Abstract
Abnormal regulation of tau phosphorylation and/or alternative splicing is associated with the development of a large (>20) group of neurodegenerative disorders collectively known as tauopathies, the most common being Alzheimer's disease. Despite intensive research, little is known about the molecular mechanisms that participate in the transcriptional and posttranscriptional regulation of endogenous tau, especially in neurons. Recently, we showed that mice lacking Dicer in the forebrain displayed progressive neurodegeneration accompanied by disease-like changes in tau phosphorylation and splicing. Dicer is a key enzyme in the biogenesis of microRNAs (miRNAs), small noncoding RNAs that function as part of the RNA-induced silencing complex (RISC) to repress gene expression at the posttranscriptional level. We identified miR-16 and miR-132 as putative endogenous modulators of neuronal tau phosphorylation and tau exon 10 splicing, respectively. Interestingly, these miRNAs have been implicated in cell survival and function, whereas changes in miR-16/132 levels correlate with tau pathology in human neurodegenerative disorders. Thus, understanding how miRNA networks influence tau metabolism and possibly other biological systems might provide important clues into the molecular causes of tauopathies, particularly the more common but less understood sporadic forms.
1. Introduction
The discovery of small noncoding miRNAs uncovered an intriguing additional level to the fine-tuning of the transcriptome and proteome. Since their discovery almost 20 years ago [1, 2], research has progressed considerably towards gaining a better understanding of the impact of the complex network of gene regulation by miRNAs on health and disease. This is well documented in the cancer field, for instance, where miRNAs are increasingly acknowledged as potential diagnostic and therapeutic agents [3, 4]. Like protein-coding genes, miRNA genes are transcribed from the genome mainly from RNA polymerase II (reviewed in [5]). To date, more than 1000 miRNA genes have been identified in humans and 750 in mice, some of which are specifically expressed in the brain [6–10]. In the cytoplasm, the endonuclease Dicer cleaves miRNA precursors (approximately 70 nucleotides in length) to generate mature (approximately 21 nucleotides in length) double-stranded RNAs. These are loaded as single-stranded RNAs into the RNA-induced silencing complex (RISC), composed of Argonaute (Ago) proteins, to negatively regulate gene expression, albeit some exceptions have been documented [11, 12]. This regulation is achieved through binding with imperfect complementarity mainly to the 3′ untranslated region (3′UTR) of target messenger RNAs (mRNAs), leading to translation inhibition or degradation. Both in vitro and in vivo studies have shown that alterations of a single miRNA (or miRNA family) could have profound effects on hundreds of target genes [13, 14], thus possibly implicating multiple biological pathways.
Abnormal phosphorylation and insoluble deposition of tau are observed in more than 20 neurodegenerative disorders, collectively known as tauopathies (reviewed in [15]). In Alzheimer's disease (AD), the most common tauopathy, hyperphosphorylated tau accumulates in the somatodendritic compartment of neurons, aggregates, and finally forms neurofibrillary tangles (NFTs). Other tauopathies include frontotemporal lobar degeneration (FTLD), Pick's disease, progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), and progressive aphasia, all of which are characterized by neuronal and/or glial tau inclusions.
Although the exact physiological role of tau remains under scrutiny, it is proposed to function to promote microtubule assembly, stabilization, spacing, and parallel-ordered organization, which are necessary for axonal transport and neurite outgrowth (reviewed in [16]). Tau binds to several proteins (reviewed in [17]) and therefore could participate in various other paradigms, including targeting the Src kinase Fyn to dendrites [18]. In the central nervous system, tau is expressed as six isoforms, resulting from inclusion or exclusion of alternative exons 2, 3, and 10 [19] (reviewed in [20]). Mutations in or surrounding tau exon 10, leading to an imbalance in tau isoforms with 3 or 4 microtubule-binding domains, can cause familial FTLD-17. Thus, tau missplicing can cause neurodegeneration and dementia in adulthood. Changes in tau isoforms have also been observed in various other tauopathies, including PSP and Pick's disease or myotonic dystrophy (reviewed in [21]); however, the underlying mechanisms involved in abnormal tau splicing and neurodegeneration remain ill-defined. Moreover, whether splicing abnormalities function upstream or concomitantly with tau hyperphosphorylation to promote neurodegeneration remains an open debate.
