Summary
Melanoma, the most aggressive form of skin cancer, has increased in incidence more rapidly than any other cancer. The completion of the human genome project and advancements in genomics technologies has allowed us to investigate genetic alterations of melanoma at a scale and depth that is unprecedented. Here, we survey the history of the different approaches taken to understand the genomics of melanoma – from early candidate genes, to gene families, to genome-wide studies. The new era of whole-exome and wholegenome sequencing has paved the way for an in-depth understanding of melanoma biology, identification of new therapeutic targets, and development of novel personalized therapies for melanoma.
Keywords: melanoma, somatic mutation, sequencing
Introduction to melanoma
Melanoma, the most fatal form of skin cancer, has increased in incidence more rapidly than any other cancer. In 2010, this disease killed approximately 8700 people and was diagnosed in 69 000 individuals in the United States (Jemal et al., 2010). Melanoma originates in melanocytes whose genetic alterations may enable their continuous proliferation and evasion of the body’s tumor surveillance mechanisms, ultimately resulting in the development of malignant melanomas (Bastian, 2003). Somatic mutations, the focus of this review, cause 90% of melanomas and are acquired during an individual’s lifespan through environmental factors such as ultraviolet (UV) light (Fernandez-Pol and Douglas, 2000; Polsky and Cordon-Cardo, 2003). Inherited mutations and epigenetic factors known to contribute to the pathophysiology of melanoma have been reviewed previously (Howell et al., 2009; Sigalotti et al., 2010).
Patterns of mutations
Compared to other tumor types, melanoma harbors a larger number of genomic changes. Existing genomic data support that UV light is the primary arbiter of higher mutation rate in melanoma as it induces point mutation causing nucleotide substitutions of thymine with cytosine (C>T) (Pleasance et al., 2010; Wei et al., 2011b). Using novel methods of high-throughput screening, numerous mutated genes and pathways have been identified. Broadly, the genes affected by mutations have been classified as oncogenes or tumor suppressors, and mutations identified in these genes have been classified as passenger or driver mutations.
Oncogenes and tumor suppressors
Melanoma causing genes, as in other cancer genes, are generally classified as oncogenes, tumor suppressors, and stability genes. A classic analogy compares the functions of oncogenes to car accelerators and tumor suppressors to car brakes. In this comparison, mutations in oncogenes can lead to acceleration of growth while mutations in tumor suppressors release the inhibition that prevents tumorigenesis (Kinzler and Vogelstein, 1996). Oncogenes are typically mutated at specific hotspots, which render similar codon changes in different tumors or cluster in particular protein domains. Additionally, only one allele is usually mutated in an oncogene, leading to a heterozygous genotype. In contrast, mutations occur throughout the gene in tumor suppressor genes, which often induce loss or truncation of the encoded protein. Mutations in tumor suppressors usually become apparent when both alleles are affected, resulting in a homozygous genotype, although some evidence for significance of heterozygous loss is also emerging.
Passenger versus driver mutations
Melanoma cells contain numerous genetic changes, some of which promote tumorigenesis and some ofwhich have no functional role (Greenman et al., 2007). ‘Driver’ mutations are genetic alterations that lead to the development of cancer cells and are pathogenic in nature. In contrast, ‘passenger’ mutations can occur along with or after driver mutations, but possess no effect on tumor growth, while non-pathogenic, passenger mutations are valuable for documenting changes that cells have undergone in route to becoming malignant (Bozic et al., 2010). Because passenger and driver mutations may arise with similar frequencies, accurately categorizing cancer mutations remains challenging (Kaminker et al., 2007; Parmigiani et al., 2009). The task of differentiating driver mutations from passenger mutations is central to cancer genomics and specifically to melanoma genomics, because of its particularly high mutation frequency. Combined use of genetic, bioinformatic, and functional assays may help to identify the nature of genetic mutations.
Overall, there are four general characteristics that can be used to distinguish passenger from driver mutations: (1) recurrence, (2) non-synonymous to synonymous ratio (N:S ratio), (3) bioinformatics analyses, and (4) biochemical / biological analysis.
1 Recurrence
When multiple cancer samples have identical mutations at the exact same amino acid or in neighboring amino acids, there is a greater likelihood that the mutation is pathogenic. For example, the activating RAS mutations occur in 30% of all human cancers and its isomer NRAS is mutated in nearly 13–25% of all malignant melanomas (Ball et al., 1994; Curtin et al., 2005; Davies et al., 2002; Goel et al., 2006; Hocker and Tsao, 2007; Schubbert et al., 2007; Van ’T Veer et al., 1989). Albino et al. (1989), nearly 22 years ago, showed that 24% of cultured malignant melanomas have an activated RAS gene. Among them, NRAS is ten times more frequently mutated than another isomer HRAS. More specifically, 15–33% of metastatic melanomas bear Q61R as the main hotspot NRAS mutation (Goydos et al., 2005). BRAF, downstream to RAS, is mutated in nearly 45% of malignant melanomas, and most mutations reside on the kinase domain (Hocker and Tsao, 2007). Notably, 80% of mutations in BRAF result in the V600E mutation (Davies et al., 2002). In addition, recurring nonsense mutations that change an amino acid to a stop codon thereby forming truncated proteins are also useful in characterizing driver mutations.
2 N:S Ratio
While non-synonymous mutations produce amino acid changes, synonymous changes do not, which lower the likelihood that the mutation causes functional effect. For a particular gene or pathway, an observed N:S ratio greater than the predicted background suggests more of the alterations in the gene are likely to be deleterious and thus more likely to be driver mutations (Greenman et al., 2007).
3 Bioinformatic analyses
Numerous bioinformatics analyses could be applied to predict whether a mutation may be a driver. For example, if the protein function and structure is known, mutations in the key positions within the functional domains may predict functional effects of the mutation. Furthermore, functional impact may be predicted based on the spectrum of the mutations and their positional configuration. Evolutionary conservation is also a useful predictor of function. Positions that are highly conserved evolutionarily are more likely to have a greater functional effect when mutated. Therefore, mutations in an evolutionarily conserved domain are more likely to be driver mutations. Algorithms such as the Scale-Invariant Feature Transform (SIFT) are helpful in predicting the effect of a mutation. SIFT calculates how likely an amino acid substitution is to contribute to protein function based on sequence homology and amino acid characteristics (Ng and Henikoff, 2003).
4 Biochemical and biological analysis
The gold standard for determining whether a mutation drives tumorigenesis is biochemical and biological analysis using various systems including overexpression, knockdown, knockout, and knockin model organisms as well as cell culture experiments (Kohli et al., 2004). However, because these methods are not well suited for highthroughput studies, a combination of bioinformatics and molecular biology tools is the best method available for identifying driver and passenger mutations.
Ultraviolet signatures
UV light is a major cause of somatic mutations in melanoma, as shown through epidemiological (Gilchrest et al., 1999) and experimental studies (Atillasoy et al., 1998; Berking et al., 2002). UV exposure leads to the generation of reactive oxygen species and free radicals that induce replication errors by causing base substitution and other genetic mutations. The characteristic signatures of UV-mediated damage are C>T and CC>TT changes; proposed mechanisms for this phenomenon include mutations via 6–4 photoproducts and formation of cyclobutane pyrimidine dimers (Jhappan et al., 2003). In addition to being a mutagen, UV light can also be a mitogen (Gilchrest et al., 1999), an inhibitor of immunogenic response against tumor cells (Donawho and Kripke, 1991) and an activator of tumorigenic paracrine mediators (Donawho and Kripke, 1991; Jamal and Schneider, 2002). Higher proportions of C>T mutations are commonly used as a predictor of UV-induced mutation signature in melanoma samples. Therefore, a non C>T mutation may be more likely to be a driver in melanoma. When C>T mutations are identified, they need to fulfill criteria described in section ‘Passenger versus Driver Mutations’ below to strengthen the argument that they are melanoma drivers.
Raw material for sequencing
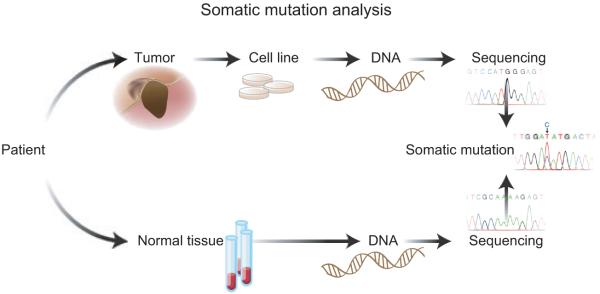
To detect somatic mutations in melanoma tumors, samples from both tumors as well as normal controls from the blood or surrounding normal tissue are needed. By comparing tumor and germline DNA, genetic changes can be identified as somatic mutations (Figure 1). Both tumor and normal DNA from the same individual are required to control for natural variations in the human genome.
Figure 1.
Representation of steps involved in the identification of somatic mutations in melanoma. Tumors are isolated from the patients and cell lines are prepared. DNA from the cell lines is used for sequencing. In parallel, DNA obtained from blood, which represents normal tissue of the patient, is sequenced. Comparison of the DNA sequences of tumor and normal tissue provides information about the somatic mutations in an individual.
To study melanoma genomics, high-quality genetic material from melanoma patients is necessary. Dissected tumors often contain normal stromal and immune cells, which dilute the ability to detect genetic changes. Thus, tumor cell DNA must be enriched before genetic analysis. Early-passage tumor cell line or mouse xenografts or microdissected tumor by a pathologist can be used for sequencing (Jones et al., 2008a) (Figure 1). Surgical resections provide larger specimens and higherquality DNA than biopsies.
