Abstract
Eukaryotic protein kinase pathways have both grown in number and changed their network architecture during evolution. We wondered if there are pivotal proteins in these pathways that have been repeatedly responsible for forming new connections through evolution, thus changing the topology of the network; and if so, whether the underlying properties of these proteins could be exploited to re-engineer and rewire these pathways. We addressed these questions in the context of the mitogen-activated protein kinase (MAPK) pathways. MAPK proteins were found to have repeatedly acquired new specificities and interaction partners during evolution, suggesting that these proteins are pivotal in the kinase network. Using the MAPKs Fus3 and Hog1 of the Saccharomyces cerevisiae mating and hyper-osmolar pathways, respectively, we show that these pivotal proteins can be re-designed to achieve a wide variety of changes in the input-output properties of the MAPK network. Through an analysis of our experimental results and of the sequence and structure of these proteins, we show that rewiring of the network is possible due to the underlying modular design of the MAPKs. We discuss the implications of our findings on the radiation of MAPKs through evolution and on how these proteins achieve their specificity.
Kinase pathways form one example of protein-protein interaction networks1. These networks have grown, changing their topology and constituent proteins, during evolution2. Whereas previous analyses have focused on the conservation properties of nodes in these networks3, it is the ability of certain nodes to consistently change connections that could allow the network topology to be malleable. Identifying such pivotal nodes in particular signalling networks may enable us to re-engineer these biological circuits with ease.
Here, we study the network of MAPK pathways that transduce different signals and regulate stress response and growth. We find that MAPK proteins are the pivotal nodes that allow their pathways to acquire new components and connections. Through a combination of sequence analysis and experiments, we show that we can continuously change the topology of the MAPK network and its signal processing capabilities by redesigning the MAPK proteins.
For our experiments, we chose two of the five MAPK pathways in S. cerevisiae4 that are involved in mating and responding to high osmolarity (Fig. 1a). These two pathways involve the MAPK proteins Fus3 and Hog1. Since another MAPK Kss1 can partially substitute for Fus3 (ref. 5), fus3Δ kss1Δ cells are sterile and hog1Δ cells die after hyper-osmolar shock6.
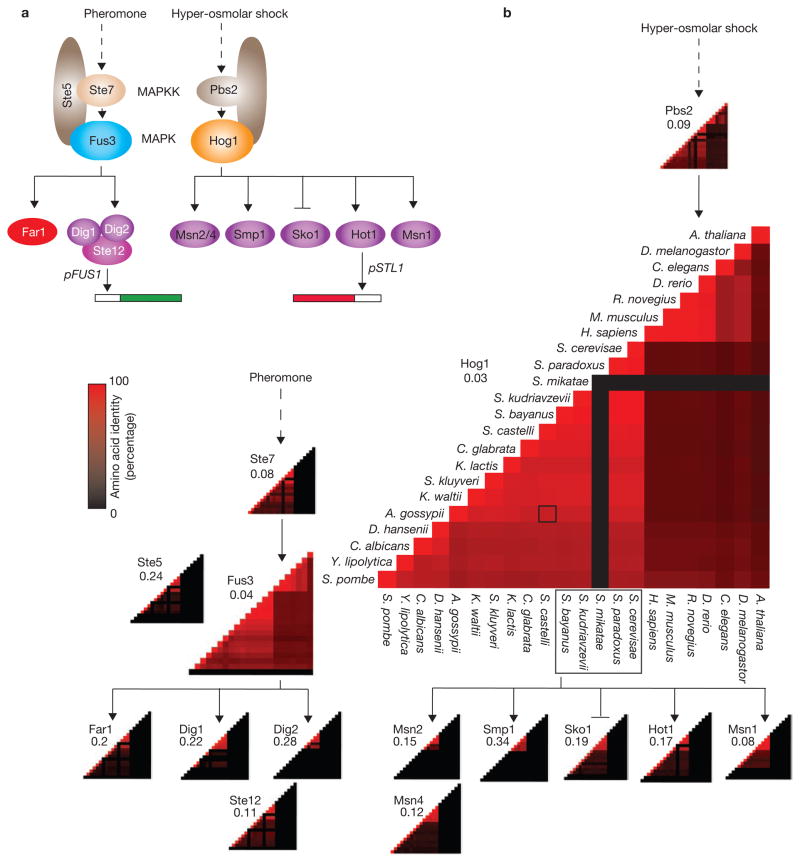
Figure 1.
