Abstract
AIM: To explore epigenetic changes in the gene encoding X chromosome-linked inhibitor of apoptosis-associated factor 1 (XAF1) during esophageal carcinogenesis.
METHODS: Methylation status of XAF1 was detected by methylation-specific polymerase chain reaction (MSP) in four esophageal cancer cell lines (KYSE30, KYSE70, BIC1 and partially methylated in TE3 cell lines), nine cases of normal mucosa, 72 cases of primary esophageal cancer and matched adjacent tissue. XAF1 expression was examined by semi-quantitative reverse transcriptional polymerase chain reaction and Western blotting before and after treatment with 5-aza-deoxycytidine (5-aza-dc), a demethylating agent. To investigate the correlation of XAF1 expression and methylation status in primary esophageal cancer, immunohistochemistry for XAF1 expression was performed in 32 cases of esophageal cancer and matched adjacent tissue. The association of methylation status and clinicopathological data was analyzed by logistic regression.
RESULTS: MSP results were as follows: loss of XAF1 expression was found in three of four esophageal cell lines with promoter region hypermethylation (completely methylated in KYSE30, KYSE70 and BIC1 cell lines and partially in TE3 cells); all nine cases of normal esophageal mucosa were unmethylated; and 54/72 (75.00%) samples from patients with esophageal cancer were methylated, and 25/72 (34.70%) matched adjacent tissues were methylated (75.00% vs 34.70%, χ2 = 23.5840, P = 0.000). mRNA level of XAF1 measured with semi-quantitative reverse transcription polymerase chain reaction was detectable only in TE3 cells, and no expression was detected in KYSE30, KYSE70 or BIC1 cells. Protein expression was not observed in KYSE30 cells by Western blotting before treatment with 5-aza-dc. After treatment, mRNA level of XAF1 was detectable in KYSE30, KYSE70 and BIC1 cells. Protein expression was detected in KYSE30 after treatment with 5-aza-dc. Immunohistochemistry was performed on 32 cases of esophageal cancer and adjacent tissue, and demonstrated XAF1 in the nucleus and cytoplasm. XAF1 staining was found in 20/32 samples of adjacent normal tissue but was present in only 8/32 samples of esophageal cancer tissue (χ2= 9.143, P = 0.002). XAF1 expression was decreased in cancer samples compared with adjacent tissues. In 32 cases of esophageal cancer, 24/32 samples were methylated, and 8/32 esophageal cancer tissues were unmethylated. XAF1 staining was found in 6/8 samples of unmethylated esophageal cancer and 2/24 samples of methylated esophageal cancer tissue. XAF1 staining was inversely correlated with XAF1 promoter region methylation (Fisher’s exact test, P = 0.004). Regarding methylation status and clinicopathological data, no significant differences were found in sex, age, tumor size, tumor stage, or metastasis with respect to methylation of XAF1 for the 72 tissue samples from patients with esophageal cancer.
CONCLUSION: XAF1 is frequently methylated in esophageal cancer, and XAF1 expression is regulated by promoter region hypermethylation.
Keywords: X chromosome-linked inhibitor of apoptosis-associated factor 1, Esophageal cancer, Methylation, Methylation-specific polymerase chain reaction, Semi-quantitative reverse transcriptional polymerase chain reaction
INTRODUCTION
Esophageal cancer is the eighth most common cancer worldwide. The incidence of esophageal cancer has increased rapidly in the past 20 years, and about 380 000 patients die each year from the disease[1-3]. Esophageal squamous cell carcinoma (ESCC) is the main type of esophageal cancer. It is highly invasive, rapidly metastatic, and results in a poor postoperative quality of life[4,5]. The mechanisms contributing to ESCC carcinogenesis are poorly understood. Recent studies have shown that aberrant promoter DNA methylation contributes to gene silencing and may participate in the carcinogenesis of human cancer. In-depth investigation of the relationship between DNA methylation and gene expression in ESCC will facilitate research in tumor pathogenesis and guide clinical practice.
