Abstract
Fused in sarcoma (FUS)-immunoreactive neuronal and glial inclusions define a novel molecular pathology called FUS proteinopathy. FUS has been shown to be a component of inclusions of familial amyotrophic lateral sclerosis with FUS mutation and three FTLD entities, including neuronal intermediate filament inclusion disease (NIFID). The pathogenic role of FUS is unknown. In addition to FUS, many neuronal cytoplasmic inclusions (NCI) of NIFID contain aggregates of α-internexin and neurofilament proteins. Herein, we have: (1) shown that FUS becomes relatively insoluble in NIFID and there are no post-translational modifications; (2) shown there are no pathogenic abnormalities in the FUS gene in NIFID; (3) performed an immunoelectron microscopy analysis of the precise localizations of FUS in NIFID, as this has not previously been described. FUS localized to euchromatin, and strongly with paraspeckles, in nuclei, consistent with its RNA/DNA-binding functions. NCI of varying morphologies were observed. Most frequent were the ‘loosely aggregated cytoplasmic inclusions’ (LACI), 81% of which had moderate or high levels of FUS-immunoreactivity. Much rarer ‘compact cytoplasmic inclusions’ (CCI) and ‘Tangled twine ball inclusions’ (TTBI) were FUS-immunoreactive at their granular peripheries, or heavily FUS-positive throughout, respectively. Thus FUS may aggregate in the cytoplasm and then admix with neuronal intermediate filament accumulations.
Keywords: Neuronal intermediate filament inclusion disease, frontotemporal lobar degeneration, FUS, neurofilament, α-internexin, immunoelectron microscopy
Introduction
Neuronal intermediate filament inclusion disease (NIFID) is a rare progressive neurological disease with a clinically heterogeneous phenotype including: frontotemporal dementia (FTD), upper motor neuron disease (MND), and extrapyramidal signs presenting, typically, at a young age (Bigio et al. 2003; Cairns et al. 2003; Josephs et al. 2003; Cairns et al. 2004a; Josephs et al. 2005; Molina-Porcel et al. 2008; Neumann et al. 2009b). Rare cases have an age at onset greater than 65 years (Molina-Porcel et al. 2008). Although the most characteristic clinical phenotype of NIFID is FTD, the clinical picture may embrace a diverse group of motor disorders and dementia: primary lateral sclerosis (upper motor neuron disease), parkinsonism, corticobasal degeneration, FTD, and combinations thereof (Bigio et al. 2003; Cairns et al. 2003; Josephs et al. 2003; Cairns et al. 2004a; Josephs et al. 2005; Molina-Porcel et al. 2008; Neumann et al. 2009b). Macroscopically, there is atrophy of the frontal and temporal lobes and often the basal ganglia are affected. Microscopically, the disease is characterized by neuronal loss and gliosis in affected areas and neuronal and glial inclusions containing fused in sarcoma (FUS) protein (Neumann et al. 2009b; Armstrong et al. 2010). In addition, there are neuronal cytoplasmic inclusions (NCI) and axonal inclusions (spheroids) that are filamentous and contain neuronal intermediate filament (IF) proteins. These latter inclusions, which are variably ubiquitinated, distinguish NIFID neuropathologically from other forms of frontotemporal lobar degeneration with FUS inclusions (FTLD-FUS) (Munoz et al. 2009; Neumann et al. 2009a and 2009b; Mackenzie et al. 2010). In about half of cases, there are neuronal intranuclear inclusions (NII), which contain epitopes of FUS and are variably ubiquitinated. The fine structure of FUS-immunoreactive neuronal inclusions of NIFID has not previously been described and the etiology of NIFID is unknown.
The FUS protein is a 526-amino acid DNA/RNA-binding protein like TAR DNA-binding protein of 43 kDa (TARDBP or TDP-43) (Buratti and Baralle 2001; Buratti et al. 2001; Kwiatkowski et al. 2009; Lagier-Tourenne and Cleveland 2009). FUS, like TDP-43, defines another molecular pathology also seen in FTLD (FTLD-FUS), but is uncommon, accounting for about 1 per cent of cases, or less, in most series (unpublished data). There are many similarities between TDP-43 and FUS: both bind RNA, through RNA recognition motifs (RRM), and both are involved in splicing and processing messenger RNA with only a few known targets (reviewed in Lagier-Tourenne and Cleveland 2009). As with TDP-43, FUS is normally a predominantly nuclear protein that is expressed at low levels in the cytoplasm. Similar to familial amyotrophic lateral sclerosis (FALS) with TARDBP mutation (Gitcho et al. 2008; Kabashi et al. 2008; Sreedharan et al. 2008; Van Deerlin et al. 2008; Benajiba et al. 2009), FALS with FUS mutation (Kwiatkowski et al. 2009; Vance et al. 2009) is characterized neuropathologically by cytoplasmic aggregates of FUS and nuclear to cytoplasmic translocation of FUS, similar to that seen in FTLD-TDP. The discovery of TARDBP mutations in FALS in about 5% of FALS established a primary genetic role for TARDBP in ALS (Gitcho et al. 2008; Kabashi et al. 2008; Van Deerlin et al. 2008; Benajiba et al. 2009). Subsequently, TARDBP variants have been reported in a few patients with FTLD-MND and FTLD in the absence of MND (Benajiba et al. 2009; Gitcho et al. 2009a). The observation that TARDBP gene defects may cause a spectrum of clinical and pathologic phenotypes (MND, FTLD-MND, and FTLD) led Van Langenhove and colleagues (2010) to investigate the genetic contribution of FUS in FTLD and FTLD-MND and they identified a missense mutation in one patient with FTLD that was predicted to be pathogenic. Though NIFID cases have no family history, these data indicate that mutations in FUS may result in a clinically heterogeneous phenotype including MND, FTLD-MND, and FTLD linked by a common molecular pathology called FUS proteinopathy. More recently, FUS has also been found in the inclusions of a number of rare neurologic disorders with overlapping features of FTLD and MND including: neuronal intermediate filament inclusion disease (NIFID) (Neumann et al. 2009b; Armstrong et al. 2010) basophilic inclusion body disease (BIBD) (Munoz et al. 2009), and atypical FTLD with ubiquitin inclusions (aFTLD-U) (Neumann et al. 2009b). FUS is also a minor component of the inclusions of several other neurodegenerative diseases including Huntington’s disease and other polyglutamine diseases (Doi et al. 2010).
It is presently unknown whether the neuronal and glial inclusions of NIFID result from toxic or protective mechanisms, but the identification and the precise ultrastructural co-localization of the range of molecules which aggregate to form inclusions will help to elucidate the mechanisms of pathogenesis in NIFID. These mechanisms may illuminate pathogenesis in other neurodegenerative diseases with abnormal neuronal protein aggregates. Here we describe, for the first time, the ultrastructural localization of FUS protein within NIFID, with particular reference to the NCI.
