Abstract
X-box binding protein 1 (XBP1) is a unique basic region leucine zipper (bZIP) transcription factor whose active form is generated by a nonconventional splicing reaction upon disruption of homeostasis in the endoplasmic reticulum (ER) and activation of the unfolded protein response (UPR). XBP1, first identified as a key regulator of major histocompatibility complex (MHC) class II gene expression in B cells, represents the most conserved signaling component of UPR and is critical for cell fate determination in response to ER stress. Here we review recent advances in our understanding of this multifaceted transcription factor in health and diseases.
Key words: X-box binding protein 1 (XBP1), Inositol-requiring enzyme 1 (IRE1α), Unfolded protein response (UPR), Splicing, Transcription, Diseases
INTRODUCTION
X-box binding protein 1 (XBP1) was initially discovered as a transcription factor critical in the regulation of human MHC class II gene expression in the early 1990s. Approximately a decade later in 2001–2002, three reports demonstrated unequivocally that XBP1 was the long sought-after mammalian homologue of HAC1 in yeast, a key transcription factor that orchestrates the unfolded protein response (UPR).
UPR, an essential arm of the quality control system designed to reestablish endoplasmic reticulum (ER) homeostasis, is initiated by the activation of three major sensors at the ER membrane: inositol-requiring enzyme 1 (IRE1), PKR-like-ER kinase (PERK), and activating transcription factor 6 (ATF6) (Fig. 1). Activation of UPR leads to the induction of chaperones and ER-associated degradation (ERAD) components as well as global translational attenuation and induction of apoptosis (if stress persists). As the activation of these three signaling pathways under pharmacological and pathophysiological conditions have been reviewed recently (40,51,73,99), we will focus primarily on the biology and role of the IRE1–XBP1 pathway in diseases as well as explore potential mechanisms to manipulate this transcription factor in therapeutic settings.
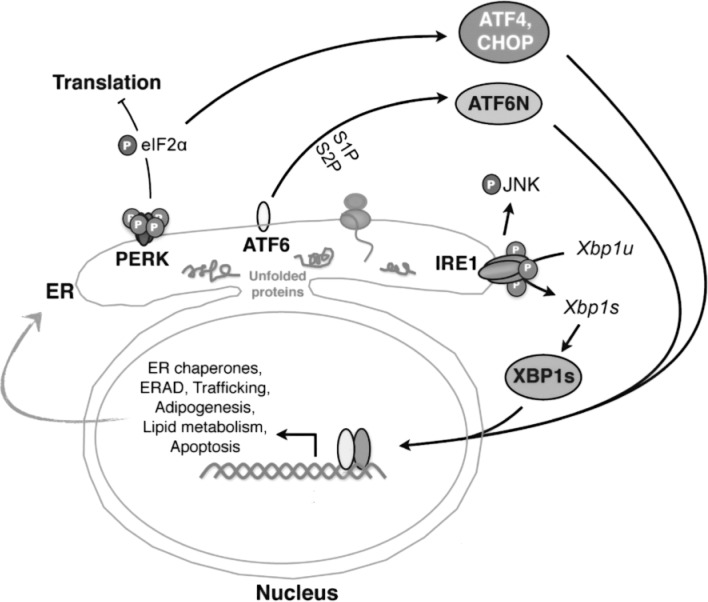
Figure 1.
Three major UPR pathways in metazoans. Misfolded proteins or homeostatic alterations in the ER activate three ER-resident sensors: IRE1α, PERK, and ATF6. Key players in each pathway are highlighted. Among the three branches, the IRE1α–XBP1 pathway, the focus of this review, is the most evolutionarily conserved. IRE1α mediates Xbp1u mRNA splicing to generate a potent transcription factor XBP1s (the spliced form of XBP1). XBP1s regulates a diverse array of genes involved in ER homeostasis, adipogenesis, lipogenesis, and cell survival. In addition, activation of IRE1α may lead to phosphorylation of JNK and hence influence the outcome of inflammatory response.
THE BIOLOGY OF XBP1
The Discovery of XBP1
The human XBP1 gene was discovered and characterized in 1990 as a basic region leucine zipper (bZIP) transcription factor present in B cells that interacted specifically with the conserved X2 boxes located in the promoters of MHC class II genes (53). XBP1 formed a stable functional heterodimer with c-fos that was critical for the expression of hXBP1 target genes (61). Further analysis of the XBP1 promoter revealed multiple regulatory cis elements, including a motif identical to the X2 sites bound by XBP1 (66,69). In situ hybridization studies revealed ubiquitous expression of XBP1 in adult tissues as well as in fetal exocrine glands and osteoblasts (12). Importantly, mice with germline XBP1 knockout died in utero from severe liver or heart hypoplasia and apoptosis (55,67).
Linking XBP1 to UPR
In 1993, two laboratories independently reported that communication between the ER and nucleus, termed UPR, was mediated by an ER transmembrane kinase ERN1/IRE1 (14,56). Subsequent work in yeast demonstrated that in addition to a kinase domain, IRE1 possessed endoribonuclease activity and Hac1 was the substrate (15). IRE1 activation led to splicing of the mRNA of Hac1u (uninduced) to generate Hac1i (induced) (82). Hac1i mRNA encoded a potent transcription factor responsible for the upregulation of many genes involved in protein folding, degradation, and trafficking (92). XBP1 was later identified as the mammalian homolog of HAC1 (6,48,100).
In metazoans, the IRE1α–XBP1 pathway is the most highly conserved and is critical for ER biogenesis and the secretory capacity of cells. Similar to yeast IRE1, metazoan IRE1α, a type I transmembrane protein, oligomerizes upon ER stress, resulting in increased activity of both its cytosolic kinase and endoribonuclease domains. Once activated, IRE1α splices 26 nucleotides from the Xbp1u mRNA (un-spliced), leading to a frameshift and the generation of XBP1s (spliced) that contains a C-terminal transactivation domain absent from XBP1u (6,48,100) (Fig. 2). Although XBP1u is very unstable, the longer half-life of XBP1s (∼22 min vs. ∼11 min for XBP1u) allows it to translocate into the nucleus and transcriptionally activate its target genes (Figs. 1 and 2).
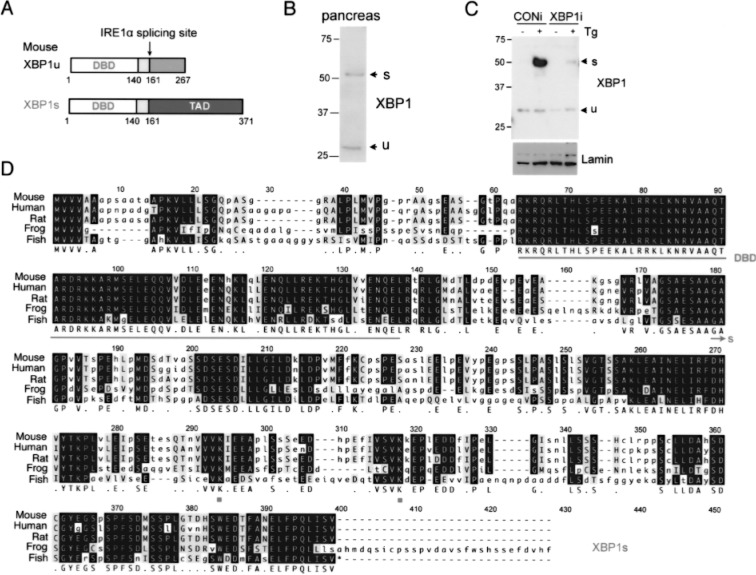
Figure 2.