Conservation of downstream components in the pheromone and hyper-osmolar glycerol pathways in S. cerevisiae. (a) The pheromone signal results in phosphorylation of the MAPKK Ste7, which phosphorylates the MAPK Fus3 aided by a scaffolding protein Ste5 (ref. 23). Phosphorylated Fus3 in turn activates the transcription factor Ste12 by relieving its repression by Dig1 and Dig2 (ref. 24), and activates Far1 leading to cell-cycle arrest. Activated Ste12 initiates the transcription of mating-specific genes including FUS1. The stimulation of cells with an osmolyte (for example, sorbitol) results in phosphorylation of the MAPKK Pbs2, which phosphorylates the MAPK Hog1, which in turn activates several transcription factors (including Hot1). This initiates the transcription of several genes, including STL1. (b) Matrices corresponding to each protein in a encode the pair-wise percentage identity between the amino-acid sequences of all orthologues of that protein. The matrix for Hog1 is enlarged and labelled. The outlined matrix element in the Hog1 matrix shows the percentage identity between the Hog1 orthologues in A.gossypii and S. castelli. As the matrices are symmetrical, only half of each is shown. Beside each matrix is the substitution ratio (N/S) measuring the evolution rate of each protein within the sensu strictu species. The MAPKs Fus3 and Hog1 are the most conserved elements of the pathways, as indicated by the predominance of red in their matrices and the low N/S values. Pbs2 is found in all the species, whereas Ste7 (a Pbs2 duplicate) is present only in yeast, as indicated by the completely black columns and rows in its matrix, corresponding to the higher eukaryotes. The remaining proteins in a only have orthologues in yeast.
To study the evolution of the MAPK pathways, we identified orthologues of their component proteins in fifteen yeast, six animal and one plant species (Figs 1b, 2; Supplementary Information Fig. S1a). From the fission yeast Schizosaccharomyces pombe7 to the higher eukaryotes, the number of MAPKs has increased from three to at least fifteen. As the MAPK family grows, many MAPK interaction partners appear de novo in different lineages. Except for Pbs2, orthologues of all the interaction partners of Fus3 and Hog1 (Fig. 1a) are found only in the yeast lineage, indicating that the specificities of these MAPKs for their partners developed uniquely in yeast (Figs 1b, 2). This includes scaffolding proteins that nucleate these multi-protein complexes aiding MAPK specificities with their appropriate upstream and downstream partners8,9. Closely related MAPK paralogues and orthologues show distinct specificities, indicating that new interactions can be acquired with little sequence divergence. For example, in S. cerevisiae the paralogues Kss1 and Fus3 can be activated by Ste7, but only Fus3 can activate Far1 causing cell-cycle arrest10. Strikingly, we also found that orthologues had switched their specificity from one MAPK in the yeast to another MAPK in the animals (Fig. 2d), providing a clear example of network topology that has changed during evolution.
Figure 2.
Evolutionary history of MAPKs and their interacting partners. (a) The phylogenetic tree of 15 yeast species and 7 higher eukaryote species used in deducing gene duplication events. Each speciation event (S0,S1,S2,S3,S4…S9) leads to the introduction of a new or duplicate gene in the upper branch. (b) MAPK-interacting partners from Fig. 1a, which are introduced de novo into the yeast lineage, are shown alongside the speciation event that created them. (c) Gene duplications accounting for all the remaining interacting partners not in b. Many of these have the new genes in b as their parent. (d) The only two MAPK-interacting partners, Pbs2 and Rlm1, found in all the species have changed their specificities towards MAPKs during evolution. In the yeast, the orthologue of the downstream transcription factor Rlm1 is activated by the orthologue of Slt2 (the MAPK of the hypo-osmolar pathway). In the higher eukaryotes, however, (for example human, mouse and rat), the Rlm1 orthologue (MEF2A) is activated by the Hog1 orthologue (p38α, refs 18, 25). A similar change occurs upstream. In yeast, the Pbs2 orthologue activates the Hog1 orthologue, whereas in human, mouse and rat the Pbs2 orthologue (MEK1/2) activates the Fus3 orthologue (ERK1/2, ref. 13). Thus despite being highly conserved, the MAPKs show flexibility in acquiring and swapping interaction partners through evolution.
Despite this plasticity, the MAPKs have been more invariant through evolution than other members of their pathways (Fig. 1b). Which molecular properties have allowed the MAPKs to remain so conserved, while still having a pivotal role in allowing their pathways to rewire and acquire new components during evolution?
The MAPKs are small, compact globular proteins. As with their linear protein sequences, their three-dimensional structures are very similar to one another (Fig. 3). To understand how these proteins achieve their specificities, we first reviewed previous biochemical11,12, structural8,13,14 and peptide10,15 analyses of the roles of certain residues in Fus3 and Hog1 (Fig. 3c, d; Supplementary Information, Section 2). We found that, other than Asp 112 and His 113 in Fus3, these residues cannot function alone as specificity determinants because they are conserved in other MAPK paralogues and their orthologues in other species.
Figure 3.