Materials and methods
Histology and immunohistochemistry
After death, the consent of the next of kin was obtained for brain removal, following local Ethical Committee procedures (Human Studies Committee, Washington University School of Medicine) and the 1964 Declaration of Helsinki (as modified in Edinburgh, 2000) (Table 1). Brain tissue was preserved in buffered 10% formalin or 4% paraformaldehyde. For this study, tissue blocks were taken from the frontal cortex at the level of the genu of the corpus callosum to study the superior frontal cortex (SFC), and temporal lobe at the level of the lateral geniculate body. Tissue was fixed in 10% phosphate buffered formal-saline and embedded in paraffin wax. Following microwave pretreatment, immunohistochemistry (IHC) was performed on 6 - 8 μm sections with either anti-FUS rabbit polyclonal antibody (1:1,500; Sigma-Aldrich, St. Louis, MO) or A300-294A (1:1,500; Bethyl Laboratories, Montgomery, TX: both generated similar staining) and sections were counterstained with haematoxylin.
Table 1.
Summary of demographic information of cases
| Case | Sex | Age at onset (years) |
Duration (years) |
Age at death (years) |
Brain weight (g) |
PMI (hours) |
References |
|---|---|---|---|---|---|---|---|
| NIFID.1 | F | 41 | 3 | 41 | 904 | 15 | [Armstrong et al. 2010; Cairns et al. 2004a; Cairns et al. 2004c] |
| NIFID.2 | M | 48 | 4 | 52 | 1,310 | ~24 | [Armstrong et al. 2010; Cairns et al. 2004a; Cairns et al. 2004c] |
| NIFID.3 | F | 52 | 2.7 | 54 | 813 | ~24 | [Armstrong et al. 2010; Cairns et al. 2004a; Cairns et al. 2004c] |
| NIFID.4 | M | 56 | 4 | 60 | 1,250 | ~24 | [Armstrong et al. 2010; Bigio et al. 2003; Cairns et al. 2004a; Cairns et al. 2004c] |
| NIFID.5 | F | 70 | 5 | 75 | 1,040 | ~24 | [Mackenzie and Feldman 2004] |
| Mean (range) | - | 53 (41-70) | 4 (2.7-5) | 56 (41-75) | 1,063 (813-1,250) | 22 (15-24) | - |
| NL.1 | F | - | - | 67 | 1,100 | 5.5 | - |
| NL.2 | M | - | - | 60 | 1,500 | 14 | - |
| NL.3 | F | - | - | 36 | 1,280 | 6 | - |
| NL.4 | M | - | - | 65 | 1,346 | 26 | - |
| Mean (range) | - | - | - | 57 (36-67) | 1,307 (1,100-1,500) | 52 (5.5-26) | - |
Abbreviations: neuronal intermediate filament inclusion disease (NIFID) and neuropathologically normal (NL) cases used in this study; PMI, post-mortem interval; M, male; F, female
Antigen retrieval of the paraffin wax-embedded tissue blocks was performed by heating sections in a solution of 0.5% ethylenediaminetetraacetic acid (EDTA) in 100 mmol/L TRIS, pH 7.6 at 100°C for 10 min. Immunohistochemistry (IHC) was undertaken on 6-10 μm thick sections prepared from formalin- (cases NIFID.1 to 4) or 4% paraformaldehyde- (NIFID.5) fixed, paraffin-embedded tissue blocks using the avidinbiotin complex detection system (Vector Laboratories, Burlingame, CA) and the chromagen 3,3′-diaminobenzidine (DAB); sections were counterstained with haematoxylin. Antibodies used included those that recognize ubiquitin, FUS, and epitopes of class IV neuronal IF proteins including phosphorylation-dependent anti-neuronal IF antibodies (Table 2). Anti-neuronal IF antibodies used in this study are well characterized and have been used previously to demonstrate epitopes of neuronal IF in NIFID (Cairns et al. 2004b).
Table 2.
Characteristics of the antibodies used in immunogold TEM
| Antigen | Poly/monoclonal | Source | Clone | Dilution (IHC-P) | Dilution (IEM) |
|---|---|---|---|---|---|
| FUS | Polyclonal | Sigma-Aldrich Inc., USA | - | 1:500 | 25μg/ml IgG |
| α-Internexin | Monoclonal | Cambridge Bioscience Ltd., UK | 2E3 | 1:400 | 5μg/ml IgG |
| Ubiquitin | Polyclonal | DakoCytomation, Denmark | - | 1:1,000 | 25μg/ml IgG |
| NEFH-phosphorylated | Monoclonal | Sternberger Monoclonals Inc., USA | SMI 31 | 1:1,000 | 1:100 |
| NEFH non-phosphorylated | Monoclonal | Sternberger Monoclonals Inc., USA | SMI 32 | 1:1,000 | 1:100 |
| Polyglutamine | Monoclonal | Chemicon International Inc., USA | 1C2 | 1:1,000 | 1:1,000 |
Abbreviations: FUS, fused in sarcoma; NEFH, neurofilament heavy polypeptide; IHC-P, immunohistochemistry using paraffin-embedded tissue; IEM, immunoelectron microscopy
Biochemistry
Fresh, snap-frozen brain tissue from neocortex was extracted and analyzed from five cases of NIFID, as previously described (Cairns et al. 2004a and 2004c; Mosaheb et al. 2005). Using an established protocol (modified), gray matter was separated from white matter and sequentially extracted with buffers of increasing strength at 2 mL/g initial wet tissue weight as follows: (1) Low-salt (LS) Tris-buffered saline (TBS; 10 mM Tris, pH 7.5, 5 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT) in 10% sucrose); (2) 1% Triton X-100 in LS-TBS, 0.5 mM NaCl; (3) Sarkosyl Buffer, LS-TBS with 1% N-lauryl-sarcosine and 0.5 mM NaCl buffer, (4) 2% SDS in 50 mM Tris, pH 7.6; and (5) Urea Buffer; 7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS), 30 mM Tris-HCl, pH 8.5. Each buffer was supplemented with a protease inhibitor cocktail (Mini Protease Inhibitor Cocktail Tablets, Roche Applied Science, Indianapolis, IN) (Gitcho et al. 2009b). Tissue was homogenized in 2 mL/g low-salt (LS) buffer and centrifuged at 25,000 x g for 30 minutes at 4°C. The supernatants were saved as the LS fraction (all extractions were repeated twice). Pellets were homogenized in 2 mL/g Triton-X (TX) buffer. Myelin precipitate was removed by suspending the pellet in Myelin Floatation Buffer (TX buffer with 30% sucrose) and centrifuged at 100,000 g for 30 minutes at 4°C. After centrifugation, the myelin layer was removed. The pellets were homogenized in 2 mL/g Sarkosyl buffer and incubated at 22°C on a shaker for 1 h before sedimentation at 180,000 g for 30 minutes at 22°C. The pellet was homogenized in 1 mL\g urea buffer and centrifuged at 25,000 g for 30 minutes at 22°C (Neumann et al. 2006). For Western blot analysis, polyvinylidene fluoride (PVDF) replicas were prepared from 4% stacking and 12% resolving SDS polyacrylamide slab gels and probed with rabbit polyclonal antibodies to FUS (C-terminal of FUS, A300-294A, 1:2,000, Bethyl Laboratories, Montgomery, TX). Primary antibodies were detected with horseradish peroxidase-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) and immunoreactive proteins were revealed using ECL chemiluminescence (ECL Advance Western Blotting Detection Kit, RPN2132, Piscataway, NJ).