Comparison of XBP1u and XBP1s proteins. (A) The domain comparison of XBP1u and XBP1s proteins. Number refers to amino acid position of mouse proteins. DBD, DNA binding domain; TAD, transactivation domain. (B) Western blot of XBP1 in nuclear extract of mouse pancreatic lysates. Note the positions of endogenous XBP1u and XBP1s proteins. (C) Western blot of XBP1s in nuclear extract of mouse macrophage RAW 264.7 cells treated with 300 nM thapsigargin (Tg) for 2 h. Cells were stably expressing either XBP1 RNAi or control RNAi. Lamin, a loading control. (D) Sequence alignment of XBP1s from different species. DBD, underlined; SUMOylation lysine (K) sites, K276 and K297; XBP1s unique sequence, arrow.
From an evolutionary perspective, the presence of eukaryotic genes with overlapping reading frames such as Xbplu and Xbpls presents an intriguing but puzzling question. Using comparative approaches, the Xbplu coding sequence (CDS) was observed to be strongly conserved and both the unspliced and spliced CDS had similar nonsynonymous substitution rates, providing evidence for a functional role for XBP1u (58). In the current model, XBP1u negatively regulates XBP1s transcriptional activity and hence UPR signaling (46,101). However, this model was challenged by the observation that overexpression of stabilized XBP1u increased the expression of XBP1s targets (89), implying a much more complicated role of XBP1u in UPR signaling. Indeed, a recent study showed that XBP1u recruited its own mRNA to the ER membrane for efficient IRE1α-mediated splicing (97). Thus, although the precise function of XBP1u remains elusive, it does appear to have a biphasic role in UPR initiation and resolution.
Transcriptional Regulation of Xbp1 Expression
In addition to the well-characterized Xbp1 mRNA splicing event, accumulating evidence suggested that transcriptional regulation of Xbp1 gene expression might also play an important role with profound pathological and therapeutic implications. Indeed, several recent studies have shown that the Xbp1 proximal promoter serves as a target for various tissue-specific and developmentally regulated transcription factors, contributing to the temporal- and spatial-specific expression of XBP1.
As the role of XBP1 in plasma cell differentiation was unraveled, studies demonstrated that Xbpl mRNA was induced by interleukin (IL)-6 in human multiple myeloma cells (96) and IL-4 in primary B cells (33). B lymphocyte-induced maturation protein, encoded by Prdml gene (BLIMP1) and interferon regulatory factor 4 (IRF4) are two major transcription factors critical for Xbpl expression and plasma cell differentiation. Microarray studies placed BLIMP1 upstream of XBP1 but as BLIMP1 acts as a transcriptional repressor, mechanistic questions arose as to how BLIMP1 induced Xbpl expression (79,80). It was discovered that BLIMP 1 repressed paired box gene 5/B cell lineage-specific activator protein (BSAP/ PAX5), a known repressor of Xbpl in B cells (69,79). IRF4 is an interferon (IFN)-regulatory family member that is expressed in B cells committed to the plasma cell lineage and required for plasmacytoid differentiation (41). XBP1s induction was ablated in IRF4-deficient cells, but BLIMP1 expression was unaffected, suggesting that IRF4 acted upstream of XBP1 in a nonredundant manner. Furthermore, other adaptive immune responses such as effector CD8+ T-cell differentiation and macrophage activation upregulated Xbpl expression through IL-2 (38) and the ligand for Toll-like receptor 4 (TLR4), lipopolysac-charide (LPS), respectively (54,71).
Various other factors contributing to the regulation of Xbpl gene expression are emerging as well. Genome-wide analysis studies identified a putative binding site for C/EBPp in the proximal promoter of Xbpl (49). Accordingly, C/EBPp was shown to directly regulate XBP1 expression in 3T3-L1 preadipocytes (78). Two myogenic transcription factors, MyoD and myogenin, were also shown to directly regulate Xbpl expression (4). In addition, Xbpl gene expression was regulated by ATF6 as well as itself, resulting in a positive feedback loop during UPR activation (100). Indeed, a human polymorphism in the proximal promoter of Xbp1 (-116C-G) that disrupts the putative binding site for XBP1s correlates with an increased risk for bipolar disorder (37). Furthermore, parathyroid hormone played a role in regulating Xbpl expression during osteoblast differentiation (103). Finally, a recent study identified XBP1 as a highly enriched white adipose gene that was repressed by PR domain containing 16 (PRDM16), a brown fat-specific transcriptional activator (36,77).
Thus, although IRE1α-mediated splicing of Xbp1 mRNA is the most well-characterized regulatory mechanism for this transcription factor, its mRNA expression is also tightly controlled by various factors in a highly dynamic manner. As UPR is constitutively active at a basal level (1,81,98), regulation through either induction or repression of Xbp1 expression may serve to fine tune ER homeostasis. Indeed, a similar situation has been identified in yeast and termed as “super-UPR” (44). Therefore, identifying novel regulators of XBP1 at the transcriptional level may provide insight into the role of “super-UPR” and may aid in the design and development of therapeutic strategies targeting human ER-associated disorders.
Posttranslational Modification of XBP1
Posttranslational modifications regulate the biological activity of many transcription factors. We recently showed that XBP1s was negatively modulated by small ubiquitin-like modifier (SUMO) (10), a transient regulatory mechanism involved in many cellular processes including transcriptional regulation, DNA damage, and signal transduction (24,74). XBP1s protein was SUMOylated at two conserved lysine residues located within the transactivation domain (Fig. 2D). Ablation of these SUMOylation events enhanced the transcriptional activity of XBP1s. In line with our study, a recent genome-wide analysis of SUMO2 modification during heat shock response also identified XBP1s as a target of SUMO2 (21). Thus, these results revealed a previously unexpected role for SUMO in the regulation of UPR activation and ER homeostasis. The role of other posttranslational modifications on XBP1 function and activity, especially phosphorylation, has yet to be studied.
XBP1-Mediated Transcriptional Events
XBP1 was reported to bind to cAMP-responsive elements (CRE) sites in the promoters of MHC class II genes (11). Further studies were performed validating that XBP1 did indeed preferentially bind to CRE-like elements in which the core “ACGT” was highly conserved (11). To identify transcriptional networks regulated by XBP1, ChIP-on-chip arrays showed that XBP1 was constitutively bound to a subset of genes involved in ER homeostasis including folding, trafficking, and ERAD (1), confirming the presence of a low level of basal or constitutive UPR (81). In most genes, XBP1 binding occurred within 200 bp of transcriptional start sites (1). In support of previous reports (11), XBP1 did indeed bind to the core “ACGT” element under physiological conditions, but XBP1 targets were also enriched in additional distinct motifs including UPR element (UPRE) and CCACG box (1).
As XBP1 is involved in various facets of biology, it is not surprising that its targets are also extremely diverse (1). Canonical XBP1 targets in UPR signaling include ER chaperones and components of ERAD including ER degradation enhance mannosidase alphalike 1 (EDEM1), DnaJ/Hsp40 homolog subfamily B member 9 (ERDJ4/DNAJB9), and DnaJ/Hsp40 homolog subfamily C member 3 (P58IPK/DNAJC3). Additional tissue-specific XBP1 targets that have been identified include IL-6 in plasma cells, C/EBPα in adipocytes, lipogenic genes in hepatocytes, proinflammatory cytokines in macrophages (see below), and Mist1 in myocytes. XBP1 was also enriched on the promoters of genes involved in a number of UPR-unrelated processes including glycolysis, gluconeogenesis, lipid metabolism, and DNA replication and repair (1). The biological relevance of these bindings requires further investigations.