Sequence analysis of MAPKs Fus3 and Hog1. (a) The degree of variability, as measured from entropy, in a multiple sequence alignment of four MAPKs and their orthologues from 22 species (Supplementary Information, Data Analysis) is shown on the structure of Fus3. High entropy (red) positions are constrained to the surface, whereas low entropy (blue) positions are in the interior. (b) Residues on Fus3 deduced computationally as putatively being responsible for the differences in Fus3 and Hog1 specificities are shown highlighted on its steric surface. The protein structure on the left has the same orientation as that in a, and the structure on the right has been rotated 180° about the vertical axis. The residues are coloured according to the segment they belong to, with the same colours as used in the six segments in c and d. Some residues, such as P80, F83, E84 and W348 shown on the structure on the right, distinguish themselves by being present on Fus3 and not on Hog1. (c, d) The segments A/a, B/b, C/c, D/d, E/e and F/f in Fus3/Hog1 used to build the hybrid kinases are shown on the structures of Fus3 and p38α (mouse Hog1 orthologue), respectively. Five regions linking these segments, shown in blue, were chosen on the basis of their strict conservation in all the MAPKs in the multiple sequence alignment used in a. Specific residues previously identified in the literature are highlighted on both structures (Supplementary Information, Section 2). Most of these residues are common to both of the MAPKs and their orthologues from other species; however, they are shown highlighted on one or the other to reflect the MAPK in which they were identified. Thus, except for D112 and H113, these residues cannot be important for specificity.
We extracted functional information on residues using a multiple sequence alignment consisting of orthologues of four S. cerevisiae MAPKs (16 Kss1, 15 Fus3, 15 Hog1 and 15 Slt2). The variable residues were almost exclusively on the surface of the proteins (Fig. 3a). Of these residues, we computationally identified a subset putatively responsible for the differences in specificities of Fus3 and Hog1 (Supplementary Information, Figs S2, S3a and Methods). Our sequence analysis suggests two characteristics of MAPKs: first, they are structurally robust to changes in most of their surface residues; second, distinct groups of surface residues stand out on each MAPK as being responsible for specific interactions. The latter finding was unexpected as it may be thought that structurally similar proteins of the same family will use the same surface regions to interact with their corresponding upstream or downstream partners. For instance, we found a group of residues (Pro 80, Phe 83, Glu 84 and Trp 348) that is conserved in Fus3, but absent in Hog1 (Fig. 3b). Other residue positions are well conserved in Hog1 but show great variability in Fus3, signifying neutral drift. Thus distinct patches may be important for the specific interactions of each MAPK with other members of its pathway (Supplementary Information, Figs S2, S3a). If MAPK plasticity stemmed from the underlying flexibility of these patches, then synthetic proteins containing different combinations of these patches should be able to deform the signalling pathways. A chimaera has been generated16 that, when expressed in mammalian cell lines, directs a stress signal into mitotic output (Supplementary Information, Section 2), suggesting that such deformations may indeed be possible.
To investigate this possibility, we constructed proteins that contained residues from Fus3 and Hog1 in various combinations. We divided both Fus3 and Hog1 into six segments (A, B, C, D, E and F for Fus3 and a, b, c, d, e and f for Hog1) linked by regions conserved in all the MAPKs (blue, Fig. 3c, d), and joined the genes coding for the 64 possible proteins composed of these segments (Supplementary Information, Methods Fig. S2). This achieved a combinatorial redistribution of the surface patches, while also ensuring that internal contacts along the sequence were preserved. The genes were driven by a FUS3 promoter. To test for pheromone output in response to either pheromone or osmolyte (sorbitol) exposure, the 64 hybrids on both low-copy (showing an expression within threefold of the native chromosomal gene; data not shown) and high-copy (showing an expression within eightfold of the native chromosomal gene; data not shown) plasmids were transformed into a haploid fus3Δ kss1Δ strain (Supplementary Information, Table S1). A fluorescent (GFP) reporter was placed under control of the native FUS1 promoter, a faithful reporter of the pheromone pathway (Supplementary Information, Fig. S3b, c). We measured the fold change of fluorescent protein after 2 h of pheromone or sorbitol exposure (Fig. 4; Supplementary Information, Fig. S5a). The strains expressing the hybrids were further assayed for their ability to mate (Fig. 4; Supplementary Information, Fig. S4a and Methods) and arrest their growth on pheromone exposure (Supplementary Table 2). To test for osmolar output in response to either input, hybrids on both low- and high-copy plasmids were transformed into a haploid hog1Δ strain with a fluorescent reporter placed under the native STL1 promoter, a faithful reporter of the osmolar pathway (Supplementary Information, Fig. S3b, d). We measured the fold change of fluorescent protein under the STL1 promoter after 2 h of pheromone or sorbitol exposure (Fig. 4; Supplementary Information, Fig. S5b). The strains with the hybrids were assayed for their ability to grow under high osmolarity conditions (Fig. 4; Supplementary Information, Fig. S4b, c). Hybrids were also assayed in a fus3Δ kss1Δ hog1Δ strain with distinct fluorescent reporters (YFP and CFP) under the FUS1 and STL1 promoters. Without the hybrids, this strain does not show any response to pheromone or sorbitol (Fig. S3e). We measured the time-series of the response for hybrids on low-copy plasmids (Supplementary Information, Figs S6, S7a), and when integrated under the FUS3 promoter in the chromosome (Supplementary Information, Fig. S7b).
