This study reveals the basis for how temporal phosphoregulation of Orm protein controls sphingolipid production in response to stress. Orm protein phosphorylation is highly responsive to sphingoid bases, and Ypk1 protein kinase transmits heat stress signals to the sphingolipid biosynthesis pathway via Orm phosphorylation.
Abstract
Sphingoid intermediates accumulate in response to a variety of stresses, including heat, and trigger cellular responses. However, the mechanism by which stress affects sphingolipid biosynthesis has yet to be identified. Recent studies in yeast suggest that sphingolipid biosynthesis is regulated through phosphorylation of the Orm proteins, which in humans are potential risk factors for childhood asthma. Here we demonstrate that Orm phosphorylation status is highly responsive to sphingoid bases. We also demonstrate, by monitoring temporal changes in Orm phosphorylation and sphingoid base production in cells inhibited for yeast protein kinase 1 (Ypk1) activity, that Ypk1 transmits heat stress signals to the sphingolipid biosynthesis pathway via Orm phosphorylation. Our data indicate that heat-induced sphingolipid biosynthesis in turn triggers Orm protein dephosphorylation, making the induction transient. We identified Cdc55–protein phosphatase 2A (PP2A) as a key phosphatase that counteracts Ypk1 activity in Orm-mediated sphingolipid biosynthesis regulation. In total, our study reveals a mechanism through which the conserved Pkh-Ypk kinase cascade and Cdc55-PP2A facilitate rapid, transient sphingolipid production in response to heat stress through Orm protein phosphoregulation. We propose that this mechanism serves as the basis for how Orm phosphoregulation controls sphingolipid biosynthesis in response to stress in a kinetically coupled manner.
INTRODUCTION
Sphingoid intermediates, including sphingoid bases, sphingoid base phosphates, and ceramides (Figure 1), play important roles in regulation of cell growth, differentiation, senescence, and apoptosis (Hannun and Obeid, 2008; Dickson, 2010; Nikolova-Karakashian and Rozenova, 2010). Serine palmitoyltransferase (SPT) mediates the rate-limiting first step in sphingolipid biosynthesis (Figure 1). Despite the importance of sphingoid intermediates as bioactive molecules, the regulation of sphingolipid biosynthesis through SPT is not well understood (Cowart and Hannun, 2007). A recent study revealed that yeast Orm proteins, encoded by ORM1 and ORM2, form a conserved complex with SPT and that their phosphorylation status affects sphingolipid production (Breslow et al., 2010; Figure 1). The authors proposed that sphingolipid levels feedback regulate Orm protein phosphorylation, thus mediating sphingolipid homeostasis (Breslow et al., 2010). However, several important questions related to this model need answers. For example, whether and which sphingolipid species affect Orm phosphorylation are not known. In addition, how temporal regulation of Orm phosphorylation relates to dynamic changes in sphingolipid biosynthesis is not known. Answers to these questions are required to better understand Orm-mediated sphingolipid homeostasis.
FIGURE 1:
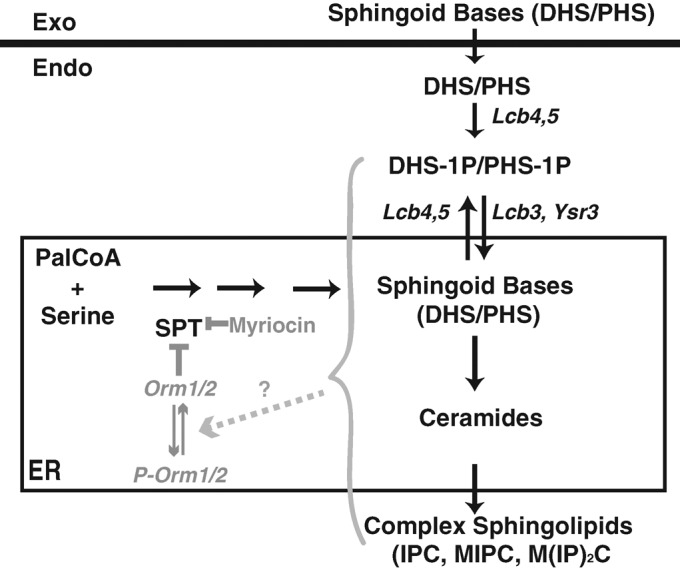
Schematic diagram of yeast sphingolipid biosynthesis from endogenous and exogenous precursors. Myriocin is a potent inhibitor of SPT (Sun et al., 2000), which is the first and rate-limiting enzyme of the sphingolipid biosynthesis pathway (Buede et al., 1991). Previous studies proposed that Orm proteins are dynamic negative regulators of SPT and their activities are inversely proportional to sphingolipid levels (Breslow et al., 2010). Exogenous sphingoid bases (DHS and PHS) can be converted into ceramides in the endoplasmic reticulum after phosphorylation and dephosphorylation by the indicated enzymes and can then be incorporated into complex sphingolipids (Qie et al., 1997; Nagiec et al., 1998; Funato et al., 2003).
Another important question concerns how Orm-mediated sphingolipid homeostasis may function in a physiological context. Increasing evidence suggests that various stimuli trigger accumulation of sphingoid intermediates, which in turn function as bioactive molecules mediating cellular responses (Hannun and Obeid, 2008; Dickson, 2010). For instance, heat stress–induced sphingoid intermediates act as signaling molecules to induce cellular responses such as translation initiation of heat shock proteins, gene regulation, and cell cycle arrest (Dickson et al., 1997; Mao et al., 1999; Jenkins and Hannun, 2001; Cowart and Hannun, 2005; Cowart et al., 2010; Meier et al., 2006; Han et al., 2010). It is striking that several groups demonstrated that heat stress induces sphingolipid biosynthesis in a rapid and transient manner (Dickson et al., 1997; Jenkins et al., 1997; Jenkins, 2003; Wells et al., 1998; Mao et al., 1999; Skrzypek et al., 1999), suggesting that biosynthesis of sphingoid intermediates in response to stresses requires precise temporal regulation. We hypothesize that the dynamic changes in sphingoid intermediate levels upon heat stress may be caused by and/or may lead to changes of Orm phosphorylation. If this is the case, heat stress could serve as a model system to address the mechanism of how Orm phosphoregulation functions in the response of sphingolipid biosynthesis to stress in general.
A recent study showed that yeast protein kinases 1 and 2 (Ypk1/2), the homologues of mammalian serum- and glucocorticoid-inducible kinase (Casamayor et al., 1999), directly phosphorylate Orm proteins in vitro (Roelants et al., 2011). This result suggests that Ypk kinase may regulate sphingolipid homeostasis through its phosphorylation of the Orm proteins. However, whether Ypk kinase activity indeed affects sphingolipid production has not been examined. Of interest, the Ypk kinases were previously considered to be downstream of heat-induced sphingolipid base accumulation (Sun et al., 2000; Friant et al., 2001; Liu et al., 2005; Hannun and Obeid, 2008; Dickson, 2010). Thus, testing for a possible role for Ypk kinase activity in the sphingolipid biosynthesis response to heat stress is an important goal.
Of importance, the Orm phosphorylation state is set not only by protein kinases, but also by phosphatases. A reasonable prediction is that the phosphatases acting on the Orm proteins are likely to be involved in and/or regulated by sphingolipid levels. Protein phosphatase 2A (PP2A) was identified as an attractive candidate for a ceramide-activated protein phosphatase in yeast (Nickels and Broach, 1996). Inactivation of the regulatory subunits (such as Cdc55) or the catalytic subunits of PP2A was reported previously to suppress the endocytic defects of a mutant with impaired SPT activity (Friant et al., 2000), suggesting that PP2A may be involved in sphingolipid production. Because PP2A was suggested previously to be involved in multiple cellular signaling pathways (Jiang, 2006), the challenge is to identify a specific relationship between Cdc55-PP2A and the Orm proteins for regulation of sphingolipid biosynthesis.
In this article, by addressing the questions just raised, we demonstrate how Orm phosphorylation regulation controls sphingolipid biosynthesis in response to heat stress in a kinetically coupled manner. We identify a signaling pathway by which the conserved Pkh-Ypk signaling cascade and Cdc55-PP2A facilitate rapid, transient sphingolipid production upon heat stress through precise regulation of Orm protein phosphorylation. Our study will therefore provide a foundation for future studies of sphingolipid-related responses to other stimuli.