Key quality control tests that confirm the identity of a tumor are also essential prior to any analysis. It should be verified that at least 75% of the cells in the melanoma tumor samples are melanoma cells and that no more than 20% of the cells are necrotic as described previously (Kairiyama et al., 1995; Quentmeier et al., 2001). This allows for efficient detection of homozygous and hemizygous deletions, amplification, duplication, and loss of heterozygosity. Hematoxylin and eosin staining can be used to verify percentage of tumor nuclei and necrotic cells, and staining with melanoma-specific markers can specifically predict the percentage of melanoma cells in a tumor sample. Genetic testing should contain cells positive for melanoma markers. Importantly, normal and tumor samples need to be matched by genotyping single nucleotide polymorphisms (SNPs) to confirm that they are from the same individual before tumor-specific somatic mutations in candidate genes can be derived. In addition, previously discovered highly mutated oncogenes and tumor suppressor genes in melanoma can serve as controls in a panel of melanoma samples when analyzing genetic alterations. Mutational analysis of melanoma genetics necessitates high-grade samples, reagents, and controls to produce reliable data.
Candidate gene sequencing
Initial studies in melanoma genetics focused on individual genes, also known as the candidate gene sequencing approach. This classic sequencing technique focused on sets of genes traditionally associated with cancer. Mutations in genes like p16, p14, and CDK4 / 6 were identified but were later found to be accountable for only a portion of sporadic and familial melanomas (Chin, 2003). Subsequent studies have identified new candidate genes, such as BRAF, NRAS, CKIT, BAP1, ERBB4, MITF, GNAQ, GNA11, MMP8, ADAMTS18, MAP3K5, MAP3K9, MAP2K1, and protein tyrosine phosphatase receptor type-D (PTPRD). For a majority of candidate genes, direct pathogenic evidence causing melanoma has yet to be validated. However, some exciting data indicating the role of a number of genes involved in melanoma tumorigenesis exist and will be described in this section.
BRAF
The RAS–RAF signaling proteins are known regulators of cell growth and drivers of multiple tumors, making them strong candidates for systemic mutational profiling in melanoma. The RAF family of serine / threonine kinases includes ARAF, BRAF, and CRAF, which activate mitogen-activated protein kinase (MAPK) kinase (MKK) that phosphorylates and induces activation of ERK / MAPK (Castellano and Downward, 2011). Candidate gene sequencing led to the seminal discovery that BRAF is mutated in 42–60% of melanoma cell lines and tumor samples (Davies et al., 2002; Hocker and Tsao, 2007). Strikingly, about 80% of the mutations in BRAF are clustered at a kinase-activation domain in which the V600E point mutation induces constitutive BRAF activation. Importantly, mutant BRAF has recently become a successful target for an FDA-approved small-molecule inhibitor, vemurafenib (PLX-4032) (Bollag et al., 2010; Chapman et al., 2011; Flaherty et al., 2010; Halaban et al., 2010).
NRAS
Upstream of BRAF in the RAS–RAF signaling pathway is the RAS subfamily, which includes HRAS, KRAS, and NRAS. Among these, NRAS and KRAS are the most relevant genes in human cancer as they are mutated in 20–30% of all cancers (Bos, 1989; Vidwans et al., 2011). Functionally, these isomers are similar but they show somewhat distinct tumor specificity (Quinlan and Settleman, 2009). KRAS-specific aberrations are mainly seen in pancreatic cancers (Hezel et al., 2006), whereas NRAS shows mutation rate up to 30% in cutaneous melanomas (Albino et al., 1989; Chin, 2003; Padua et al., 1984; Tsao et al., 2004), HRAS on the other hand is a key player in the maintenance of murine melanomas (Chin et al., 1999). Although activated NRAS has been accepted as the key oncogene in human melanomas, it requires another stimulus to yield truly transformed melanoma cells such as by inactivation of p53 or CDKN2A (Bardeesy et al., 2001; Chin et al., 1997).
C-KIT
c-KIT or CD117 belongs to the receptor tyrosine kinase (RTK) family whose downstream effectors include MAPK, phospholipase C, MITF, and Src and is activated by a ligand called stem cell factor (SCF) or KIT-ligand (Fecher et al., 2007; Grichnik, 2006; Hemesath et al., 1998). In melanocyte development, c-KIT-SCF signaling is required for differentiation, proliferation, and migration (Fecher et al., 2007; Grichnik, 2006). Both inactivating and activating mutations have been discovered in c-KIT. Germline-inactivating mutations cause loss of c-KIT kinase activity and lead to a loss of melanocytes and other abnormalities associated with piebaldism (Alexeev and Yoon, 2006). In contrast, activating mutations of c-KIT are associated with several cancer subtypes and are known to dysregulate diverse mechanisms providing survival advantages to tumor cells. In a seminal paper by Curtin et al. (2005), analysis of 102 primary melanomas found c-KIT mutations in 21% of mucosal, 11% of acral, and 17% of sun-damaged skin. In contrast, no KIT mutations were identified in melanomas without longterm sun damage. Furthermore, this study also included an analysis of DNA copy number alterations showing that KIT gene amplification was present in 6% of chronic sun-damaged, 7% of acral lentiginous, and 8% of mucosal melanomas. These genetic data were validated by numerous subsequent studies (Antonescu et al., 2007; Ashida et al., 2009; Beadling et al., 2008; Handolias et al., 2010; Rivera et al., 2008; Torres-Cabala et al., 2009). Recently, a study by Monsel et al. (2009) investigated the signaling pathway regulated by c-KIT mutants K642E and L576P. They found that under normoxic conditions, these mutations strongly activate the phosphatidyl-inositol-3 kinase (PI3K) pathway and weakly activate the Ras / Raf / Mek / Erk pathway. However, this induction was insufficient to instigate melanocyte transformation and growth. In contrast, under hypoxic conditions that mimic tumor microenvironment or with constitutive expression of hypoxia-inducible factor 1-α (HIF-1α), these mutations strongly activated the Ras / Raf / Mek / Erk pathway and stimulated melanocyte transformation and proliferation (Monsel et al., 2009). In another study, Phung et al. found that wild-type c-KIT stimulation leads to activation of MITF transcription factor because of c-KIT phosphorylation at the PI3K-binding site (Y721) and Src-binding sites (Y568 and Y570). In addition, c-KIT-induced activation of MITF is dependent on PI3K-, Akt-, Src-, p38- or Mek kinases and mutation of c-KIT phosphorylation sites restrict proliferation. However, this study utilized human embryonic kidney cells 293T cells, so additional studies are needed to confirm the role of c-KIT signaling in melanocyte transformation and melanoma progression (Phung et al., 2011). Overall, c-KIT is an important regulator of melanocyte fate, and its activating mutations favor melanoma formation by dysregulating various cell-signaling pathways.
Importantly, recent Phase II clinical trials of the c-KIT inhibitor Imatinib, which is FDA approved for gastrointestinal stromal tumors, reported an overall durable response rate of 16–23%, specifically in patients who harbor mutations in exons 11 or 13 in c-Kit or with an amplification of c-KIT. These studies therefore proved a significant clinical response in a subset of melanoma patients (Carvajal et al., 2011; Guo et al., 2011).
GNAQ/ GNA11
Unlike other types of melanoma, uveal melanomas lack BRAF and NRAS mutations (Cruz et al., 2003). The Bastian laboratory identified a novel mutation in a G-protein family of proteins called guanine nucleotide-binding protein G(q) subunit α (GNAQ) that is mutated in 83% of blue nevi and in 46% of uveal melanomas. They discovered the hotspot Q209L mutation in the Ras-like domain of GNAQ that produces a dominant oncogenic phenotype. This GNAQ Q209L mutation accelerates anchorage-independent growth and causes melanocytic transformation by inducing MAP kinase activation (Van Raamsdonk et al., 2009). Functionally, the α subunit of the heterotrimeric G-protein is activated only when bound to GTP and is inactivated when the bound GTP (guanosine-5′-triphosphate) is hydrolyzed (Landis et al., 1989). The glutamine residue at 209 is essential for conformation switching; mutations at this position cause the a subunit to lock in a constitutively active conformation, which produces oncogenic pro-survival signaling (Kalinec et al., 1992; Landis et al., 1989; Sondek et al., 1994). The Bastian laboratory further extended their studies to 713 melanocytic neoplasms of different subtypes and sequenced exon 5 of both GNAQ and its paralogue GNA11. They found that mutations in GNAQ and GNA11 are mutually exclusive and reported that 55% of blue nevi, 45% of uveal melanomas, and 22% of metastatic cells of uveal melanoma have GNAQ mutations at the glutamine in position 209. In addition, 7% of blue nevi, 32% of primary uveal melanomas, and 57% of metastatic uveal melanoma have GNA11 mutations affecting glutamine 209 (Van Raamsdonk et al., 2010). Xenograft studies in mice with cells bearing GNA11 Q209L mutations produced large tumors that spontaneously metastasized. However, the study did not include a comparison with GNAQ Q209L, which would be an interesting hypothesis to test in the future. Overall, both GNAQ and GNA11 are two important drivers in melanomas that lack Ras / Raf mutations.
MITF and SOX10
Candidate gene studies have recently also revealed genetic alterations in microphthalmia-associated transcription factor (MITF), which is involved in pigmentation, differentiation, and growth of melanocytes. Garraway et al. identified MITF amplifications that were correlated with patient survival, metastatic events, chemotherapeutic resistance, BRAF mutations, and inactivation of p16. These investigators showed that MITF has the ability to transform primary melanocytes in conjunction with BRAF, which further implicates MITF as a melanoma oncogene (Garraway et al., 2005).