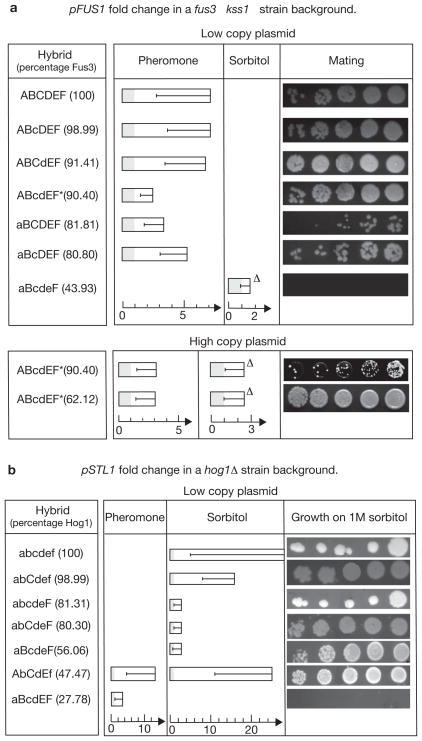
Figure 4.
Several hybrid MAPKs function in vivo to faithfully transduce and cross-wire the pheromone and hyper-osmolar signals. (a) The mean fold change in pFUS1 activity (upper panel, measured by a fluorescent reporter) in ~100 induvidual fus3Δ kss1Δ haploid cells containing the hybrid MAPKs expressed from a low-copy plasmid, after stimulation for 2 h by 0.6 μM pheromone or 1 M sorbitol. The percentage of residues in the hybrid that belong to Fus3 and differ from Hog1 is shown in parentheses. Serial dilutions show the mating efficiency of fus3Δ kss1Δ MATa haploids containing the indicated hybrids. Error bars reflect the intrinsically large range of response in the cells and not the measurement of uncertainty in a single cell. One hybrid expressed in low-copy plasmid (upper panel), and two hybrids expressed in high-copy plasmid (lower panel) mediate a cross-wiring whereby sorbitol evokes pFUS1 activity. The persistence of this cross-wiring after deleting STE7 is indicated by ‘Δ’. Hybrids showing constitutive pFUS1 activity are marked with an asterisk. Sorbitol-induced pFUS1 activity of aBcdeF was measured in fus3Δkss1Δhog1Δ cells. (b) The mean fold change in pSTL1 activity measuring an osmolar response, in ~100 single hog1Δ haploid cells containing the hybrid MAPKs and stimulated in the same manner as in a. The axes for fold change are scaled differently for the pheromone and sorbitol stimuli. Serial dilutions show the efficient growth of hog1Δ strains carrying the hybrid on plates containing 1 M sorbitol. Two hybrids mediated a cross-wiring showing pSTL1 activity on pheromone exposure (Supplementary Information, Data Analysis). The pheromone induced pSTL1 activity of aBcdEF was measured in fus3Δ kss1Δ cells.
Several hybrids show FUS1 promoter (pFUS1) activity in response to either stimulus (Fig. 4a). Another nine (ABcdeF, ABCDeF, aBCdEF, abCDeF, AbCDEF, abCDEf, abCdEF, aBcdEF, AbcDEF) do so when expressed from high-copy plasmids (Supplementary Information, Figs S5a, S8). All low-copy hybrids that showed pFUS1 activity upon pheromone exposure also rescued the cells ability to mate (Fig. 4a). Unstimulated cells carrying ABcdEF and ABcdeF had constitutively active pFUS1 showing about 50% and 20%, respectively, of the fluorescence shown by pheromone stimulated wild-type cells. (Supplementary Information, Fig. S8 and Section 3). This is noteworthy, as producing phospho-mimicking mutations in the activation loop does not constitutively activate MAPKs as it does for the activating MAPK Kinases (MAPKK)17, 18, 19.