RESULTS
Orm2 is the major Orm species in budding yeast
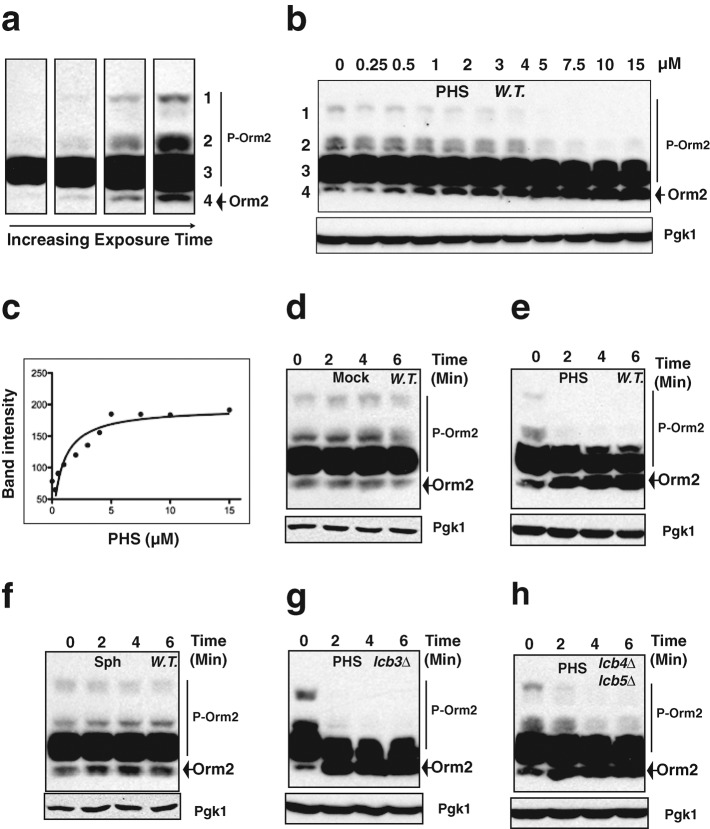
A 3XFLAG sequence was inserted at the 5′ end of the genes ORM1 and ORM2, which encode the two homologous yeast Orm proteins. Because Orm2 expression was at least 10 times higher than Orm1 expression (Supplemental Figure S1a) and because 3XFLAG-Orm1 in an orm2∆ strain, but not 3XFLAG-Orm2 in an orm1∆ strain, caused slow cell growth (Supplemental Figure S1, b and c), we concluded that Orm2 is the major Orm protein. Consistently, orm2∆ cells but not orm1∆ cells showed perturbation in sphingolipid homeostasis (Han et al., 2010). These results suggested that Orm2 provides the majority of the Orm protein function. Thus, we decided to monitor Orm2 phosphorylation in our studies. As shown in Figure 2a, four groups of Orm2 bands were observed when resolved by phosphate-affinity gel, which is widely used for the analysis of phosphoprotein isotypes (Kinoshita et al., 2006). As previously reported (Breslow et al., 2010), the slower-migrating bands in the top three groups were confirmed to be phosphorylated forms of Orm2 (Supplemental Figure S2). On the basis of the principle responsible for protein separation on phosphate-affinity gels (Kinoshita et al., 2006), the bands in the top three groups are likely to be phosphorylated to different extents, with the slowest-migrating species being the most highly phosphorylated. The bands in the third group observed by Western blotting are often overexposed under the conditions that allow us to observe bands in all four groups (Figure 2a). Thus, we monitored changes in intensity of groups 1, 2, and 4 as clear indicators of changes in Orm2 phosphorylation status.
FIGURE 2:
Exogenously provided sphingoid bases are sufficient to induce rapid Orm2 dephosphorylation. (a, b, d–h), Western blots showing Orm2 phosphorylation patterns after separation of the indicated yeast cell extracts on phosphate-affinity gels (top). P-Orm2 indicates phosphorylated forms of Orm2. We used 3-phosphoglycerate kinase 1 (Pgk1) as a loading control (bottom). All cells were grown to early log phase in YPD at 25°C before the indicated treatments. Cell extracts were prepared as described in Materials and Methods. (a) The majority of the Orm2 is moderately phosphorylated. Western blotting analysis of cell exacts from wild-type cells expressing 3XFLAG-Orm2 at its endogenous locus with different exposure times: 30 s and 1, 3, and 6 min (left to right). (b, c) Exogenous phytosphingosine (PHS) induces Orm2 dephosphorylation in a concentration-dependent manner. Wild-type cells expressing 3XFLAG-Orm2 at its endogenous locus were cultured in media containing the indicated concentrations of PHS (dissolved in methanol) for 10 min. The intensities of the fastest-migrating 3XFLAG Orm2 bands (the fourth group) in each lane (b) were analyzed and plotted in the graph shown in c. (d, e) Exogenous PHS induces rapid Orm2 dephosphorylation. Wild-type cells expressing 3XFLAG-Orm2 from its endogenous locus were cultured in media containing methanol alone (mock) or 5 μM PHS dissolved in methanol for the indicated times. (f) Exogenous sphingosine (Sph) does not affect Orm2 phosphorylation. Wild-type cells expressing 3XFLAG-Orm2 from its endogenous locus were cultured in the presence of 5 μM Sph (dissolved in methanol) for the indicated times. (g, h) Exogenous PHS induces rapid Orm2 dephosphorylation in lcb3∆ cells or in lcb4∆ lcb5∆ cells. lcb3∆ cells or lcb4∆ lcb5∆ cells expressing 3XFLAG-Orm2 from its endogenous locus were cultured in the presence of 5 μM PHS for the indicated times.
Exogenously provided sphingoid bases are sufficient to induce rapid Orm2 dephosphorylation
We examined how exogenously provided sphingoid bases, which are early sphingoid intermediates (Figure 1), affect Orm2 phosphorylation status. As shown in Figure 2, b and c, dephosphorylated Orm2, seen as the fastest-migrating bands (the fourth group), increased in response to a 10-min treatment at 25°C with a natural yeast sphingoid base, phytosphingosine (PHS), in a concentration-dependent manner. PHS at 5 μM is sufficient to cause maximal effects on Orm2 dephosphorylation (Figure 2c), whereas 20 μM of exogenous dihydrosphingosine (DHS), another sphingoid base, had a similar effect on Orm2 dephosphorylation (unpublished data). Dephosphorylated Orm2 greatly increased within 2 min of treatment with 5 μM PHS at 25°C (Figure 2, d and e). In contrast, adding 5 μM sphingosine (Sph), which is a mammalian sphingoid base that cannot rescue a yeast sphingolipid deficiency (Wells and Lester, 1983), did not affect Orm2 phosphorylation (Figure 2, d and f), indicating that the effect of PHS is highly specific. Furthermore, stearylamine (Jenkins and Hannun, 2001), which is a long-chain primary amine, also did not induce Orm2 dephosphorylation (unpublished data), providing further evidence for chemical specificity.
The rapid response to the exogenous addition of PHS (Figure 2e) suggests that the sphingoid base itself, rather than its downstream sphingolipid metabolites (Nagiec et al., 1997; Figure 1), triggers Orm2 dephosphorylation. To explore this possibility further, we used a knockout mutant of the LCB3 gene, which encodes the sphingosine-1P phosphatase required for ceramide synthesis from exogenous PHS (Mao et al., 1997, 1999; Qie et al., 1997; Figure 1). In the absence of Lcb3, exogenous PHS cannot be incorporated into ceramides (Qie et al., 1997; Funato et al., 2003). However, exogenous PHS still rapidly induced Orm2 dephosphorylation in the lcb3∆ strain (Figure 2g), suggesting that exogenous PHS induces Orm2 dephosphorylation without being converted to ceramides. A previous study indicated that lack of Orm2 results in increased PHS but decreased ceramides in vivo (Han et al., 2010). Here we found that Orm1 phosphorylation is greatly reduced in a strain lacking Orm2 (Supplemental Figure S1a), in agreement with PHS being sufficient to induce Orm dephosphorylation. In addition, exogenous PHS still triggered Orm2 dephosphorylation in mutant cells lacking both Lcb4 and Lcb5 (Figure 2h), which are sphingoid base kinases required for phosphorylation of exogenously provided PHS (Nagiec et al., 1998; Funato et al., 2003; Figure 1). Thus, exogenous PHS itself, without being converted to phosphorylated sphingoid bases (Figure 1), can induce Orm2 dephosphorylation. However, compared with the wild-type cells (Figure 2e), Orm2 dephosphorylation in lcb4∆ lcb5∆ cells responds to PHS to a lesser extent (Figure 2h), suggesting that phosphorylated sphingoid bases may also contribute to Orm2 phosphoregulation.
Taken together, our data demonstrate that exogenous addition of PHS is sufficient to rapidly induce Orm2 dephosphorylation, indicating that Orm phosphorylation status is highly responsive to levels of sphingoid bases. Although we cannot exclude the possibility that ceramide and complex sphingolipids also contribute to Orm phosphoregulation, the results with lcb3∆ mutants (Figure 2g) support the conclusion that neither of these sphingolipids is required for Orm phosphoregulation.