Cronin et al. compared MITF genetic sequences among patients with primary and metastatic lesions. Overall, over 20% of metastatic melanoma cases had alterations in the MITF pathway. Of the primary melanomas, 2 / 26 had mutations in MITF and 6 / 55 showed mutations in SOX10, a binding partner of MITF (Cronin et al. 2009). While no amplifications of MITF were observed in the primary tumor samples, 4 / 50 metastatic samples had MITF amplifications, suggesting that MITF amplification may be a late-stage change in cancer progression. Moreover, the range of mutations observed in SOX10 was characteristic of a tumor suppressor gene. Of the metastatic melanoma samples, three frameshift mutations were predicted to result in protein truncations before the DNA-binding domain or at the C-terminus. Functional analysis of SOX10 mutations showed reduced activity on target promoters and association with loss of heterozygosity (Cronin et al., 2009). As expected, MITF and SOX10 mutations were mutually exclusive, indicative of a disruption in a common genetic pathway.
Functional analysis of the MITF 4TΔ2B mutation, a splice variant mutation that causes loss of exon 2B of MITF, in the presence of wild-type SOX10, showed significantly greater activation of human dopachrome tautomerase (HuDct) and tyrosinase promoter but not of cell cycle arrest marker p21. This suggests that mutated MITF allows cells to escape p21-mediated cell cycle arrest. Dopachrome tautomerase (Dct) gene is expressed early and is required for the melanocyte pigmentation and development (Ludwig et al., 2004). Furthermore, mutations resulting in early truncation of SOX10 abolished activity of MITF-mediated HuDct activation, suggesting that SOX10 is indispensable for MITF activity. Further validation of the mutated MITF functional effects was recently published by Taylor et al. (2011) who used a zebrafish model to show mutant MITF 4TΔ2B potentiated melanocyte cell division and differentiation compared with wild-type human MITF. This again supports finding by Cronin et al. that MITF 4TΔ2B is important in melanoma genesis.
Two recent Nature articles identified a germline mutation leading to a substitution of glutamic acid 318 with lysine (E318K) in MITF that was associated with both sporadic melanoma and renal cell carcinoma (RCC). The E318K mutation is located in a small-ubiquitin-like modifier (SUMO) consensus site in MITF, which has been found to interrupt sumoylation and increase susceptibility to cancer. Bertolotto et al. (2011) sequenced MITF in 62 patients with both melanoma and renal cell cancer and observed a higher occurrence of E318K in cases over controls. This study found MITF E318K increased the risk of either melanoma or RCC by more than fivefold. Chromatin immunoprecipitation analysis of the genome-wide effects of E318K on MITF-occupied loci showed greater promoter occupancy by MITF E318K and an increase in MITF E318K occupancy at the HIF1A promoter. Bertolotto et al. propose that the E318K mutation induces hypoxia-inducible factor 1α (HIF1α) transcription, a pathway previously found to be dysregulated in RCC. The study concludes that MITF E318K expression augmented in vitro migration, invasion, and colony formation of both melanoma and RCC cell lines. In a separate study, Yokoyama et al. (2011) sequenced the genomes of a number of melanoma families and found a SNP in MITF that led to independent identification of the E318K substitution. Researchers determined E318K had a log of odds (lod) score of 2.7, suggesting E318K represents an intermediate risk variant. This variant allele was linked with higher nevus counts and nonblue eye color. The group performed functional analysis to confirm E318K led to impaired sumoylation and dysregulation of specific MITF targets. The Bertolotto and Yokoyama papers used different methods to characterize the E318K mutation and its association with both melanoma and RCC.
PTPRD
Another gene identified using candidate gene studies to be genetically altered in melanoma is PTPRD. Because activation of tyrosine phosphorylation signaling pathways (e.g. via EGFR mutation or HER2 / Neu amplification) are well-characterized hallmarks of cancer, inactivation of protein tyrosine phosphatases (PTPs) that regulate tyrosine phosphorylation and growth signaling was expected to produce a similar effect.
In a genome-wide loss of heterozygosity and copy number alteration study, Stark and Hayward (2007) found that the PTPRD locus underwent homozygous deletions in 9% of melanoma cell lines, while candidate gene sequencing of PTPRD in malignant melanomas revealed somatic mutations in 12% of melanoma samples (Solomon et al., 2008). Similarly, a paper recently accepted to Nature Genetics by Hayward et al. also found PTPRD to be highly mutated in melanoma (Stark et al., 2011). Functional studies have shown that reconstitution of PTPRD inhibited growth and induced apoptosis in cells harboring its deletions or mutations. However, constitutive expression of somatic mutations alleviated the effect of wild-type PTPRD expression (Solomon et al., 2008).
In addition to melanoma, PTPRD has been found to be deleted in a fraction of glioblastomas (Solomon et al., 2008), adenocarcinomas of the colon and lung (Sjoblom et al., 2006; Weir et al., 2007), and squamous carcinomas of the head and neck (Veeriah et al., 2009). Together, these findings indicate that PTPRD plays an important role as a tumor suppressor gene in neoplasms of neuroectodermal origin.
AKT
The Akt / PKB family is a serine / threonine protein kinase that has been linked to tumor cell survival, proliferation, and metastasis. The family includes Akt1, Akt2, and Akt3. Stahl et al. (2004) were the first to identify that AKT3 is deregulated in melanoma by copy number increases. Furthermore, this group reported that this amplification activates the AKT3 function that promotes cell survival. Later, Carpten et al. (2007) described a novel point mutation in AKT1, namely an E17K substitution in breast, colon, and ovarian cancer. Functionally, E17K translocates to the membrane and induces constitutive phosphorylation of AKT1 that is sufficient to produce fibroblast transformation and leukemia in transgenic mice (Carpten et al., 2007). Using the candidate gene approach, Davies et al. investigated the significance of the AKT E17K mutation in 137 melanoma samples and 65 human melanoma cell lines. They found only one E17K mutation in AKT1 in all samples. Surprisingly, they discovered activating E17K mutation in AKT3 in two melanoma samples obtained from one patient and two commercially available melanoma cell lines, WM46 and D40 (Davies et al., 2008). These findings suggest that the AKT3 E17K mutation may serve as another potential target for melanoma therapy.
ETV1
The Garraway laboratory identified several tumor samples containing amplifications on chromosome 7p, one of the most common sites of melanoma driver cancer gene mutations (Jane-Valbuena et al., 2010). The minimal common overlapping region among the tumor samples bridged the oncogene ETV1 (Ets Variant Gene 1). The authors used fluorescence in situ hybridization analysis to show that over 40% of samples contained copy gains in the ETV1 locus. In melanoma cell lines, ETV1 expression was essential for proliferation and growth independent of anchorage. The combination of ETV1 overexpression and oncogenic NRASG12D transformed primary melanocytes that led to tumor growth in mouse models. In immortalized melanocytes, ETV1 also increased MITF expression, which was necessary for ETV1-induced oncogenicity. This study implicates ETV1 deregulation in melanoma genesis.
Candidate gene family sequencing
With the completion of the Human Genome Project, cancer gene identification progressed from studying candidate genes to candidate families. Combined with the advent of new sequencing technologies and capture methods, hundreds of targeted genes can be investigated at once. Tyrosine kinase family The first gene families to be comprehensively sequenced were the tyrosine kinase families. Proteins associated with protein and lipid phosphorylation are central to physiologic and pathologic signaling and proliferation, and several members of the protein kinase families had already been correlated with tumorigenesis. In addition, kinases serve as potential drug targets as they are susceptible to pharmacologic inhibition.
One of these seminal studies describes a pattern of somatic mutations in 518 protein kinase genes in a sample set of 210 diverse human cancers (Greenman et al., 2007). In this study, the investigators identified substantial variation in the mutation spectrum of distinct cancer subtypes (Greenman et al., 2007). To differentiate between passenger and driver mutations, Greenman et al. (2007) analyzed several conserved domains and concluded that driver mutations are located in approximately 120 of 518 protein kinases sequenced. Melanoma and glioma were found to contain the largest incidences of mutations per megabase pairs of DNA, 18.54 and 22.37, respectively. Furthermore, a unique spectra of mutations were found in melanomas because of C:G>T:A, again implicating the role of UV light as a potential carcinogen. Of the 518 genes encoding protein kinases, the top three most likely to carry at least one driver mutation based on selection pressure estimates were Titin (TTN), BRAF, and ATM. With an N:S ratio of 4.8, TTN is the largest polypeptide encoded by the human genome and is a component of many cell types with a known function in muscle contractile machinery. Whether TTN mutations play a role in oncogenesis has yet to be biologically evaluated.
Because of the diversity of human cancer samples used in this study and the small number of samples investigated per cancer type, further studies were needed to identify the candidate genes for specific malignancies. For example, among known tyrosine kinases mutated in melanoma, only BRAF ranked in the top 50. Other mutated kinases identified in melanoma such as AKT3, ERBB4, and FLT1 ranked 185th, 214th, and 88th of 518, respectively (Greenman et al., 2007). Nevertheless, this study established the platform for large-scale systemic family sequencing studies and identification of potential drivers in cancer progression.
Prickett et al. recently performed a mutational analysis of the protein tyrosine kinome in 79 cutaneous malignant melanomas. An initial genetic screen of the kinase domain-encoding exons revealed somatic mutations in 19 of 86 tyrosine kinases. Subsequent complete gene sequencing of the 19 PTKs showed 19% of the mutations were in ERBB4, 10% in FLT1, and 10% in PTK2B (Prickett et al., 2009). Furthermore, a recent study by Wagle et al. (2011) identified an ERBB4 mutation in a melanoma patient found to be resistant to BRAF treatment. Importantly, ERBB4 somatic alterations, some of which overlap with the initial Prickett et al. study, were recently also identified by Hayward et al. in their melanoma samples (Stark et al., 2011). Importantly, two of the identified alterations in the study by Prickett et al. had substitutions in the same residue (E452K). This particular alteration was also identified by Hayward et al. in an additional three melanoma samples (personal communication). Moreover, another of the alterations identified in ERBB4 (E872K) has previously been discovered in lung adenocarcinoma forming a ‘mini-hotspot’ (Ding et al., 2008).