One hybrid expressed from a low-copy plasmid, and several hybrids expressed from a high-copy plasmid (Fig. 4a; Supplementary Information, Fig. S5a), also showed pFUS1 activity in response to sorbitol, in fus3Δ kss1Δ cells. Such cross-wiring could occur by either a direct or an indirect mechanism. A direct mechanism would involve the hybrid MAPK being directly activated by Pbs2 and in turn activating Ste12. There are two possible indirect mechanisms: the first is where the hybrid inhibits native Hog1 or Pbs2 while also performing the function of Fus3. This is because strains in which the osmolar pathway is interrupted at the level of Hog1 or Pbs2 (either by deletion or inhibition of these proteins) promiscuously channel the hyper-osmolar signal into a pheromone output (Supplementary Information, Fig. S3d). This promiscuous channelling happens upstream of the MAPK and is abolished by deleting Ste7 (ref. 20). The second indirect mechanism is one in which Hog1 fails to inhibit the hybrid, allowing the osmolar signal to leak through the pheromone pathway, again through Ste7. To discriminate between the direct and indirect mechanisms of cross-wiring, we assayed the hybrid MAPKs in a ste7Δ hog1Δ strain. Without the hybrids, a sorbitol stimulus does not get cross-wired into pheromone output in this strain (Information, Fig. S9a). Three hybrids (ABcdEF, ABcdeF and aBcdeF) showed pFUS1 activity on hyper-osmolar shock in the ste7Δ hog1Δ strain, suggesting that they were activated directly by Pbs2 (Fig. 4a; Supplementary Information, Fig. S9b). For the remaining hybrids, cross-activation was abolished in the ste7Δ hog1Δ strain, implying that the cross-wiring occurred indirectly (Supplementary Information, Fig. S9c).
When expressed from a low-copy plasmid in fus3Δ kss1Δ hog1Δ cells, only the hybrid aBcdeF is capable of cross-wiring a sorbitol input into a pheromone output. Cells carrying aBcdeF are insensitive to pheromone (Fig. 5a, b), implying that this hybrid is directly activated by Pbs2 but not by Ste7. The cross-wiring persisted in the ste7Δ hog1Δ strain. The cross-wiring only occurred in the absence of native HOG1 when aBcdeF was expressed from a low-copy plasmid. Over-expression from a high-copy plasmid rescued this cross-talk in the presence of native HOG1, suggesting either competitive binding to Pbs2 between aBcdeF and Hog1 or direct inhibition of aBcdeF by Hog1.
Figure 5.
Modular design allows some hybrid MAPKs to be activated by either input and others capable of activating either output. The protein space between extant Fus3 and Hog1 is rich in function. (a, b) Differential interference contrast (DIC) and fluorescence images of cells containing three hybrids: aBcdeF and AbCdEf on a low-copy plasmid and ABcdEF on a high-copy plasmid. Cells were stimulated with pheromone (left) and sorbitol (right). Green fluorescence (a) shows pFUS1 activity in fus3Δ kss1Δ cells (which, except for aBcdeF, have an intact osmolar pathway) mediated by the hybrid on pheromone or sorbitol stimulation. Red fluorescence (b) shows the pSTL1 activity in hog1Δ cells (which have an intact pheromone pathway) mediated by the hybrid on stimulation by pheromone or sorbitol. (c) aBcdeF interacts upstream only with the MAPKK Pbs2 of the osmolar pathway, but can interact downstream with transcription factors of both pathways (Ste12, Hot1). Thus sufficiently non-overlapping groups of residues on the original MAPKs must be responsible for these downstream interactions. Analogously, AbCdEf interacts downstream only with Hot1 but interacts upstream with the MAPKKs Ste7 and Pbs2. Therefore sufficiently non-overlapping groups of residues are also responsible for these upstream interactions. (d) Functional properties of each hybrid are summarized schematically in a 2 × 2 matrix. The hybrids are arranged from Hog1 (top) to Fus3 (bottom). Hybrids in each successive row below Hog1 have one more segment taken from Fus3. The hybrids discussed in a, b and c are underlined. Each element of the matrix denotes the hybrids ability to mediate one of four distinct input-output characteristics in a cell. Top left (green) indicates pheromone response to pheromone stimulus. Top right (green) indicates pheromone response to sorbitol stimulus. Bottom left (red) indicates sorbitol response to pheromone stimulus. Bottom right (red) indicates sorbitol response to sorbitol stimulus. The bottom right is grey for three hybrids rescuing growth on 1 M sorbitol plates without showing a transcriptional response (Supplementary Information, Fig. S4b, c). Our sampling of the intervening MAPK protein space between Hog1 and Fus3 reveals several continuous paths via functioning intermediates.
