs used for PCR analysis were as follows: TBX3 (forward, 5′-CTCAGCTCTATCCCCCAGCA-3′; reverse, 5′-CGCTCTTTCGCTCTCGAGGAG-3′) and GAPDH (forward, 5′-GAAGGCTGGGGCTCATTT-3′; reverse, 5′-CAGGAGGCATTGCTGATGAT-3′).
Lentiviral stable knockdown cell lines
ME1402 cells were transduced with vesicular stomatitis virus G–pseudotyped lentiviral particles essentially as described in Aagaard et al. (2007). Briefly, 5 × 10 cm dishes of HEK293T cells at ∼60% confluency were transfected using CaPO4 with 13 μg of the pTIG-shTBX3 lentiviral vector, 3.75 μg of the Gag–Pol packaging plasmid (pRRE), 3 μg of the Rev packaging plasmid (pRSV-Rev), and 3.75 μg of vesicular stomatitis virus G envelope plasmid (pVSVG) per dish. Forty-eight hours after transfection, virus supernatants were harvested and concentrated by ultracentrifugation at 25,000 rpm at 4°C for 2 h (SW 55 Ti; Optima L-80 XP; Beckman Coulter, Brea, CA) and the viral pellets resuspended in sterile PBS. ME1402 cells were transduced with concentrated viral particles in the presence of 8 μg/ml polybrene (hexadimethrine bromide; Sigma-Aldrich) overnight. A pure population of positively transduced cells was obtained by fluorescence-activated cell sorting for enhanced green fluorescent protein and successful knockdown of TBX3 in the shTBX3-ME1402 cells confirmed by Western blotting.
Growth curves
Short-term growth of the ME1402 and shTBX3-ME1402 cell lines was compared as described previously (Prince et al., 2003). Cells were seeded in triplicate at 104 cells/well in 12-well plates, collected by trypsinization, and counted on a hemocytometer at 2- to 3-d intervals.
In situ hybridization and embryo culture
ICR female mice housed with male mice overnight were checked daily for vaginal plugs, with noon of the day a plug appeared taken as E0.5. Raldh2−/+ female mice (mated with Raldh2−/+ males) were supplied with all-trans retinoic acid in 1g of applesauce from E7.5 to E9.5 of gestation. On the first day the mouse received 150 μg RA, and on E8.5 and E9.5 the mouse received 400 μg. Pregnant females were killed on the appropriate day by cervical dislocation, embryos were collected in cold PBS containing BSA (Sigma-Aldrich), and extraembryonic tissues were removed. Mouse organ cultures were performed essentially as described previously, using a method shown to support limb bud outgrowth (Naiche and Papaioannou, 2003). Briefly, limb buds were dissected from E10.5 to E12.5 ICR embryos and cultured ventral side down on a 0.3 μM isopore membrane filter (Millipore, Billerica, MA) in Opti-MEM Reduced Serum Medium (Life Technologies, Carlsbad, CA) supplemented with 10% FBS or 5′ MITO+ Serum Extender (BD Biosciences, San Diego, CA) and 1% penicillin/streptomycin for 72 h at 37°C in 5% CO2. Genital ridges were dissected from E11.5 embryos and cultured dorsal side down as described for limb buds. For RA supplementation, 200 nM all-trans and 9-cis RA (Sigma-Aldrich) were added to the medium. Organ cultures were subsequently fixed and processed for in situ hybridization using standard protocols as previously described (Wilkinson and Nieto, 1993). Digoxigenin-labeled RNA probes were made using standard protocols. The mouse Tbx3 template plasmid has been described previously (Chapman et al., 1996), and the pBS-cRet and pBS-RARβTotal plasmids were kindly provided by F. Costantini and C. Mendelsohn, respectively (Columbia University, New York, NY).
Statistical analysis
Statistical analysis was performed by using the two-sample t test (Excel; Microsoft, Redmond, WA).