Through a series of functional studies, Prickett et al. showed that ERBB4 mutations imparted cells with an increased kinase activity and an ability to transform mouse fibroblast NIH3T3 cells as well as melanoma cells. Using shRNA, the study showed that ERBB4 knockdown in ERBB4 mutant melanoma cell lines inhibited growth compared with cells bearing wild-type ERBB4. Pharmacological inhibition of ERBB4 by lapatinib, a pan-ERBB kinase inhibitor, preferentially killed more ERBB4 mutant melanoma cell lines than cells expressing wild-type ERBB4. Interestingly, distinct ERBB4 mutant cell lines showed variable sensitivity to lapatinib, suggesting that ERBB4 mutants contribute to drug sensitivity. However, as lapatinib is known to also inhibit EGFR and ERBB2, there is a need for the development of specific ERBB4 kinase inhibitors to exclude the role of EGFR and / or ERBB2 in addressing this issue. Furthermore, recent extension of the mutational data shows significant mutual exclusivity between mutations in ERBB4 and BRAF (P > 0.02 Fisher’s exact test). These provocative discoveries reveal an exciting new melanoma gene and elucidate an interesting but complex role of growth factor receptor signaling in melanoma.
To further characterize the role of RTKs in melanoma, the Stern laboratory conducted a phosphoproteomic screen of 58 human RTKs in a panel of earlypassage melanoma cell lines. Functional analysis of 42 RTKs showed the insulin receptor family (IGF1R, IR), MET family (MET, MST1R), EGFR family (ERBB1, ERBB3, and ERBB4), and TAM family (TYRO3, AXL, and MERTK) to be among the most frequently activated RTKs in melanoma. Subsequent shRNA-mediated knockdown of AXL, HER3, and IGFR1R resulted in decreased melanoma cell proliferation, and AXL knockdown decreased melanoma cell migration (Tworkoski et al., 2011). Importantly, the expression and activation of RTKs was unrelated to mutation status, and signaling through wild-type RTKs may be a mechanism of resistance in tumor cells treated with pathway-directed therapeutics (Mcdermott et al., 2010). The study identified novel RTK targets for pharmacological inhibition and drug-resistance prevention in melanoma.
Tyrosine phosphatases
Since the identification of PTPRD to be mutated in melanoma, researchers have performed genetic investigation of the entire family of PTPs. Genomic analysis showed that PTP receptor type-T (PTPRT) and PTPRD are the most and second most highly mutated PTPs in melanoma, respectively. Biochemical functional assays suggest that PTPRT is a tumor suppressor, whose genetic inactivation results in survival advantage in tumor cells. Recent studies on PTPRT validated two interacting proteins STAT3 (Zhang et al., 2007) and paxillin (Zhao et al., 2010) and underscored the role of PTPRT in tumorigenesis.
As mentioned earlier, Solomon et al. (2008) identified PTPRD mutations in 12% of melanomas and glioblastomas. Ectopic expression of PTPRD in glioblastoma and malignant melanoma cell lines bearing deletions or mutations induced growth inhibition, whereas expression of mutant PTPRD alleviated this phenotype. However, little is known about the binding partners of PTPRD and how its expression affects melanoma cell biology. Because stable expression of PTPRD is growth inhibitory, Walia et al. created a novel doxycycline-inducible single plasmid-based lentiviral plasmid with 3X-Flag tag attached to PTPRD and expressed it in a melanoma cell line. Currently, investigators are screening and validating its interacting partners by mass spectrometry experiments (Walia et al., unpublished). PTPs such as PTPRT and PTPRD clearly play an important role in melanoma, and further investigation of their biology might identify their corresponding substrates and potential interacting tyrosine kinases.
Proteases
Proteases are generally classified into five major classes: metalloproteinase, serine, cysteine, threonine, and aspartic proteinases (Rivera et al., 2010). Metalloproteinases, the largest protease class, includes matrix metalloproteinases (MMPs), a disintegrin and metalloproteinase (ADAMs), and ADAMs with thrombospondin domain (ADAMTS). Matrix metalloproteinases, ADAMs, and ADAMTS have recently been investigated for somatic mutations in melanoma (Palavalli et al., 2009; Stocker and Bode, 1995).
MMP
Matrix metalloproteinases are signal-peptide containing proteins that are predominantly secreted and function extracellularly to degrade basement membranes and extracellular matrix (Brinckerhoff and Matrisian, 2002). They are constitutively expressed until activated by enzymes cleaving prodomains or releasing cysteine bonds (Balbin et al., 2003; Egeblad and Werb, 2002; Freije et al., 2003). Matrix metalloproteinases have been linked both to cancer metastasis and tumor suppression, and mouse experiments revealed that MMPs possess anticancer properties (Balbin et al., 2003; Gutierrez-Fernandez et al., 2008; Montel et al., 2004). MMP8-deficient mice showed a significantly elevated incidence of skin tumors and metastasis, suggesting that MMP8 hinders tumorigenesis and suppresses metastasis (Balbin et al., 2003; Gutierrez-Fernandez et al., 2008). Unfortunately, small-molecule inhibitors of MMPs developed as promising anticancer agents showed no significant benefit in human clinical trials. Instead, MMP inhibitors accelerated tumor growth in some cases (Coussens et al., 2002; Lopez-Otin and Matrisian, 2007).
To further analyze the role of MMPs in human melanoma, Palavalli et al. performed a comprehensive mutational analysis of the MMP gene family. The study showed that in 6% of cases, MMP-8 was mutated. Most of these mutations were accompanied by loss of heterozygosity suggesting that MMP-8 acts as a tumor suppressor gene. Functional studies showed that in contrast to wild-type MMP-8, mutant MMP-8 possesses lower MMP enzymatic activity, promotes cell growth in soft agar, and forms tumor ulceration in xenograft mouse models. A whole-genome analysis of a melanoma tumor also identified an MMP somatic mutation (Pleasance et al., 2010); however, no functional evaluation of this mutation exists as of yet. Interestingly, a recent study testing the association of MMP-8 variation and increased risk of melanoma showed that particular MMP-8 polymorphisms are indeed associated with the risk of developing melanoma (Debniak et al., 2011).
ADAM
Somatic mutations identified in ADAM family members suggest their role in tumorigenesis of distinct subtypes (Dalgliesh et al., 2010; Parsons et al., 2008; Pleasance et al., 2010; Sjoblom et al., 2006; TCGA, 2008). Systematic analysis of the entire ADAM family in a large set of human melanoma samples identified ADAM29 and ADAM7 as mutated genes in melanoma (Wei et al., 2011a). Similarly, the Pleasance et al. (2010) paper as well as the Stark et al. (2011) paper identified a ADAM29 and ADAM7 somatic mutations in their melanoma whole-genome and melanoma whole-exome analysis. ADAMs are mostly membrane-anchored glycoproteins that regulate cell–cell fusion, cell migration, and cell–cell adhesion (Mochizuki and Okada, 2007). Approximately half of all ADAMs are involved in cell adhesion and the remaining half possess proteolytic activity that causes shedding of surface proteins.
Functionally, ADAM29 mutations promote melanoma cell adhesion to collagen I, II, and IV, whereas mutations in ADAM7 reduce melanoma cell adhesion to collagen IV and laminin and increase cell migration compared with cells expressing wild-type ADAM7 (Wei et al., 2011a). Furthermore, studies showed ADAM7 harbors three mini-hotspots within the same functional reprolysin domain. This indicates that mutated ADAM genes regulate migration and adhesion of melanoma cells to different extracellular matrix proteins. Future studies are needed to explore whether ADAM7 mutations promote intravasation and ADAM29 mutations promote extravasation during melanoma metastasis.
ADAMTS
The ADAMTS family, which includes metalloproteinases with a thrombospondin domain, encodes extracellular proteins and functions as proteolytic proteins (Rocks et al., 2008). Their extracellular topology makes them excellent candidates for systemic drug targeting. In general, ADAMTS family members are thought to possess tumor suppressive activities (Li et al., 2010; Rocks et al., 2008; Viloria et al., 2009). However, their detailed role in cancer is still under study. Several papers have reported that ADAMTS15 and ADAMTS18 are somatically mutated in colorectal cancers (Sjoblom et al., 2006; Viloria et al., 2009; Wood et al., 2007). A follow-up study investigating the mutation spectrum of ADAMTS family in melanoma identified ADAMTS18 as a mutated ADAMTS gene among melanoma patients (Wei et al., 2010). In melanoma xenograft models, mutated ADAMTS18 promotes growth, cell migration, and metastasis. It is known that proteolysis of the C-terminal domain of ADAMTS18 modifies its non-catalytic ancillary domain that regulates interactions with its substrates and other proteins. Interestingly, several of the truncation mutations of ADAMTS18 lie near the C-terminal domain suggesting that this may affect its interaction with physiological substrates. Indeed, the mutations identified in this region affected substrate recognition and enzyme localization in immunoprecipitation and immunofluorescence staining assays (Wei et al., 2010). Thus, future identification of specific substrates of ADAMTS18 will provide useful information about its mechanism of action and how its mutations modulate melanoma cell phenotypes.