To date, several groups have identified factors involved in the regulation of tau splicing. These include regulatory sequences (cis-elements) within or around tau exon 10 as well as specific regulatory proteins (trans-acting factors) (reviewed in [20]). Similarly, a number of enzymes have been proposed to regulate tau phosphorylation (reviewed in [22]). Although these studies have been insightful, they were mostly based on artificial and/or overexpression paradigms, which makes it difficult to extrapolate these observations to endogenous tau.
In 2006, Bilen et al. [23] showed a remarkable enhancement of tau-mediated cell death in Drosophila cells upon suppression of miRNA maturation. Specifically, the authors showed that retinal degeneration caused by the expression of normal or mutant (R406W) human tau in vivo was enhanced by loss of R3D1/loquacious, a double-stranded RNA binding protein that is required for the activity of Dcr-1 (Dicer homologue) in miRNA processing. Interestingly, upregulation of bantam suppressed tau-induced degeneration, suggesting that this miRNA could mitigate tau-induced neurotoxicity. However, as bantam is not conserved in humans, it is tempting to speculate that other miRNAs play a similar role in the mammalian brain. More recently, a neuronal miRNA, miR-128, was shown to modulate the expression of BAG2, a cochaperone potentially involved in tau degradation and aggregation in cultured COS-7 cells and in primary neurons [24]. Our recently published data indicate that conditional knockout (cKO) of Dicer (resulting in a global reduction in miRNA production) in the adult mouse brain results in disease-like hyperphosphorylation of endogenous tau [25]. Moreover, the Dicer mutant mice display changes in tau exon 10 splicing [26], as seen in various tauopathies including PSP and Pick's disease. In this paper, we highlight salient observations with regard to these studies and highlight outstanding questions related to miRNA research in tauopathies.
2. Tau Phosphorylation Regulation by the miR-15 Family
Tau is a phosphoprotein that contains more than 80 potential phosphorylation sites (reviewed in [27]). It is generally well accepted that tau phosphorylation is important for microtubule binding, whereas phosphorylation causes tau to detach from microtubules. Hyperphosphorylation, defined as increased phosphorylation of physiological sites and additional phosphorylation at pathological sites, characterizes insoluble aggregates of tau. However, in its unphosphorylated form, under thermal stress, tau localizes to the nucleus, where it protects DNA from double-stranded DNA damage [28]. A phosphorylated pool of tau could also localize to somatodendritic compartment as well as the dendritic spine to modulate neuronal plasticity and glutamatergic transmission [18]. Tau phosphorylation is very sensitive to intrinsic and extrinsic changes (e.g., heat, cold, stress, and starvation) [29–32]. Thus, any changes in the delicate balance between the tau kinases and phosphatases could have serious biological consequences, and the identification of these regulatory enzymes in vivo is of particular importance. It is noteworthy that some of those central key kinases also regulate indirectly tau splicing through phosphorylation of splicing factors [33–36]. Together, deregulation of kinase and/or phosphatase activity could be dually detrimental towards tau splicing and phosphorylation, which synergistically would promote tau aggregation.
As general posttranslational regulation of gene expression, miRNAs are potential modulators of kinase, phosphatase, and/or splicing factor expression. While studying the effects of Dicer loss in the brain, we observed significant changes in endogenous tau phosphorylation and splicing [26]. This was demonstrated at the RNA and protein levels using, RT-PCR, 2D electrophoresis and tau phospho-specific antibodies. Because of using the Cre-LoxP system, Dicer inactivation was limited to neurons, and in particular postmitotic pyramidal neurons. It is noteworthy that only a few studies have documented changes in endogenous tau phosphorylation in vivo, as most biological models express exogenous and/or mutated human tau. Remarkably, several phosphoepitopes related to disease, including serine 422, were increased in the Dicer mutant mice when compared with controls [25]. Unfortunately, given the rather quick lethality associated with Dicer loss (approximately 4 weeks), we could not determine whether tau hyperphosphorylation concurred before or after neurodegeneration. Nevertheless, these results provide a proof of concept that miRNA haploinsufficiency causes abnormal tau hyperphosphorylation and missplicing.