Acknowledgments
This work was supported by grants from the SA Medical Research Council, the National Research Foundation, the Cancer Association of South Africa, the Cancer Research Initiative of South Africa, the University of Cape Town, and the National Institutes of Health (HD033082). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Abbreviations used:
- RA
retinoic acid
- Raldh
retinaldehyde dehydrogenase
- RAR
retinoic acid receptor
- RARE
retinoic acid response element
- RXR
retinoid X receptor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0790) on April 25, 2012.
REFERENCES
- Aagaard L, Amarzguioui M, Sun G, Santos LC, Ehsani A, Prydz H, Rossi JJ. A facile lentiviral vector system for expression of doxycycline-inducible shRNAs: knockdown of the pre-miRNA processing enzyme Drosha. Mol Ther. 2007;15:938–945. doi: 10.1038/sj.mt.6300118. [DOI] [PubMed] [Google Scholar]
- Aström A, Pettersson U, Chambon P, Voorhees JJ. Retinoic acid induction of human cellular retinoic acid-binding protein-II gene transcription is mediated by retinoic acid receptor-retinoid X receptor heterodimers bound to one far upstream retinoic acid-responsive element with 5-base pair spacing. J Biol Chem. 1994;269:22334–22339. [PubMed] [Google Scholar]
- Bakker ML, Boukens BJ, Mommersteeg MTM, Brons JF, Wakker V, Moorman AFM, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Balmer JE, Blomhoff R. Gene expression regulation by retinoic acid. J Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- Bamshad M, et al. The spectrum of mutations in TBX3: genotype/phenotype relationship in ulnar-mammary syndrome. Am J Hum Genet. 1999;64:1550–1562. doi: 10.1086/302417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Behesti H, Holt JKL, Sowden JC. The level of BMP4 signaling is critical for the regulation of distinct T-box gene expression domains and growth along the dorso-ventral axis of the optic cup. BMC Dev Biol. 2006;6:62. doi: 10.1186/1471-213X-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, Lohuizen van M, Bernards R. TBX-3, the gene mutated in ulnar-mammary syndrome, is a negative regulator of p19ARF and inhibits senescence. J Biol Chem. 2002;277:6567–6572. doi: 10.1074/jbc.M110492200. [DOI] [PubMed] [Google Scholar]
- Carlson H, Ota S, Campbell CE, Hurlin PJ. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum Mol Genet. 2001;10:2403. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- Chapman DL, et al. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206:379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Chen H, Zhang H, Lee J, Liang X, Wu X, Zhu T, Lo P-K, Zhang X, Sukumar S. HOXA5 acts directly downstream of retinoic acid receptor beta and contributes to retinoic acid-induced apoptosis and growth inhibition. Cancer Res. 2007;67:8007–8013. doi: 10.1158/0008-5472.CAN-07-1405. [DOI] [PubMed] [Google Scholar]
- Chia I, Grote D, Marcotte M, Batourina E, Mendelsohn C, Bouchard M. Nephric duct insertion is a crucial step in urinary tract maturation that is regulated by a Gata3-Raldh2-Ret molecular network in mice. Development. 2011;138:2089–2097. doi: 10.1242/dev.056838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Hu JK-H, Berge ten D, Fernandez-Teran M, Ros MA, Tabin CJ. Initiation of proximal-distal patterning in the vertebrate limb by signals and growth. Science. 2011;332:1083–1086. doi: 10.1126/science.1199499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- Dhandapani L, Yue P, Ramalingam SS, Khuri FR, Sun S. Retinoic acid enhances TRAIL-induced apoptosis in cancer cells by upregulating TRAIL receptor 1 expression. Cancer Res. 2011;71:5245–5254. doi: 10.1158/0008-5472.CAN-10-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato LJ, Suh JH, Noy N. Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res. 2007;67:609–615. doi: 10.1158/0008-5472.CAN-06-0989. [DOI] [PubMed] [Google Scholar]