Whole-exome sequencing
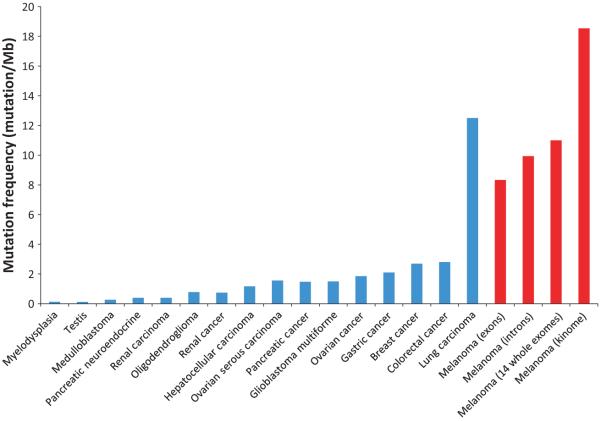
While sequencing gene families has been invaluable in identifying cancer genes, entire exomes can now be queried using increasingly high-throughput sequencing technologies to provide an unbiased profiling of the genomic basis of melanoma. Wei et al. performed the first comprehensive exome-wide genetic study of melanoma by sequencing the exomes of 14 untreated malignant melanoma samples along with their matched normal DNA. As this was the first comprehensive analysis of the melanoma exome, it allowed an approximate calculation of the frequency of mutations per megabase of melanoma DNA, which was determined to be 11.4 mutations / Mb. This fits well with the frequency identified by Pleasance et al. as well as Greenman et al. who identified the frequency to be 18.54 mut / Mb and 9.9 mut / Mb, respectively (Greenman et al., 2007; Pleasance et al., 2010). Importantly, this is about an order of magnitude higher than frequencies reported in other solid cancers, apart for lung carcinoma (Figure 2).
Figure 2.
Prevalence of somatic mutations in different cancers. Number of somatic mutations detected per megabasepair of genes sequenced (y-axis) is plotted against different types of tumors. Mb: megabasepairs of DNA. References, Myelodysplasia (Papaemmanuil et al., 2011; assuming 38 Mb captured / exome), Testis (Greenman et al., 2007), Medulloblastoma (Parsons et al., 2011), Pancreatic Neuroendocrine (Jiao et al., 2011), Renal Carcinoma (Varela et al., 2011), Oligodendroglioma (Bettegowda et al., 2011; assuming 38 Mb captured / exome), Renal cancer (Greenman et al., 2007), Hepatocellular Carcinoma (Li et al., 2011; assuming 38 Mb captured / exome), Ovarian Serous Carcinoma (TCGA, 2011, assuming 38 Mb captured / exome), Pancreatic Cancer (Jones et al., 2008b), Glioblastoma Multiforme (Parsons et al., 2008), Ovarian cancer (Greenman et al., 2007), Gastric cancer (Greenman et al., 2007), Breast cancer (Greenman et al., 2007), Colorectal cancer (Sjoblom et al., 2006), Lung Carcinoma, protein-coding genes (Lee et al., 2010), Melanoma (exons) (Pleasance et al., 2010), Melanoma (introns) (Pleasance et al., 2010), Melanoma (14 whole exomes) (Wei et al., 2011b), Melanoma (kinome) (Greenman et al., 2007).
Filtering through approximately 4000 candidate somatic mutations, Wei et al. (2011b) found seven novel recurrent non-synonymous mutations, in addition to the known BRAF recurrent mutation. Scale-up studies of these candidates in a prevalence panel of 153 melanoma samples revealed that the majority of the recurrent mutations only scaled up to a total of 1–4% of cases. However, calculation of the likelihood of acquiring this number of identical mutations at the same position in each case was significant. The recurrent alteration that scaled up the most was the S722F substitution in TRRAP that occurred in six different melanoma samples. Importantly, this exact somatic alteration was also identified by Halaban et al. as well as Meltzer et al. in their melanoma samples (unpublished data; personal communication). Recurrent mutations of a single amino acid suggested that TRRAP acts as a melanoma oncogene, which was further validated by functional data described below.
TRRAP, a multi-function protein complex with histone acetyltransferase activity, serves as a transcription co-activator of several genes such as Mdm-2, MYC, and E2F. Recurrent mutations and transformation assays suggest that TRRAP acts as an oncogene. The serine-722 in TRRAP is generally conserved in other species indicating an essential role of this amino acid segment in development. However, no known functional domain was found to encompass the S722F mutation. Although some functional work on the identified hotspot mutation has shown TRRAP mutations to be transforming in NIH3T3 fibroblast cells and to be involved in apoptosis, further understanding of its effect of on melanoma biology has yet to be determined. Importantly, the identification of TRRAP recurrent mutations adds melanoma to the growing list of cancers driven by mutations in proteins with histone modifying activity.
This same whole-exome analysis also identified 68 genes that were mutated in more than two independent samples (Wei et al., 2011b). Sixteen candidate driver genes that harbored a higher number of somatic mutations beyond what would be expected by chance were identified and re-sequenced in 38 untreated melanoma samples. Most significantly, GRIN2A, which encodes a subunit of the ionotropic glutamate receptor, was found mutated in the largest fraction of melanoma cases. A screen of 135 total melanomas using different cohorts of validation samples revealed 34 distinct mutations and indicated that GRIN2A is mutated in 25.2% of melanomas (33, 28.2, and 15.6% in three respective melanoma cohorts). Overall, the investigators found two mutation clusters, three recurrent alterations, and five nonsense mutations in the full-length GRIN2A subunit, suggesting a tumor suppressive role of GRIN2A in melanoma. Importantly, GRIN2A mutations were identified at a similar frequency of 17% (14 somatic mutations in 80 melanoma samples) by Halaban et al. in their melanoma samples (unpublished data) as well as by Stark et al. (2011). Further investigation on exactly how GRIN2A mutations affect melanoma biology is under investigation.
The comprehensive nature of the whole-exome study allowed the identification of the glutamate signaling pathway to be significantly altered in melanoma. Glutamate activates two distinct types of receptors: ionotropic glutamate receptors (iGluRs) and metabotropic glutamate receptors (mGluRs). iGluRs are ligand-gated ion channels that permit cations such as calcium and potassium to traverse the plasma cell membrane following glutamate and receptor binding. There are three types of iGluRs: N-methyl-D-aspartate (NMDA), Kainate, and α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors. GRIN2A that was described as highly mutated in the screen encodes a glutamate receptor subunit that binds NMDA (Wei et al., 2011b). Of the genes encoding mGluRs, GRM1, GRM3, and GRM5 have been found to play a role in melanoma genesis as further described below (Arcella et al., 2005; Choi et al., 2011; Pollock et al., 2003).
In contrast with iGluRs, mGluRs are G-protein-coupled metabotropic receptors. Pharmacological blocking of mGLuRs has been shown to inhibit growth in cancer cells (Arcella et al., 2005). Seminal work by Pollock et al. (2003) has found that expression of GRM1 coding for mGluR1 correlates with hyper-proliferation of mouse melanocytes. In humans, GRM1 expression was detected in melanoma cell lines and biopsies but was absent in normal melanocytes and benign nevi (Pollock et al., 2003). Another study from the same group showed that ectopic expression of GRM1 possesses oncogenic activities and alone is sufficient to induce melanocytes transformation (Shin et al., 2008). Treatment of GRM1-expressing melanoma cells with glutamate release inhibitors and GRM1 antagonists such as Rizudole decreases extracellular levels of glutamate and inhibits melanoma cell growth (Namkoong et al., 2007).
Choi et al. linked GRM5 to melanoma via finding that the majority of the founders of GRM5 transgenic lines had severe, early-onset phenotypes of melanoma. A majority of the GRM5 transgene-positive mice possessed hyperpigmentation of the pinnae and tail 3–5 days after birth. Examination of tumor samples revealed a considerable increase in the phosphorylation of ERK (Choi et al., 2011). The study demonstrated the role of GRM5-mediated glutamatergic signaling in the development of an aggressive phenotype of melanoma in vivo.
Finally, GRM3, which encodes another glutamate receptor, mGluR3, was recently identified through exomic sequencing to be mutated in melanoma (Prickett et al., 2011) (Stark et al., 2011). Prickett et al. (2011) found that GRM3 was mutated in 16% of melanomas. GRM3 mutations include 18 non-synonymous mutations and two hotspot mutations M547K (identified in two samples) and E870K (identified in four samples derived from two independent melanoma cohorts). Functional studies reveal that GRM3 alterations regulate MEK phosphorylation, thereby increasing anchorage-independent growth and migration. Moreover, shRNA-mediated knockdown of mutant GRM3 or treatment of cells with MEK inhibitor AZD-6244 significantly reduced melanoma cell growth, migration, and growth in vivo. As previous studies showed that pharmacological blocking of mGLuRs inhibits the growth of cancer cells (Arcella et al., 2005), these results show possible therapeutic relevance.
In addition to GRM1, 3 and 5 a GRM-signaling downstream effector called phospholipase C, beta 4 (PLCB4) was also shown to be mutated through whole-exome sequencing (Wei et al., 2011b). The protein encoded by PLCB4 catalyzes the formation of second messenger inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAG) from phosphatidylinositol 4,5-bisphosphate (PIP3). IP3 diffuses into the cytoplasm and acts as an agonist for calcium-sensitive IP3 receptors in endoplasmic reticulum that causes a release of intracellular calcium. DAG in the membrane activates protein kinase C, which regulates phosphorylation of various proteins inside the cell.