As stated above, several tau kinases and phosphatases have been identified, some of which are believed to contribute significantly to tau hyperphosphorylation in vivo. In attempt to identify such enzymes in the Dicer cKO mice, we performed whole-genome microarrays and western blot analyses. These experiments led to the identification of ERK1/MAPK3 (and possibly ERK2/MAPK1) as a major regulator of neuronal tau phosphorylation in vivo, at least in this model. In line with this observation, several ERK1-dependent epitopes were hyperphosphorylated in the Dicer mutant mice. Of course, several other enzymes can potentially contribute to tau phosphorylation misregulation in this and other models, and in particular disease conditions in humans. In line with this hypothesis, a number of proposed tau kinases, including GSK3β and JNK/MAPK8, are prone to miRNA regulation in various cell types [37, 38]. Nevertheless, it is interesting to observe that ERK phosphorylation is increased in tau-positive neurons in AD and other tauopathies [39–41]. In addition, ERK is essential for brain development and involved in neuronal death [42, 43]. Specific gene knockout mouse models are required to assess the role of these proteins in the regulation of tau phosphorylation and neurodegeneration in vivo.
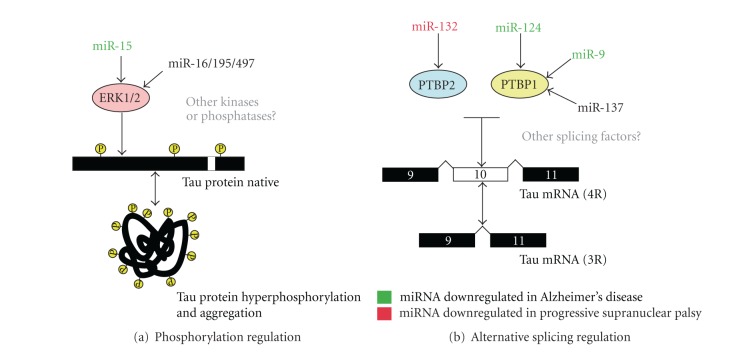
Although it is a conceptually crude experimental approach, the Dicer cKO mice provide a unique and unbiased model to study global miRNA function in the brain. To identify miRNAs involved in the regulation of ERK1 (and consequently tau phosphorylation), we used several prediction programs that are available online, including TargetScan (http://targetscan.org/). This program identified a number of potential miRNA-binding sites in the 3′UTR of ERK1. Our functional assays provided the validation that several members of the miR-16 family (miR-16, -15, -195, -497) could directly modulate endogenous ERK1 and tau phosphorylation in neuronal cells in vitro, including rat primary neurons. Of mention, both endogenous miR-16 and tau are enriched in distal axons of sympathetic neurons [44]. Intriguingly, miR-15a and miR-15b are downregulated in AD brain and cerebrospinal fluid, respectively [25, 45, 46], providing clinical relevance for these observations. Moreover, miR-15 targets the proapoptotic protein Bcl-2, whose protein levels are increased in AD [47–49]. In addition, miR-16 overexpression could regulate APP expression in vivo in the mouse brain [50]. Taken together, these observations highlight the potential importance of the miR-16 family in AD development by regulating cell survival, amyloid production, and tau phosphorylation (Figure 1). Interestingly, TargetScan predicts more than 1000 human target genes for miR-16, several of which are associated with networks related to cell death, cellular organization, and molecular transport (S. S. Hébert, unpublished observations). Notably, among the high-scoring predicted targets are miRNA-processing regulators such as TNRC6B [51]. It will be interesting to see whether loss of miR-16 family members in vivo recapitulates, at least in past, the observed effects on ERK and, most importantly, tau phosphorylation in vivo.
Figure 1.
Potential role of miRNAs in tau metabolism regulation. (a, b) Two models are shown demonstrating how specific miRNAs could be involved in the regulation of tau phosphorylation (and aggregation) and/or tau exon 10 alternative splicing. Note that some miRNAs are affected in disease conditions (in green, downregulated in AD; in red, downregulated in PSP). Whether other miRNA target effectors are involved in the physiological and/or pathological regulation of tau metabolism remains to be explored. Any changes in the level or function of these miRNAs could have serious biological consequences, including tau hyperphosphorylation and aggregation and an imbalance in tau microtubule-binding repeats (encoded in tau exon 10, giving rise to either four 4R-tau or 3R-tau). Regulation of tau exon 10 splicing by PTBP1 or PTBP2 may be direct or may implicate other coregulators such as TDP-43 or PSF, both of which have been shown to either regulate PTBP2 expression or regulate PTBP splicing and repress tau exon 10 inclusion [26, 52–54].