THE ROLE OF XBP1 IN HEALTH AND DISEASES
This section highlights and addresses recent reports on the relevance and importance of the IRE1α–XBP1 pathway in multiple pathophysiological conditions such as pathogen defense and immunity, obesity and type 2 diabetes, circadian rhythm regulation, cancer, and neurodegeneration and aging.
XBP1 in Adaptive Immunity
XBP1 is required for plasma cell differentiation (68), but does not influence memory B cell commitment (90,91). XBP1-deficient B cells exhibited normal proliferation and activation, but expressed decreased levels of J chain, a component required for Ig assembly. Consequently, these animals were more susceptible to infections, but restoration of XBP1s expression rescued Ig production (68). Furthermore, Xbp1 mRNA splicing (i.e., IRE1α activity) was attenuated in mice lacking Ig heavy chains, suggesting that IRE1α activity and UPR was modulated and induced by Ig synthesis and production (33,104). XBP1-mediated ER expansion was required for adoption of a “professional secretory cell” phenotype characteristic of plasma cells (80) (Fig. 3). In addition, XBP1s induced the expression of terminal differentiation factor IL-6 in splenic B cells (33). Thus, XBP1 in professional secretory cells may have evolved additional functions allowing these cells to respond to ‘physiological” UPR (80).
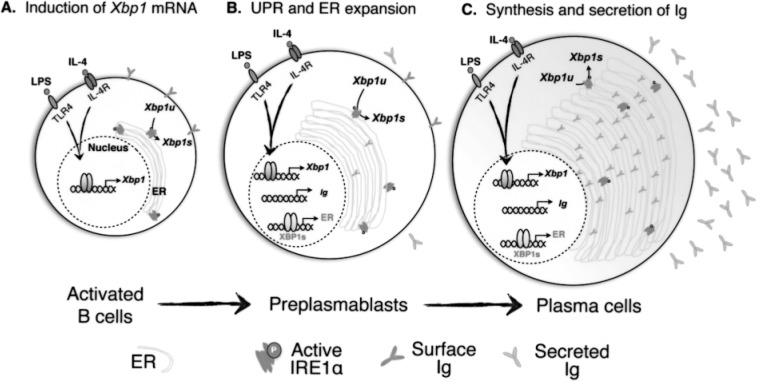
Figure 3.
XBP1 in B cell differentiation to plasma cells. LPS and IL-4 stimulate the expression of Xbp1 mRNA and lead to the elevation of XBP1s protein in activated B cells (A). XBP1s is responsible for expanding the ER capacity in preparation for upcoming waves of immunoglobulin (Ig) biosynthesis in preplasmablasts (B). In fully differentiated plasma cells, ER capacity reaches a new set point of homeostasis to accommodate Ig biosynthesis (C). The question of whether and when UPR activation occurs during this process remains open.
In contrast to previous reports (19,20,33,104), a more recent study reported that Xbp1 induction was independent of differentiation as B cells lacking IgM still maintained Xbp1 activation. This discovery suggested that Xbp1 activation may be required for normal plasma cell differentiation rather than as a consequence of massive Ig synthesis and secretion (30). Furthermore, Tirosh et al. (88) showed that while XBP1 was required for IgM synthesis and secretion, glycoprotein degradation was unaffected by loss of XBP1. Hence, the timing and mechanism of UPR and XBP1 activation during plasma cell differentiation remain an interesting and open question.
XBP1 in Innate Immunity
The IRE1α–XBP1 signaling pathway of UPR is also critical for the development and survival of another immune population, dendritic cells (DCs). Loss of XBP1 in DCs reduced IFN-α production upon stimulation with CpG, an agonist of TLR2, and rendered cells prone to ER stress-induced or differentiation-associated cell death (34). Indeed, both conventional and plasmacytoid DCs in XBP1-deficient animals exhibited decreased survival at basal levels and in response to TLR signaling. Conversely, forced expression of XBP1s enhanced DC development (34).
Most recently, XBP1 was shown to have a critical role in regulating the expression of key proinflammatory cytokines in macrophages (54). Both TLR2 and TLR4 signaling specifically activated the IRE1α–XBP1 branch, which in turn increased the expression and secretion of IL-6, tumor necrosis factor-α (TNF-α), and IFN-β without inducing canonical UPR genes. Mice with macrophage-specific deficiency of XBP1 exhibited increased bacterial burden postinfection (54). The function of XBP1 in innate immunity seemed to be highly conserved as similar observations were made in worms; XBP1-deficient worms were hypersensitive to pathogen infection (70). Therefore, XBP1 plays a critical and protective role in both innate and adaptive immunity. This is not surprising given that the RNase domain of IRE1, both α and β, shares unique homology with RNase L (87), a critical component of the antiviral system that cleaves single-stranded RNA (84).
XBP1 in Inflammatory Bowel Disease (IBD)
In line with the role of the IRE1α–XBP1 pathway in immunity, XBP1 has been implicated in IBD, a common chronic human disorder. Mice with specific Xbp1 deletion in intestinal epithelial cells were more susceptible to developing spontaneous small intestinal enteritis (39). Patients with Crohn’s disease and ulcerative colitis, two forms of IBD, exhibited decreased XBP1s levels. In addition, several genome-wide linkage studies hinted at an association between IBD and a region of the genome physically close to the Xbp1 gene (2,23,94) and the IRE1β gene (5,31). Moreover, deep sequencing identified novel rare single nucleotide polymorphisms in Xbp1 that, along with other environmental and genetic risk factors, might confer susceptibility to IBD (39). Further supporting a key role of XBP1 in IBD, loss of IRE1β, the isoform expressed predominantly in the gastroin-stestinal track, resulted in hypersensitivity to dextran sodium sulfate (DSS)-induced colitis in mice (3). It is quite interesting that IRE1α expression alone in intestinal epithelial cells failed to protect IRE1β−/− animals from induced colitis. Thus, these studies suggested that the IRE1β-XBP1 pathway likely played an important role in the pathogenesis of IBD.
XBP1 in Obesity and Insulin Resistance
ER stress, particularly the IRE1α-XBP1 branch, has been implicated in obesity-induced insulin resistance and type 2 diabetes (62-64,105). Initial reports demonstrated a link between IRE1α activation and JNK-dependent inhibitory serine phosphorylation of insulin receptor substrate 1 (IRS-1) at serine 307 (Ser307) (63,93). In line with the role of IRE1α activation in attenuating insulin signaling, XBP1s over-expression in mouse embryonic fibroblast (MEF) cells suppressed ER-stress-induced JNK activation and IRS-1 phosphorylation on Ser307, whereas XBP1+/− tissues showed opposite effects (63). Furthermore, XBP1+/− mice exhibited increased ER stress and more severe insulin resistance upon high-fat diet (HFD)-induced obesity accompanied with elevated p-Ser307 on IRS1 (63). Conversely, reduction of ER stress via the administration of chemical chaperones such as 4-phenyl butyric acid (PBA) and tauroursodeoxycholic acid (TUDCA) attenuated phosphorylation of IRS1 at Ser307 and improved the insulin sensitivity of obese animals (64). More recently, it was shown that compromised insulin signaling during obesity might decrease the levels of functional nuclear XBP1s in the liver of obese mice (65). The interaction between the heterodimer p85α and p85β, the regulatory subunits of phosphoinositide 3-kinase (PI3K) and XBP1 was disrupted in obese animals, leading to defects in the nuclear entry of XBP1s and elevated ER stress. Over-expression of p85α or p85β in the liver improved glucose tolerance in obese animals (65). Hence, ER stress has been proposed to be a prime culprit in linking obesity with insulin resistance (29).