- Dupé V, Ghyselinck NB, Thomazy V, Nagy L, Davies PJ, Chambon P, Mark M. Essential roles of retinoic acid signaling in interdigital apoptosis and control of BMP-7 expression in mouse autopods. Dev Biol. 1999;208:30–43. doi: 10.1006/dbio.1998.9176. [DOI] [PubMed] [Google Scholar]
- Fan W, Huang X, Chen C, Gray J, Huang T. TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res. 2004;64:5132–5139. doi: 10.1158/0008-5472.CAN-04-0615. [DOI] [PubMed] [Google Scholar]
- Frasch M, Chen X, Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995;121:957–974. doi: 10.1242/dev.121.4.957. [DOI] [PubMed] [Google Scholar]
- Ghyselinck NB, Dupé V, Dierich A, Messaddeq N, Garnier JM, Rochette-Egly C, Chambon P, Mark M. Role of the retinoic acid receptor beta (RARbeta) during mouse development. Int J Dev Biol. 1997;41:425–447. [PubMed] [Google Scholar]
- Gould A, Itasaki N, Krumlauf R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron. 1998;21:39–51. doi: 10.1016/s0896-6273(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91:212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- Han J, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Ito A, Asamoto M, Hokaiwado N, Takahashi S, Shirai T. Tbx3 expression is related to apoptosis and cell proliferation in rat bladder both hyperplastic epithelial cells and carcinoma cells. Cancer Lett. 2005;219:105–112. doi: 10.1016/j.canlet.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jerome-Majewska LA, Jenkins GP, Ernstoff E, Zindy F, Sherr CJ, Papaioannou VE. Tbx3, the ulnar-mammary syndrome gene, and Tbx2 interact in mammary gland development through a p19Arf/p53-independent pathway. Dev Dyn. 2005;234:922–933. doi: 10.1002/dvdy.20575. [DOI] [PubMed] [Google Scholar]
- Lee KA, Green MR. Small-scale preparation of extracts from radiolabeled cells efficient in pre-mRNA splicing. Methods Enzymol. 1990;181:20–30. doi: 10.1016/0076-6879(90)81108-7. [DOI] [PubMed] [Google Scholar]
- Liberatore CM, Searcy-Schrick RD, Yutzey KE. Ventricular expression of tbx5 inhibits normal heart chamber development. Dev Biol. 2000;223:169–180. doi: 10.1006/dbio.2000.9748. [DOI] [PubMed] [Google Scholar]
- Lingbeek ME, Jacobs JL, Lohuizen van M. The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. J Biol Chem. 2002;277:26120–26127. doi: 10.1074/jbc.M200403200. [DOI] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dolle P, Decimo D, Lemeur M, Dierich A, Gorry P, Chambon P. Development roles of the retinoic acid receptors. J Steroid Biochem Mol Biol. 1995;53:475–486. doi: 10.1016/0960-0760(95)00094-g. [DOI] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dollé P, Dierich A, Gorry P, Gansmuller A, Chambon P. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- Lomnytska M, Dubrovska A, Hellman U, Volodko N, Souchelnytskyi S. Increased expression of cSHMT, Tbx3 and utrophin in plasma of ovarian and breast cancer patients. Int J Cancer. 2006;118:412–421. doi: 10.1002/ijc.21332. [DOI] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–1497. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Lucas PC, O'Brien RM, Mitchell JA, Davis CM, Imai E, Forman BM, Samuels HH, Granner DK. A retinoic acid response element is part of a pleiotropic domain in the phosphoenolpyruvate carboxykinase gene. Proc Natl Acad Sci USA. 1991;88:2184–2188. doi: 10.1073/pnas.88.6.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M, Canoun C, Ma C, Sank A, Shuler C. Interdigital soft tissue separation induced by retinoic acid in mouse limbs cultured in vitro. Int J Dev Biol. 1993;37:555–564. [PubMed] [Google Scholar]
- Lyng H, et al. Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer. BMC Genomics. 2006;7:268. doi: 10.1186/1471-2164-7-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader S, Thiele K, Breuer B, Alonso A. The promoter of the H1zero histone gene contains a DNA element bound by retinoic acid receptors. J Mol Biol. 1994;242:37–44. doi: 10.1006/jmbi.1994.1555. [DOI] [PubMed] [Google Scholar]