DNA extraction and PCR
Genomic DNA was isolated from frozen brain tissue. Briefly, approximately 100 mg of frozen brain tissue was pulverized in liquid nitrogen. DNA was extracted using the Qiagen DNeasy Blood & Tissue Kit (Quiagen, Valencia, CA). Primers used were previously reported and conditions optimized for PCR (Kwiatkowski et al. 2009). The purified PCR products (QIAquick PCR Purification Kit, Qiagen, Valencia, CA) were directly sequenced using BigDye version 3.1 (Applied Biosystems, Carlsbad, CA) on an ABI Prism 3100 automated DNA sequencer (AME Bioscience A/S Toroed, Norway) with PCR primers. Sequencher 4.9 software (Gene Codes Corp., Ann Arbor, MI) was used for sequence analysis.
Tissue preparation for transmission electron microscopy
Frozen brain tissue stored at −70°C was brought to −20°C. The grey matter was identified, dissected, and samples placed directly into a cold (4°C) solution of 4% (w/v) formaldehyde and 0.1% (v/v) glutaraldehyde (vacuum-distilled) in phosphate-buffered saline (PBS). The following procedures were carried out at 4°C. After 18 h of fixation, the samples were rinsed thoroughly in PBS then dehydrated in an ethanol series and embedded in Unicryl resin (British BioCell International, Cardiff, UK) as previously described (Thorpe et al. 1999) and as used in our previous transmission electron microscope (TEM) study of NIFID (Mosaheb et al. 2005). Details of the antibodies and the dilutions used for immunogold labeling TEM studies are described in Table 2; for the TEM study, 5 and 10nm gold particle-conjugated goat anti-rabbit (GaR5, GaR10) and 10nm goat anti-mouse IgG (GaM10) secondary probes were obtained from British BioCell International (Cardiff, UK).
Immunogold labeling transmission electron microscopy
We used established methods to perform immunoelectron microscopy (Thorpe, 1999). Briefly, modified phosphate-buffered saline, pH 8.2 containing 1% BSA, 500μl/L Tween-20, 10mM NaEDTA and 0.2g/L NaN3 (henceforward termed PBS+) was used throughout all the following procedures for all dilutions of antibodies and gold probes. Thin sections were initially blocked in normal goat serum (1:10 in PBS+) for 30 min at ambient temperature, then either single-labeled with anti-FUS and anti-ubiquitin antibodies (in initial labeling experiments we used normal rabbit serum as a specificity control and all dilutions were performed at 25μg/ml IgG final concentration) or double-labeled with anti-FUS and SMI31 or SMI32 (both at 1:100) or α-internexin (INA) (5μg/ml IgG final concentration).
After 3 × 2 min PBS+ rinses, the sections were immunolabeled in GaR10 for the single labeling or, for the double-labeling, a mixture of GaR5/GaM10 secondary gold probes (all 1:10 in PBS+) for 1 h at ambient temperature. Sections were subsequently rinsed in PBS+ (3 × 10 min) and distilled water (4 × 5 min). All immunogold-labeled thin sections were subsequently post-stained in 2% (w/v) aqueous, 0.22μm-filtered uranyl acetate for 1h and examined in a Hitachi-7100 TEM at 100kV. The sections were systematically and thoroughly scanned for pathology and images acquired digitally with an axially-mounted (2K x 2K pixel) Gatan Ultrascan 1000 CCD camera (Gatan UK, Oxford, UK).
Results
Neuropathology
The histological changes in NIFID included the stereotypical lesions of frontotemporal lobar degeneration (FTLD): neuronal loss, microvacuolation, particularly in superficial laminae, and astrocytosis in temporal and frontal isocortex. However, the most striking feature of the NIFID brains was the presence of inclusions containing FUS (Fig. 1a to c). At the light microscope level, abnormal FUS-immunoreactivity was detected at four sites: neuronal cytoplasmic inclusions (NCI) (Fig. 1), neuronal intranuclear inclusions (NII), dystrophic neurites (DN), and in sparse glial cytoplasmic inclusions (GI) (Neumann et al. 2009b; Armstrong et al. 2010). As previously reported, NCI were present in neurons that lacked nuclear FUS-immunoreactivity, indicating a redistribution of the normally predominantly nuclear protein to abnormal cytoplasmic aggregates (Fig. 3a). In the cases examined, the NCI was the most frequent morphological type, whilst NII were rare in the sections available. Thus, for the TEM studies, we present data relating to NCI only; the infrequency of NII, DN and GI in thin sections precluded a sufficiently detailed analysis of these inclusions.
Fig. 1.
Fused in sarcoma (FUS)-immunoreactive neuronal cytoplasmic inclusions (NCI) in NIFID. Neuronal loss, gliosis, microvacuolation and FUS-immunoreactive neuronal cytoplasmic inclusions (NCI) are present in the superficial laminae of the parahippocampal gyrus (a). NCI in the dentate gyrus (b). Globose and skein-like NCI in motor neurons of the precentral gyrus; one motor neuron contains a compact cytoplasmic/hyaline conglomerate inclusion (CCI) with eccentric FUS-immunoreactivity (c). FUS immunohistochemistry (a-c); x400 (a, c); x200 (b).
Fig. 3.