Inhibitors of apoptosis (IAPs) are antiapoptotic factors in cancer cells that render cells resistant to apoptosis by inhibition of core death executioners, the caspases, or by neutralizing antagonists[6]. In the IAP family, X chromosome-linked IAP (XIAP) has been recognized as the most versatile caspase inhibitor[7,8]. In many models of cancer, XIAP is overexpressed[9]. XIAP-associated factor 1 (XAF1) is one of the antagonists that has been identified as a mediator of XIAP by rescuing XIAP-suppressed caspase activity[10]. XAF1 is also a new candidate tumor suppressor. Recent studies have suggested that loss of XAF1 expression may occur in different human cancers because of aberrant DNA methylation[11-17]. Zou et al[15] found that loss of XAF1 expression is associated with tumor progression in human gastric and colon cancers. Lee et al[14] also discovered that downregulation of XAF1 expression is correlated with human urogenital malignancies. However, the relationship between the expression level of XAF1 and the methylation status of XAF1 in esophageal cancer has not been demonstrated.
In this study, we investigated whether promoter region methylation was associated with the progression of esophageal cancer and analyzed the relationship between XAF1 expression and promoter region methylation. We identified XAF1 as a potential esophageal cancer biomarker for prognosis and a target for future therapeutic agents. Detection of the methylation status of XAF1 appears to be promising as a predictive factor in primary ESCC.
MATERIALS AND METHODS
Human tissue samples and cell lines
Tissue samples taken from 72 cases of ESCC and 72 matched adjacent normal tissues were used in this study. Nine cases of normal esophageal epithelia were removed during endoscopy biopsy and then snap frozen. All samples were collected from the Chinese PLA General Hospital under the guidelines approved by the Institutional Review Board of the Chinese PLA General Hospital.
Four esophageal cancer cell lines (KYSE30, KYSE70, BIC1 and TE3) were examined in this study. All esophageal cancer cell lines were previously established from primary esophageal cancer and were maintained in 90% RPMI 1640 (Invitrogen, Carlsbad, CA, United States) supplemented with 10% fetal bovine serum. Cells were passaged once at a ratio of 1:3. Cells were then allowed to grow to total confluence (about 106 cells) on a 75-cm2 culture flask (NEST Biotechnology, Jiangsu, China).
Esophageal cancer cell lines were split to low density (30% confluence) 12 h before treatment. Cells were treated with 2 mol/L 5-aza-2’-deoxycytidine (5-aza-dc) (Sigma, St. Louis, MO, United States), a demethylating agent, which was added fresh every 24 h for a total of 96 h. At the end of the treatment, RNA and protein were extracted from the cells (see below).
RNA isolation and reverse transcriptional polymerase chain reaction
Total RNA was isolated with the Trizol reagent (Life Technologies, Gaithersburg, MD, United States). Agarose gel (1%) electrophoresis and spectrophotometric analysis (A260 nm/A280 nm ratio) were used to evaluate RNA quality and quantity. RNA was stored at -80 °C prior to use. First-strand cDNA was synthesized from 5 μg total RNA with random 6-mer primers and a Superscript II reverse transcriptional kit (Invitrogen). The reaction mixture was then diluted to 100 μL with water. Subsequently, 2.5 μL of this diluted cDNA mixture was used for polymerase chain reaction (PCR) amplification in a 25 μL reaction (final volume). PCR amplification of XAF1 was carried out using primers 5′-GAGCATGCAGAAGTCCTCGCT-3′ (forward) and 5′-CCTGTTCACTGCGACAGACATCT-3′ (reverse). The primer set for XAF1 was designed to span intronic sequences between exons to exclude amplification of genomic DNA. A total of 32 cycles of amplification was performed for each reverse transcriptional polymerase chain reaction (RT-PCR) experiment. As an internal control, glyceraldehyde-3-phosphate dehydrogenase was amplified with 25 cycles to ensure cDNA quality and quantity for each RT-PCR reaction. Amplified products were analyzed on 1.5% agarose gels.