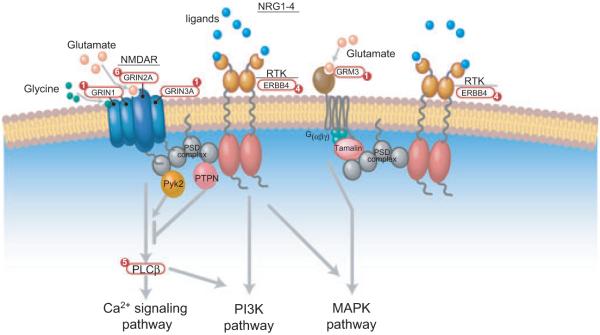
Taken together, these studies underscore the importance of glutamate signaling including both iGluRs (AMPA and NMDA receptors), as well as mGluRs (GRM1, GRM3, PLCB4, and GRM5) in melanoma. Importantly, Ozaki et al. showed the link between glutamate signaling and its modulation by the ERBB4 receptor and its ligand NRG1, which suggest a crucial crosstalk between these pathways (Anton et al., 2004; Dalva et al., 2000; Ozaki et al., 1997; Rieff et al., 1999) (Figure 3). Recently, the use of combined therapy of Riluzole, a benzothiazole derivative that blocks glutamate release and Sorafenib, a small-molecule, multi-kinase inhibitor, has shown promising results both in vitro and in vivo (Lee et al., 2011). Single agent therapy with either Riluzole or sorafenib has been proven to be ineffective in the treatment of melanoma (Lee et al., 2011; Mehnert et al., 2011), suggesting that both kinase and glutamate pathways need to be targeted for an effective therapy. Additionally, the effect of combined Riluzole and Sorafenib therapy was independent of the Raf V600E mutation spectrum of the patients suggesting a novel viable therapeutic option (Lee et al., 2011). Thus, further investigation into the role of the glutamate pathway in melanoma and its therapeutic intervention is highly warranted.
Figure 3.
The mutations in glutamate signaling pathway and its associated proteins in melanoma. Proteins that play an important functional role in glutamate signaling are shown. The number of somatic mutations in each gene is labeled in the circle. RTK, receptor tyrosine kinase; NMDAR, N-methyl-d-aspartic acid (NMDA) receptor; PSD complex, postsynaptic density complex; NRG, neuregulin; GRIN, glutamate receptor, ionotropic N-methyl D-aspartate; ERBB, epidermal growth factor receptor; PTPN, Protein tyrosine phosphatases, nonreceptor type.
An additional whole-exome sequencing study by the Bowcock laboratory showed BRCA1-associated protein 1 (BAP1), which encodes a ubiquitin carboxy-terminal hydrolase, harbors inactivating somatic mutations in 84% (26 of 31) of metastasizing uveal melanoma (Harbour et al., 2010). Of note, 15 of 31 mutations caused premature protein termination and five altered its ubiquitin carboxyl terminal hydroxylase domain. The authors concluded that loss of BAP1 might increase susceptibility to uveal melanoma metastasis and serve as a valuable therapeutic target. In an independent study, the Bastian laboratory showed germline SNPs observed in BAP1 conferred a predisposition to melanocytic neoplasms (Wiesner et al., 2011). The investigators describe two families with multiple skin-colored, elevated melanocytic tumors that ranged from epithelioid nevi to atypical melanocytic proliferations sharing features with melanoma. Several individuals in the families developed uveal or cutaneous melanomas and were found to have inactivating germline mutations of BAP1. Inactivating somatic mutations of the remaining wildtype BAP1 allele in these patients led to the development of melanocytic neoplasms. In addition, the authors detected BAP1 mutations in a fraction of sporadic melanocytic tumors with similar histology as the familial tumors. Taken together, these findings suggest loss of BAP1 is associated with a distinct type of melanocytic neoplasm in both clinical and morphologic presentation.
Two recent papers also used exome sequencing to examine variations in melanoma exonic sequences, with specific focus on the MAPK family. Nikolaev et al. (2011) used seven melanoma cell lines containing characteristic UV-induced DNA repair alongside donormatched germline samples. Interesting, two melanoma samples with BRAF mutations contained gain-offunction MAP2K1 (MEK1) and MAP2K2 (MEK2) mutations, which led to constitutive ERK phosphorylation and encoded proteins with greater resistance to BRAF or MEK inhibitors. The authors analyzed a larger group of melanoma patients and found 8% harbored somatic MAP2K1 and MAP2K2 mutations. In a separate study, Stark et al. (2011) sequenced eight melanoma exomes to detect novel somatic mutations and concentrated on the MAP3K family. Overall, 24% of melanoma cell lines contained mutations in the protein-coding regions of either MAP3K5 or MAP3K9. Using structural modeling, the investigators predicted that mutations in the kinase domain might affect the function and regulation of these protein kinases. The MAPK mutations are likely inactivating given their location and resultant loss of heterozygosity in 85% of the MAP3K5 and 67% of the MAP3K9 melanoma samples. The authors also used in vitro kinase assays to show MAP3K5 I780F and MAP3K9 W333 variants had decreased kinase activity. Moreover, when MAP3K5 or MAP3K9 mutations were overexpressed in HEK293T cells, the phosphorylation of downstream MAP kinases was reduced. The authors used siRNA to attenuate MAP3K9 function in melanoma cells and showed increased cell viability after temozolomide treatment. Altogether, the studies by Nikolaev et al. and Stark et al. used whole-exome sequencing to evaluate the possible role of MAPK mutations in melanoma genesis and chemoresistance.
Whole-genome sequencing
While whole-exome sequencing provides an unbiased genome-wide investigation of the coding regions in melanoma, whole-genome studies provide an even more indepth understanding, including regulatory regions undergoing somatic alterations and structural changes that drive melanoma tumorigenesis. The first comprehensive genomic catalog of somatic mutations in melanoma was performed on a malignant melanoma cell line COLO-829 and lymphoblastoid line derived from the same individual (Pleasance et al., 2010). This study identified several putative cancer genes such as SPDEF, a member of the ETS transcription factor family (Oettgen et al., 2000); MMP28, a MMPs, ADAM29; and UVRAG, a gene involved in UV light sensitivity, autophagy and tumor suppression (Liang et al., 2006). Genomic analysis also revealed traces of DNA repair initiated preferentially at transcribed regions such as exons compared with untranscribed DNA regions such as introns.
In the future, generation and analysis of high-quality genomic data from a large number of samples promises to provide greater insight into the processes of DNA damage, mutation, and repair mechanisms that contribute to the evolution of melanoma. Furthermore, higher sensitivity and specificity is already helping to identify structural variants and regulatory region alterations.
Molecular definition of melanoma subclasses
With a heterogeneous array of manifestations, melanoma has undergone various subclassifications. Formerly, melanoma was categorized according to its associated precursor lesion. In 1967, Dr. Wallace Clark developed a melanoma classification based on the histopathologic in situ component of the tumor adjacent to any invasive component. In this system, the disease was subdivided into superficial spreading melanoma, lentigo maligna melanoma, and nodular melanoma. These categories along with later additions remain in the WHO melanoma classification. Criticism of the current classification scheme centers on the overlap between clinical and histopathological features, poor predictability of patient outcome, and lack of applicability to patient care. However, melanoma subtypes emphasize the various clinical and pathological patterns of melanoma. Recent studies of somatic mutations in melanomas may also be integrated into later versions of the melanoma classification system. In the future, the mechanistic understanding of identified melanoma mutations will be enhanced using some of the approaches described in this review. Melanoma will then likely be categorized by clinical, pathologic, and melanoma characteristics that define unique biological subsets of melanoma that have similar somatic alterations, clinical presentation, and treatment options (Vidwans et al., 2011). Use of molecular alterations to define melanoma subclasses might allow for the scaling up of the various mini-hotspot alterations described in this review as well as categorizing the various driver genes discussed in this review.
Conclusion
Clearly, melanoma possesses a complex biological behavior resulting from a diverse range of genetic mutations. Advancements in DNA sequencing technologies from candidate gene, to candidate family, to exome, to whole-genome sequencing continue to revolutionize our ability to identify and classify mutations. The exciting discoveries of BRAF, CKIT, GNAQ, GRIN2A, TRRAP, MAP3K5, and other cancer genes and families set the stage for identifying more novel biomarkers and genomic alterations that may help us develop and select the best treatment for future melanoma patients. In the future, it will be important to develop inducible cell lines and animal models with driver mutants to test whether the genotype indeed drives the melanoma phenotype and to determine the efficacy of small molecular inhibitors. Ultimately, classification of melanoma by histologic and genetic subtypes will revolutionize the design of personalized melanoma therapies.
Acknowledgements
We thank Mike Davies and Jared Gartner for insightful comments. This work was supported by the Intramural Research Programs of the National Human Genome Research Institute, National Institutes of Health, USA.