Several studies, however, have suggested that this model may not be all-inclusive. First, liver-specific XBP1-null mice failed to exhibit overt changes in ER ultrastructure or activation of two other UPR braches PERK and ATF6 (47). This is in line with another report showing that Xbp1 expression and the active form of ATF6 were reduced in the hepatocytes of obese mice, indicative of decreased ER stress (95). Furthermore, ER stress was not detectable in white adipose tissues upon 12 weeks HFD in XBP1-splic-ing reporter mice (102), questioning the notion that ER stress in adipose tissues played a causal role in obesity-associated insulin resistance. Moreover, a recent study demonstrated that p-Ser307 of IRS1 was not critical for the development of insulin resistance, but rather promoted insulin sensitivity in mice (13). The IRS1 Ser307Ala knockin mice exhibited increased insulin resistance upon HFD-induced obesity. Finally, liver-specific disruption of p85α improved systemic glucose tolerance and insulin sensitivity in both lean and obese mice while overexpression of p85α in the liver had the opposite effect (85,86). Hence, the role of ER stress and the IRE1α–XBP1 pathway in obesity and diabetes warrants further studies.
XBP1 has also been implicated in hepatic lipid metabolism and adipocyte differentiation. Using hepatocyte-specific conditional XBP1 knockout mice, it was shown that XBP1-deficient hepatocytes exhibited reduced de novo lipid biosynthesis (47). XBP1 played an unexpected role in regulating hepatic lipogenesis by directly binding to the promoters of key lipogenic factors including diacylglycerol acetyltransferase 2 (DGAT2), stearoyl-CoA reductase 1 (SCD1), and acetyl-CoA carboxylase (ACC2) (47). In addition, we recently showed that XBP1-deficient preadipocytes and MEF cells showed dramatic defects in adipogenesis as XBP1s interacted with the promoter of Cebpα, a master regulator of adipogenesis, and activated its expression (78) (Fig. 4). Thus, XBP1 played a critical regulatory role during adipogenesis by integrating into the transcriptional cascade underlying adipogenic differentiation. This finding was consistent with reports of an absence of fat depot in XBP1-/-neonates rescued with hepatic XBP1s overexpression (45).
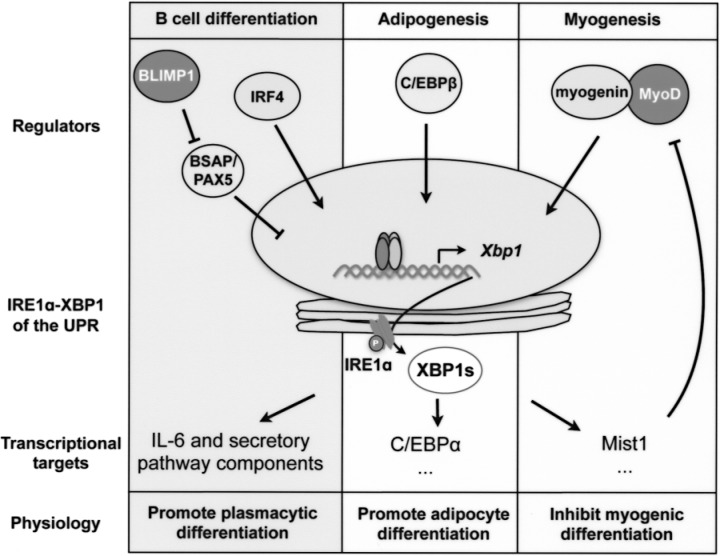
Figure 4.
Simplified schematic outline of IRE1α–XBP1-mediated signaling cascades during B cell differentiation, adipogenesis, and myogenesis. The key transcription factors (regulators) that are responsible for Xbp1 mRNA induction, XBP1s targets, and physiological effects of the signaling cascades are shown for each event.
XBP1 in Circadian Rhythm
Circadian rhythms allow organisms to synchronize environmental inputs such as light and nutrient availability with biological processes with a periodicity of 24 h (42). In 1972, Chedid and Nair (8) showed that the morphology and amount of hepatic smooth ER structures were regulated by a diurnal rhythm, which coincided with the rhythmic patterns of drug-metabolizing enzymes in the ER membranes. A recent study, the first to examine the relationship between UPR activation and the circadian clock, showed that the IRE1α–XBP1 pathway was activated rhythmically every 12 h in the liver and influenced hepatic lipid metabolism (16). Animals lacking a functional circadian clock exhibited constitutive activation of the IRE1α–XBP1 pathway, which was proposed to be responsible for asynchronous expression of enzymes involved in liver metabolism and leading to perturbed lipid metabolism and triglyceride accumulation in the liver (16). It remains unclear how the IRE1α–XBP1 pathway fits into the canonical clock network of transcriptional and translational feedback loops.
XBP1 in Cancer
Genome-wide profiling along with association studies demonstrated that XBP1 expression was induced in a variety of cancers including lymphoid malignancies such as multiple myeloma and acute myeloid leukemia (17,35,50,57) as well as breast cancers (18,22,43). Moreover, multiple myeloma cells with overexpression of superoxide dismutase (SOD2), an enzyme that eliminates free superoxide radicals, exhibited decreased proliferation correlated with decreased Xbp1 expression (32). In support of a direct role for XBP1 in tumorigenesis, the loss of XBP1 was shown to severely inhibit tumor growth (72). Indeed, transformed cells with XBP1 deficiency were more sensitized to hypoxia and underwent apoptosis, implicating XBP1 as a survival factor (72). In addition, mice with ectopic expression of XBP1s in B cells exhibited enhanced B cell proliferative potential along with development of multiple myeloma that recapitulated many critical aspects of the human disease (7). Finally, it was shown that XBP1 was activated in primary mammary tumors with its expression correlating with enhanced tumor growth (83).
Thus, the role of XBP1 as a survival factor deems it a very attractive therapeutic target. However, UPR can also initiate apoptosis in the face of persistent ER stress. A study demonstrated that acute myeloid leukemia (AML) patients with UPR activation actually merited better prognosis as indicated by lower relapse rates, and better overall and disease-free survival (76). Therefore, to fully understand the involvement of XBP1 in cancer development and progression, future studies that can carefully monitor UPR activation and delineate the respective roles of all three UPR branches are required.
XBP1 in Neurodegeneration and Aging
The role of XBP1 in neurodegeneration remains controversial and appears to be disease-specific. Toxic intracellular protein aggregates, one of the primary underlying causes of neurodegenerative pathologies, induce ER stress and activate UPR (52). Indeed, cellular and animal models of Huntington’s (59,60) and Parkinson’s (28) diseases are reportedly associated with activation of the IRE1α–XBP1 pathway. However, it remains unclear whether UPR activation in these models represents a direct cause of the diseases or a secondary effect associated with tissue damage.
XBP1 occupancy was observed on the promoters of genes linked to neurodegenerative pathologies including Alzheimer’s disease (1), although the relevance of these events remains speculative. Ectopic expression of XBP1s played a protective role in cells against chemical-induced cell death and significantly attenuated the degeneration of dopaminergic neurons in a mouse model of Parkinson’s disease (75). In contrast, SOD1 transgenic mice with XBP1 deficiency specifically in the nervous system were more resistant to the development of familial amyotrophic lateral sclerosis (ALS) (27). These animals exhibited increased macroautophagy concomitant with reduced accumulation of mutant SOD1, providing further evidence on an intimate link between UPR and autophagy. In contrast, XBP1 did not appear to influence prion pathogenesis as loss of XBP1 had no effect on prion aggregation, neuronal survival, or overall animal survival (26). Consistently, many UPR markers were unaffected in the brains of prion-infected XBP1-deficient mice when compared to the wild-type cohort (26).