Methylation-specific PCR
Genomic DNA from all types of samples was prepared using the proteinase K method. After phenol/chloroform extraction, DNA was precipitated in ethanol, dissolved in low TE buffer, and stored at -20 °C. Genomic DNA from esophageal cancer, adjacent tissues, and cell lines was bisulfite modified as described before[18]. Methylation-specific PCR (MSP) was carried out using primers XAF1-ML: 5′-TTTGTAAGAAACGAAATTTAATCGA-3′ and XAF1-MR: 5′-CCTACCCTTAAAACCCACGAT-3′ and XAF1-UL: 5′-TTTGTAAGAAATGAAATTTAATTGA-3′ and XAF1-UR: 5′-CTCCTACCCTTAAAACCCACAAT-3′[10]. Each MSP reaction included about 200 ng bisulfite-treated DNA, 25 pmol each primer, 100 pmol dNTPs, 2.5 μL 10 × PCR buffer, and 1 U Taq Polymerase (Invitrogen) in a final reaction volume of 25 μL. Cycle conditions were as follows: 95 °C for 10 min; 35 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s; and 72 °C for 10 min. MSP products were analyzed using 2% agarose gel electrophoresis.
Immunohistochemistry
Immunohistochemistry was performed on 4-μm-thick serial sections cut from paraffin blocks containing formaldehyde-fixed esophageal cancer tissue and paired adjacent tissue. After deparaffinization and rehydration, endogenous peroxidase activity was blocked for 30 min in methanol containing 0.3% hydrogen peroxide. Antigen retrieval was performed in target retrieval solution for 45 min at 96 °C, which was followed by a cooling-off period of 20 min. The primary rabbit antibody (anti-XAF1, 1:200; OriGene Technologies, MD, United States) was then incubated overnight at 4 °C. Then, the catalyzed signal amplification system (ZSGB Biotech., Beijing, China) was used to detect XAF1 staining.
Protein preparation and Western blotting
KYSE30 cells were treated with 5-aza-dc (as described above), harvested, and lysed in ice-cold Tris buffer (20 mmol/L Tris, pH 7.5) containing 137 mmol/L NaCl, 2 mmol/L ethylene diamine tetraacetic acid, 1% Triton X-100, 10% glycerol, 50 mmol/L NaF, 1 mmol/L dithiothreitol, and a protease inhibitor cocktail (Roche Applied Science). Cell lysate (35 μg) was loaded into each lane, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and electroblotted onto PVDF membranes (Hybond-P; Amersham, United States). After being blocked with 5% nonfat milk and 0.1% Tween-20 in TBS, the membranes were incubated with primary rabbit anti-XAF1 (1:1000; OriGene Technologies). Rabbit anti-actin (Beyotime Biotech., China) was used as a loading control. The blots were visualized using enhanced chemiluminescence (Pierce Bioscience, Rockford, IL, United States).
Statistical analysis
Statistical analysis was carried out using the χ2 test and Fisher’s exact test. The relationship between methylation status and clinicopathological data was carried out using multiple logistic regression. P < 0.05 was considered statistically significant.
RESULTS
XAF1 promoter methylation status in esophageal cancer
To ascertain whether the XAF1 promoter methylation status was associated with esophageal carcinogenesis, MSP was performed on nine cases of normal esophageal mucosa, 72 samples taken from patients with esophageal cancer, and 72 paired adjacent normal tissue samples. All nine cases of normal esophageal mucosa were unmethylated; 54 of 72 (75%) samples taken from patients with esophageal cancer were methylated; and 25 of 72 (34.7%) matched adjacent normal tissues were methylated (χ2 = 23.5840, P = 0.000; Figure 1B). These results suggest that methylation of the XAF1 promoter region is a potential early detection marker of esophageal cancer.
Figure 1.