- Mahlamäki EH, Barlund M, Tanner M, Gorunova L, Hoglund M, Karhu R, Kallioniemi A. Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer. 2002;35:353–358. doi: 10.1002/gcc.10122. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín M, Gallego-Llamas J, Ribes V, Kedinger M, Niederreither K, Chambon P, Dollé P, Gradwohl G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol. 2005;284:399–411. doi: 10.1016/j.ydbio.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Lohnes D, Décimo D, Lufkin T, LeMeur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Ruberte E, LeMeur M, Morriss-Kay G, Chambon P. Developmental analysis of the retinoic acid-inducible RAR-beta 2 promoter in transgenic animals. Development. 1991;113:723–734. doi: 10.1242/dev.113.3.723. [DOI] [PubMed] [Google Scholar]
- Mercader N, Leonardo E, Piedra ME, Martínez-A C, Ros MA, Torres M. Opposing RA and FGF signals control proximodistal vertebrate limb development through regulation of Meis genes. Development. 2000;127:3961–3970. doi: 10.1242/dev.127.18.3961. [DOI] [PubMed] [Google Scholar]
- Mesbah K, Harrelson Z, Théveniau-Ruissy M, Papaioannou VE, Kelly RG. Tbx3 is required for outflow tract development. Circ Res. 2008;103:743–750. doi: 10.1161/CIRCRESAHA.108.172858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mic FA, Haselbeck RJ, Cuenca AE, Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development. 2002;129:2271–2282. doi: 10.1242/dev.129.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowla S, Pinnock R, Leaner VD, Goding CR, Prince S. PMA-induced up-regulation of TBX3 is mediated by AP-1 and contributes to breast cancer cell migration. Biochem J. 2011;433:145–153. doi: 10.1042/BJ20100886. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development. 2003;130:2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Fraulob V, Garnier J-M, Chambon P, Dollé P. Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech Dev. 2002a;110:165–171. doi: 10.1016/s0925-4773(01)00561-5. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dollé P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Vermot J, Fraulob V, Chambon P, Dollé P. Retinaldehyde dehydrogenase 2 (RALDH2)-independent patterns of retinoic acid synthesis in the mouse embryo. Proc Natl Acad Sci USA. 2002b;99:16111–16116. doi: 10.1073/pnas.252626599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Vermot J, Messaddeq N, Schuhbaur B, Chambon P, Dollé P. Embryonic retinoic acid synthesis is essential for heart morphogenesis in the mouse. Development. 2001;128:1019–1031. doi: 10.1242/dev.128.7.1019. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nat Lett. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Ogura T, Evans RM. A retinoic acid-triggered cascade of HOXB1 gene activation. Proc Natl Acad Sci USA. 1995;92:387–391. doi: 10.1073/pnas.92.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AI, Crotty DA, Elwell VA, Wolgemuth DJ. Expression of the murine Hoxa4 gene requires both autoregulation and a conserved retinoic acid response element. Development. 1998;125:1991–1998. doi: 10.1242/dev.125.11.1991. [DOI] [PubMed] [Google Scholar]
- Peres J, Davis E, Mowla S, Bennett DC, Li JA, Wansleben S, Prince S. The highly homologous T-box transcription factors, TBX2 and TBX3, have distinct roles in the oncogenic process. Genes Cancer. 2010;1:272–282. doi: 10.1177/1947601910365160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince S, Carreira S, Abrahams A, Goding CR. Tbx2 directly represses the expression of the p21WAF1 cyclin-dependent kinase inhibitor. Cancer Res. 2004;64:1669–1674. doi: 10.1158/0008-5472.can-03-3286. [DOI] [PubMed] [Google Scholar]