FUS is present in the nucleus of unaffected cells (a-c). Light microscopy reveals normal nuclear staining (arrow) in the superficial layers of the parahippocampal cortex; in affected neurons there is an apparent translocation of FUS-immunoreactivity from the nucleus to the cytoplasm to form a compact neuronal cytoplasmic inclusion (double arrows). In some neurons, there is less intense immunoreactivity in the cytoplasm, possibly representing an intermediate form, or pre-inclusion (double arrowheads); in the same neuron there is a vermiform intranuclear inclusion (arrowhead). Within the neuropil, some FUS-immunoreactive grains are seen (asterisk). TEM reveals FUS immunogold labeling of nuclear structures (b, c). Epitopes of FUS are preferentially localized to euchromatin, rather than (the denser) heterochromatin (asterisk) regions, within the nucleus; the nuclear envelope is marked by an arrow (b). Paraspeckles (solid arrow), have the appearance of dense round areas in the nucleus and are strongly FUS-immunoreactive (c). The paraspeckles were, most often, located next to interchromatin granules (dashed arrow). FUS immunohistochemistry (a), FUS immunogold TEM (b, c). Scale bars 50 μm (a), 100 nm (b) and 200 nm (c)
Biochemistry
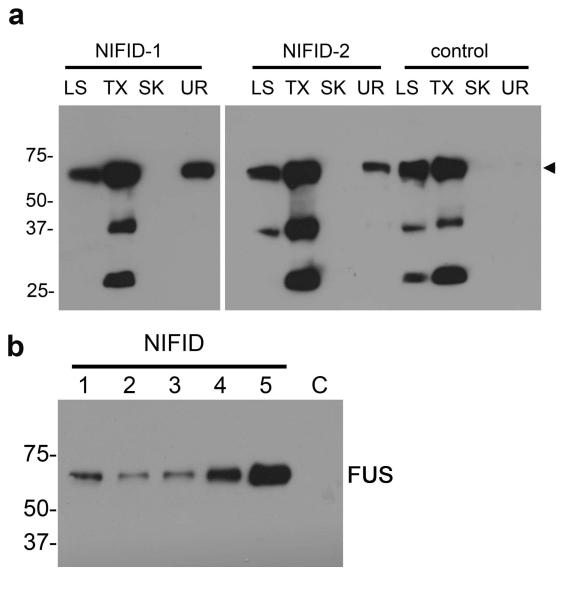
To determine if NIFID is a protein misfolding/aggregating disease we investigated the solubility of FUS protein using sequential fractionation methods. We extracted protein from gray matter of frontal cortex from patients with NIFID as well as age-matched controls. Immunoblotting showed variability in the amount of FUS detected in all brains analyzed; however, there was a consistent alteration in FUS solubility in NIFID (Fig. 2a). In addition, we detected only a single major protein band of approximately 73 kDa, consistent with the predicted molecular weight (Fig. 2a arrowhead). We did not identify higher molecular weight species in the soluble or urea fractions, but lower molecular weight species were seen in the soluble fractions, possibly indicating splice variants or cleavage products of the normal protein. As FUS was present in the urea-soluble fraction only in the NIFID cases (Fig. 2), further investigation is needed to examine the possible role of post-translational and structural modifications associated with FUS and, possibly, other urea-soluble proteins in NIFID.
Fig. 2.
FUS in NIFID is relatively insoluble. Gray matter from the frontal cortex of five NIFID cases (lanes 1 to 5) and a control patient (C) was sequentially extracted with buffers of increasing extraction strength. Each extract, corresponding to 20 mg of initial wet tissue weight, was resolved by SDS-PAGE and immunoblotted with antibodies to FUS. There is a consistent increase in insolubility of FUS in NIFID: normal FUS was present in the soluble fractions of NIFID and control (arrowhead), but insoluble FUS (urea fraction) was only present in NIFID cases and not the control (a). Urea-soluble FUS was present in all NIFID.1-5 cases but not in the control brain tissue (NL) (b).
Molecular genetics
In this study, we sequenced the complete coding region and the flanking intronic regions of FUS. A single nucleotide polymorphism (SNP) was detected at position 4327 in two cases (C/T genotype, SNP: rs1052352). However, this SNP has also been reported in normal controls. We did not detect any novel polymorphisms in the FUS gene.
Immunogold labeling transmission electron microscopy
Neuronal nuclear localization of FUS
Within normal neuronal nuclei, FUS-immunogold particles were preferentially localized to euchromatin regions (Fig. 3b). Additionally, there was a strong association of FUS-immunogold particles with discrete, round subnuclear structures, called ‘paraspeckles’ (Bond and Fox, 2009) (Fig. 3c, solid arrow). These paraspeckles were, most often, located next to interchromatin granules (Fig. 3c, dashed arrow). The presence of FUS in nuclear structures in unaffected cells is consistent with its normal DNA/RNA-processing function. Initial specificity control labeling with normal rabbit serum at matched IgG concentration to that used for the anti-FUS antibody generated acceptably very minimal labeling (data not shown).
Neuronal cytoplasmic inclusions
The number of neuronal cytoplasmic FUS-immunogold particles was, generally, very low in unaffected neurons of NIFID (data not shown). In affected (NCI-bearing) neurons, FUS-labeling was strongly and specifically associated with certain inclusions, of which three major morphological subtypes were identified: (1) ‘Loosely Aggregated Cytoplasmic Inclusion’ (LACI), (2) ‘Compact Cytoplasmic Inclusion’ (CCI), consistent with the morphological subtypes previously described (Mosaheb et al. 2005), and (3) ‘Tangled-Twine-Ball Inclusion’ (TTBI).
The LACI was the most common type of inclusion, as was previously reported in our ultrastructural studies of the NCI with neuronal IF proteins in NIFID (Cairns et al. 2004a; Mosaheb et al. 2005). In thin sections, LACI appeared predominantly as round, oval (Fig. 4a), or elongated (Fig. 4b) inclusions. Discrete inclusions were adjacent to the nucleus while others were annular. None of these inclusions had a limiting membrane around the border of the inclusion. FUS labeling of the LACI was prominent in granular regions (Fig. 4d), while filamentous regions had much lower levels of immunoreactivity. Visual inspection and semi-quantitative assessment of all LACI (n = 88) revealed that 81% had moderate or high levels of immunogold labeling. In the LACI, α-internexin and phosphorylated neurofilament immunolabeling was low, as we have previously described (Mosaheb et al. 2005). INA and phosphorylated NEFH epitopes did not, in most inclusions, co-localize with FUS. Conversely, ubiquitin labeling of the LACI was high as previously reported (Mosaheb et al. 2005) and was localized, largely, to areas of FUS-immunogold labeling.
Fig. 4.
Ultrastructural analysis of the NCI in NIFID. Neuronal cytoplasmic inclusions have varying morphologies: a round, neuronal, loosely aggregated cytoplasmic inclusion (LACI) (arrow) adjacent to the nucleus (a), a crescentic LACI (arrow) (b), and annular filaments of a LACI (arrows) surrounding the neuronal nucleus (c). FUS labeling is preferentially over granular structures within the LACI (d). FUS immunogold TEM. Scale bars = 2 μm (a-c), 200 nm (d)
The CCI, corresponding to the hyaline conglomerate inclusion identified by light microscopy (Fig. 1c) (Bigio et al. 2003; Cairns et al. 2004a) and that we have previously shown to contain neuronal IF proteins (Mosaheb et al. 2005), was much less frequent than LACI. They were usually round or oval in shape and contained more densely packed parallel filaments than was seen in the LACI (Fig. 5a and 6a [CCI]). Some of these CCI had transitional zones in which the filaments were orientated in different directions. Typically, surrounding the CCI was a layer of more dense and more granular material (Fig. 5a and 5b, arrows). The dense core of the CCI was not, or only modestly, FUS-immunoreactive (Fig. 5b and 5c, regions with asterisks), corresponding to the FUS-negative regions of CCI seen by IHC (Fig. 1c). Eccentric or surrounding areas were moderately FUS-immunolabeled (Fig. 5b, arrows [single-labeled for FUS] and Fig. 5c [serial section of same CCI double-labeled for FUS and phosphorylated neurofilament). Previously, we showed that the core of the CCI contains variable amounts of INA, ubiquitin and phosphorylated NEFH (Mosaheb et al. 2005).