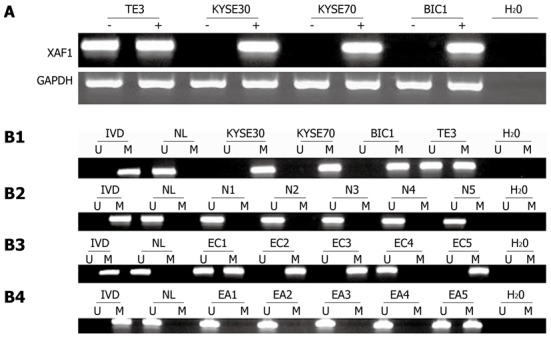
X chromosome-linked inhibitor of apoptosis-associated factor 1 expression was silenced by DNA methylation. A: X chromosome-linked inhibitor of apoptosis-associated factor 1 (XAF1) expression was analyzed by semi-quantitative reverse transcriptional polymerase chain reaction before and after 5-aza-dc treatment (2 mol/L, 96 h) of the esophageal cancer cell lines (KYSE30, KYSE70, BIC1 and TE3). Methylation status of XAF1 CpG islands in esophageal cancer cell lines, esophageal normal mucosa, esophageal cancer tissue, and matched adjacent normal tissue. Primer efficiency was verified with a positive control (in vitro methylated DNA, IVD) and a negative control (normal blood lymphocyte DNA, NL). “U” indicates the presence of unmethylated alleles; “M” indicates the presence of methylated alleles; B1: Methylation of XAF1 in esophageal cancer cell lines (KYSE30, KYSE70, BIC1 and TE3); B2: Methylation of XAF1 in normal esophageal mucosa (NE1, NE2, NE3, NE4 and NE5); B3: Representative methylation-specific polymerase chain reaction (MSP) results for XAF1 in esophageal primary cancer tissue samples (EC); B4: Representative MSP results for XAF1 in esophageal matched adjacent normal tissue (EA). GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
XAF1 promoter methylation and XAF1 expression in esophageal cancer cell lines
To determine whether XAF1 is a tumor suppressor in esophageal cancer tumorigenesis, XAF1 expression levels were detected with semi-quantitative RT-PCR and Western blotting. The mRNA level of XAF1 was detectable only in the TE3 cell line, and no expression was detected in the KYSE30, KYSE70 or BIC1 cell lines (Figure 1A). Protein expression was not observed in the KYSE30 cell line before treatment with 5-aza-dc (Figure 2). To examine if these findings were due to promoter region methylation of XAF1, the methylation status of XAF1 was analyzed in these cells lines with MSP. XAF1 was completely methylated in the KYSE30, KYSE70 and BIC1 cell lines and partially methylated in TE3 cells (Figure 1B).
Figure 2.
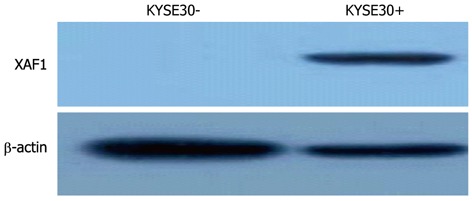
Western blotting analysis of X chromosome-linked inhibitor of apoptosis-associated factor 1 protein expression in the KYSE30 cell line before and after treatment with 2 mol/L 5-aza-deoxycytidine (+) for 96 h. XAF1: X chromosome-linked inhibitor of apoptosis-associated factor 1.
To demonstrate further whether XAF1 expression was restored with promoter region methylation, these cell lines were treated with 5-aza-dc. As shown in Figure 1, XAF1 was expressed in KYSE30, KYSE70 and BIC1 cells after 5-aza-dc treatment, and we detected protein expression in KYSE30 after treatment with 5-aza-dc (Figure 2), suggesting that XAF1 expression was regulated by promoter region methylation in esophageal cancer.
Correlation of XAF1 expression and methylation status in esophageal cancer tissue
To explore the expression of XAF1, immunohistochemistry was performed on 32 cases of esophageal cancer and adjacent tissue paraffin samples. XAF1 expression was found in the nucleus and cytoplasm as previously reported[19,20]. XAF1 staining was found in 50% (16/32) of adjacent tissue samples and only in 25% (8/32) of cancer tissue samples (χ2 = 9.143, P = 0.002). Reduced expression of XAF1 was found in cancer tissues as compared with the adjacent tissue samples (Figure 3).
Figure 3.