Temporal association between Orm phosphorylation dynamics and de novo sphingoid base production in response to heat stress
Previous studies suggested that heat stress induces rapid transient de novo accumulation of sphingoid intermediates, including PHS. We next asked whether and how Orm protein phosphorylation may be involved in heat-induced sphingolipid biosynthesis.
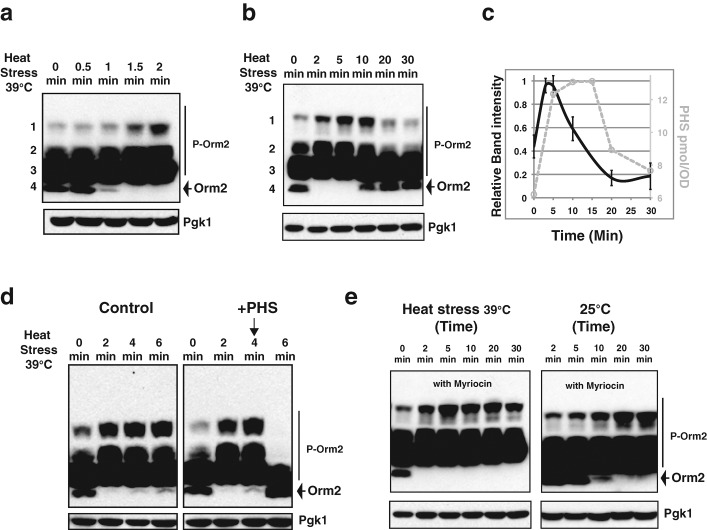
We monitored Orm2 phosphorylation after cells were shifted from 25 to 39°C. Within 1 min after the temperature shift, dephosphorylated Orm2, which is the fastest-migrating band on phosphate-affinity gels, started to disappear (Figure 3a). Correspondingly, Orm2 phosphorylation levels increased, as evidenced by the increased abundance of the slower-migrating bands in the top two groups (Figure 3a). Orm2 phosphorylation further increased at 2 min (Figure 3a), with the maximal effect observed at ∼5 min, followed by a decrease (Figure 3, b and c). Orm2 phosphorylation was further reduced by 20 min of heat stress and then stabilized (Figure 3, b and c). Several independent studies demonstrated that heat stress induces rapid de novo accumulation of various sphingoid intermediates, including sphingoid bases (Dickson et al., 1997; Jenkins et al., 1997; Jenkins, 2003; Wells et al., 1998; Mao et al., 1999; Skrzypek et al., 1999). The sphingoid base levels peak by 10–15 min and decrease to near-basal levels by 30 min of heat stress (Dickson et al., 1997; Jenkins et al., 1997). Our measurements of PHS levels confirmed these previous conclusions (Figure 3c, gray, broken line). Thus, our results demonstrate a striking temporal association between Orm phosphorylation dynamics and de novo sphingoid base production in response to heat stress (Figure 3c).
FIGURE 3:
Orm2 phosphorylation dynamics and sphingoid base production upon heat shock. (a, b, d, e) Western blots showing Orm2 phosphorylation patterns after separation of the indicated yeast cell extracts on phosphate-affinity gels (top). P-Orm2 indicates phosphorylated forms of Orm2. Pgk1 was used as a loading control (bottom). Wild-type cells expressing 3XFLAG-Orm2 from its endogenous locus were used in all experiments and cultured to early log phase at 25°C before the indicated treatments. Cell extracts were prepared as described in Materials and Methods. (a–c) Temporal association between Orm phosphorylation dynamics and sphingoid base production upon heat stress. Cell extracts were prepared after cell cultures were shifted from 25 to 39°C for the indicated times. The relative intensity of the two slowest-migrating 3XFLAG Orm2 bands (groups 1 and 2) shown in b was quantified. Time-course experiments shown in b were performed an additional four times. Data collected from the five independent experiments were plotted in the graph shown in c (black line). Data shown represent the means with standard deviations. The C18-PHS concentration (c, gray, broken line) was determined by HPLC as described in Materials and Methods. (d), Orm2 phosphorylation status upon heat stress in the presence of exogenous sphingoid bases. Cells were shifted from 25 to 39°C for the indicated times. Methanol alone (control) or 5 μM PHS dissolved in methanol was added exogenously 4 min after cells were shifted from 25 to 39°C. (e) Orm2 phosphorylation upon heat stress in the presence of myriocin. Left, 0.5 μg/ml myriocin was added to cells upon shift from 25 to 39°C. Right, cells were treated with 0.5 μg/ml myriocin at 25°C for the indicated times.
High levels of Orm phosphorylation induced by heat stress decrease when the sphingoid bases (PHS) reach their peak levels (Figure 3c, time points between 5 and 15 min), suggesting that heat-induced sphingoid base accumulation and Orm dephosphorylation may be mechanistically linked. To investigate this possibility further, we altered the timing of the sphingoid base accumulation by adding exogenous PHS 4 min after the temperature shift, when de novo–synthesized sphingolipids normally just begin to accumulate (Figure 3c; Dickson et al., 1997; Jenkins et al., 1997; Wells et al., 1998; Mao et al., 1999; Skrzypek et al., 1999). In control cells, increased Orm2 phosphorylation peaks after 4 min of heat stress and is only slightly diminished after 6 min of heat stress (Figure 3d, left). However, addition of exogenous PHS at 4 min reduced Orm2 phosphorylation to the basal level at 6 min (Figure 3d, right).
To further test whether de novo synthesis of sphingoid intermediates in response to heat stress is required to trigger Orm dephosphorylation, we monitored Orm2 phosphorylation in response to heat stress in the presence of myriocin, a potent SPT inhibitor (Sun et al., 2000; Figure 1). No obvious changes in Orm2 phosphorylation were observed up to 5 min after myriocin treatment at 25°C (Figure 3e), and 2 min of heat stress induced Orm2 phosphorylation irrespective of myriocin presence (Figure 3, b and e), indicating that heat stress affects Orm2 phosphorylation before the myriocin-sensitive step. Of interest, in the presence of myriocin, the increased Orm2 phosphorylation did not decline even after 30 min of heat stress (compare Figure 3e with Figure 3b), indicating that de novo synthesis of sphingoid intermediates is required for reducing the increased Orm2 phosphorylation caused by heat stress. Together, these results provide strong evidence that increased levels of sphingoid intermediates trigger Orm dephosphorylation during the heat stress response. Thus, our results suggest that both exogenously provided sphingoid intermediates (such as PHS) and the accumulation of physiologically induced sphingoid intermediates (by heat stress), cause rapid Orm2 dephosphorylation (Figures 2 and 3).
Ypk kinase transmits heat stress signals to the sphingolipid biosynthesis pathway via Orm phosphorylation
Another example of a striking temporal association between Orm phosphorylation and sphingolipid intermediate production is the observation that heat stress triggers Orm phosphorylation in <1 min (Figure 3a), with phosphorylation reaching its maximum before the level of sphingoid bases reaches its peak (Figure 3, b and c). This temporal association raises the important question of determining how the heat stress signal is transmitted to induce rapid Orm phosphorylation.
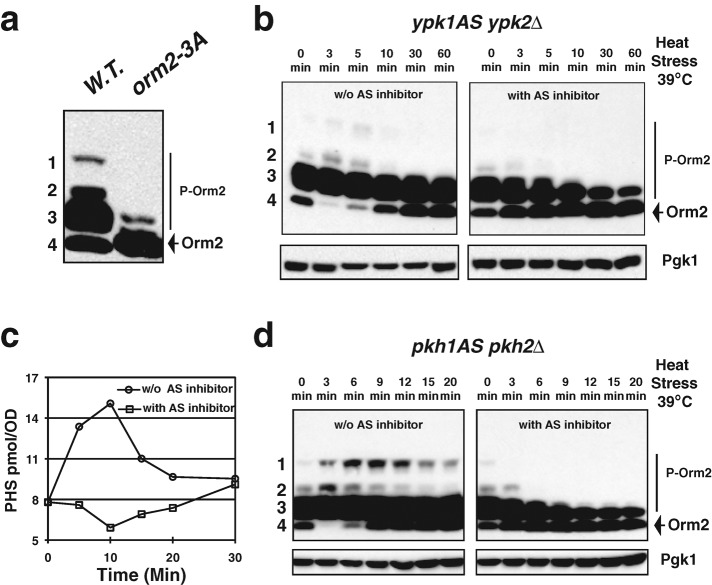
A recent study suggested that Ypk1, a homologue of mammalian serum- and glucocorticoid-inducible kinase (Casamayor et al., 1999), specifically phosphorylates Orm2 residues S46, S47, and S48 in vitro (Roelants et al., 2011). To examine the role of phosphorylation of these serine residues on Orm function, we mutated S46, S47, and S48 of Orm2 to alanine or aspartic acid to generate orm2-3A (mimicking the dephosphorylated form) or orm2-3D (mimicking the phosphorylated form), respectively. The level of sphingoid bases was greatly reduced in orm2-3A cells and was greatly increased orm2-3D cells (Supplemental Figure S3), providing evidence that phosphorylation of these serines plays a role in sphingolipid production. Furthermore, as shown in Figure 4a, the majority of orm2-3A protein is in dephosphorylated forms, demonstrating that phosphorylation of Orm2 residues S46, S47, and S48 is responsible for the slower migration of phosphorylated Orm2 (Figure 4a; Breslow et al., 2010). Together, these results strongly suggest that phosphorylation of Orm2 by Ypk1 is involved in Orm-mediated sphingolipid biosynthesis regulation.