Recent studies have implicated the IRE1–XBP1 pathway in aging in worms (9,25). First, loss of hypoxia inducible factor 1 (HIF1) extended life span in part via the activation of the IRE1 pathway. Defects in IRE1 signaling significantly reduced the life span of the long-lived hif1 loss-of-function mutant (9). This effect appeared to be IRE1 specific as a PERK deletion mutant had no effect. A similar observation was recently reported in insulin/IGF-1 pathway mutant worms (25). Loss of IRE1 or XBP1 shortened the life span of long-lived daf-2 mutants to a much lesser extent than in wild-type worms, suggesting that the effect of XBP1 on life span may depend on one of these factors in the insulin/IGF-1 pathway. Nonetheless, IRE1 activity and Xbp1s mRNA were unexpectedly very low in the daf-2 mutant, indicative of improved overall ER homeostasis. Mechanistically, it was proposed that XBP1 might regulate the expression of a conserved Zn-finger protein downstream of Xbp1 (DOX-1) in a DAF-16/FOXO-dependent manner. The effect of IRE1–XBP1 in the aging process of higher organisms merits further studies.
CONCLUDING REMARKS
The role of XBP1 as a critical transcription factor and mediator of UPR signaling has been very well documented in the literature, but in addition to this vital role, the history of XBP1 discovery as well as recent insights into immune regulation has demonstrated that it is also required for various aspects of immunity including B cell and effector CD8+ T-cell differentiation, dendritic cell survival, and TLR-induced macrophage responses. In addition, SNPs in the hXBP1 gene rendered individuals susceptible to IBD. Collectively, reports on XBP1 in immunity have revealed novel roles for this transcription factor in both innate and adaptive immune responses although interestingly, none are directly related to the function of MHC class II genes. Thus, increased understanding of the molecular actions and transcriptional networks regulated by XBP1 in immune cells may aid in the development of potential therapeutics targeting immune disorders.
Given its unique regulatory mechanisms and short half-life, the XBP1 protein has been touted as an important regulator that can quickly integrate transient environmental cues with downstream gene activation. Indeed, XBP1 is important for differentiation in various cell types including myocytes, adipocytes, and plasma cells. Of note, it is interesting to compare the roles of XBP1 in these differentiation events, all of which involve complex regulatory circuits (Fig. 4). From these insights, the IRE1α–XBP1 branch of UPR appears to be acting as a key component of a differentiation switch to control diverse cellular fates. One outstanding question remaining from these studies is whether the signals responsible for activating XBP1 in these various developmental processes arise from accumulation of unfolded proteins in the ER lumen or from ER-independent differentiation signals.
As disease progression is normally manifested as a collective outcome involving many tissues and signaling pathways, simply using downstream targets of UPR signaling to assess the status of ER stress and UPR activation is unlikely to be sufficient and may not be reliable in certain pathophysiological settings. Cross-talk among various signaling pathways (e.g., insulin and TLR) and other stress responses (e.g., amino acid starvation) may confound assessment of ER stress and UPR activation, thus making it critical to assess ER stress and UPR activation at the level of UPR sensors (98). Moreover, as XBP1 activity is regulated at multiple levels, XBP1 may modulate ER homeostasis independently of classical UPR activation. Therefore, it is highly conceivable that these super-UPR-like events may be critical for maintenance of ER homeostasis under physiological conditions to circumvent the deleterious consequences of prolonged UPR activation. As ER stress has been implicated in an increasing number of human diseases (40,99), novel methods to assess and accurately quantitate UPR sensor activation and ER stress under physiological and pathological conditions will be instrumental to the future of the field and its therapeutic implications.
ACKNOWLEDGMENTS
Due to space limitations, we apologize for being unable to cite all relevant works. The work in the laboratory is supported by the American Federation for Aging Research (AFAR), the American Diabetes Association (ADA), and NIH NIDDK RO1DK082582.
REFERENCES
- 1. Acosta-Alvear D.; Zhou Y.; Blais A.; Tsikitis M.; Lents N. H.; Arias C.; Lennon C. J.; Kluger Y.; Dynlacht B. D. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell 27:53–66; 2007. [DOI] [PubMed] [Google Scholar]
- 2. Barmada M. M.; Brant S. R.; Nicolae D. L.; Achkar J. P.; Panhuysen C. I.; Bayless T. M.; Cho J. H.; Duerr R. H. A genome scan in 260 inflammatory bowel disease-affected relative pairs. Inflamm. Bowel Dis. 10:513–520; 2004. [DOI] [PubMed] [Google Scholar]
- 3. Bertolotti A.; Wang X.; Novoa I.; Jungreis R.; Schlessinger K.; Cho J. H.; West A. B.; Ron D. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. J. Clin. Invest. 107:585–593; 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blais A.; Tsikitis M.; Acosta-Alvear D.; Sharan R.; Kluger Y.; Dynlacht B. D. An initial blueprint for myogenic differentiation. Genes Dev. 19:553–569; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brant S. R.; Fu Y.; Fields C. T.; Baltazar R.; Ravenhill G.; Pickles M. R.; Rohal P. M.; Mann J.; Kirschner B. S.; Jabs E. W.; Bayless T. M.; Hanauer S. B.; Cho J. H. American families with Crohn’s disease have strong evidence for linkage to chromosome 16 but not chromosome 12. Gastroenterology 115:1056–1061; 1998. [DOI] [PubMed] [Google Scholar]
- 6. Calfon M.; Zeng H.; Urano F.; Till J. H.; Hubbard S. R.; Harding H. P.; Clark S. G.; Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96; 2002. [DOI] [PubMed] [Google Scholar]
- 7. Carrasco D. R.; Sukhdeo K.; Protopopova M.; Sinha R.; Enos M.; Carrasco D. E.; Zheng M.; Mani M.; Henderson J.; Pinkus G. S.; Munshi N.; Horner J.; Ivanova E. V.; Protopopov A.; Anderson K. C.; Tonon G.; DePinho R. A. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 11:349–360; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chedid A.; Nair V. Diurnal rhythm in endoplasmic reticulum of rat liver: Electron microscopic study. Science 175:176–179; 1972. [DOI] [PubMed] [Google Scholar]
- 9. Chen D.; Thomas E. L.; Kapahi P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans . PLoS Genet. 5: e1000486; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen H.; Qi L. SUMO modification regulates transcriptional activity of XBP1. Biochem. J. 429:95–102; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clauss I. M.; Chu M.; Zhao J. L.; Glimcher L. H. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 24:1855–1864; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clauss I. M.; Gravallese E. M.; Darling J. M.; Shapiro F.; Glimcher M. J.; Glimcher L. H. In situ hybridization studies suggest a role for the basic region-leucine zipper protein hXBP-1 in exocrine gland and skeletal development during mouse embryogenesis. Dev. Dyn. 197:146–156; 1993. [DOI] [PubMed] [Google Scholar]