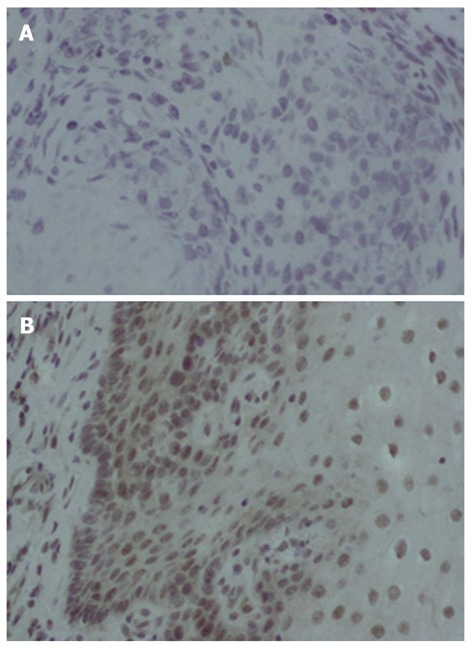
Immunohistochemistry analysis of X chromosome-linked inhibitor of apoptosis-associated factor 1 in esophageal cancer tissue and adjacent tissue. Esophageal cancer and adjacent normal tissue samples were immunohistologically analyzed with anti-X chromosome-linked inhibitor of apoptosis-associated factor 1 (XAF1) (1:200 dilution; × 400). A: XAF1 was not detected in esophageal cancer tissue; B: XAF1 was localized in the nucleus and cytoplasm in adjacent normal esophageal tissue.
We then analyzed the relationship between the methylation status and expression of XAF1 in 32 esophageal cancer samples. Twenty-four esophageal cancer samples (24/32, 75.00%) were methylated, and eight esophageal cancer tissue (8/32, 25.00%) were unmethylated. XAF1 staining was found in six samples (6/8, 75.00%) of unmethylated esophageal cancer tissue and two samples (2/24, 8.33%) of methylated esophageal cancer tissue. XAF1 staining was inversely correlated with XAF1 promoter region methylation (Fisher’s exact test, P = 0.004).
Correlation of XAF1 methylation status with clinicopathological factors
Regarding methylation status and clinicopathological data, we found no significant differences in sex, age, tumor size, tumor stage, or metastasis with respect to the methylation status of XAF1 for the 72 primary esophageal cancer patients (Table 1).
Table 1.
Clinicopathological characteristics and XAF1 methylation status of 40 patients with esophageal cancer n (%)
| Clinical parameter | n |
XAF1 methylation status |
P value | |
| Methylatedn = 30 (75%) | Unmethylatedn = 10 (25%) | (χ2test) | ||
| Age (yr) | ||||
| < 65 | 40 | 30 (75) | 10 (25) | 1.0 |
| ≥ 65 | 32 | 24 (75) | 8 (25) | |
| Gender | ||||
| Male | 53 | 40 (75.5) | 13 (24.5) | 0.8773 |
| Female | 19 | 14 (73.7) | 5 (26.3) | |
| Tumor size (cm) | ||||
| < 5 | 48 | 33 (68.8) | 15 (31.2) | |
| ≥ 5 | 24 | 21 (87.5) | 3 (12.5) | 0.0833 |
| Tumor stage | ||||
| I | 12 | 6 (50.0) | 6 (50.0) | 0.081 |
| II | 21 | 17 (81.0) | 4 (19.0) | |
| III | 6 | 6 (100.0) | 0 (0) | |
| IV | 1 | 1 (100) | 0 (0) | |
| Metastasis | ||||
| Negative | 45 | 33 (73.3) | 12 (26.7) | 0.6733 |
| Positive | 27 | 21 (77.8) | 6 (22.2) | |
In this 72 patients, only 32 cases with tumor stage history.
DISCUSSION
The development of cancer is influenced by many factors and many genes and progresses through many stages[21]. In some ways, cancer is considered an epigenetic disease as well as a genetic disease. Epigenetics is the inheritance of information based on gene expression levels, whereas genetics refers to information transmitted according to the gene sequence. Epigenetic changes may lead to chromosomal instability, activation of parasitic endogenous sequences, loss of imprinting, illegitimate expression, aneuploidy, and mutations, and it may contribute to the transcriptional silencing of tumor suppressor genes[22]. In the human genome, CpG dinucleotides are inconsonantly distributed, resulting in CpG-rich regions[23]. The main epigenetic modification is aberrant CpG island methylation, which is tissue-specific but not species-specific. Methylation affects many pathways in cellular networks, such as the cell cycle and apoptosis[22].