Fig. 5.
Ultrastructural analysis of the compact cytoplasmic inclusion (CCI) in NIFID. A low magnification electron micrograph showing one CCI (asterisks) (a). The core of the CCI (asterisk) shows no, or minimal, FUS-immunoreactivity (single antigen labeling), whereas the peripheral, more granular, regions (arrows) are moderately labeled by anti-FUS antibodies (b). Serial section of the same CCI as in (b) showing a zone of phosphorylated neurofilament immunoreactivity (asterisks) and a peripheral zone of FUS-immunoreactivity (circles) and a transitional zone double-labeled for FUS and phosphorylated NEFH (circle with white asterisk). FUS (5nm gold particles) and phosphorylated NEFH (10nm gold particles) double-labeled immunogold TEM. Scale bars 2 μm (a, b) and 200 nm (c)
Fig. 6.
Filaments of NCI in NIFID contain FUS protein. NCI with the appearance of tangled twine balls (TTBI) reveal a dense central core with radiating bundles of filaments (a, b). Immunoelectron double-labeling with FUS and phosphorylated NEFH reveals that the thick radiating filament bundles of the TTBI are strongly FUS-immunolabeled (5nm gold particles within circles) and there is moderate labeling with anti-phosphorylated NEFH antibodies (10nm gold particles) (c). In addition to containing FUS and phosphorylated NEFH, TTBI contain epitopes of ubiquitin; the thick radiating filament bundles showed the most intense labeling (d). FUS, phosphorylated NEFH, and ubiquitin immunogold TEM. Scale bars 2 μm (a, b) and 200 nm (c, d)
The tangled twine ball inclusion (TTBI), like the CCI, was much less numerous than LACI (Mosaheb et al. 2005). These TTBI were first found and described by Mosaheb (2008 thesis, unpublished data) and are composed of a dense, central, tightly tangled ball of filaments with invading/radiating filaments, resembling loose ends which have escaped a tangled twine ball. Moderate levels of FUS labeling were identified in the central dense region and high levels in the peripheral filament bundles (Fig. 6c). Epitopes of phosphorylated NEFH were present to a moderate degree (Fig. 6c) and more numerous epitopes of ubiquitin were seen (Fig. 6d). Labeling of FUS, neuronal IF, and ubiquitin was localized to similar regions of the TTBI indicating proximate co-localization of these proteins in the TTBI of NIFID. FUS, neuronal IF, and ubiquitin immuno-labeling of filamentous structures in NIFID is summarized in Table 3.
Table 3.
Immunolabelling and filament dimensions of three types of neuronal cytoplasmic inclusions in NIFID
| Antigen | Neuronal cytoplasmic inclusion | ||
|---|---|---|---|
| LACI | TTBI | CCI | |
| FUS | ++ | +++ | − |
| α-Internexin* | +/− | − | (+++) |
| Phosphorylated NEFH* | + | ++ | (+++) |
| Ubiquitin* | +++ | +++ | (++) |
| Polyglutamine* | (−) | (−) | (−) |
| Filament width mean ± s.e.m. (range) |
8.9 ± 0.2 (5.9-15.0) nm | 9.6 ± 0.5 (5.6-12.4) nm | 10.1 ± 0.7 (6.1-14.2) nm |
Abbreviations: LACI, loosely aggregated cytoplasmic inclusion; TTBI, ‘tangled twine ball inclusion; CCI, compact cytoplasmic inclusion; FUS, fused in sarcoma; NEFH, neurofilament heavy polypeptide. Semi-quantitative assessment of the density of immunogold particles/filament: +++ high; ++ moderate; + low; +/− negligible; − unlabelled.
Data previously reported (Mosaheb et al. 2005) are included here (in parentheses) for comparison with FUS immunoelectron microscopy
Discussion
FUS has recently been added to the list of protein misfolding diseases which are characterized by relatively insoluble aggregates which form either within neurons, glial cells, or both. FUS was first identified as the pathological protein in about 5% of cases of familial ALS with FUS gene mutation (Kwiatkowski et al. 2009; Vance et al. 2009). So far, only one FUS variant has been reported in a case of FTD (Van Langenhove et al. 2010). Subsequently, it has been found in the inclusions of a diverse group of FTLD entities including: NIFID, BIBD, and atypical FTLD with ubiquitin-immunoreactive inclusions (Munoz et al. 2009; Neumann et al. 2009 and 2009b; Armstrong et al. 2010). Together, this group of diseases is linked by a common molecular pathology called FUS proteinopathy. The pathological signature of FUS in FTLD-FUS remains to be determined, but preliminary data indicate that FUS becomes relatively insoluble in aFTLD-U, a similarity shared with other protein misfolding diseases including: tauopathy, synucleinopathy, and prionopathy (Forman et al. 2004; Winklhofer et al. 2008). The specificity of FUS within FTLD is uncertain as FUS is also a component of the neuronal intranuclear inclusions of Huntington’s disease, spinocerebellar ataxias 1 and 3, and neuronal intranuclear inclusion body disease (Woulfe et al. 2010).
FUS protein is an RNA/DNA binding protein that is localized predominantly to the nucleus (Andersson et al. 2008). It is involved in many diverse cellular processes, including cell proliferation (Bertrand et al. 1999), DNA repair (Baechtold et al. 1999) transcriptional regulation and RNA splicing (Yang et al. 1998), and nucleo-cytoplasmic shuttling (Zinszner et al. 1997). It has been suggested that the non-coding RNAs (ncRNAs) present in paraspeckles may act as a ligand for FUS causing an allosteric change resulting in its release from an inactive conformation (Wang et al. 2008). The accumulation of FUS in paraspeckles may indicate a partial loss of function. The observation that FUS co-localizes with paraspeckles indicates a specific function(s) for this protein distinct from those of TDP-43 and deserves further investigation.