- 13. Copps K. D.; Hancer N. J.; Opare-Ado L.; Qiu W.; Walsh C.; White M. F. Irs1 serine 307 promotes insulin sensitivity in mice. Cell Metab. 11:84–92; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox J. S.; Shamu C. E.; Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197–1206; 1993. [DOI] [PubMed] [Google Scholar]
- 15. Cox J. S.; Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391–404; 1996. [DOI] [PubMed] [Google Scholar]
- 16. Cretenet G.; Le Clech M.; Gachon F. Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 11:47–57; 2010. [DOI] [PubMed] [Google Scholar]
- 17. Davies F. E.; Dring A. M.; Li C.; Rawstron A. C.; Shammas M. A.; O’Connor S. M.; Fenton J. A.; Hideshima T.; Chauhan D.; Tai I. T.; Robinson E.; Auclair D.; Rees K.; Gonzalez D.; Ashcroft A. J.; Dasgupta R.; Mitsiades C.; Mitsiades N.; Chen L. B.; Wong W. H.; Munshi N. C.; Morgan G. J.; Anderson K. C. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood 102:4504–4511; 2003. [DOI] [PubMed] [Google Scholar]
- 18. Doane A. S.; Danso M.; Lal P.; Donaton M.; Zhang L.; Hudis C.; Gerald W. L. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 25:3994–4008; 2006. [DOI] [PubMed] [Google Scholar]
- 19. Gass J. N.; Gifford N. M.; Brewer J. W. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J. Biol. Chem. 277:49047–49054; 2002. [DOI] [PubMed] [Google Scholar]
- 20. Gass J. N.; Jiang H. Y.; Wek R. C.; Brewer J. W. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol. Immunol.:45:1035–1043;2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Golebiowski F.; Matic I.; Tatham M. H.; Cole C.; Yin Y.; Nakamura A.; Cox J.; Barton G. J.; Mann M.; Hay R. T. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2: ra24; 2009. [DOI] [PubMed] [Google Scholar]
- 22. Gomez B. P.; Riggins R. B.; Shajahan A. N.; Klimach U.; Wang A.; Crawford A. C.; Zhu Y.; Zwart A.; Wang M.; Clarke R. Human X-box binding protein-1 confers both estrogen independence and antiestrogen resistance in breast cancer cell lines. FASEB J. 21:4013–4027; 2007. [DOI] [PubMed] [Google Scholar]
- 23. Hampe J.; Shaw S. H.; Saiz R.; Leysens N.; Lantermann A.; Mascheretti S.; Lynch N. J.; MacPherson A. J.; Bridger S.; van Deventer S.; Stokkers P.; Morin P.; Mirza M. M.; Forbes A.; Lennard-Jones J. E.; Mathew C. G.; Curran M. E.; Schreiber S. Linkage of inflammatory bowel disease to human chromosome 6p. Am. J. Hum. Genet. 65:1647–1655; 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hay R. T. SUMO: A history of modification. Mol. Cell 18:1–12; 2005. [DOI] [PubMed] [Google Scholar]
- 25. Henis-Korenblit S.; Zhang P.; Hansen M.; McCormick M.; Lee S. J.; Cary M.; Kenyon C. Insulin/ IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl. Acad. Sci. USA 107:9730–9735; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hetz C.; Lee A. H.; Gonzalez-Romero D.; Thielen P.; Castilla J.; Soto C.; Glimcher L. H. Unfolded protein response transcription factor XBP-1 does not influence prion replication or pathogenesis. Proc. Natl. Acad. Sci. USA 105:757–762; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hetz C.; Thielen P.; Matus S.; Nassif M.; Court F.; Kiffin R.; Martinez G.; Cuervo A. M.; Brown R. H.; Glimcher L. H. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 23:2294–2306; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holtz W. A.; O’Malley K. L. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J. Biol. Chem. 278:19367–19377; 2003. [DOI] [PubMed] [Google Scholar]
- 29. Hotamisligil G. S. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140:900–917; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu C. C.; Dougan S. K.; McGehee A. M.; Love J. C.; Ploegh H. L. XBP-1 regulates signal transduction, transcription factors and bone marrow colonization in B cells. EMBO J. 28:1624–1636; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hugot J. P.; Laurent-Puig P.; Gower-Rousseau C.; Olson J. M.; Lee J. C.; Beaugerie L.; Naom I.; Dupas J. L.; Van Gossum A.; Orholm M.; Bonaiti-Pellie C.; Weissenbach J.; Mathew C. G.; Lennard-Jones J. E.; Cortot A.; Colombel J. F.; Thomas G. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature 379:821–823; 1996. [DOI] [PubMed] [Google Scholar]
- 32. Hurt E. M.; Thomas S. B.; Peng B.; Farrar W. L. Integrated molecular profiling of SOD2 expression in multiple myeloma. Blood 109:3953–3962; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iwakoshi N. N.; Lee A. H.; Vallabhajosyula P.; Otipoby K. L.; Rajewsky K.; Glimcher L. H. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321–329; 2003. [DOI] [PubMed] [Google Scholar]
- 34. Iwakoshi N. N.; Pypaert M.; Glimcher L. H. The transcription factor XBP-1 is essential for the development and survival of dendritic cells. J. Exp. Med. 204:2267–2275; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Juric D.; Lacayo N. J.; Ramsey M. C.; Racevskis J.; Wiernik P. H.; Rowe J. M.; Goldstone A. H.; O’Dwyer P. J.; Paietta E.; Sikic B. I. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J. Clin. Oncol. 25:1341–1349; 2007. [DOI] [PubMed] [Google Scholar]
- 36. Kajimura S.; Seale P.; Tomaru T.; Erdjument-Bromage H.; Cooper M. P.; Ruas J. L.; Chin S.; Tempst P.; Lazar M. A.; Spiegelman B. M. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 22:1397–1409; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kakiuchi C.; Iwamoto K.; Ishiwata M.; Bundo M.; Kasahara T.; Kusumi I.; Tsujita T.; Okazaki Y.; Nanko S.; Kunugi H.; Sasaki T.; Kato T. Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat. Genet. 35:171–175; 2003. [DOI] [PubMed] [Google Scholar]
- 38. Kamimura D.; Bevan M. J. Endoplasmic reticulum stress regulator XBP-1 contributes to effector CD8+ T cell differentiation during acute infection. J. Immunol. 181:5433–5441; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaser A.; Lee A. H.; Franke A.; Glickman J. N.; Zeissig S.; Tilg H.; Nieuwenhuis E. E.; Higgins D. E.; Schreiber S.; Glimcher L. H.; Blumberg R. S. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134:743–756; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kim I.; Xu W.; Reed J. C. Cell death and endoplasmic reticulum stress: Disease relevance and therapeutic opportunities. Nat. Rev. Drug Discov. 7:1013–1030; 2008. [DOI] [PubMed] [Google Scholar]
- 41. Klein U.; Casola S.; Cattoretti G.; Shen Q.; Lia M.; Mo T.; Ludwig T.; Rajewsky K.; Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782; 2006. [DOI] [PubMed] [Google Scholar]
- 42. Kovac J.; Husse J.; Oster H. A time to fast, a time to feast: The crosstalk between metabolism and the circadian clock. Mol. Cell 28:75–80; 2009. [DOI] [PubMed] [Google Scholar]
- 43. Lacroix M.; Leclercq G. About GATA3, HNF3A, and XBP1, three genes co-expressed with the oestrogen receptor-alpha gene (ESR1) in breast cancer. Mol. Cell. Endocrinol. 219:1–7; 2004. [DOI] [PubMed] [Google Scholar]