In life and death decisions at the cellular level, there is a balance between pro- and antiapoptotic factors, and a variety of pathological conditions such as cancer and autoimmune and neurodegenerative diseases can result from disruption of this balance[6]. Apoptosis is crucial for eliminating defective or potentially dangerous cells and provides a defense against malignant transformation and autoimmunity[11]. IAPs are a new family of intrinsic cell death proteins that work as endogenous caspase inhibitors and participate in cell cycle regulation and modulation of receptor-mediated signal transduction. A recent study has reported that the level of XIAP mRNA is relatively high in many human cancers, suggesting that XIAP is one of the most potent and versatile inhibitors of caspases and apoptosis[15].
XAF1 has been previously identified as a binding partner of XIAP. In contrast to Smac/DIABLO and HtrA2, which promote caspase activation, XAF1 reverses XIAP-mediated inhibition of caspase-3 activity. Furthermore, XAF1 induces cell cycle arrest during G2/M phase and mitotic catastrophe, and the restoration of XAF1 expression induces cancer cell apoptosis and inhibits tumor growth in many types of human cancers[24]. XAF1 is ubiquitously expressed in all normal adult and fetal tissues but is drastically decreased in many cancer cell lines[11]. Loss of XAF1 expression is associated with methylation in its promoter region in many cancers. For example, XAF1 is present at very low or undetectable levels in gastric cancer, colorectal cancer[15], and cervical carcinoma[25].
In this study, we found that XAF1 was frequently methylated in esophageal cancer. Moreover, expression of XAF1 was inversely correlated with its methylation status. XAF1 was methylated in three esophageal cancer cell lines and 54 samples of esophageal cancer tissue. XAF1 methylation resulted in loss of expression in esophageal cancer cell lines, and the expression of XAF1 was restored in KYSE30, KYSE70 and BIC1 cells after treatment with 5-aza-dc. Furthermore, we observed the expression of XAF1 in the methylated cell line KYSE30 after treatment with 5-aza-dc. The results indicated that promoter region methylation regulated the expression of XAF1. XAF1 was frequently methylated in esophageal cancer tissue (75%) but was methylated only in 34.7% of matched adjacent normal tissues and not at all in normal esophageal mucosa, indicating that promoter region methylation of XAF1 was likely to be related to esophageal carcinogenesis. In addition, XAF1 protein expression was decreased in cancer tissues as compared with adjacent normal samples, and low expression of XAF1 was significantly correlated with promoter region methylation. XAF1 expression and promoter region methylation status have been reported to be useful for identifying poorly differentiated cancer or patients with a poor disease outcome[16,17,26].
XAF1 is frequently methylated in esophageal cancer, and XAF1 expression is regulated by promoter region methylation. The loss of XAF1 expression may play an important role in tumor growth, and methylation of XAF1 may serve as an early detection marker for esophageal cancer.
COMMENTS
Background
Esophageal cancer is the eighth most common cancer worldwide. The incidence of esophageal cancer has increased rapidly in the past 20 years. Esophageal squamous cell carcinoma (ESCC) is the main type of esophageal cancer. It is highly invasive, rapidly metastatic, and results in a poor postoperative quality of life. The mechanisms contributing to ESCC carcinogenesis are poorly understood. Recent studies showed that aberrant promoter DNA methylation contributes to gene silencing and may participate in the carcinogenesis of human cancer. X chromosome-linked inhibitor of apoptosis (XIAP)-associated factor 1 (XAF1) is a new candidate tumor suppressor gene. Recent studies have suggested that loss of XAF1 expression may occur in different human cancers because of aberrant DNA methylation. However, the relationship between the expression level of XAF1 and the methylation status of XAF1 in esophageal cancer has not been demonstrated.
Research frontiers
XAF1 is a new candidate tumor suppressor gene. Recent studies have suggested that loss of XAF1 expression may occur in human gastric cancer, colon cancer and urogenital malignancies because of aberrant DNA methylation. However, the relationship between expression level of XAF1 and methylation status of XAF1 in esophageal cancer has not been demonstrated. In this study, the authors demonstrated that expression level of XAF1 was inversely correlated with methylation status of XAF1, and XAF1 expression was regulated by promoter region methylation.