FIGURE 4:
Ypk kinase transmits heat stress signals to the sphingolipid biosynthesis pathway via Orm phosphorylation. (a, b, d) Western blots showing Orm2 phosphorylation patterns after separation of the indicated yeast cell extracts on phosphate affinity gels (top). P-Orm2 indicates phosphorylated forms of Orm2. Pgk1 was used as a loading control (bottom). All cells were grown to early log phase in YPD at 25°C before the indicated treatments. Cell extracts were prepared as described in Materials and Methods. (a) Orm2 residues S46, S47, and S48 are responsible for slower migration of phosphorylated Orm2. (b) Orm2 phosphorylation in response to heat stress in the absence of Ypk activity. Dimethyl sulfoxide (DMSO) alone or 50 μM of 3-MOB-PP1 dissolved in DMSO, which specifically inhibits ypk1-as kinase activity, was added to ypk1as ypk2∆ cells upon shift from 25 to 39°C. (c) Heat-induced sphingoid base accumulation requires Ypk kinase activity. The C18-PHS concentration was determined in the ypk1as ypk2∆ cells cultured in the absence (line with circle) or the presence (line with square) of 50 μM 3-MOB-PP1 shifted from 25 to 39°C for the indicated times. (d) Orm2 phosphorylation status upon heat stress in the absence of Pkh activity. DMSO alone or 100 μM CZ21 dissolved in DMSO, which specifically inhibits pkh1-as kinase activity, was added to pkh1as pkh2∆ cells upon shift from 25 to 39°C.
To determine whether the rapid Orm2 phosphorylation induced by heat stress (Figure 3c, time points between 0 and 4 min) requires Ypk kinase activity in vivo, we generated an analogue-sensitive (Bishop et al., 2000) ypk1-as allele in ypk2∆ background. Deletion of YPK2 has no apparent phenotypic defect, but loss of YPK1 results in slow growth (Chen et al., 1993). Knocking out both YPK1 and YPK2 causes lethality (Chen et al., 1993). Orm2 phosphorylation in response to heat stress in the ypk1-as ypk2∆ mutant was monitored in the presence of 3-MOB-PP1, which specifically inhibits ypk1-as kinase activity. Heat stress–induced phosphorylation no longer occurs when Ypk kinase activity is abolished (Figure 4b and Supplemental Figure S4a), indicating that heat stress facilitates Orm phosphorylation through Ypk kinase activity. More important, heat induced-sphingoid base accumulation no longer occurs when Ypk kinase activity is inhibited (Figure 4c and Supplemental Figure S4b). These results not only provide strong evidence that Orm phosphorylation positively regulates sphingolipid production in vivo, but they also indicate that Ypk kinase transmits the heat stress signals to sphingoid intermediate production through phosphorylation of the Orm proteins.
Ypk1/2 are phosphorylated and activated by Pkh1/2, the homologues of mammalian PKD1 (Casamayor et al., 1999). Neither Pkh1 nor Pkh2 alone is required for cell growth, but loss of both proteins causes lethality (Casamayor et al., 1999). Consistently, heat stress–induced phosphorylation is abolished when Pkh kinase activity is inhibited (Figure 4d).
A recent study proposed that Orm2 expression levels regulate sphingolipid synthesis (Liu et al., 2012). However, as shown in Figure 3a, Orm phosphorylation starts to increase within 1 min after introduction of heat stress, reaching its maximum at around 5 min, and then decreases. The rapid time scale of these events strongly suggests that regulation of sphingolipid synthesis in response to heat is primarily due to the changes in Orm phosphorylation, not to changes in Orm expression.
Together, the results shown in Figures 3 and 4 establish a physiologically relevant context for the association between sphingolipid homeostasis and Orm protein phosphorylation dynamics and identify a novel feedback pathway for temporal regulation of sphingolipid biosynthesis during the heat stress response: heat stress rapidly activates the Pkh-Ypk signaling pathway to induce Orm phosphorylation, which in turn promotes sphingolipid intermediate production. The accumulated sphingoid intermediates then trigger Orm dephosphorylation, which in turn down regulates sphingolipid biosynthesis.
Cdc55-PP2A is a key phosphatase that counteracts Ypk1 activity in Orm-mediated sphingolipid biosynthesis regulation
Orm phosphorylation state is set not only by kinases but also by phosphatases. We next sought to identify the phosphatase involved in Orm dephosphorylation. We hypothesized that the phosphatase acting on the Orm proteins is likely to be involved in and/or regulated by sphingolipid levels. PP2A (Figure 5a) was identified as an attractive candidate for a ceramide-activated protein phosphatase in yeast (Nickels and Broach, 1996). Inactivation of Cdc55 or the catalytic subunits of PP2A was reported previously to suppress the endocytic defects of a mutant with impaired SPT activity (Friant et al., 2000), suggesting that PP2A may be involved in sphingolipid production. We explored the specific relationship between Cdc55-PP2A activity and Orm regulation at standard growth temperatures (25 or 30°C).
FIGURE 5:
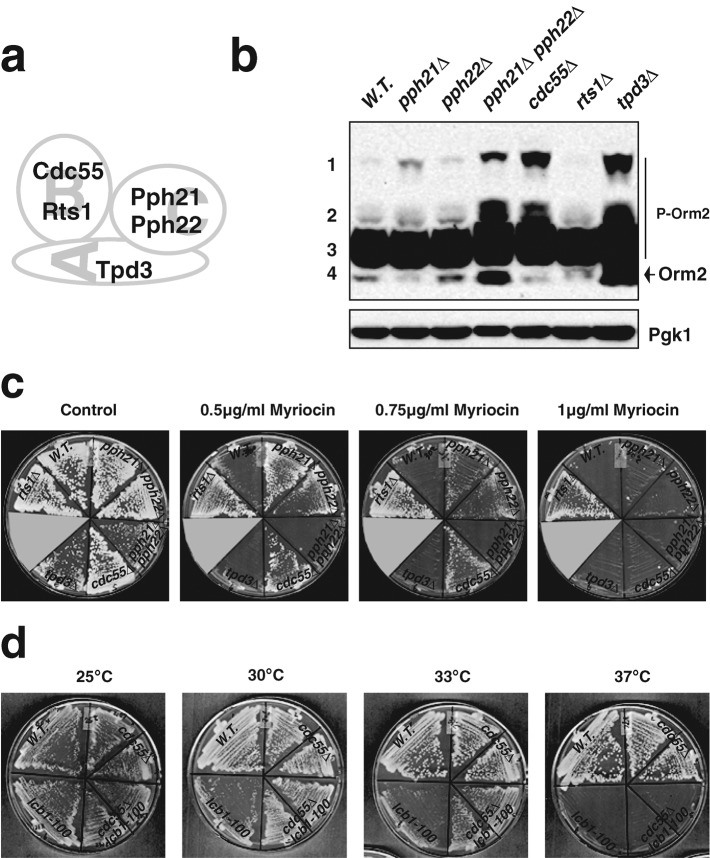
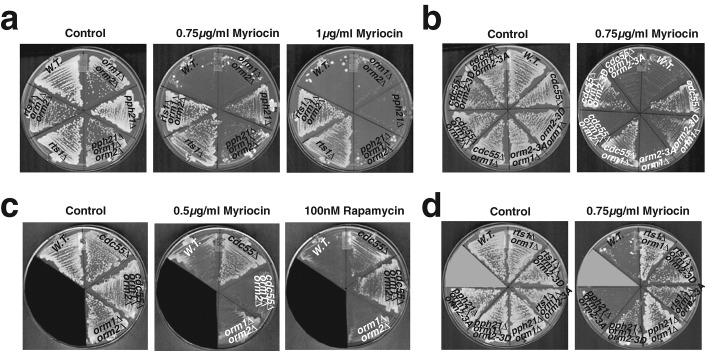
Orm2 phosphorylation status and myriocin resistance in various PP2A mutants. (a) Subunit composition of yeast PP2A. (b) Orm2 phosphorylation status in cells lacking the indicated PP2A subunits. Western blots showing the phosphorylation patterns for Orm2 after separation on phosphate-affinity gels. All cells were grown to early log phase in YPD at 25°C. Cell extracts were prepared as described in Materials and Methods. Pgk1 was used as a loading control. (c) Growth of PP2A mutants in the presence of myriocin. Various mutants were grown on plates containing the indicated concentrations of myriocin at 30°C for 3 d. (d) Cdc55 absence rescued the growth of an lcb1-100 ts mutant at semirestrictive temperature but not restrictive temperature. Mutants were grown at the indicated temperature for 3 d. Note that cdc55∆ lcb1-100 cells grow better than lcb1-100 at 30 and 33°C.