- Prince S, Wiggins T, Hulley PA, Kidson SH. Stimulation of melanogenesis by tetradecanoylphorbol 13-acetate (TPA) in mouse melanocytes and neural crest cells. Pigment Cell Res. 2003;16:26–34. doi: 10.1034/j.1600-0749.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- Renard C-A, et al. Tbx3 is a downstream target of the Wnt/beta-catenin pathway and a critical mediator of beta-catenin survival functions in liver cancer. Cancer Res. 2007;67:901–910. doi: 10.1158/0008-5472.CAN-06-2344. [DOI] [PubMed] [Google Scholar]
- Ribeiro I, Kawakami Y, Büscher D, Raya A, Rodríguez-León J, Morita M, Rodríguez Esteban C, Izpisúa Belmonte JC. Tbx2 and Tbx3 regulate the dynamics of cell proliferation during heart remodeling. PloS One. 2007;2:e398. doi: 10.1371/journal.pone.0000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V, Wang Z, Dollé P, Niederreither K. Retinaldehyde dehydrogenase 2 (RALDH2)-mediated retinoic acid synthesis regulates early mouse embryonic forebrain development by controlling FGF and sonic hedgehog signaling. Development. 2006;133:351–361. doi: 10.1242/dev.02204. [DOI] [PubMed] [Google Scholar]
- Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- Roberts C, Ivins SM, James CT, Scambler PJ. Retinoic acid down-regulates Tbx1 expression in vivo and in vitro. Dev Dyn. 2005;232:928–938. doi: 10.1002/dvdy.20268. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, Aladowicz E, Lanfrancone L, Goding CR. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 2008;68:7872–7881. doi: 10.1158/0008-5472.CAN-08-0301. [DOI] [PubMed] [Google Scholar]
- Roselló-Díez A, Ros MA, Torres M. Diffusible signals, not autonomous mechanisms, determine the main proximodistal limb subdivision. Science. 2011;332:1086–1088. doi: 10.1126/science.1199489. [DOI] [PubMed] [Google Scholar]
- Rowley M, Grothey E, Couch FJ. The role of Tbx2 and Tbx3 in mammary development and tumorigenesis. J Mammary Gland Biol Neoplasia. 2004;9:109–118. doi: 10.1023/B:JOMG.0000037156.64331.3f. [DOI] [PubMed] [Google Scholar]
- Sucov HM, Murakami KK, Evans RM. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc Natl Acad Sci USA. 1990;87:5392–5396. doi: 10.1073/pnas.87.14.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Sekiya S, Büscher D, Izpisúa Belmonte JC, Taniguchi H. Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development. 2008;135:1589–1595. doi: 10.1242/dev.016634. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Takeuchi J, Koshiba-Takeuchi K, Ogura T. Tbx genes specify posterior digit identity through Shh and BMP signaling. Dev Cell. 2004;6:43–53. doi: 10.1016/s1534-5807(03)00401-5. [DOI] [PubMed] [Google Scholar]
- Tang X, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345–364. doi: 10.1146/annurev-pathol-011110-130303. [DOI] [PubMed] [Google Scholar]
- Tickle C, Alberts B, Wolpert L, Lee J. Local application of retinoic acid to the limb bond mimics the action of the polarizing region. Nature. 1982;296:564–566. doi: 10.1038/296564a0. [DOI] [PubMed] [Google Scholar]
- Tümpel S, Sanzezquerro J, Isaac A, Eblaghie M, Dobson J, Tickle C. Regulation of Tbx3 expression by anteroposterior signalling in vertebrate limb development. Dev Biol. 2002;250:251–262. [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridisation to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Zhao X, Uehara M, Yamashita K, Nishijima M, Nishino J, Saijoh Y, Sakai Y, Hamada H. Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev Cell. 2004;6:411–422. doi: 10.1016/s1534-5807(04)00062-0. [DOI] [PubMed] [Google Scholar]
- Zhao X, Brade T, Cunningham TJ, Duester G. Retinoic acid controls expression of tissue remodeling genes Hmgn1 and Fgf18 at the digit-interdigit junction. Dev Dyn. 2010;239:665–671. doi: 10.1002/dvdy.22188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Sirbu IO, Mic FA, Molotkova N, Molotkov A, Kumar S, Duester G. Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning. Curr Biol. 2009;19:1050–1057. doi: 10.1016/j.cub.2009.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]