We have extended these observations and speculations by (1) undertaking DNA sequencing of the FUS gene in NIFID, (2) determining the relative solubility of FUS protein by biochemical sequential fractionation methods, and (3) performing immunogold electron microscopic studies of the inclusions to elucidate their fine structure in order to illuminate their evolution, as far as this is possible in a static section of brain tissue. Specifically, we have used immunogold labeling TEM to localize FUS protein within unaffected neurons and within the various morphological subtypes of NCI seen in NIFID (LACI, CCI, and the TTBI) (Mosaheb et al. 2005). In this TEM study, we focused on FUS-immunogold labeling and undertook double-labeling of FUS and phosphorylated NEFH and FUS and INA, together with (single) ubiquitin labeling of serial sections and compared these data with our previous ultrastructural studies of these inclusions.
In unaffected NIFID neurons, nuclear FUS immunolabeling was preferentially associated with regions of euchromatin, which is consistent with its role in transcription, and similar to our previous ultrastructural study of nuclear TDP-43 in normal brain neurons (Thorpe et al. 2008). A novel observation of this study is that FUS is strongly associated with subnuclear structures called paraspeckles in adult human brain (Bond and Fox, 2009). These structures are only found in mammalian nuclei in cells which have undergone differentiation (Fox et al. 2002); they are known to contain proteins involved in transcription and RNA processing, with active RNA polymerase II and newly transcribed RNA present in their structures (Xie et al. 2006). Paraspeckles are thought to regulate gene expression by the retention of RNA (Zhang and Carmichael, 2001) and they and their functions are very dynamic (Bond and Fox, 2009). For example, a virus-infected cell increases paraspeckle diameter either as a result of viral processing or through activation of cell defense mechanisms (Zolotukhin et al. 2003). Specific labeling of FUS within paraspeckles correlates with its well-described functions as an RNA/DNA binding protein (Crozat et al. 1993; Perrotti et al. 1998) involved in transcription regulation, RNA splicing (Yang et al. 1998), and nucleo-cytoplasmic shuttling (Zinszner et al. 1997). The observation that FUS co-localizes with paraspeckles indicates a specific function(s) for this protein distinct from those of TDP-43 and deserves further scrutiny.
Initial observations at the light microscope level show that NCI-bearing neurons exhibit a lack of nuclear FUS staining (Figs. 1 and 3); this redistribution of a usually predominantly nuclear protein to the cytoplasm in affected neurons is similar to our previous findings, at the ultrastructural level, in regard to TDP-43 in FTLD with TDP-43 proteinopathy (Thorpe et al. 2008) and to Pin1 protein in a range of FTLD entities with tauopathy and Alzheimer’s disease (Thorpe et al. 2004). Thus, the mis-localization of important regulators of nuclear function appears to be a common feature in a spectrum of neurodegenerative diseases and may lead to cell death by common pathogenic mechanisms leading to a toxic gain of function and/or loss of function.
With regards to FUS localization in affected NIFID neurons, this study provides further evidence that it is an important component of the pathological inclusions, supporting the initial evidence provided by Neumann et al. (2009b). Three types of NCI were identified, the most common being the LACI (Mosaheb et al. 2005); many were round or oval in shape (Pick body-like), though some were neurofibrillary tangle-like, crescentic, or annular, as previously described and more recently described using FUS IHC (Bigio et al. 2003; Josephs et al. 2003; Cairns et al. 2004a; Mackenzie and Feldman, 2004; Neumann et al. 2009b; Armstrong et al. 2010) at the light microscope level. Immunogold labeling with anti-phosphorylated neurofilament antibodies was low, α-internexin labeling was negligible, whilst ubiquitin epitopes in LACI were numerous both in this and our previous study (Mosaheb et al. 2005). Eighty-one percent of LACI were labeled, either moderately or heavily, with anti-FUS antibodies, indicating an important role for FUS in the pathogenesis of NIFID. The much rarer TTBI and the CCI/hyaline conglomerate inclusion had variable proportions of molecular components. There was more abundant phosphorylated NEFH in TTBI, suggesting a potentially more significant role for this protein in this form of inclusion compared to a LACI. The distribution of FUS labeling around the granular peripheries of CCI/hyaline conglomerate inclusions suggests an evolution in the aggregation of proteins including neuronal IF. Previously, we showed very high levels of INA labeling in the CCI from the same NIFID cases as used in the present study (Mosaheb et al. 2005). Reproducibility between studies was confirmed by comparing the mean width of filaments containing INA (mean filament width: 10.1 nm) of this study with that calculated from our original study by a different observer (mean filament width: 9.9 nm) of our previous study (Mosaheb et al. 2005).
Previously, we used a candidate gene approach to identify potential gene defects in several neuronal IF genes and found none in NIFID (Momeni et al. 2006). Similarly, in this study, in agreement with Neumann et al. (2009b), we found no pathogenic mutations in the FUS gene. Interestingly, a transgenic mouse model with a deletion in gigaxonin (GAN), a cytoskeletal regulating protein, and the genetic cause of giant axonal neuropathy developed a phenotype reminiscent of the INA-immunoreactive inclusions of NIFID. Although the NCIs of NIFID and GANDex1/Dex1 mice were immunohistochemically similar, no GAN variant was identified in DNA obtained from well-characterized cases of NIFID (Dequen et al. 2009). To date, no pathogenic gene defect has been reported in NIFID.
Using post-mortem brain tissue, it is a challenge to determine the pathogenic chronology of the disease and in particular the formation and evolution of the inclusions. The apparent alternatives are that either the three forms of NCI reported here and previously (LACI, CCI, and TTBI; Bigio et al. 2003; Mosaheb et al. 2005; Uchikado et al. 2006) represent unique structures reflective of differing molecular pathogenic cascades operating within different neurons, or they represent different appearances of the same inclusion at different stages in its evolution towards maturity. This latter would be analogous to the proposed evolution of the ‘diffuse’ beta amyloid plaque into a ‘classical’ ring-with-core structure and, ultimately, a ‘burned-out’ plaque in the pathogenesis of AD. Previous studies (Neumann et al. 2009b; Armstrong et al. 2010), suggest that as there is often diffuse, but intense, staining of neuronal IF in the cytoplasm of neurons with FUS-immunoreactive NCI, an initial defect in FUS metabolism, either as a consequence of a reduction in normal function or toxic accumulation of aggregates, may cause a secondary abnormality in the processing and regulation of intermediate filament trafficking. This sequence of events might result from the malfunction of FUS as a transcriptional regulator. Alternatively, the apparent accumulation of neuronal IF in the cytoplasm may be a consequence of the formation and growth of the FUS-immunoreactive inclusion (Neumann et al. 2009b). The observation that some separate and distinct inclusions of neuronal IF and FUS can occur in the same neuron indicates that both of these processes may operate within a single neuron.