3. Tau Alternative Splicing Regulation by miR-132
As discussed above, abnormal regulation of tau exon 10 splicing can cause disease. It is interesting to note that Dicer deficiency in the adult brain is also associated with changes in tau splicing [26]. Using a similar strategy as above (e.g., bioinformatics, microarrays, literature search, western blot analysis, etc.), we identified miR-132 and the neuronal splicing regulator PTBP2 as potential regulators of endogenous tau exon 10 splicing in neurons. These results are consistent with previous findings linking tau exon 10 splicing regulation by PTBP1 in vitro [55].
While not discussed in detail in our study, other miRNAs, including miR-124 and miR-9, could also regulate endogenous tau exon 10 splicing in neuronal cells by targeting specific regulatory and/or splicing factors [26]. Both miRNAs are downregulated in AD [56–58], which could have important consequences for tau metabolism, at least in certain biological contexts (Figure 1). For instance, downregulation of miR-9 is observed in the presence of exogenous Aβ in mouse primary neurons [59]. Whether this or other miRNAs function as intermediates between Aβ peptides, tau missplicing and hyperphosphorylation remain an exciting possibility. Interestingly, differential splicing of the tau transcript has also been reported in AD [60, 61].
PSP is a cause of late-onset atypical parkinsonism described by Steele et al. [62]. Dementia is also a common feature at the end stage of the disease. Neuropathologically, PSP is characterized by neuronal loss, gliosis, and NFT formation. Glial fibrillary tangles have also been described. In these patients, tau aggregates are mainly composed of tau with 4 microtubule-binding domains (4R-tau) [13, 15]. Using PSP as a model disease, we identified miR-132 to be selectively downregulated in pathological conditions. Interestingly, PTBP2 protein (but not mRNA) levels were increased in PSP patients and correlated significantly with miR-132 expression [26]. These experiments provide unprecedented molecular links among abnormal tau splicing, hyperphosphorylation, and sporadic tauopathies. Interestingly, changes in PTBP1 and PTBP2 levels, and by extension alternative splicing patterns, have been documented in human diseases, including neurodegenerative disorders [63, 64]. On the basis of these observations, it is tempting to speculate that miRNAs could contribute significantly to several aspects of tau metabolism and neuronal dysfunction in various diseases.
4. Outstanding Questions
Although the above-mentioned studies are interesting, many questions remain unanswered. For instance, what are the biological and clinical significance of these findings? Are other miRNAs involved in tau metabolism regulation? Can miRNAs be used as diagnostic and possibly therapeutic agents for sporadic tauopathies? Without a doubt, these and other questions will require extensive followup studies in various models, from cells to animals to humans.
Conflict of Interests
The authors declared that there is no conflict of interests.
Acknowledgments
This work was supported by the Alzheimer Society of Canada, the French National Research Agency, and the Research on Alzheimer's Disease and Related Disorders Québec-France-Canada Collaboration grant (TAUMIRNA). This work has been developed through the LABEX (Laboratories of Excellence, investing for the future program) and DISTALZ (Development of Innovative Strategies for a Transdisciplinary Approach to Alzheimer's Disease).