Several hybrids, when expressed from a low-copy plasmid, show STL1 promoter (pSTL1) activity in response to either input (Fig. 4b; Supplementary Information, Fig. S9d). The hybrid AbCdEf, which faithfully transduces the sorbitol signal almost as efficiently as wild-type Hog1, also cross-wires the pathways by showing pSTL1 activity in response to pheromone exposure (Fig. 5a, b). Another hybrid, aBcdEF, achieves similar cross-wiring but is insensitive to sorbitol itself (Supplementary Information, Fig. S9d). The cross-wiring by aBcdEF occurred only in the absence of native Fus3 and Kss1, suggesting a competitive binding to Ste7 between aBcdEF and Fus3 or Kss1. All low-copy hybrids that showed pSTL1 activity on sorbitol exposure also rescued the ability of the cells to grow under high osmolar conditions (Fig. 4b). Interestingly, certain hybrids rescued the growth of the cells on a high osmolar medium (Supplementary Information, Fig. S4b, c), but did not mediate reporter activity in the cell (Fig. 4b; Supplementary Information, Fig. S5b).
Besides demonstrating the flexibility of MAPKs, our results allow us to draw some conclusions about the modularity and specificity of these proteins.
We find that the MAPK proteins are modular. One in three hybrids were functional, some with as many of their distinguishing residues taken from Fus3 as from Hog1. This strongly suggests that the common conserved residues at the core are sufficient for folding, and the variable exterior residues, where most of the evolution has occurred, control specificity. However, despite their having similar structures and symmetrical roles in their respective pathways, the patches of surface residues used by the paralogues Fus3 and Hog1 to interact with their up and downstream factors seem substantially different. This is highlighted by the hybrids (Fig. 5a, b, c) that are able to interact with both upstream partners, such as ABcdEF (which is one of three hybrids in which sorbitol evoked a pheromone response through Pbs2), or both downstream partners (aBcdeF). The hybrid aBcdeF yields both an osmolar and pheromone response to an osmolar input, but is non-responsive to pheromone (Fig. 5a), implying that it can be activated by Pbs2 alone but can activate both Ste12 and Hot1. As every segment is either from Fus3 or Hog1, the regions on Fus3 and Hog1 that interact with Ste12 and Hot1 must be considerably non-overlapping. Not only do we find that these MAPKs show a modular design, we also note that this design is implemented differently in the two MAPKs in spite of their great structural similarity. Thus, conformational change, which commonly underlies enzyme promiscuity in other synthetic proteins21, is unlikely to be the mechanism by which our hybrid kinases achieved their various cross-wirings and specificities.
We can uncover how MAPK proteins achieve specificity. Our results indicate that the segment BEF is required in conjunction with the segments A or D for pheromone input to invoke pheromone output (Fig. 4a). However, as aBcdeF activates the pheromone pathway only on osmolar input, and not on pheromone input, it appears that BF alone is sufficient for pheromone output and that E is important for pheromone input. The segment BF contains all the residues associated with the ‘docking domain’ (Fig. 3c; Supplementary Information, Section 2). These were identified and studied mainly in the context of Fus3 binding to Ste7 and to phosphatases10. However, that BF appears sufficient for pheromone output suggests it is more important invivo for the Fus3–Ste12 interaction, whereas E is more important for the Fus3–Ste7 interaction. The patch of residues (Pro 80, Phe 83, Glu 84 and Trp 348) identified by our sequence analysis to be unique to Fus3 is contained in BF. The hybrid aBCDEF transduces the pheromone signal to induce high pFUS1 activity but fails to mediate cell-cycle arrest (Supplementary Table 2). Consistently, the strains carrying aBCDEF have a low mating efficiency (Supplementary Information, Fig. S4a). This implicates segment A in Fus3–Far1 interaction. Segment d is necessary for transducing a high osmolar signal into any output (Fig. 4ab). In Fus3, D has a disordered structure and most of its residues undergo neutral drift. This again illustrates how members of this family have specialized different residue patches on their surfaces to achieve analogous specificities rather than refining the residues on some common catalytic loop22.
Our data show that these proteins can find new specificities with relatively few changes in their sequence (as few as seven point-mutations). They can retain their original function while acquiring new interaction partners. This might be why they have increased in number, having repeatedly found new functions after duplication (Supplementary Information, Fig. S10). Indeed our results suggest promiscuous intermediates through which duplicated enzymes could evolve, en route to their new specificities22. We have also gained insights into the specificity determinants of extant proteins. Exploration building on our hybrids can complement traditional biochemical techniques, which are challenged by the transient nature of kinase–substrate interactions. The implication for synthetic design is that pivotal proteins such as these may serve as the best templates, or scaffolds, with which to design new specificities to create new connections in existing pathways.