In this study, we examined 88 LACI using FUS immunogold TEM and make some very cautious suggestions regarding their possible evolution. We observed a small number of LACI with significant filamentous areas that were not labeled with anti-FUS antibodies, but which were labelled for α-internexin and neurofilament. However, the granular regions around these inclusions labeled moderately for FUS. Therefore, it may be that LACI evolve composed of intermediate filaments, with the subsequent accumulation of FUS protein as a downstream event. Previously, we demonstrated high levels of INA and neurofilament labeling in the central filamentous core of CCI in the same NIFID cases (Mosaheb et al. 2005). In this study, we have observed that the granular regions surrounding the CCI is FUS-immunoreactive (Fig. 5), similarly to those surrounding some LACI. Thus, the CCI may represent a precursor LACI. Granular FUS-immunoreactive material may be deposited early at the periphery of the filamentous INA- and neurofilament-containing inclusion (CCI) and then invade and admix with the inclusion to form a LACI. The mature inclusion (LACI) would eventually assume the morphology of a more loosely-aggregated filamentous form, concomitant with lowered levels of reactivity to INA and neurofilament proteins. This and the disaggregation of the inclusion may reflect a degree of protein degradation; the higher levels of ubiquitin within the LACI, compared with the CCI, support this hypothesis (see Table 3 for a summary of these labeling data).
In summary, we provide the first ultrastructural analysis of the association of FUS protein within NCI of NIFID. Allied to our previous data on neuronal IF proteins in NIFID, these data support an important role for FUS, alongside neuronal IF proteins, and ubiquitin in the molecular pathogenic cascade leading to NCI in NIFID. These unique inclusions of NIFID distinguish it from other FUS proteinopathies including ALS with FUS mutation, BIBD and aFTLD-U.
Acknowledgements
A preliminary report of these data was presented at the 7th International Conference on Frontotemporal Dementias 2010, Indianapolis, Indiana, USA. We thank the families of patients whose generosity made this research possible. We acknowledge the staff of the Charles F. and Joanne Knight Alzheimer’s Disease Research Center Neuropathology Laboratory, Departments of Neurology and Pathology & Immunology, Washington University School of Medicine, St. Louis, Missouri, USA, for technical assistance. Support for this work was provided by grants from the Wellcome Trust, UK, (GR066166AIA) to JRT and NJC, and the National Institute on Aging of the National Institutes of Health (P50 AG05681 and P01 AG03991), and the Friedman Award to NJC.
References
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong RA, Gearing M, Bigio EH, Cruz-Sanchez FF, Duyckaerts C, Mackenzie IR, Perry RH, Skullerud K, Yokoo H, Cairns NJ. The spectrum and severity of FUS-immunoreactive inclusions in the frontal and temporal lobes of ten cases of neuronal intermediate filament inclusion disease. Acta Neuropathol. 2010;121:219–228. doi: 10.1007/s00401-010-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechtold H, Kuroda M, Sok J, Ron D, Lopez BS, Akhmedov AT. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- Benajiba L, Le Ber I, Camuzat A, Lacoste M, Thomas-Anterion C, Couratier P, Legallic S, Salachas F, Hannequin D, Decousus M, Lacomblez L, Guedj E, Golfier V, Camu W, Dubois B, Campion D, Meininger V, Brice A. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- Bertrand P, Akhmedov AT, Delacote F, Durrbach A, Lopez BS. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene. 1999;18:4515–4521. doi: 10.1038/sj.onc.1203048. [DOI] [PubMed] [Google Scholar]
- Bigio EH, Lipton AM, White CL, III, Dickson DW, Hirano A. Frontotemporal and motor neurone degeneration with neurofilament inclusion bodies: additional evidence for overlap between FTD and ALS. Neuropathol Appl Neurobiol. 2003;29:239–253. doi: 10.1046/j.1365-2990.2003.00466.x. [DOI] [PubMed] [Google Scholar]
- Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Grossman M, Arnold SE, Burn DJ, Jaros E, Perry RH, Duyckaerts C, Stankoff B, Pillon B, Skullerud K, Cruz-Sanchez FF, Bigio EH, Mackenzie IR, Gearing M, Juncos JL, Glass JD, Yokoo H, Nakazato Y, Mosaheb S, Thorpe JR, Uryu K, Lee VM, Trojanowski JQ. Clinical and neuropathologic variation in neuronal intermediate filament inclusion disease. Neurology. 2004a;63:1376–1384. doi: 10.1212/01.wnl.0000139809.16817.dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Perry RH, Jaros E, Burn D, McKeith IG, Lowe JS, Holton J, Rossor MN, Skullerud K, Duyckaerts C, Cruz-Sanchez FF, Lantos PL. Patients with a novel neurofilamentopathy: dementia with neurofilament inclusions. Neurosci Lett. 2003;341:177–180. doi: 10.1016/s0304-3940(03)00100-9. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Uryu K, Bigio EH, Mackenzie IR, Gearing M, Duyckaerts C, Yokoo H, Nakazato Y, Jaros E, Perry RH, Arnold SE, Lee VM, Trojanowski JQ. α-Internexin aggregates are abundant in neuronal intermediate filament inclusion disease (NIFID) but rare in other neurodegenerative diseases. Acta Neuropathol (Berl) 2004b;108:213–223. doi: 10.1007/s00401-004-0882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Zhukareva V, Uryu K, Zhang B, Bigio E, Mackenzie IR, Gearing M, Duyckaerts C, Yokoo H, Nakazato Y, Jaros E, Perry RH, Lee VM, Trojanowski JQ. α-Internexin is present in the pathological inclusions of neuronal intermediate filament inclusion disease. Am J Pathol. 2004c;164:2153–2161. doi: 10.1016/s0002-9440(10)63773-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- Dequen F, Cairns NJ, Bigio EH, Julien JP. Gigaxonin mutation analysis in patients with NIFID. 2009. doi:10.1016/j.neurobiolaging.2009.08.018. [DOI] [PMC free article] [PubMed]