References
- Albino AP, Nanus DM, Mentle IR, Cordon-Cardo C, Mcnutt NS, Bressler J, Andreeff M. Analysis of ras oncogenes in malignant melanoma and precursor lesions: correlation of point mutations with differentiation phenotype. Oncogene. 1989;4:1363–1374. [PubMed] [Google Scholar]
- Alexeev V, Yoon K. Distinctive role of the cKit receptor tyrosine kinase signaling in mammalian melanocytes. J. Invest. Dermatol. 2006;126:1102–1110. doi: 10.1038/sj.jid.5700125. [DOI] [PubMed] [Google Scholar]
- Anton ES, Ghashghaei HT, Weber JL, et al. Receptor tyrosine kinase ErbB4 modulates neuroblast migration and placement in the adult forebrain. Nat. Neurosci. 2004;7:1319–1328. doi: 10.1038/nn1345. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Busam KJ, Francone TD, et al. L576P KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int. J. Cancer. 2007;121:257–264. doi: 10.1002/ijc.22681. [DOI] [PubMed] [Google Scholar]
- Arcella A, Carpinelli G, Battaglia G, D’onofrio M, Santoro F, Ngomba RT, Bruno V, Casolini P, Giangaspero F, Nicoletti F. Pharmacological blockade of group II metabotropic glutamate receptors reduces the growth of glioma cells in vivo. Neuro Oncol. 2005;7:236–245. doi: 10.1215/S1152851704000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida A, Takata M, Murata H, Kido K, Saida T. Pathological activation of KIT in metastatic tumors of acral and mucosal melanomas. Int. J. Cancer. 2009;124:862–868. doi: 10.1002/ijc.24048. [DOI] [PubMed] [Google Scholar]
- Atillasoy ES, Seykora JT, Soballe PW, Elenitsas R, Nesbit M, Elder DE, Montone KT, Sauter E, Herlyn M. UVB induces atypical melanocytic lesions and melanoma in human skin. Am. J. Pathol. 1998;152:1179–1186. [PMC free article] [PubMed] [Google Scholar]
- Balbin M, Fueyo A, Tester AM, Pendas AM, Pitiot AS, Astudillo A, Overall CM, Shapiro SD, Lopez-Otin C. Loss of collagenase-2 confers increased skin tumor susceptibility to male mice. Nat. Genet. 2003;35:252–257. doi: 10.1038/ng1249. [DOI] [PubMed] [Google Scholar]
- Ball NJ, Yohn JJ, Morelli JG, Norris DA, Golitz LE, Hoeffler JP. Ras mutations in human melanoma: a marker of malignant progression. J. Invest. Dermatol. 1994;102:285–290. doi: 10.1111/1523-1747.ep12371783. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Bastian BC, Hezel A, Pinkel D, Depinho RA, Chin L. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol. Cell. Biol. 2001;21:2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian BC. The longer your telomeres, the larger your nevus? Am. J. Dermatopathol. 2003;25:83–84. [PubMed] [Google Scholar]
- Beadling C, Jacobson-Dunlop E, Hodi FS, et al. KIT gene mutations and copy number in melanoma subtypes. Clin. Cancer Res. 2008;14:6821–6828. doi: 10.1158/1078-0432.CCR-08-0575. [DOI] [PubMed] [Google Scholar]
- Berking C, Takemoto R, Binder RL, Hartman SM, Ruiter DJ, Gallagher PM, Lessin SR, Herlyn M. Photocarcinogenesis in human adult skin grafts. Carcinogenesis. 2002;23:181–187. doi: 10.1093/carcin/23.1.181. [DOI] [PubMed] [Google Scholar]
- Bertolotto C, Lesueur F, Giuliano S, et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat. Rev. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- Carvajal RD, Antonescu CR, Wolchok JD, et al. KIT as a therapeutic target in metastatic melanoma. JAMA. 2011;305:2327–2334. doi: 10.1001/jama.2011.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Downward J. RAS interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011;2:261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L. The genetics of malignant melanoma: lessons from mouse and man. Nat. Rev. Cancer. 2003;3:559–570. doi: 10.1038/nrc1145. [DOI] [PubMed] [Google Scholar]
- Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner JW, II, Depinho RA. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- Choi KY, Chang K, Pickel JM, Badger JD, II, Roche KW. Expression of the metabotropic glutamate receptor 5 (mGluR5) induces melanoma in transgenic mice. Proc. Natl Acad. Sci. U.S.A. 2011;108:15219–15224. doi: 10.1073/pnas.1107304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- Cronin JC, Wunderlich J, Loftus SK, et al. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;22:435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz F, III, Rubin BP, Wilson D, Town A, Schroeder A, Haley A, Bainbridge T, Heinrich MC, Corless CL. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003;63:5761–5766. [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, Lazar AJ, Gershenwald JE, Mills GB. A novel AKT3 mutation in melanoma tumours and cell lines. Br. J. Cancer. 2008;99:1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debniak T, Jakubowska A, Serrano-Fernandez P, Kurzawski G, Cybulski C, Chauhan SR, Laxton RC, Maleszka R, Lubinski J, Ye S. Association of MMP8 gene variation with an increased risk of malignant melanoma. Melanoma Res. 2011;21:464–468. doi: 10.1097/CMR.0b013e3283485fdd. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donawho CK, Kripke ML. Evidence that the local effect of ultraviolet radiation on the growth of murine melanomas is immunologically mediated. Cancer Res. 1991;51:4176–4181. [PubMed] [Google Scholar]
- Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J. Clin. Oncol. 2007;25:1606–1620. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- Fernandez-Pol JA, Douglas MG. Molecular interactions of cancer and age. Hematol. Oncol. Clin. North Am. 2000;14:25–44. doi: 10.1016/s0889-8588(05)70276-8. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije JM, Balbin M, Pendas AM, Sanchez LM, Puente XS, Lopez-Otin C. Matrix metalloproteinases and tumor progression. Adv. Exp. Med. Biol. 2003;532:91–107. doi: 10.1007/978-1-4615-0081-0_9. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Gilchrest BA, Eller MS, Geller AC, Yaar M. The pathogenesis of melanoma induced by ultraviolet radiation. N. Engl. J. Med. 1999;340:1341–1348. doi: 10.1056/NEJM199904293401707. [DOI] [PubMed] [Google Scholar]
- Goel VK, Lazar AJ, Warneke CL, Redston MS, Haluska FG. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J. Invest. Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- Goydos JS, Mann B, Kim HJ, Gabriel EM, Alsina J, Germino FJ, Shih W, Gorski DH. Detection of B-RAF and N-RAS mutations in human melanoma. J. Am. Coll. Surg. 2005;200:362–370. doi: 10.1016/j.jamcollsurg.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grichnik JM. Kit and melanocyte migration. J. Invest. Dermatol. 2006;126:945–947. doi: 10.1038/sj.jid.5700164. [DOI] [PubMed] [Google Scholar]
- Guo J, Si L, Kong Y, et al. Phase II, open-label, singlearm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J. Clin. Oncol. 2011;29:2904–2909. doi: 10.1200/JCO.2010.33.9275. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Fernandez A, Fueyo A, Folgueras AR, et al. Matrix metalloproteinase-8 functions as a metastasis suppressor through modulation of tumor cell adhesion and invasion. Cancer Res. 2008;68:2755–2763. doi: 10.1158/0008-5472.CAN-07-5154. [DOI] [PubMed] [Google Scholar]
- Halaban R, Zhang W, Bacchiocchi A, Cheng E, Parisi F, Ariyan S, Krauthammer M, Mccusker JP, Kluger Y, Sznol M. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res. 2010;23:190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handolias D, Salemi R, Murray W, Tan A, Liu W, Viros A, Dobrovic A, Kelly J, Mcarthur GA. Mutations in KIT occur at low frequency in melanomas arising from anatomical sites associated with chronic and intermittent sun exposure. Pigment Cell Melanoma Res. 2010;23:210–215. doi: 10.1111/j.1755-148X.2010.00671.x. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, Bowcock AM. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Hocker T, Tsao H. Ultraviolet radiation and melanoma: a systematic review and analysis of reported sequence variants. Hum. Mutat. 2007;28:578–588. doi: 10.1002/humu.20481. [DOI] [PubMed] [Google Scholar]
- Howell PM, Jr, Liu S, Ren S, Behlen C, Fodstad O, Riker AI. Epigenetics in human melanoma. Cancer Control. 2009;16:200–218. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- Jamal S, Schneider RJ. UV-induction of keratinocyte endothelin-1 downregulates E-cadherin in melanocytes and melanoma cells. J. Clin. Invest. 2002;110:443–452. doi: 10.1172/JCI13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jane-Valbuena J, Widlund HR, Perner S, et al. An oncogenic role for ETV1 in melanoma. Cancer Res. 2010;70:2075–2084. doi: 10.1158/0008-5472.CAN-09-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Noonan FP, Merlino G. Ultraviolet radiation and cutaneous malignant melanoma. Oncogene. 2003;22:3099–3112. doi: 10.1038/sj.onc.1206450. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, et al. DAXX / ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Chen WD, Parmigiani G, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. U.S.A. 2008a;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008b;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kairiyama C, Slavutsky I, Larripa I, Morvillo V, Bravo AI, Bover L, Podhajcer OL, Mordoh J. Biologic, immunocytochemical, and cytogenetic characterization of two new human melanoma cell lines: IIB-MEL-LES and IIB-MEL-IAN. Pigment Cell Res. 1995;8:121–131. doi: 10.1111/j.1600-0749.1995.tb00652.x. [DOI] [PubMed] [Google Scholar]
- Kalinec G, Nazarali AJ, Hermouet S, Xu N, Gutkind JS. Mutated alpha subunit of the Gq protein induces malignant transformation in NIH 3T3 cells. Mol. Cell. Biol. 1992;12:4687–4693. doi: 10.1128/mcb.12.10.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Zhang Y, Waugh A, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007;67:465–473. doi: 10.1158/0008-5472.CAN-06-1736. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colon cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Kohli M, Rago C, Lengauer C, Kinzler KW, Vogelstein B. Facile methods for generating human somatic cell gene knockouts using recombinant adeno-associated viruses. Nucleic Acids Res. 2004;32:e3. doi: 10.1093/nar/gnh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis CA, Masters SB, Spada A, Pace AM, Bourne HR, Vallar L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature. 1989;340:692–696. doi: 10.1038/340692a0. [DOI] [PubMed] [Google Scholar]