METHODS
Orthology and duplication
To construct the phylogeny and search for duplications involving a gene extant in S. cerevisiae, we found the best reciprocal BLAST hit in each species. The orthology relationship was established by confirming that orthologues in species sharing the same nearest common ancestor with S.cerevisiae had similar distances from the S. cerevisiae protein, and by noting the progressive increase in this distance for orthologues in species whose nearest common ancestor was further away (Supplementary Information, Fig. 1a). For the genes, putative orthologues were found in most species in one branch of a speciation event and not in the other, thereby making it obvious if, and in which, species this gene was lost. A simple indicator of the possibility of a duplication event was two proteins in S. cerevisiae having separate orthologues in all species on one side of a speciation event, but BLASTed to the same protein in species on the other side. Rather than multiple gene-loss events, a single duplication at the speciation event is the most parsimonious explanation. Again, the duplication was verified by viewing the protein sequences and BLAST scores in relation to the species tree (see Supplementary Information for more details).
Computational identification of variable and putative specificity residues in MAPKs
To identify variable residues, the entropy at a given position j (Fig. 3a; Supplementary Information, Fig. S1c) was computed from the multiple sequence alignment using . Here, i indexes the 20 types of amino acid, and is the normalized frequency with which amino acid i occurs at position j.
To identify the specificity patches of Fus3 and Hog1 in S. cerevisiae, we performed a multiple alignment of Fus3 and Hog1 orthologues from the yeast species. Higher eukaryotic orthologues were deliberately excluded because the MAPK specificities in this lineage are different (Fig. 2d). Fus3 (Hog1) orthologue residues appearing directly above or below each other at a given position in the alignment were scored pair-wise by the BLOSUM50 matrix, and the sum of these scores was taken as indicative of the level of conservation at that position in Fus3 (Hog1). The linear ordering of the orthologues, on which this number depends, was taken to be that best representing the evolutionary relationships of the underlying phylogenetic tree. A plot of the score versus positions on Fus3 (Supplementary Information, Fig. 1b; sorted by increasing scores) shows a conspicuous plateau at a score of 42, indicating a natural cut-off point above which a position is declared to be ‘well conserved’. For a position to qualify as putatively endowing specificity to Fus3 it also had to be different at the corresponding position in Hog1. This difference was quantified by summing the pair-wise BLOSUM50 scores for the Fus3 and Hog1 residues taken from orthologues belonging to the same species. A negative score was taken to signify a difference.
Yeast strains, plasmids and growth conditions
See Supplementary Table 1 for the list of strains and plasmids used in this study. Cells transformed with plasmids were grown overnight at 30 °C in a selective medium of SC-URA, re-inoculated into fresh medium for 8 h of exponential growth and then incubated with pheromone (0.6 μM) and sorbitol (1 M). The time-dependence of the reporter activity was measured using fluorescence microscopy (Fig. 4 reports the mean fluorescence after 2 h; Supplementary Information, Figs S6, S7a).
Construction of hybrid MAPKs
See Supplementary Information, Fig. S2 for the amino-acid sequences of the six segments A/a, B/b, C/c, D/d, E/e and F/f in Fus3/Hog1. The base-pair sequences of the hybrids were designed ab initio as follows: Codons for the common conserved regions separating the segments were selected to create unique restriction sites recognized by five different restriction enzymes. Achieving this in the region separating E/e and F/f, where the amino-acid sequences of Fus3 and Hog1 are not identical, effectively introduced two mutations in the amino-acid sequence of Fus3: Gln 287, Arg 288 to Glu 287, Lys 288, forcing them to agree with the corresponding residues of Hog1. The substitution of Gln to Glu is seen in the closely related sensu strictu yeast Saccharomyces kudriavzevii, and the substitution Arg to Lys is extremely conservative as both Arg and Lys are positively charged. All other codons in the genes were optimized for expression in S. cerevisiae.
Microscopy
Cells were observed using a Zeiss 200M fluorescent microscope and Orca-II-ER and Hamamatsu EM-CCD cameras. Objectives used were: ×100 with N.A. = 1.45 and ×63 with N.A. = 1.4. Emission of CFP was visualized at 470 nm (30-nm bandwidth) on excitation at 430 nm (25-nm bandwidth). Emission from YFP was collected at 535 nm (30-nm bandwidth) on excitation at 500 nm (20-nm bandwidth). Emission from GFP was collected at 528 nm (38-nm bandwidth) on excitation at 490 nm (20-nm bandwidth).
Mating assay
fus3Δ kss1Δ MATa strains containing the hybrid MAPKs were grown in a selective medium to exponential phase, and then plated on YPD simultaneously with a MATα strain, also grown to exponential phase. By measuring the cell density of the two cultures before plating, an equal number of cells of each mating type was plated. After 36 h, cells scraped off the resulting lawn were inoculated in YPD for 2 h before plating a threefold serial dilution of 10 μl spots on plates selected for diploids (Supplementary Information, Fig. 4a). As only the MATa strains were autotrophic for the amino acids histidine and tryptophan, and only the MATα strain contained a drug-resistant gene (resistant to Kanamycin), plates lacking histidine and tryptophan and containing the drug were used to select for diploids. Again, the cell density was carefully measured to ensure that the same initial density of cells was used for plating the first spot.