The yeast PP2A, similar to its mammalian counterparts, is a heterotrimer composed of three distinct subunits, namely A (the structural subunit, encoded by TPD3), B (the regulatory subunit, encoded by two distinct genes, CDC55 and RTS1), and C (the catalytic subunit, also encoded by two distinct genes, PPH21 and PPH22) (Jiang, 2006). If PP2A does dephosphorylate Orm2, lack of PP2A activity would be expected to result in increased Orm2 phosphorylation. tpd3∆ and pph21∆ pph22∆ showed increases in both phosphorylated (the two slowest-migrating bands) and dephosphorylated forms (the fastest-migrating band) of Orm2 (Figure 5b). The total Orm2 expression levels in these two mutants also appeared to be much higher than in wild-type cells (Figure 5b). Moreover, both of these strains have severe growth defects (Figure 5c). Thus, we conclude that it is not possible to address PP2A function, particularly in Orm phosphoregulation, using these two mutants; the elevated Orm2 expression in these mutants may affect sphingolipid synthesis (Liu et al., 2012). In contrast, pph21∆, pph22∆, cdc55∆, and rts1∆ single mutants grow relatively normally (Figure 5c). Compared to wild-type cells, Orm2 phosphorylation levels increased dramatically in cdc55∆ mutants (Figure 5b). pph21∆ cells also showed higher Orm2 phosphorylation (Figure 5b). In contrast, the Orm2 phosphorylation patterns in rts1∆ and wild-type cells were indistinguishable (Figure 5b). These results indicate that Cdc55-PP2A activity, rather than Rts1-PP2A activity, mediates Orm dephosphorylation.
We next examined how PP2A mutants respond to the SPT inhibitor myriocin. Previous studies suggested that Orm proteins negatively regulate SPT activity and that Orm phosphorylation relieves their inhibition of SPT (Breslow et al., 2010; Figure 1). Thus, impaired PP2A activity, which increases Orm2 phosphorylation, might lead to enhanced myriocin resistance. Unfortunately, tpd3∆ and pph21∆ pph22∆ mutants show severe growth defects even without myriocin (Figure 5c), making assessment of their myriocin sensitivity difficult. However, pph21∆, pph22∆, and cdc55∆ single mutants showed resistance to 0.75 μg/ml myriocin, a concentration that inhibits growth of wild-type cells (Figure 5c). The same set of PP2A mutants could not survive 1 μg/ml myriocin (Figure 5c), a concentration that likely causes more complete inhibition of SPT activity. In addition, cdc55∆ rescued the growth of the lcb1-100 ts mutant at a semirestrictive temperature but not at the restrictive temperature (Figure 5d). Lcb1 is an essential component of yeast SPT (Buede et al., 1991). Thus, it appears that the myriocin resistance of the Cdc55-PP2A–deficient mutants is only observable when SPT activity is not completely blocked and thus is still regulated by Orm proteins. The rts1∆ mutant shows resistance to even 1 μg/ml myriocin (Figure 5c), implying that the rts1∆ mutant cells and cdc55∆ mutant cells are resistant to myriocin through a different mechanism.
To further explore the relationship between PP2A, the Orm proteins, and SPT regulation, we examined the myriocin sensitivity of PP2A mutants in an orm-null mutant background. We found that pph21∆ orm1∆ orm2∆ and cdc55∆ orm1∆ orm2∆ triple mutants were not resistant to 0.75 μg/ml myriocin (Figure 6, a and b), whereas rts1∆ orm1∆ orm2∆ mutants were resistant to even 1 μg/ml myriocin (Figure 6a), establishing that Orm proteins are required for the myriocin resistance of Cdc55-PP2A–deficient mutants but not of the rts1∆ mutant.
FIGURE 6:
PP2A-deficient mutants confer myriocin resistance specifically through effects on the Orm proteins. (a, b, d) Impaired Cdc55-PP2A activity results in myriocin resistance dependent on Orm phosphorylation status. Various mutants were grown at 30°C for 3 d on plates containing the indicated myriocin concentrations. (c) Orm proteins are required for myriocin resistance but not rapamycin resistance in cdc55∆. Various mutants were grown on plates containing 0.5 μg/ml myriocin or 100 nM rapamycin at 30°C for 3 d.
We next tested the role of Ypk phosophorylation of the three Orm2 serines (Roelants et al., 2011) in the myriocin resistance of Cdc55-PP2A deficient mutants. As shown in Figure 6, b and d, the pph21∆ orm2-3D orm1∆ and cdc55∆ orm2-3D orm1∆ strains show myriocin resistance, whereas the pph21∆ orm2-3A orm1∆ and cdc55∆ orm2-3A orm1∆ strains do not. These results support the conclusion that Cdc55-PP2A counteracts Ypk1 phosphorylation of the three indicated Orm2 serines in regulation of sphingolipid production. In contrast, the rts1∆ orm2-3A orm1∆ strain is resistant to myriocin (Figure 6d), which further confirms our conclusion that myriocin resistance of rts1∆ is independent of the Orm proteins.
A previous study demonstrated that cdc55∆ confers resistance to rapamycin (Jiang and Broach, 1999), which is an inhibitor of TOR signaling. Strikingly, we found that Orm proteins are required for myriocin resistance of cdc55∆ cells but not for their rapamycin resistance (Figure 6c). This result establishes that whereas PP2A may be involved in multiple cellular signaling pathways (Jiang, 2006), Cdc55-PP2A and Orm protein activities are linked specifically for sphingolipid biosynthesis. Furthermore, sphingoid base levels were substantially increased in cdc55∆ cells (Supplemental Figure S3), indicating that Cdc55-PP2A activity is involved in sphingolipid production.
PP2A activity contributes to the regulation of Orm phosphorylation dynamics in response to heat stress
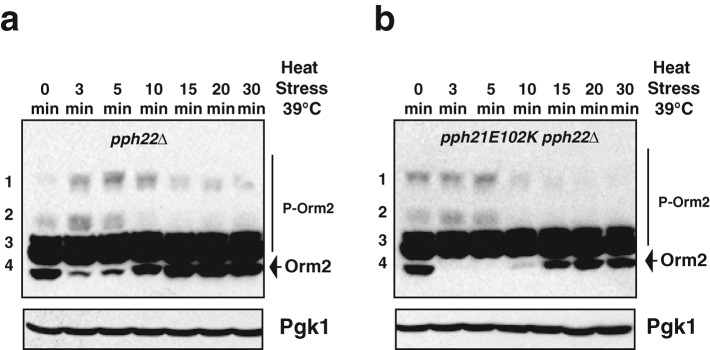
We next examined the role of Cdc55-PP2A in Orm2 dephosphorylation dynamics in response to heat stress. We generated a PP2A ts mutant, pph21 E102K pph22∆ (Lin and Arndt, 1995), which allowed us to simultaneously introduce heat stress and turn off PP2A activity. As shown in Figure 7, substantially less dephosphorylated Orm2, seen as the fastest-migrating bands on a phosphate-affinity gel, appears in the PP2A ts mutant at both the 10- and 15-min time points compared with pph22∆ cells. The observation that Orm dephosphorylation was not completely abolished in the PP2A ts mutant suggests that other phosphatases may also contribute to Orm protein dephosphorylation in response to heat stress. It is also possible that PP2A activity was not completely lost in the PP2A ts mutant after the short temperature shift. Nevertheless, our results indicate that PP2A activity contributes to regulation of Orm phosphorylation dynamics in response to heat stress.
FIGURE 7:
Orm phosphorylation status in PP2A-deficient mutants upon heat shock. Western blots showing the phosphorylation patterns for Orm2 after separation on phosphate-affinity gels. P-Orm2 indicates phosphorylated forms of Orm2. Pgk1 was used as a loading control. Cell extracts were prepared from cells shifted from 25 to 39°C for the indicated times as described in Materials and Methods. The pph21E102K pph22∆ mutant is temperature sensitive.
DISCUSSION
A model for how Orm phosphorylation regulation controls sphingolipid biosynthesis in response to stress in a kinetically coupled manner
As bioactive molecules, sphingoid intermediates transmit signals when their cellular levels change in response to various stresses (Hannun and Obeid, 2008; Dickson, 2010; Nikolova-Karakashian and Rozenova, 2010). However, both lack of sphingolipids and constitutive high levels of sphingolipids compromise cell viability (Buede et al., 1991; Chung et al., 2001). Thus, biosynthesis of sphingoid intermediates in response to stresses requires precise temporal control. Previously, very little was known about the regulatory mechanism.