Abbreviation
- 3′UTR:
3′ Untranslated region.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116:S89–S92. doi: 10.1016/s0092-8674(04)00035-2. [DOI] [PubMed] [Google Scholar]
- 3.Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends in Genetics. 2008;24(9):448–456. doi: 10.1016/j.tig.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Nana-Sinkam SP, Croce CM. MicroRNAs as therapeutic targets in cancer. Translational Research. 2011;157(4):216–225. doi: 10.1016/j.trsl.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 5.Hébert SS, de Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends in Neurosciences. 2009;32(4):199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Miska EA, Alvarez-Saavedra E, Townsend M, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biology. 2004;5(9):p. R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barad O, Meiri E, Avniel A, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Research. 2004;14(12):2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome biology. 2004;5(3):p. R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shao NY, Hu HY, Yan Z, et al. Comprehensive survey of human brain microRNA by deep sequencing. BMC Genomics. 2010;11:p. 409. doi: 10.1186/1471-2164-11-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babiarz JE, Hsu R, Melton C, Thomas M, Ullian EM, Blelloch R. A role for noncanonical microRNAs in the mammalian brain revealed by phenotypic differences in Dgcr8 versus Dicer1 knockouts and small RNA sequencing. RNA. 2011;17(8):1489–1501. doi: 10.1261/rna.2442211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 12.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(5):1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 15.Buée L, Bussière T, Buée-Scherrer V, Delacourte A, Hof PR. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Research Reviews. 2000;33(1):95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 16.Morris M, Maeda S, Vossel K, Mucke L. The Many faces of tau. Neuron. 2011;70(3):410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brandt R, Leschik J. Functional interactions of tau and their relevance for Alzheimer’s disease. Current Alzheimer Research. 2004;1(4):255–269. doi: 10.2174/1567205043332054. [DOI] [PubMed] [Google Scholar]
- 18.Ittner LM, Ke YD, Delerue F, et al. Dendritic function of tau mediates amyloid-β toxicity in Alzheimer’s disease mouse models. Cell. 2010;142(3):387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- 19.Chapple PJ, Anthony K, Martin TR, Dev A, Cooper TA, Gallo JM. Expression, localization and tau exon 10 splicing activity of the brain RNA-binding protein TNRC4. Human Molecular Genetics. 2007;16(22):2760–2769. doi: 10.1093/hmg/ddm233. [DOI] [PubMed] [Google Scholar]
- 20.Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochimica Et Biophysica Acta. 2005;1739(2):91–103. doi: 10.1016/j.bbadis.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 21.Sergeant N, Bretteville A, Hamdane M, et al. Biochemistry of Tau in Alzheimer’s disease and related neurological disorders. Expert Review of Proteomics. 2008;5(2):207–224. doi: 10.1586/14789450.5.2.207. [DOI] [PubMed] [Google Scholar]
- 22.Buée L, Troquier L, Burnouf S, et al. From tau phosphorylation to tau aggregation: what about neuronal death? Biochemical Society Transactions. 2010;38(4):967–972. doi: 10.1042/BST0380967. [DOI] [PubMed] [Google Scholar]
- 23.Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Molecular Cell. 2006;24(1):157–163. doi: 10.1016/j.molcel.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 24.Carrettiero DC, Hernandez I, Neveu P, Papagiannakopoulos T, Kosik KS. The cochaperone BAG2 sweeps paired helical filament-insoluble tau from the microtubule. Journal of Neuroscience. 2009;29(7):2151–2161. doi: 10.1523/JNEUROSCI.4660-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hébert SS, Papadopoulou AS, Smith P, et al. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Human Molecular Genetics. 2010;19(20):3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 26.Smith PY, Delay C, Girard J, et al. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Human Molecular Genetics. 2011;20(20):4016–4024. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- 27.Hanger DP, Noble W. Functional implications of glycogen synthase kinase-3-mediated tau phosphorylation. International Journal of Alzheimer's Disease. 2011;2011:11 pages. doi: 10.4061/2011/352805. Article ID 352805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sultan A, Nesslany F, Violet M, et al. Nuclear Tau, a key player in neuronal DNA protection. Journal of Biological Chemistry. 2011;286(6):4566–4575. doi: 10.1074/jbc.M110.199976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittington RA, Papon MA, Chouinard F, Planel E. Hypothermia and Alzheimer's disease neuropathogenic pathways. Current Alzheimer Research. 2010;7(8):717–725. doi: 10.2174/156720510793611646. [DOI] [PubMed] [Google Scholar]