- Lee W, Jiang Z, Liu J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Wall BA, Wangari-Talbot J, Shin SS, Rosenberg SA, Chan JL, Namkoong J, Goydos JS, Chen S. Glutamatergic pathway targeting in melanoma; single agent and combinatorial therapies. Clin. Cancer Res. 2011;17:7080–7092. doi: 10.1158/1078-0432.CCR-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang W, Shao Y, et al. High-resolution melting analysis of ADAMTS18 methylation levels in gastric, colorectal and pancreatic cancers. Med. Oncol. 2010;27:998–1004. doi: 10.1007/s12032-009-9323-8. [DOI] [PubMed] [Google Scholar]
- Li M, Zhao H, Zhang X, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat. Rev. Cancer. 2007;7:800–808. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- Ludwig A, Rehberg S, Wegner M. Melanocyte-specific expression of dopachrome tautomerase is dependent on synergistic gene activation by the Sox10 and Mitf transcription factors. FEBS Lett. 2004;556:236–244. doi: 10.1016/s0014-5793(03)01446-7. [DOI] [PubMed] [Google Scholar]
- Mcdermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res. 2010;70:1625–1634. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehnert JM, Wen Y, Lee JH, Dudek L, Pruski-Clark L, Shih W, Chen S, Goydos JS. A phase II trial of riluzole, an antagonist of metabotropic glutamate receptor (GRM1) signaling, in advanced melanoma; 2011 ASCO Annual Meeting; 2011; abstr. 8557. [Google Scholar]
- Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsel G, Ortonne N, Bagot M, Bensussan A, Dumaz N. c-Kit mutants require hypoxia-inducible factor 1alpha to transform melanocytes. Oncogene. 2009;29:227–236. doi: 10.1038/onc.2009.320. [DOI] [PubMed] [Google Scholar]
- Montel V, Kleeman J, Agarwal D, Spinella D, Kawai K, Tarin D. Altered metastatic behavior of human breast cancer cells after experimental manipulation of matrix metalloproteinase 8 gene expression. Cancer Res. 2004;64:1687–1694. doi: 10.1158/0008-5472.can-03-2047. [DOI] [PubMed] [Google Scholar]
- Namkoong J, Shin SS, Lee HJ, Marin YE, Wall BA, Goydos JS, Chen S. Metabotropic glutamate receptor 1 and glutamate signaling in human melanoma. Cancer Res. 2007;67:2298–2305. doi: 10.1158/0008-5472.CAN-06-3665. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev SI, Rimoldi D, Iseli C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat. Genet. 2011;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- Oettgen P, Finger E, Sun Z, et al. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J. Biol. Chem. 2000;275:1216–1225. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- Ozaki M, Sasner M, Yano R, Lu HS, Buonanno A. Neuregulin-beta induces expression of an NMDA-receptor subunit. Nature. 1997;390:691–694. doi: 10.1038/37795. [DOI] [PubMed] [Google Scholar]
- Padua RA, Barrass N, Currie GA. A novel transforming gene in a human malignant melanoma cell line. Nature. 1984;311:671–673. doi: 10.1038/311671a0. [DOI] [PubMed] [Google Scholar]
- Palavalli LH, Prickett TD, Wunderlich JR, et al. Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nat. Genet. 2009;41:518–520. doi: 10.1038/ng.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Eng. J. Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmigiani G, Boca S, Lin J, Kinzler KW, Velculescu V, Vogelstein B. Design and analysis issues in genome-wide somatic mutation studies of cancer. Genomics. 2009;93:17–21. doi: 10.1016/j.ygeno.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung B, Sun J, Schepsky A, Steingrimsson E, Ronnstrand L. C-KIT signaling depends on microphthalmiaassociated transcription factor for effects on cell proliferation. PLoS ONE. 2011;6:e24064. doi: 10.1371/journal.pone.0024064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock PM, Cohen-Solal K, Sood R, et al. Melanoma mouse model implicates metabotropic glutamate signaling in melanocytic neoplasia. Nat. Genet. 2003;34:108–112. doi: 10.1038/ng1148. [DOI] [PubMed] [Google Scholar]
- Polsky D, Cordon-Cardo C. Oncogenes in melanoma. Oncogene. 2003;22:3087–3091. doi: 10.1038/sj.onc.1206449. [DOI] [PubMed] [Google Scholar]
- Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA, Samuels Y. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat. Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prickett TD, Wei X, Cardenas-Navia I, et al. Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat. Genet. 2011;43:1119–1126. doi: 10.1038/ng.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentmeier H, Osborn M, Reinhardt J, Zaborski M, Drexler HG. Immunocytochemical analysis of cell lines derived from solid tumors. J. Histochem. Cytochem. 2001;49:1369–1378. doi: 10.1177/002215540104901105. [DOI] [PubMed] [Google Scholar]
- Quinlan MP, Settleman J. Isoform-specific ras functions in development and cancer. Future Oncol. 2009;5:105–116. doi: 10.2217/14796694.5.1.105. [DOI] [PubMed] [Google Scholar]
- Rieff HI, Raetzman LT, Sapp DW, Yeh HH, Siegel RE, Corfas G. Neuregulin induces GABA(A) receptor subunit expression and neurite outgrowth in cerebellar granule cells. J. Neurosci. 1999;19:10757–10766. doi: 10.1523/JNEUROSCI.19-24-10757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera RS, Nagatsuka H, Gunduz M, Cengiz B, Gunduz E, Siar CH, Tsujigiwa H, Tamamura R, Han KN, Nagai N. C-kit protein expression correlated with activating mutations in KIT gene in oral mucosal melanoma. Virchows Arch. 2008;452:27–32. doi: 10.1007/s00428-007-0524-2. [DOI] [PubMed] [Google Scholar]
- Rivera S, Khrestchatisky M, Kaczmarek L, Rosenberg GA, Jaworski DM. Metzincin proteases and their inhibitors: foes or friends in nervous system physiology? J. Neurosci. 2010;30:15337–15357. doi: 10.1523/JNEUROSCI.3467-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks N, Paulissen G, El Hour M, Quesada F, Crahay C, Gueders M, Foidart JM, Noel A, Cataldo D. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90:369–379. doi: 10.1016/j.biochi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Shin SS, Namkoong J, Wall BA, Gleason R, Lee HJ, Chen S. Oncogenic activities of metabotropic glutamate receptor 1 (Grm1) in melanocyte transformation. Pigment Cell Melanoma Res. 2008;21:368–378. doi: 10.1111/j.1755-148X.2008.00452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigalotti L, Covre A, Fratta E, Parisi G, Colizzi F, Rizzo A, Danielli R, Nicolay HJ, Coral S, Maio M. Epigenetics of human cutaneous melanoma: setting the stage for new therapeutic strategies. J. Transl. Med. 2010;8:56. doi: 10.1186/1479-5876-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Kim JS, Cronin JC, et al. Mutational inactivation of PTPRD in glioblastoma multiforme and malignant melanoma. Cancer Res. 2008;68:10300–10306. doi: 10.1158/0008-5472.CAN-08-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- Stark MS, Woods SL, Gartside MG, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat. Genet. 2011;44:165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker W, Bode W. Structural features of a superfamily of zinc-endopeptidases: the metzincins. Curr. Opin. Struct. Biol. 1995;5:383–390. doi: 10.1016/0959-440x(95)80101-4. [DOI] [PubMed] [Google Scholar]
- Taylor KL, Lister JA, Zeng Z, Ishizaki H, Anderson C, Kelsh RN, Jackson IJ, Patton EE. Differentiated melanocyte cell division occurs in vivo and is promoted by mutations in Mitf. Development. 2011;138:3579–3589. doi: 10.1242/dev.064014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cabala CA, Wang WL, Trent J, et al. Correlation between KIT expression and KIT mutation in melanoma: a study of 173 cases with emphasis on the acral-lentiginous / mucosal type. Mod. Pathol. 2009;22:1446–1456. doi: 10.1038/modpathol.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN / MMAC1 inactivation in melanoma. J. Invest. Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworkoski K, Singhal G, Szpakowski S, Zito CI, Bacchiocchi A, Muthusamy V, Bosenberg M, Krauthammer M, Halaban R, Stern DF. Phosphoproteomic screen identifies potential therapeutic targets in melanoma. Mol. Cancer Res. 2011;9:801–812. doi: 10.1158/1541-7786.MCR-10-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van ’T Veer LJ, Burgering BM, Versteeg R, Boot AJ, Ruiter DJ, Osanto S, Schrier PI, Bos JL. N-ras mutations in human cutaneous melanoma from sun-exposed body sites. Mol. Cell. Biol. 1989;9:3114–3116. doi: 10.1128/mcb.9.7.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’brien JM, Simpson EM, Barsh GS, Bastian BC. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI / SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeriah S, Brennan C, Meng S, et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidwans SJ, Flaherty KT, Fisher DE, Tenenbaum JM, Travers MD, Shrager J. A melanoma molecular disease model. PLoS ONE. 2011;6:e18257. doi: 10.1371/journal.pone.0018257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viloria CG, Obaya AJ, Moncada-Pazos A, Llamazares M, Astudillo A, Capella G, Cal S, Lopez-Otin C. Genetic inactivation of ADAMTS15 metalloprotease in human colorectal cancer. Cancer Res. 2009;69:4926–4934. doi: 10.1158/0008-5472.CAN-08-4155. [DOI] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Prickett TD, Viloria CG, et al. Mutational and functional analysis reveals ADAMTS18 metalloproteinase as a novel driver in melanoma. Mol. Cancer Res. 2010;8:1513–1525. doi: 10.1158/1541-7786.MCR-10-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Moncada-Pazos A, Cal S, Soria-Valles C, Gartner J, Rudloff U, Lin JC, Rosenberg SA, Lopez-Otin C, Samuels Y. Analysis of the disintegrin-metalloproteinases family reveals ADAM29 and ADAM7 are often mutated in melanoma. Hum. Mutat. 2011a;32:E2148–E2175. doi: 10.1002/humu.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 2011b;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Woods SL, Boyle GM, et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo A, Yu J, et al. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao Y, Zhang X, Guda K, et al. Identification and functional characterization of paxillin as a target of protein tyrosine phosphatase receptor T. Proc. Natl Acad. Sci. U.S.A. 2010;107:2592–2597. doi: 10.1073/pnas.0914884107. [DOI] [PMC free article] [PubMed] [Google Scholar]