Here we provided evidence supporting a model in which the conserved Pkh-Ypk signaling cascade and Cdc55-PP2A facilitate/ensure rapid, transient sphingolipid production upon heat stress through regulation of Orm protein phosphorylation (Figure 8): 1) A Pkh-Ypk cascade is rapidly activated upon heat stress. 2) Orm phosphorylation rapidly increases. 3) Orm phosphorylation releases inhibition of SPT activity. 4) Activated SPT promotes de novo synthesis of sphingoid intermediates. 5) Accumulated sphingoid intermediates act as signaling molecules to initiate the cellular heat shock responses, as described in the Introduction. Sphingoid intermediate accumulation also feeds back to dephosphorylate Orm proteins, possibly by inhibiting Pkh-Ypk kinase activity or/and activating Cdc55-PP2A phosphatase activity. 6) Dephosphorylated Orm proteins inhibit SPT activity.
FIGURE 8:
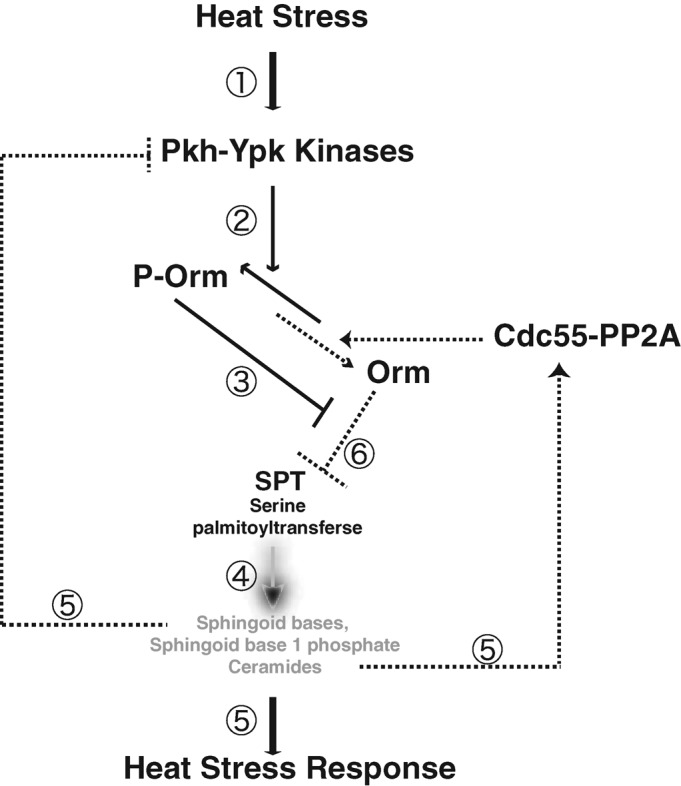
A feedback regulation pathway in which Orm protein phosphorylation dynamics rapidly and precisely regulate sphingolipid biosynthesis in response to heat stress. See the text for a description.
The Pkh-Ypk cascade was previously proposed to be downstream of heat-induced sphingolipid accumulation (Dickson, 2010). However, our data indicate that the Pkh-Ypk cascade is activated in response to heat stress and induces de novo sphingolipid biosynthesis (Figure 4). Of interest, a previous in vitro study indicated that Pkh1/Pkh2 kinases are inhibited by micromolar concentrations of sphingoid bases (Friant et al., 2001). Thus, the most harmonious interpretation of our results and previous findings is that the Pkh-Ypk cascade is also regulated by heat-induced sphingolipid intermediate accumulation via a negative feedback mechanism by which increased sphingolipids inhibit Pkh-Ypk kinase–mediated phosphorylation of Orm proteins, thereby restoring SPT inhibition.
How could heat stress activate Ypk kinases to trigger Orm2 phosphorylation in such a rapid manner (Figure 3a)? Breslow et al. (2010) showed that disruption of sphingolipid biosynthesis by myriocin results in an increase in Orm phosphorylation. However, the increase in Orm phosphorylation in response to heat stress apparently is not due to low levels of sphingolipids, because both the levels of Orm phosphorylation and the levels of sphingolipid intermediates actually continue to increase at these early time points after heat stress (Figure 2C, time points between 0 and 4 min; Dickson et al., 1997; Jenkins et al., 1997; Jenkins, 2003; Wells et al., 1998; Mao et al., 1999; Skrzypek et al., 1999). Moreover, our data suggest that heat stress affects Orm phosphorylation before the myriocin-sensitive step (Figure 3e). Ypk1 activation is known to require its phosphorylation by both Pkh kinases and the TORC2 complex (Casamayor et al., 1999; Kamada et al., 2005). Recently Slm1/2 was demonstrated to play important roles in recruiting Ypk1 to the plasma membrane for activation by the TORC2 complex (Niles et al., 2012). Of interest, both Pkh kinases and Slm1/2 localize in punctate eisosome plasma membrane domains (Walther et al., 2007; Grossmann et al., 2008), and the TORC2 complex localizes in small foci on the plasma membrane but not in eisosomes (Berchtold and Walther, 2009). Another recent study suggested that plasma membrane stress might induce relocalization of Slm1/2 and activation of TORC2 (Berchtold et al., 2012). Thus, it is possible that heat stress may rapidly decrease the rigidity of plasma membrane, resulting in release of Pkh kinase and Slm1/2 from eisosomes, thereby triggering Ypk kinase activation.
Both the lipids and the proteins involved in this regulatory circuit (Figure 8) are highly conserved between yeast and mammalian cells. Thus, the mechanism we proposed here may serve a general basis for how Orm phosphoregulation controls sphingolipid biosynthesis in response to stress in a kinetically coupled manner.
Orm phosphorylation status is highly responsive to sphingoid bases
In this study, we demonstrated that the high levels of Orm phosphorylation induced by heat stress decline while the sphingoid bases (PHS) reach their peak levels (Figure 3c, time points between 5 and 15 min). Several previous studies established that sphingoid bases and sphingoid base phosphates accumulate with similar timing (Dickson et al., 1997; Jenkins et al., 1997). However, the heat-induced ceramides peak ∼10 min after the sphingoid bases reach their peak (Dickson et al., 1997; Jenkins et al., 1997), and no obvious changes in the levels of complex sphingolipids were observed during 2 h of heat stress (Jenkins et al., 1997). Thus, the decrease of Orm phosphorylation induced by heat stress is kinetically coupled to changes in sphingoid base and sphingoid base phosphate levels but not ceramide or complex sphingolipid levels, suggesting that Orm phosphorylation is primarily regulated by sphingoid bases and sphingoid base phosphates. In agreement with these conclusions, our experiments indicate that exogenously provided sphingoid bases are sufficient to induce Orm dephosphorylation without being converted to ceramide or to complex sphingolipids (Figure 2).
A recent study proposed that Orm1 and Orm2 may regulate sphingolipid synthesis via two different mechanisms (Liu et al., 2012). We observed the phosphorylation dynamics of Orm1 and Orm2 upon heat stress to be very similar (unpublished data), suggesting that both of the Orm proteins function similarly in regulation of the sphingolipid biosynthesis response to heat stress. However, we found that Orm2 expression was at least 10 times higher than Orm1 expression (Supplemental Figure S1a). It is also worth mentioning that neither C-terminally tagged Orm1 nor Orm2 is functional (unpublished data) and that Orm1 tagged at its N-terminus with 3XFLAG cannot fulfill Orm1 function in the orm2∆ background (Supplemental Figure S1A). Our studies indicate that Orm function is preserved in N-terminally tagged Orm2.
Phosphate-affinity gels separated Orm2 protein into four mobility groups. Western blotting analysis revealed that the majority of Orm proteins are moderately phosphorylated (bands in the third group, Figure 2a) under standard growth conditions. This large pool of moderately phosphorylated Orm protein may explain how cells rapidly respond to various environmental stresses in a graded manner. In addition, the majority of orm2-3A proteins were detected as dephosphorylated forms, and levels of sphingoid bases were greatly reduced in orm2-3A cells, further supporting the conclusion that dephosphorylated Orm proteins negatively regulate sphingolipid biosynthesis. Given that the reduction of dephosphorylated Orm2 and the increase in hyperphosphorylated Orm2 occur at the same time in response to heat stress (Figure 3b, time points 2 and 5 min), SPT activation in response to heat stress could be caused by release of SPT inhibition by dephosphorylated Orm2, by positive regulation of SPT by hyperphosphorylated Orm2, or both.