- 44. Leber J. H.; Bernales S.; Walter P. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2:E235; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee A. H.; Chu G. C.; Iwakoshi N. N.; Glimcher L. H. XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J. 24:4368–4380; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee A. H.; Iwakoshi N. N.; Anderson K. C.; Glimcher L. H. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA 100:9946–9951; 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee A. H.; Scapa E. F.; Cohen D. E.; Glimcher L. H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320:1492–1496; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee K.; Tirasophon W.; Shen X.; Michalak M.; Prywes R.; Okada T.; Yoshida H.; Mori K.; Kaufman R. J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452–466; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lefterova M. I.; Zhang Y.; Steger D. J.; Schupp M.; Schug J.; Cristancho A.; Feng D.; Zhuo D.; Stoeckert C. J. Jr.; Liu X. S.; Lazar M. A. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 22:2941–2952; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leleu X.; Hunter Z. R.; Xu L.; Roccaro A. M.; Moreau A. S.; Santos D. D.; Hatjiharissi E.; Bakthavachalam V.; Adamia S.; Ho A. W.; Soumerai J.; Patterson C. J.; Manning R. J.; Hamilton S.; Verselis S.; Fox E.; Carrasco R.; Ghobrial I. M.; Treon S. P. Expression of regulatory genes for lymphoplasmacytic cell differentiation in Waldenstrom Macroglobulinemia. Br. J. Haematol. 145:59–63; 2009. [DOI] [PubMed] [Google Scholar]
- 51. Lin J. H.; Walter P.; Yen T. S. Endoplasmic reticulum stress in disease pathogenesis. Annu. Rev. Pathol. 3:399–425; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lindholm D.; Wootz H.; Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 13:385–392; 2006. [DOI] [PubMed] [Google Scholar]
- 53. Liou H. C.; Boothby M. R.; Finn P. W.; Davidon R.; Nabavi N.; Zeleznik-Le N. J.; Ting J. P.; Glimcher L. H. A new member of the leucine zipper class of proteins that binds to the HLA DR alpha promoter. Science 247:1581–1584; 1990. [DOI] [PubMed] [Google Scholar]
- 54. Martinon F.; Chen X.; Lee A. H.; Glimcher L. H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 11:411–418; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Masaki T.; Yoshida M.; Noguchi S. Targeted disruption of CRE-binding factor TREB5 gene leads to cellular necrosis in cardiac myocytes at the embryonic stage. Biochem. Biophys. Res. Commun. 261:350–356; 1999. [DOI] [PubMed] [Google Scholar]
- 56. Mori K.; Ma W.; Gething M. J.; Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74:743–756; 1993. [DOI] [PubMed] [Google Scholar]
- 57. Munshi N. C.; Hideshima T.; Carrasco D.; Shammas M.; Auclair D.; Davies F.; Mitsiades N.; Mitsiades C.; Kim R. S.; Li C.; Rajkumar S. V.; Fonseca R.; Bergsagel L.; Chauhan D.; Anderson K. C. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood 103:1799–1806; 2004. [DOI] [PubMed] [Google Scholar]
- 58. Nekrutenko A.; He J. Functionality of unspliced XBP1 is required to explain evolution of overlapping reading frames. Trends Genet. 22:645–648; 2006. [DOI] [PubMed] [Google Scholar]
- 59. Nishitoh H.; Kadowaki H.; Nagai A.; Maruyama T.; Yokota T.; Fukutomi H.; Noguchi T.; Matsuzawa A.; Takeda K.; Ichijo H. ALS-linked mutant SOD1 induces ER stress- and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 22:1451–1464; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nishitoh H.; Matsuzawa A.; Tobiume K.; Saegusa K.; Takeda K.; Inoue K.; Hori S.; Kakizuka A.; Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 16:1345–1355; 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ono S. J.; Liou H. C.; Davidon R.; Strominger J. L.; Glimcher L. H. Human X-box-binding protein 1 is required for the transcription of a subset of human class II major histocompatibility genes and forms a heterodimer with c-fos. Proc. Natl. Acad. Sci. USA 88:4309–4312; 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ozcan L.; Ergin A. S.; Lu A.; Chung J.; Sarkar S.; Nie D.; Myers M. G. Jr.; Ozcan U. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell Metab. 9:35–51; 2009. [DOI] [PubMed] [Google Scholar]
- 63. Ozcan U.; Cao Q.; Yilmaz E.; Lee A. H.; Iwakoshi N. N.; Ozdelen E.; Tuncman G.; Gorgun C.; Glimcher L. H.; Hotamisligil G. S. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science 306:457–461; 2004. [DOI] [PubMed] [Google Scholar]
- 64. Ozcan U.; Yilmaz E.; Ozcan L.; Furuhashi M.; Vaillancourt E.; Smith R. O.; Gorgun C. Z.; Hotamisligil G. S. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313:1137–1140; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Park S. W.; Zhou Y.; Lee J.; Lu A.; Sun C.; Chung J.; Ueki K.; Ozcan U. The regulatory sub-units of PI3K, p85alpha and p85beta, interact with XBP-1 and increase its nuclear translocation. Nat. Med. 16:429–437; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ponath P. D.; Fass D.; Liou H. C.; Glimcher L. H.; Strominger J. L. The regulatory gene, hXBP-1, and its target, HLA-DRA, utilize both common and distinct regulatory elements and protein complexes. J. Biol. Chem. 268:17074–17082; 1993. [PubMed] [Google Scholar]
- 67. Reimold A. M.; Etkin A.; Clauss I.; Perkins A.; Friend D. S.; Zhang J.; Horton H. F.; Scott A.; Orkin S. H.; Byrne M. C.; Grusby M. J.; Glimcher L. H. An essential role in liver development for transcription factor XBP-1. Genes Dev. 14:152–157; 2000. [PMC free article] [PubMed] [Google Scholar]
- 68. Reimold A. M.; Iwakoshi N. N.; Manis J.; Vallabhajosyula P.; Szomolanyi-Tsuda E.; Gravallese E. M.; Friend D.; Grusby M. J.; Alt F.; Glimcher L. H. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412:300–307; 2001. [DOI] [PubMed] [Google Scholar]
- 69. Reimold A. M.; Ponath P. D.; Li Y. S.; Hardy R. R.; David C. S.; Strominger J. L.; Glimcher L. H. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J. Exp. Med. 183:393–401; 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Richardson C. E.; Kooistra T.; Kim D. H. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463:1092–1095; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Roach J. C.; Smith K. D.; Strobe K. L.; Nissen S. M.; Haudenschild C. D.; Zhou D.; Vasicek T. J.; Held G. A.; Stolovitzky G. A.; Hood L. E.; Aderem A. Transcription factor expression in lipopolysaccharide-activated peripheral-blood-derived mononuclear cells. Proc. Natl. Acad. Sci. USA 104:16245–16250; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Romero-Ramirez L.; Cao H.; Nelson D.; Hammond E.; Lee A. H.; Yoshida H.; Mori K.; Glimcher L. H.; Denko N. C.; Giaccia A. J.; Le Q. T.; Koong A. C. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 64:5943–5947; 2004. [DOI] [PubMed] [Google Scholar]
- 73. Ron D.; Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell. Biol. 8:519–529; 2007. [DOI] [PubMed] [Google Scholar]
- 74. Rytinki M. M.; Kaikkonen S.; Pehkonen P.; Jaaskelainen T.; Palvimo J. J. PIAS proteins: Pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci. 66:3029–3041; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sado M.; Yamasaki Y.; Iwanaga T.; Onaka Y.; Ibuki T.; Nishihara S.; Mizuguchi H.; Momota H.; Kishibuchi R.; Hashimoto T.; Wada D.; Kitagawa H.; Watanabe T. K. Protective effect against Parkinson’s disease-related insults through the activation of XBP1. Brain Res. 1257:16–24; 2009. [DOI] [PubMed] [Google Scholar]