Image processing
The mean fold changes in fluorescence (Fig. 4) were computed by dividing the increase in fluorescence levels averaged over ~100 stimulated cells by the mean fluorescence levels averaged over unstimulated cells. The pheromone-induced pSTL1 fold change in cells carrying AbCdEf and aBcdEF was computed (Fig. 4b) using data from a selected fraction of the cells imaged, as approximately 50% of the cells carrying AbCdEf and about 5% of the cells carrying aBcdEF showed a response.
Supplementary Material
Acknowledgments
We thank B. Stern for critical readings and suggestions, H. Dohlman, A. Drummond, M. DePristo, A. Murray, D. Huse, D. Fisher, M. McClean, M. Thomson and I. Nachman for discussions and comments, and R.e Hellmiss for help with figures. Work was supported by an NIH grant (2P50GM068763).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
AUTHOR CONTRIBUTIONS
A. M. and S. R. conceived and planned the project and wrote the manuscript. A.M. designed and implemented the computational aspects of the project, performed the microscopy and analysed the data. J.W. and A.M. performed the strain and plasmid construction.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 2.Stelling J, Sauer U, Szallasi Z, Doyle FJ, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Wuchty S, Oltvai ZN, Barabasi AL. Evolutionary conservation of motif constituents in the yeast protein interaction network. Nature Genet. 2003;35:176–179. doi: 10.1038/ng1242. [DOI] [PubMed] [Google Scholar]
- 4.Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabbagh W, Jr, Flatauer LJ, Bardwell AJ, Bardwell L. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell. 2001;8:683–691. doi: 10.1016/s1097-2765(01)00322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 7.Sipiczki M. Where does fission yeast sit on the tree of life? Genome Biol. 2000;1:R1011. doi: 10.1186/gb-2000-1-2-reviews1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharyya RP, et al. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 10.Remenyi A, Good MC, Bhattacharyya RP, Lim WA. The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol Cell. 2005;20:951–962. doi: 10.1016/j.molcel.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J. Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol. 1996;16:3637–3650. doi: 10.1128/mcb.16.7.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nature Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 13.Chang CI, Xu BE, Akella R, Cobb MH, Goldsmith EJ. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- 14.Goldsmith EJ, Cobb MH, Chang CI. Structure of MAPKs. Methods Mol Biol. 2004;250:127–144. doi: 10.1385/1-59259-671-1:127. [DOI] [PubMed] [Google Scholar]
- 15.Bardwell AJ, Abdollahi M, Bardwell L. Docking sites on mitogen-activated protein kinase (MAPK) kinases, MAPK phosphatases and the Elk-1 transcription factor compete for MAPK binding and are crucial for enzymic activity. Biochem J. 2003;370:1077–1085. doi: 10.1042/BJ20021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet A, Pouyssegur J. Identification of MAP kinase domains by redirecting stress signals into growth factor responses. Science. 1996;272:1652–1655. doi: 10.1126/science.272.5268.1652. [DOI] [PubMed] [Google Scholar]
- 17.Mansour SJ, Candia JM, Gloor KK, Ahn NG. Constitutively active mitogen-activated protein kinase kinase 1 (MAPKK1) and MAPKK2 mediate similar transcriptional and morphological responses. Cell Growth Differ. 1996;7:243–250. [PubMed] [Google Scholar]
- 18.Zhang J, Zhang F, Ebert D, Cobb MH, Goldsmith EJ. Activity of the MAP kinase ERK2 is controlled by a flexible surface loop. Structure. 1995;3:299–307. doi: 10.1016/s0969-2126(01)00160-5. [DOI] [PubMed] [Google Scholar]
- 19.Diskin R, Askari N, Capone R, Engelberg D, Livnah O. Active mutants of the human p38alpha mitogen-activated protein kinase. J Biol Chem. 2004;279:47040–47049. doi: 10.1074/jbc.M404595200. [DOI] [PubMed] [Google Scholar]
- 20.O’Rourke SM, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr Opin Chem Biol. 2006;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 22.Glasner ME, Gerlt JA, Babbitt PC. Evolution of enzyme superfamilies. Curr Opin Chem Biol. 2006;10:492–497. doi: 10.1016/j.cbpa.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 24.Gelli A. Rst1 and Rst2 are required for the a/alpha diploid cell type in yeast. Mol Microbiol. 2002;46:845–854. doi: 10.1046/j.1365-2958.2002.03213.x. [DOI] [PubMed] [Google Scholar]
- 25.Yang SH, Galanis A, Sharrocks AD. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol Cell Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.