- Doi H, Koyano S, Suzuki Y, Nukina N, Kuroiwa Y. The RNA-binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neurosci Res. 2010;66:131–133. doi: 10.1016/j.neures.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, Levitch D, Hatanpaa KJ, White CL, III, Bigio EH, Caselli R, Baker M, Al Lozi MT, Morris JC, Pestronk A, Rademakers R, Goate AM, Cairns NJ. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Bigio EH, Mishra M, Johnson N, Weintraub S, Mesulam M, Rademakers R, Chakraverty S, Cruchaga C, Morris JC, Goate AM, Cairns NJ. TARDBP 3′-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 2009a;118:633–645. doi: 10.1007/s00401-009-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitcho MA, Strider J, Carter D, Taylor-Reinwald L, Forman MS, Goate AM, Cairns NJ. VCP mutations causing frontotemporal lobar degeneration disrupt localization of TDP-43 and induce cell death. J Biol Chem. 2009b;284:12384–12398. doi: 10.1074/jbc.M900992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephs KA, Holton JL, Rossor MN, Braendgaard H, Ozawa T, Fox NC, Petersen RC, Pearl GS, Ganguly M, Rosa P, Laursen H, Parisi JE, Waldemar G, Quinn NP, Dickson DW, Revesz T. Neurofilament inclusion body disease: a new proteinopathy? Brain. 2003;126:2291–2303. doi: 10.1093/brain/awg231. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Uchikado H, McComb RD, Bashir R, Wszolek Z, Swanson J, Matsumoto J, Shaw G, Dickson DW. Extending the clinicopathological spectrum of neurofilament inclusion disease. Acta Neuropathol. 2005;109:427–32. doi: 10.1007/s00401-004-0974-4. [DOI] [PubMed] [Google Scholar]
- Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, Vande VC, Bouchard JP, Lacomblez L, Pochigaeva K, Salachas F, Pradat PF, Camu W, Meininger V, Dupre N, Rouleau GA. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, Valdmanis P, Rouleau GA, Hosler BA, Cortelli P, de Jong PJ, Yoshinaga Y, Haines JL, Pericak-Vance MA, Yan J, Ticozzi N, Siddique T, McKenna-Yasek D, Sapp PC, Horvitz HR, Landers JE, Brown RH., Jr Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Feldman H. Neurofilament inclusion body disease with early onset frontotemporal dementia and primary lateral sclerosis. Clin Neuropathol. 2004;23:183–193. [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, Ince PG, Kamphorst W, Revesz T, Rozemuller AJ, Kumar-Singh S, Akiyama H, Baborie A, Spina S, Dickson DW, Trojanowski JQ, Mann DM. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119:1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Porcel L, Llado A, Rey MJ, Molinuevo JL, Martinez-Lage M, Esteve FX, Ferrer I, Tolosa E, Blesa R. Clinical and pathological heterogeneity of neuronal intermediate filament inclusion disease. Arch Neurol. 2008;65:272–275. doi: 10.1001/archneurol.2007.37. [DOI] [PubMed] [Google Scholar]
- Momeni P, Cairns NJ, Perry RH, Bigio EH, Gearing M, Singleton AB, Hardy J. Mutation analysis of patients with neuronal intermediate filament inclusion disease (NIFID) Neurobiol Aging. 2006;27:778–778. doi: 10.1016/j.neurobiolaging.2005.03.030. [DOI] [PubMed] [Google Scholar]
- Mosaheb S, Thorpe JR, Hashemzadeh-Bonehi L, Bigio EH, Gearing M, Cairns NJ. Neuronal intranuclear inclusions are ultrastructurally and immunologically distinct from cytoplasmic inclusions of neuronal intermediate filament inclusion disease. Acta Neuropathol (Berl) 2005;110:360–368. doi: 10.1007/s00401-005-1057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz DG, Neumann M, Kusaka H, Yokota O, Ishihara K, Terada S, Kuroda S, Mackenzie IR. FUS pathology in basophilic inclusion body disease. Acta Neuropathol. 2009;118:629–631. doi: 10.1007/s00401-009-0598-9. [DOI] [PubMed] [Google Scholar]
- Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009a;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Roeber S, Kretzschmar HA, Rademakers R, Baker M, Mackenzie IRA. Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol. 2009b;118:605–616. doi: 10.1007/s00401-009-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Lozzo RV, Cooper DR, Calabretta B. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de Belleroche J, Mitchell JD, Leigh PN, Al Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe JR. The application of LR gold resin for immunogold labeling. Methods Mol Biol. 1999;117:99–110. doi: 10.1385/1-59259-201-5:99. [DOI] [PubMed] [Google Scholar]
- Thorpe JR, Mosaheb S, Hashemzadeh-Bonehi L, Cairns NJ, Kay JE, Morley SJ, Rulten SL. Shortfalls in the peptidyl-prolyl cis-trans isomerase protein Pin1 in neurons are associated with frontotemporal dementias. Neurobiol Dis. 2004;17:237–249. doi: 10.1016/j.nbd.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Thorpe JR, Rulten SL, Kay JE. The binding of a putative and a known chaperone protein revealed by immunogold labelling transmission electron microscopy: A suggested use of chaperones as probes for the distribution of their target proteins. J Histochem Cytochem. 1999;47:1633–1640. doi: 10.1177/002215549904701215. [DOI] [PubMed] [Google Scholar]
- Thorpe JR, Tang H, Atherton J, Cairns NJ. Fine structural analysis of the neuronal inclusions of frontotemporal lobar degeneration with TDP-43 proteinopathy. J Neural Transm. 2008;115:1661–1671. doi: 10.1007/s00702-008-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikado H, Li A, Lin WL, Dickson DW. Heterogeneous inclusions in neurofilament inclusion disease. Neuropathology. 2006;26:417–21. doi: 10.1111/j.1440-1789.2006.00709.x. [DOI] [PubMed] [Google Scholar]
- Van Deerlin VM, Leverenz JB, Bekris LM, Bird TD, Yuan W, Elman LB, Clay D, Wood EM, Chen-Plotkin AS, Martinez-Lage M, Steinbart E, McCluskey L, Grossman M, Neumann M, Wu IL, Yang WS, Kalb R, Galasko DR, Montine TJ, Trojanowski JQ, Lee VM, Schellenberg GD, Yu CE. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Langenhove T, van der Zee J, Sleegers K, Engelborghs S, Vandenberghe R, Gijselinck I, Van den Broeck M, Mattheijssens M, Peeters K, De Deyn PP, Cruts M, Van Broeckhoven C. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–371. doi: 10.1212/WNL.0b013e3181ccc732. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al Saraj S, Al Chalabi A, Leigh PN, Blair IP, Nicholson G, de Belleroche J, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklhofer KF, Tatzelt J, Haass C. The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27:336–349. doi: 10.1038/sj.emboj.7601930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woulfe J, Gray DA, Mackenzie IR. FUS-immunoreactive intranuclear inclusions in neurodegenerative disease. Brain Pathol. 2010;20:589–597. doi: 10.1111/j.1750-3639.2009.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie SQ, Martin S, Guillot PV, Bentley DL, Pombo A. Splicing speckles are not reservoirs of RNA polymerase II, but contain an inactive form, phosphorylated on serine2 residues of the C-terminal domain. Mol Biol Cell. 2006;17:1723–1733. doi: 10.1091/mbc.E05-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Embree LJ, Tsai S, Hickstein DD. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem. 1998;273:27761–27764. doi: 10.1074/jbc.273.43.27761. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci. 1997;110(Pt 15):1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- Zolotukhin AS, Michalowski D, Bear J, Smulevitch SV, Traish AM, Peng R, Patton J, Shatsky IN, Felber BK. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol Cell Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]