- 30.Planel E, Bretteville A, Liu L, et al. Acceleration and persistence of neurofibrillary pathology in a mouse model of tauopathy following anesthesia. The FASEB Journal. 2009;23(8):2595–2604. doi: 10.1096/fj.08-122424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okawa Y, Ishiguro K, Fujita SC, Avila J. Stress-induced hyperphosphorylation of tau in the mouse brain. FEBS Letters. 2003;535(1–3):183–189. doi: 10.1016/s0014-5793(02)03883-8. [DOI] [PubMed] [Google Scholar]
- 32.Yanagisawa M, Planel E, Ishiguro K, Fujita SC. Starvation induces tau hyperphosphorylation in mouse brain: implications for Alzheimer’s disease. FEBS Letters. 1999;461(3):329–333. doi: 10.1016/s0014-5793(99)01480-5. [DOI] [PubMed] [Google Scholar]
- 33.Qian W, Liang H, Shi J, et al. Regulation of the alternative splicing of tau exon 10 by SC35 and Dyrk1A. Nucleic Acids Research. 2011;39(14):6161–6171. doi: 10.1093/nar/gkr195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J, Qian W, Yin X, et al. Cyclic AMP-dependent protein kinase regulates the alternative splicing of tau exon 10: a mechanism involved in tau pathology of Alzheimer disease. Journal of Biological Chemistry. 2011;286(16):14639–14648. doi: 10.1074/jbc.M110.204453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glatz DC, Rujescu D, Tang Y, et al. The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer’s disease. Journal of Neurochemistry. 2006;96(3):635–644. doi: 10.1111/j.1471-4159.2005.03552.x. [DOI] [PubMed] [Google Scholar]
- 36.Hernández F, Pérez M, Lucas JJ, Mata AM, Bhat R, Avila J. Glycogen synthase kinase-3 plays a crucial role in tau exon 10 splicing and intranuclear distribution of SC35: implications for Alzheimer’s disease. Journal of Biological Chemistry. 2004;279(5):3801–3806. doi: 10.1074/jbc.M311512200. [DOI] [PubMed] [Google Scholar]
- 37.Mohamed JS, Lopez MA, Boriek AM. Mechanical stretch up-regulates microRNA-26a and induces human airway smooth muscle hypertrophy by suppressing glycogen synthase kinase-3β . Journal of Biological Chemistry. 2010;285(38):29336–29347. doi: 10.1074/jbc.M110.101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Chen S, Luan X, et al. MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of hela cells. IUBMB Life. 2009;61(11):1075–1082. doi: 10.1002/iub.252. [DOI] [PubMed] [Google Scholar]
- 39.Ferrer I, Blanco R, Carmona M, et al. Phosphorylated map kinase (ERK1, ERK2) expression is associated with early tau deposition in neurones and glial cells, but not with increased nuclear DNA vulnerability and cell death, in Alzheimer disease, Pick’s disease, progressive supranuclear palsy and corticobasal degeneration. Brain Pathology. 2001;11(2):144–158. doi: 10.1111/j.1750-3639.2001.tb00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iqbal K, Alonso Adel C, El-Akkad E, et al. Significance and mechanism of Alzheimer neurofibrillary degeneration and therapeutic targets to inhibit this lesion. Journal of Molecular Neuroscience. 2002;19(1-2):95–99. doi: 10.1007/s12031-002-0017-3. [DOI] [PubMed] [Google Scholar]
- 41.Perry G, Roder H, Nunomura A, et al. Activation of neuronal extracellular receptor kinase (ERK) in Alzheimer disease links oxidative stress to abnormal phosphorylation. NeuroReport. 1999;10(11):2411–2415. doi: 10.1097/00001756-199908020-00035. [DOI] [PubMed] [Google Scholar]
- 42.Satoh Y, Kobayashi Y, Takeuchi A, Pagès G, Pouysségur J, Kazama T. Deletion of ERK1 and ERK2 in the CNS causes cortical abnormalities and neonatal lethality: Erk1 deficiency enhances the impairment of neurogenesis in Erk2-deficient mice. Journal of Neuroscience. 2011;31(3):1149–1155. doi: 10.1523/JNEUROSCI.2243-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramaniam S, Unsicker K. ERK and cell death: ERK1/2 in neuronal death. The FEBS Journal. 2010;277(1):22–29. doi: 10.1111/j.1742-4658.2009.07367.x. [DOI] [PubMed] [Google Scholar]