Innovations and breakthroughs
Recent reports have highlighted that the loss of XAF1 expression or downregulation of XAF1 expression may occur in different human cancers because of aberrant DNA methylation. This is the first study to report that XAF1 is also loss of expression because of aberrant DNA methylation in esophageal cancer.
Applications
By understanding the relationship between loss of XAF1 expression and methylation status of XAF1 in esophageal cancer, and by inducing its expression with 5-aza-deoxycytidine, this study may represent a future strategy for therapeutic intervention in the treatment of patients with esophageal cancer.
Terminology
Inhibiters of apoptosis (IAPs) are antiapoptotic factors in cancer cells that render cells resistant to apoptosis by inhibition of core death executioners, the caspases, or by neutralizing antagonists. In the IAP family, XIAP has been recognized as the most versatile caspase inhibitor. In many models of cancer, XIAP is overexpressed. XAF1 is one of the antagonists that has been identified as a mediator of XIAP by rescuing XIAP-suppressed caspase activity. XAF1 is a new candidate tumor suppressor gene. Recent studies have suggested that loss of XAF1 expression may occur in different human cancers because of aberrant DNA methylation.
Peer review
The study is very interesting and throws light on future studies and confirms increased methylation of XAF1 in squamous cell carcinoma.
Footnotes
Supported by Grants from the National Basic Research Program (973 Program), No. 2012CB934002, 2010CB912802; National Key Scientific Instrument Special Programme of China, No. 2011YQ03013405; and National Science Foundation of China, No. 81121004, 81071953 and 81161120432
Peer reviewer: Dr. Subbaramiah Sridhar, Medical College of Georgia, BBR 2544, Medical College of Georgia,15th Street, Augusta, GA 30912, United States
S- Editor Cheng JX L- Editor Kerr C E- Editor Zhang DN
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 3.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 4.Naidoo R, Ramburan A, Reddi A, Chetty R. Aberrations in the mismatch repair genes and the clinical impact on oesophageal squamous carcinomas from a high incidence area in South Africa. J Clin Pathol. 2005;58:281–284. doi: 10.1136/jcp.2003.014290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 6.Wright CW, Duckett CS. Reawakening the cellular death program in neoplasia through the therapeutic blockade of IAP function. J Clin Invest. 2005;115:2673–2678. doi: 10.1172/JCI26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y, Nakabayashi Y, Nakata K, Reed JC, Takahashi R. X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase-3 and -7 in distinct modes. J Biol Chem. 2001;276:27058–27063. doi: 10.1074/jbc.M102415200. [DOI] [PubMed] [Google Scholar]
- 9.Karikari CA, Roy I, Tryggestad E, Feldmann G, Pinilla C, Welsh K, Reed JC, Armour EP, Wong J, Herman J, et al. Targeting the apoptotic machinery in pancreatic cancers using small-molecule antagonists of the X-linked inhibitor of apoptosis protein. Mol Cancer Ther. 2007;6:957–966. doi: 10.1158/1535-7163.MCT-06-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liston P, Fong WG, Kelly NL, Toji S, Miyazaki T, Conte D, Tamai K, Craig CG, McBurney MW, Korneluk RG. Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat Cell Biol. 2001;3:128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- 11.Byun DS, Cho K, Ryu BK, Lee MG, Kang MJ, Kim HR, Chi SG. Hypermethylation of XIAP-associated factor 1, a putative tumor suppressor gene from the 17p13.2 locus, in human gastric adenocarcinomas. Cancer Res. 2003;63:7068–7075. [PubMed] [Google Scholar]