- 76. Schardt J. A.; Weber D.; Eyholzer M.; Mueller B. U.; Pabst T. Activation of the unfolded protein response is associated with favorable prognosis in acute myeloid leukemia. Clin. Cancer Res. 15:3834–3841; 2009. [DOI] [PubMed] [Google Scholar]
- 77. Seale P.; Bjork B.; Yang W.; Kajimura S.; Chin S.; Kuang S.; Scime A.; Devarakonda S.; Conroe H. M.; Erdjument-Bromage H.; Tempst P.; Rudnicki M. A.; Beier D. R.; Spiegelman B. M. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454:961–967; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sha H.; He Y.; Chen H.; Wang C.; Zenno A.; Shi H.; Yang X.; Zhang X.; Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 9:556–564; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shaffer A. L.; Lin K. I.; Kuo T. C.; Yu X.; Hurt E. M.; Rosenwald A.; Giltnane J. M.; Yang L.; Zhao H.; Calame K.; Staudt L. M. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 17:51–62; 2002. [DOI] [PubMed] [Google Scholar]
- 80. Shaffer A. L.; Shapiro-Shelef M.; Iwakoshi N. N.; Lee A. H.; Qian S. B.; Zhao H.; Yu X.; Yang L.; Tan B. K.; Rosenwald A.; Hurt E. M.; Petroulakis E.; Sonenberg N.; Yewdell J. W.; Calame K.; Glimcher L. H.; Staudt L. M. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21:81–93; 2004. [DOI] [PubMed] [Google Scholar]
- 81. Shen X.; Ellis R. E.; Sakaki K.; Kaufman R. J. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 1:e37; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sidrauski C.; Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 90:1031–1039; 1997. [DOI] [PubMed] [Google Scholar]
- 83. Spiotto M. T.; Banh A.; Papandreou I.; Cao H.; Galvez M. G.; Gurtner G. C.; Denko N. C.; Le Q. T.; Koong A. C. Imaging the unfolded protein response in primary tumors reveals microenvironments with metabolic variations that predict tumor growth. Cancer Res. 70:78–88; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stark G. R.; Kerr I. M.; Williams B. R.; Silverman R. H.; Schreiber R. D. How cells respond to interferons. Annu. Rev. Biochem. 67:227–264; 1998. [DOI] [PubMed] [Google Scholar]
- 85. Taniguchi C. M.; Aleman J. O.; Ueki K.; Luo J.; Asano T.; Kaneto H.; Stephanopoulos G.; Cantley L. C.; Kahn C. R. The p85alpha regulatory subunit of phosphoinositide 3-kinase potentiates c-Jun N-terminal kinase-mediated insulin resistance. Mol. Cell. Biol. 27:2830–2840; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Taniguchi C. M.; Tran T. T.; Kondo T.; Luo J.; Ueki K.; Cantley L. C.; Kahn C. R. Phosphoinositide 3-kinase regulatory subunit p85alpha suppresses insulin action via positive regulation of PTEN. Proc. Natl. Acad. Sci. USA 103:12093–12097; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tirasophon W.; Lee K.; Callaghan B.; Welihinda A.; Kaufman R. J. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 14:2725–2736; 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tirosh B.; Iwakoshi N. N.; Glimcher L. H.; Ploegh H. L. XBP-1 specifically promotes IgM synthesis and secretion, but is dispensable for degradation of glycoproteins in primary B cells. J. Exp. Med. 202:505–516; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tirosh B.; Iwakoshi N. N.; Glimcher L. H.; Ploegh H. L. Rapid turnover of unspliced Xbp-1 as a factor that modulates the unfolded protein response. J. Biol. Chem. 281:5852–5860; 2006. [DOI] [PubMed] [Google Scholar]
- 90. Todd D. J.; Lee A. H.; Glimcher L. H. The endoplasmic reticulum stress response in immunity and autoimmunity. Nat. Rev. Immunol. 8:663–674; 2008. [DOI] [PubMed] [Google Scholar]
- 91. Todd D. J.; McHeyzer-Williams L. J.; Kowal C.; Lee A. H.; Volpe B. T.; Diamond B.; McHeyzer-Williams M. G.; Glimcher L. H. XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. J. Exp. Med. 206:2151–2159; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Travers K. J.; Patil C. K.; Wodicka L.; Lockhart D. J.; Weissman J. S.; Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258; 2000. [DOI] [PubMed] [Google Scholar]
- 93. Urano F.; Wang X.; Bertolotti A.; Zhang Y.; Chung P.; Harding H. P.; Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664–666; 2000. [DOI] [PubMed] [Google Scholar]
- 94. Vermeire S.; Rutgeerts P.; Van Steen K.; Joossens S.; Claessens G.; Pierik M.; Peeters M.; Vlietinck R. Genome wide scan in a Flemish inflammatory bowel disease population: Support for the IBD4 locus, population heterogeneity, and epistasis. Gut 53:980–986; 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang Y.; Vera L.; Fischer W. H.; Montminy M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature 460:534–537; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wen X. Y.; Stewart A. K.; Sooknanan R. R.; Henderson G.; Hawley T. S.; Reimold A. M.; Glimcher L. H.; Baumann H.; Malek L. T.; Hawley R. G. Identification of c-myc promoter-binding protein and X-box binding protein 1 as interleukin-6 target genes in human multiple myeloma cells. Int. J. Oncol. 15:173–178; 1999. [DOI] [PubMed] [Google Scholar]
- 97. Yanagitani K.; Imagawa Y.; Iwawaki T.; Hosoda A.; Saito M.; Kimata Y.; Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell 34:191–200; 2009. [DOI] [PubMed] [Google Scholar]
- 98. Yang L.; Xue Z.; He Y.; Sun S.; Chen H.; Qi L. A Phostag-based method reveals the extent of physiological endoplasmic reticulum stress. PLoS ONE 5: e11621; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yoshida H. ER stress and diseases. FEBS J. 274:630–658; 2007. [DOI] [PubMed] [Google Scholar]
- 100. Yoshida H.; Matsui T.; Yamamoto A.; Okada T.; Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891; 2001. [DOI] [PubMed] [Google Scholar]
- 101. Yoshida H.; Oku M.; Suzuki M.; Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172:565–575; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yoshiuchi K.; Kaneto H.; Matsuoka T. A.; Kohno K.; Iwawaki T.; Nakatani Y.; Yamasaki Y.; Hori M.; Matsuhisa M. Direct monitoring of in vivo ER stress during the development of insulin resistance with ER stress-activated indicator transgenic mice. Biochem. Biophys. Res. Commun. 366:545–550; 2008. [DOI] [PubMed] [Google Scholar]
- 103. Zambelli A.; Mongiardini E.; Villegas S. N.; Carri N. G.; Boot-Handford R. P.; Wallis G. A. Transcription factor XBP-1 is expressed during osteoblast differentiation and is transcriptionally regulated by parathyroid hormone (PTH). Cell Biol. Int. 29:647–653; 2005. [DOI] [PubMed] [Google Scholar]
- 104. Zhang K.; Wong H. N.; Song B.; Miller C. N.; Scheuner D.; Kaufman R. J. The unfolded protein response sensor IRE1alpha is required at 2 distinct steps in B cell lymphopoiesis. J. Clin. Invest. 115:268–281; 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhang X.; Zhang G.; Zhang H.; Karin M.; Bai H.; Cai D. Hypothalamic IKKbeta/NF-kappaB and ER stress link overnutrition to energy imbalance and obesity. Cell 135:61–73; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]