Cdc55-PP2A counteracts Ypk1 activity in Orm-mediated sphingolipid biosynthesis regulation
In comparison to kinases, phosphatases often function in a less specific manner. In this study, we performed a series of experiments to demonstrate a specific relationship between Cdc55-PP2A and the Orm proteins in regulating sphingolipid biosynthesis. First, sphingolipid biosynthesis increased in cdc55∆ mutants. Second, Orm proteins were required for myriocin resistance but not for rapamycin resistance of cdc55∆ cells. Finally, genetic analysis suggested that Cdc55-PP2A functions to counteract Ypk1 kinase–mediated Orm phosphorylation.
In contrast, we found that Rts1, another PP2A regulatory subunit, is involved in sphingolipid regulation in an Orm protein–independent manner. Previous studies suggested that Sac1 binds and modulates SPT activity in a pathway independent of Orm proteins (Breslow et al., 2010). Future investigations are needed to test whether Rts1-PP2A is involved in Sac1 modulation of SPT activity.
PP2A was previously identified as an attractive candidate for a ceramide-activated protein phosphatase in yeast (Nickels and Broach, 1996), although direct evidence that ceramides (or other sphingoid intermediates) activate PP2A has not been reported. It is not clear whether other phosphatases are also involved in Orm phosphoregulation. Loss of Ypk kinase activity abolished sphingolipid biosynthesis in response to heat stress (Figure 4c), but Orm phosphorylation still decreased (Figure 4b, right), suggesting that Cdc55-PP2A may affect Orm phosphorylation at least partially in a constitutive manner. Because both Pkh kinases and the TORC2 complex are required for Ypk kinase activity, an alternative explanation for our results could be that Cdc55-PP2A controls Orm phosphorylation by negatively regulating these two upstream effectors of Ypk kinases, instead of directly dephosphorylating the Orm proteins. However, this possibility seems unlikely because Orm2 dephosphorylation should have no longer occurred upon loss of Ypk kinase activity (Figure 4b, right), which is contrary to our observation.
MATERIALS AND METHODS
Media and strain construction
The yeast strains used in this study are listed in Supplemental Table S1. They were grown in standard rich media (yeast extract/peptone/dextrose [YPD]). The primers used for cloning are listed in Supplemental Table S2.
To generate a strain expressing the 3XFLAG-fused Orm2 protein from its genetic locus, we first constructed a vector using following strategies. Two DNA fragments were obtained by PCR amplification, using primer pairs of HindIII-ORM2-F/FLAG-ORM2-interR and FLAG-ORM2-interF/XhoI-ORM2-R, respectively. The two DNA fragments were then converted into one by PCR amplification, using primer pairs of HindIII-ORM2-F/XhoI-ORM2-R. The resulting PCR product was digested and ligated into the pBluescript II KS(+) vector using the HindIII and XhoI restriction enzymes. In addition, a NAT1 (nourseothricin acetyltransferase) DNA fragment, amplified by primer pairs NatR-ORM2-F/NatR-ORM2-R, was inserted into the same vector using the HindIII and SpeI restriction enzymes. The resulting plasmid, named pBS-NAT1-FLAG-ORM2, was digested with HindIII and XhoI, and the fragment containing NAT1-FLAG-ORM2 was transformed into a yeast strain in which ORM2 had been replaced by Candida glabrata (Cg) URA3. We isolated transformants that no longer grow on a plate lacking uracil but do grow on a nourseothricin-dihydrogen sulfate (clonNAT)–containing plate. The pBS-NAT1-FLAG-ORM2 plasmid was also used as a template to generate NAT1-FLAG-ORM2-3A and NAT1-FLAG ORM2-3D plasmids using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) and primers ORM2-3A-F/ORM2-3A-R and ORM2-3D-F/ORM2-3D-R, respectively. A strain expressing 3XFLAG-Orm1 was generated by a similar strategy but using different primers, listed in Supplemental Table S2.
To obtain strains expressing Ypk1-as (Ypk1 L424A), two fragments were PCR amplified from genomic DNA using primers OYS227/OYS268 and OYS228/OYS267. The two fragments were then converted into one by PCR amplification, using primer pairs of OYS227/OYS228. The resulting fragment was digested and ligated into pRS306 vector. pRS306-YPK1AS (YPK1 L424A) was linearized using StuI and transferred into the URA3 locus of a ypk1∆::CgLEU2 strain.
To obtain strains expressing Pkh1-as, a fragment containing 500 base pairs upstream and downstream of PKH1 was PCR amplified from genomic DNA using primers PKH1-XhoI-F and PKH1-SacII-R. The PCR product was digested by XhoI/SacII and was ligated into pRS306 vector, creating pRS306-PKH1. pRS306-PKH1 was used as template to generate the PKH1-AS (PKH1 F187V, L203A) plasmid using the QuikChange Lightning Site-Directed Mutagenesis Kit and primers PKH1-F187V/L203A-F and PKH1-F187V/L203A-R. PKH1-AS (PKH1 F187V, L203A) plasmid was linearized using StuI and inserted into the URA3 locus of a pkh1∆::CgLEU2 strain.
PPH21 E102K plasmid was obtained using a similar strategy as for the PKH1-AS plasmid. Primers PPH21-BamHI-F and PPH21-NotI-R were used to amplify the PPH21 fragment. The PPH21 fragment was ligated into the pRS306 vector, creating pRS306-PPH21. pRS306-PPH21 was used as a template to generate PPH21 E102K plasmid using the QuikChange Lightning Site-Directed Mutagenesis Kit and primers PPH21-E102K-F and PPH21-E102K-R. Linearized pRS306-PPH21 E102K plasmid was integrated into the URA3 locus of a pph21∆::CgHIS3 strain.
Lipid reagents
Phytosphingosine purified from Saccharomyces cerevisiae was obtained from Avanti Polar Lipids (Alabaster, AL). Sphingosine was obtained from Santa Cruz Biotechnology (Santa Cruz, CA), and stearylamine was obtained from Sigma-Aldrich (St. Louis, MO). All lipids were dissolved in methanol.
Preparation of whole-cell extracts
Yeast cells were grown to early logarithmic phase in YPD. After the indicated treatments, cold trichloroacetic acid (TCA) was added to the yeast culture to a final concentration of 20% (vol/vol). The growth medium was removed after centrifugation, and cell pellets were flash frozen in liquid nitrogen. The cells were thawed, resuspended in 5% (vol/vol) TCA, and lysed by bead beating at 4°C for 10 min. Whole-cell extracts were collected by 5 min of centrifugation at 14,000 × g at 4°C. The pellets were resuspended in SDS–PAGE sample buffer containing 50 mM dithiothreitol.
Detection of protein phosphorylation
To detect phosphorylation-dependent mobility shifts of FLAG-Orm1 and FLAG-Orm2, whole-cell extracts were loaded onto 8% SDS polyacrylamide gels containing 25 μM Phos-tag acrylamide (Wako Chemicals USA, Richmond, VA) and 25 μM MnCl2. Before transfer to nitrocellulose membranes, gels were washed twice for 10 min in 2 mM EDTA containing transfer buffer (25 mM Tris-HCl pH 8.3, 192 mM glycine, 20% [vol/vol] methanol) and then once for 10 min in transfer buffer without EDTA. Membranes were then probed with 1:4000 mouse anti-FLAG (Sigma-Aldrich).
Lipid analysis by high-performance liquid chromatography
Extraction and processing of sphingoid bases from yeast cells for fluorescence high-performance liquid chromatography (HPLC) analysis using the AQC reagent (Waters, Milford, MA) was performed as described previously (Lester and Dickson, 2001). Sampling of heat-stressed cells was as described. HPLC analysis was performed using a C18 column (4.6 × 250 mm, XDB-C18; Hewlett-Packard, Palo, Alto, CA) on a Shimadzu LC10A series liquid chromatography system. Isocratic elution was carried out for 60 min at a flow rate of 1.0 ml/min. Lipid-reacted AQC reagent was excited with 244-nm UV radiation, and the resultant emission signal at 398 nm was detected. C18-PHS reacted with the AQC reagent was used as a standard for quantification.
Supplementary Material
Acknowledgments
We are grateful to Françoise Roelants and Jeremy Thorner for sharing information before publication on the identity of the kinase that phosphorylates the Orm proteins. We thank Howard Riezman for providing the lcb1-100 strain. We gratefully acknowledge Betsy Wong and Casey Drubin for the preparation of constructs and strains. The manuscript was improved by the critical comments of Georjana Barnes, Christa Cortesio, Jonathan Wong, and Nathaniel Krefman. This work was supported by National Institutes of Health Grant R01 GM 50399 to D.G.D. Y.M. acknowledges a Human Frontiers Science Program Long-term Fellowship.
Abbreviations used:
- Cg
Candida glabrata
- DHS
dihydrosphingosine
- DMSO
dimethyl sulfoxide
- HPLC
high-performance liquid chromatography
- PHS
phytosphingosine
- PP2A
phosphatase 2A
- Sph
sphingosine
- SPT
serine palmitoyltransferase
- TCA
trichloroacetic acid
- Ypk1
yeast protein kinase 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0209) on April 25, 2012.