- 44.Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16(8):1516–1529. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cogswell JP, Ward J, Taylor IA, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. Journal of Alzheimer’s Disease. 2008;14(1):27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 46.Nunez-Iglesias J, Liu C-C, Morgan TE, Finch CE, Zhou XJ. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer’s disease cortex reveals altered miRNA regulation. PloS One. 2010;5(2):p. e8898. doi: 10.1371/journal.pone.0008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cimmino A, Calin GA, Fabbri M, et al. MiR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu G, Kwong WH, Li Q, Wang X, Feng Z, Yew DT. Bcl2, bax, and nestin in the brains of patients with neurodegeneration and those of normal aging. Journal of Molecular Neuroscience. 2005;27(2):167–174. doi: 10.1385/JMN:27:2:167. [DOI] [PubMed] [Google Scholar]
- 49.Jordan-Sciutto KL, Malaiyandi LM, Bowser R. Altered distribution of cell cycle transcriptional regulators during Alzheimer disease. Journal of Neuropathology and Experimental Neurology. 2002;61(4):358–367. doi: 10.1093/jnen/61.4.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu W, Liu C, Zhu J, et al. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiology of Aging. 2012;33:522–534. doi: 10.1016/j.neurobiolaging.2010.04.034. [DOI] [PubMed] [Google Scholar]
- 51.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-Terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15(5):781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ray P, Kar A, Fushimi K, Havlioglu N, Chen X, Wu JY. PSF suppresses tau exon 10 inclusion by interacting with a stem-loop structure downstream of exon 10. Journal of Molecular Neuroscience. 2011;45(3):453–466. doi: 10.1007/s12031-011-9634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sephton CF, Cenik C, Kucukural A, et al. Identification of neuronal RNA targets of TDP-43-containing ribonucleoprotein complexes. Journal of Biological Chemistry. 2011;286(2):1204–1215. doi: 10.1074/jbc.M110.190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei ML, Memmott J, Screaton G, Andreadis A. The splicing determinants of a regulated exon in the axonal MAP tau reside within the exon and in its upstream intron. Molecular Brain Research. 2000;80(2):207–218. doi: 10.1016/s0169-328x(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Gao QS, Wang Y, Lafyatis R, Stamm S, Andreadis A. Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. Journal of Neurochemistry. 2004;88(5):1078–1090. doi: 10.1046/j.1471-4159.2003.02232.x. [DOI] [PubMed] [Google Scholar]
- 56.Hébert SS, Horré K, Nicolaï L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/β-secretase expression. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith P, Al Hashimi A, Girard J, Delay C, Hébert SS. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. Journal of Neurochemistry. 2011;116(2):240–247. doi: 10.1111/j.1471-4159.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- 58.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18(3):297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 59.Schonrock N, Ke YD, Humphreys D, et al. Neuronal microRNA deregulation in response to Alzheimer’s disease amyloid-β . PloS one. 2010;5(6):p. e11070. doi: 10.1371/journal.pone.0011070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasojima K, McGeer EG, McGeer PL. Tangled areas of Alzheimer brain have upregulated levels of exon 10 containing tau mRNA. Brain Research. 1999;831(1-2):301–305. doi: 10.1016/s0006-8993(99)01486-9. [DOI] [PubMed] [Google Scholar]
- 61.Caffrey TM, Joachim C, Paracchini S, Esiri MM, Wade-Martins R. Haplotype-specific expression of exon 10 at the human MAPT locus. Human Molecular Genetics. 2006;15(24):3529–3537. doi: 10.1093/hmg/ddl429. [DOI] [PubMed] [Google Scholar]
- 62.Steele JC, Richardson JC, Olszewski J. Progressive supranuclear palsy. A heterogeneous degeneration involving the brain stem, basal ganglia and cerebellum with vertical gaze and pseudobulbar palsy, nuchal dystonia and dementia. Archives of Neurology. 1964;10:333–359. doi: 10.1001/archneur.1964.00460160003001. [DOI] [PubMed] [Google Scholar]
- 63.Tollervey JR, Wang Z, Hortobágyi T, et al. Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Research. 2011;21(10):1572–1582. doi: 10.1101/gr.122226.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sterne-Weiler T, Howard J, Mort M, Cooper DN, Sanford JR. Loss of exon identity is a common mechanism of human inherited disease. Genome Research. 2011;21(10):1563–1571. doi: 10.1101/gr.118638.110. [DOI] [PMC free article] [PubMed] [Google Scholar]