- 12.Fang X, Liu Z, Fan Y, Zheng C, Nilson S, Egevad L, Ekman P, Xu D. Switch to full-length of XAF1 mRNA expression in prostate cancer cells by the DNA methylation inhibitor. Int J Cancer. 2006;118:2485–2489. doi: 10.1002/ijc.21636. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Peng Y, Sun YW, He H, Zhu S, An X, Li M, Lin MC, Zou B, Xia HH, et al. All-trans retinoic acid induces XAF1 expression through an interferon regulatory factor-1 element in colon cancer. Gastroenterology. 2006;130:747–758. doi: 10.1053/j.gastro.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 14.Lee MG, Huh JS, Chung SK, Lee JH, Byun DS, Ryu BK, Kang MJ, Chae KS, Lee SJ, Lee CH, et al. Promoter CpG hypermethylation and downregulation of XAF1 expression in human urogenital malignancies: implication for attenuated p53 response to apoptotic stresses. Oncogene. 2006;25:5807–5822. doi: 10.1038/sj.onc.1209867. [DOI] [PubMed] [Google Scholar]
- 15.Zou B, Chim CS, Zeng H, Leung SY, Yang Y, Tu SP, Lin MC, Wang J, He H, Jiang SH, et al. Correlation between the single-site CpG methylation and expression silencing of the XAF1 gene in human gastric and colon cancers. Gastroenterology. 2006;131:1835–1843. doi: 10.1053/j.gastro.2006.09.050. [DOI] [PubMed] [Google Scholar]
- 16.Chung SK, Lee MG, Ryu BK, Lee JH, Han J, Byun DS, Chae KS, Lee KY, Jang JY, Kim HJ, et al. Frequent alteration of XAF1 in human colorectal cancers: implication for tumor cell resistance to apoptotic stresses. Gastroenterology. 2007;132:2459–2477. doi: 10.1053/j.gastro.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 17.Kempkensteffen C, Hinz S, Schrader M, Christoph F, Magheli A, Krause H, Schostak M, Miller K, Weikert S. Gene expression and promoter methylation of the XIAP-associated Factor 1 in renal cell carcinomas: correlations with pathology and outcome. Cancer Lett. 2007;254:227–235. doi: 10.1016/j.canlet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng KC, Campos EI, Martinka M, Li G. XAF1 expression is significantly reduced in human melanoma. J Invest Dermatol. 2004;123:1127–1134. doi: 10.1111/j.0022-202X.2004.23467.x. [DOI] [PubMed] [Google Scholar]
- 20.Straszewski-Chavez SL, Visintin IP, Karassina N, Los G, Liston P, Halaban R, Fadiel A, Mor G. XAF1 mediates tumor necrosis factor-alpha-induced apoptosis and X-linked inhibitor of apoptosis cleavage by acting through the mitochondrial pathway. J Biol Chem. 2007;282:13059–13072. doi: 10.1074/jbc.M609038200. [DOI] [PubMed] [Google Scholar]
- 21.An JY, Fan ZM, Gao SS, Zhuang ZH, Qin YR, Li JL, He X, Tsao GS, Wang LD. Loss of heterozygosity in multistage carcinogenesis of esophageal carcinoma at high-incidence area in Henan Province, China. World J Gastroenterol. 2005;11:2055–2060. doi: 10.3748/wjg.v11.i14.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 23.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 24.Sun PH, Zhu LM, Qiao MM, Zhang YP, Jiang SH, Wu YL, Tu SP. The XAF1 tumor suppressor induces autophagic cell death via upregulation of Beclin-1 and inhibition of Akt pathway. Cancer Lett. 2011;310:170–180. doi: 10.1016/j.canlet.2011.06.037. [DOI] [PubMed] [Google Scholar]
- 25.Micali OC, Cheung HH, Plenchette S, Hurley SL, Liston P, LaCasse EC, Korneluk RG. Silencing of the XAF1 gene by promoter hypermethylation in cancer cells and reactivation to TRAIL-sensitization by IFN-beta. BMC Cancer. 2007;7:52. doi: 10.1186/1471-2407-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakemi R, Yano H, Ogasawara S, Akiba J, Nakashima O, Fukahori S, Sata M, Kojiro M. X-linked inhibitor of apoptosis (XIAP) and XIAP-associated factor-1 expressions and their relationship to apoptosis in human hepatocellular carcinoma and non-cancerous liver tissues. Oncol Rep. 2007;18:65–70. [PubMed] [Google Scholar]