REFERENCES
- Berchtold D, Piccolis M, Chiaruttini M, Riezman I, Riezman H, Roux A, Walther TC, Loewith R. Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat Cell Biol. 2012;14:542–547. doi: 10.1038/ncb2480. [DOI] [PubMed] [Google Scholar]
- Berchtold D, Walther TC. TORC2 plasma membrane localization is essential for cell viability and restricted to a distinct domain. Mol Biol Cell. 2009;20:1565–1575. doi: 10.1091/mbc.E08-10-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Collins SR, Bodenmiller B, Aebersold R, Simons K, Shevchenko A, Ejsing CS, Weissman JS. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–1053. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buede R, Rinker-Schaffer C, Pinto WJ, Lester RL, Dickson RC. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J Bacteriol. 1991;173:4325–4332. doi: 10.1128/jb.173.14.4325-4332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Torrance PD, Kobayashi T, Thorner J, Alessi DR. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol. 1999;9:186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- Chen P, Lee KS, Levin DE. A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol Gen Genet. 1993;236:443–447. doi: 10.1007/BF00277146. [DOI] [PubMed] [Google Scholar]
- Chung N, Mao C, Heitman J, Hannun YA, Obeid LM. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J Biol Chem. 2001;276:35614–35621. doi: 10.1074/jbc.M105653200. [DOI] [PubMed] [Google Scholar]
- Cowart LA, Gandy JL, Tholanikunnel B, Hannun YA. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem J. 2010;431:31–38. doi: 10.1042/BJ20100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowart LA, Hannun YA. Using genomic and lipidomic strategies to investigate sphingolipid function in the yeast heat-stress response. Biochem Soc Trans. 2005;33:1166–1169. doi: 10.1042/BST20051166. [DOI] [PubMed] [Google Scholar]
- Cowart LA, Hannun YA. Selective substrate supply in the regulation of yeast de novo sphingolipid synthesis. J Biol Chem. 2007;282:12330–12340. doi: 10.1074/jbc.M700685200. [DOI] [PubMed] [Google Scholar]
- Dickson RC. Roles for sphingolipids in Saccharomyces cerevisiae. Adv Exp Med Biol. 2010;688:217–231. doi: 10.1007/978-1-4419-6741-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, Nagiec EE, Skrzypek M, Tillman P, Wells GB, Lester RL. Sphingolipids are potential heat stress signals in Saccharomyces. J Biol Chem. 1997;272:30196–30200. doi: 10.1074/jbc.272.48.30196. [DOI] [PubMed] [Google Scholar]
- Friant S, Lombardi R, Schmelzle T, Hall MN, Riezman H. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 2001;20:6783–6792. doi: 10.1093/emboj/20.23.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S, Zanolari B, Riezman H. Increased protein kinase or decreased PP2A activity bypasses sphingoid base requirement in endocytosis. EMBO J. 2000;19:2834–2844. doi: 10.1093/emboj/19.12.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K, Lombardi R, Vallee B, Riezman H. Lcb4p is a key regulator of ceramide synthesis from exogenous long chain sphingoid base in Saccharomyces cerevisiae. J Biol Chem. 2003;278:7325–7334. doi: 10.1074/jbc.M209925200. [DOI] [PubMed] [Google Scholar]
- Grossmann G, Malinsky J, Stahlschmidt W, Loibl M, Weig-Meckl I, Frommer WB, Opekarova M, Tanner W. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J Cell Biol. 2008;183:1075–1088. doi: 10.1083/jcb.200806035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Lone MA, Schneiter R, Chang A. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc Natl Acad Sci USA. 2010;107:5851–5856. doi: 10.1073/pnas.0911617107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Jenkins GM. The emerging role for sphingolipids in the eukaryotic heat shock response. Cell Mol Life Sci. 2003;60:701–710. doi: 10.1007/s00018-003-2239-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Hannun YA. Role for de novo sphingoid base biosynthesis in the heat-induced transient cell cycle arrest of Saccharomyces cerevisiae. J Biol Chem. 2001;276:8574–8581. doi: 10.1074/jbc.M007425200. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J Biol Chem. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:440–449. doi: 10.1128/MMBR.00049-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- Lester RL, Dickson RC. High-performance liquid chromatography analysis of molecular species of sphingolipid-related long chain bases and long chain base phosphates in Saccharomyces cerevisiae after derivatization with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate. Anal Biochem. 2001;298:283–292. doi: 10.1006/abio.2001.5368. [DOI] [PubMed] [Google Scholar]
- Lin FC, Arndt KT. The role of Saccharomyces cerevisiae type 2A phosphatase in the actin cytoskeleton and in entry into mitosis. EMBO J. 1995;14:2745–2759. doi: 10.1002/j.1460-2075.1995.tb07275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Zhang X, Lester RL, Dickson RC. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J Biol Chem. 2005;280:22679–22687. doi: 10.1074/jbc.M502972200. [DOI] [PubMed] [Google Scholar]
- Liu M, Huang C, Polu SR, Schneiter R, Chang A. Regulation of sphingolipid synthesis via Orm1 and Orm2 in yeast. J Cell Sci. doi: 10.1242/jcs.100578. doi: 10.1242/jcs.100578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C, Saba JD, Obeid LM. The dihydrosphingosine-1-phosphate phosphatases of Saccharomyces cerevisiae are important regulators of cell proliferation and heat stress responses. Biochem J. 1999;342:667–675. [PMC free article] [PubMed] [Google Scholar]
- Mao C, Wadleigh M, Jenkins GM, Hannun YA, Obeid LM. Identification and characterization of Saccharomyces cerevisiae dihydrosphingosine-1-phosphate phosphatase. J Biol Chem. 1997;272:28690–28694. doi: 10.1074/jbc.272.45.28690. [DOI] [PubMed] [Google Scholar]
- Meier KD, Deloche O, Kajiwara K, Funato K, Riezman H. Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1164–1175. doi: 10.1091/mbc.E05-11-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC. Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene. J Biol Chem. 1997;272:9809–9817. doi: 10.1074/jbc.272.15.9809. [DOI] [PubMed] [Google Scholar]
- Nagiec MM, Skrzypek M, Nagiec EE, Lester RL, Dickson RC. The LCB4 (YOR171c) and LCB5 (YLR260w) genes of Saccharomyces encode sphingoid long chain base kinases. J Biol Chem. 1998;273:19437–19442. doi: 10.1074/jbc.273.31.19437. [DOI] [PubMed] [Google Scholar]
- Nickels JT, Broach JR. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- Nikolova-Karakashian MN, Rozenova KA. Ceramide in stress response. Adv Exp Med Biol. 2010;688:86–108. doi: 10.1007/978-1-4419-6741-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niles BJ, Mogri H, Hill A, Vlahakis A, Powers T. Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc Natl Acad Sci USA. 2012;109:1536–1541. doi: 10.1073/pnas.1117563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qie L, Nagiec MM, Baltisberger JA, Lester RL, Dickson RC. Identification of a Saccharomyces gene, LCB3, necessary for incorporation of exogenous long chain bases into sphingolipids. J Biol Chem. 1997;272:16110–16117. doi: 10.1074/jbc.272.26.16110. [DOI] [PubMed] [Google Scholar]
- Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2011;108:19222–19227. doi: 10.1073/pnas.1116948108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzypek MS, Nagiec MM, Lester RL, Dickson RC. Analysis of phosphorylated sphingolipid long-chain bases reveals potential roles in heat stress and growth control in Saccharomyces. J Bacteriol. 1999;181:1134–1140. doi: 10.1128/jb.181.4.1134-1140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Taniguchi R, Tanoue D, Yamaji T, Takematsu H, Mori K, Fujita T, Kawasaki T, Kozutsumi Y. Sli2 (Ypk1), a homologue of mammalian protein kinase SGK, is a downstream kinase in the sphingolipid-mediated signaling pathway of yeast. Mol Cell Biol. 2000;20:4411–4419. doi: 10.1128/mcb.20.12.4411-4419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Aguilar PS, Frohlich F, Chu F, Moreira K, Burlingame AL, Walter P. Pkh-kinases control eisosome assembly and organization. EMBO J. 2007;26:4946–4955. doi: 10.1038/sj.emboj.7601933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells GB, Dickson RC, Lester RL. Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J Biol Chem. 1998;273:7235–7243. doi: 10.1074/jbc.273.13.7235. [DOI] [PubMed] [Google Scholar]
- Wells GB, Lester RL. The isolation and characterization of a mutant strain of Saccharomyces cerevisiae that requires a long chain base for growth and for synthesis of phosphosphingolipids. J Biol Chem. 1983;258:10200–10203. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.