Abstract
Cancer recapitulates Darwinian evolution. Mutations acquired during life that provide cells with a growth or survival advantage will preferentially multiply to form a tumor. As a result of The Cancer Genome Atlas project we have now gathered detailed information on the nucleotide sequence changes in a number of human cancers. The sources of mutations in cancer are diverse and the complexity of those found to be clonally present in tumors has increasingly made it difficult to identify key rate-limiting genes for tumor growth that might serve as potential targets for directed therapies. The impact of DNA sequencing on future cancer research and personalized therapy is likely to be profound and merits critical evaluation.
Keywords: Evolution, Cancer Genome, DNA Sequencing
Introduction
The early historical reliance on surgical excision for treatment of cancers suggests the belief that tumorigenesis was a fundamentally irreversible process. The multiplicity of chromosomal aberrations associated with abnormal cellular morphology in many human tumors was noted by pathologists more than a century ago and has continued to serve as a means of identifying malignant cells and stratifying the aggressiveness of certain cancers (Figure 1). As early as 1902 Boveri suggested that the intercellular cooperation required between cell types during embryogenesis was disrupted in tumors as a result of chromosomal aberrations (1). We now no longer consider chromosomes, or even individual genes, as the irreducible genetic units of cancer, but instead we can identify single-base changes buried among billions of unaltered nucleotides in the human genome. Modern biochemical approaches allow us, not only to identify mutated cancer genes, but also to infer how specific mutations affect gene function. As a result, we are able to catalog unique nucleotide changes that contribute to malignant phenotypes or that increase the susceptibility of individuals to develop specific tumors.
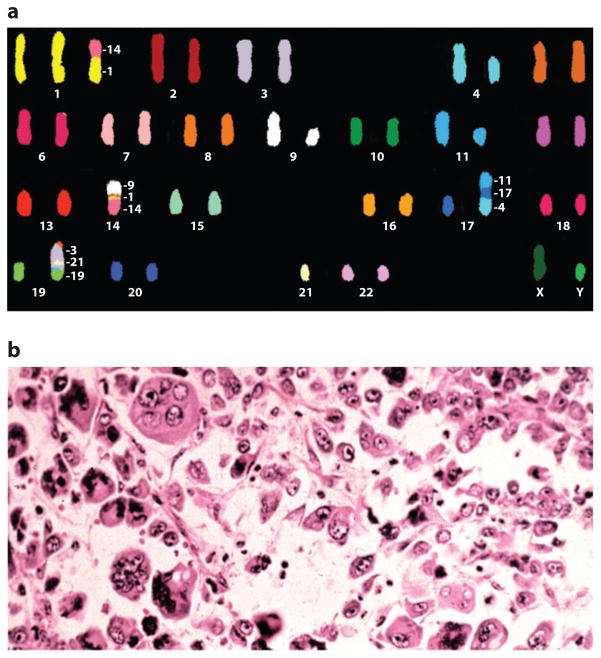
Figure 1. Heterogeneity in cancer.
A) Chromosomal heterogeneity. Spectral karyotype from an acute myelogenous leukemia (AML) cell demonstrating aneuploidy and multiple chromosomal rearrangements. Courtesy of Dr. Karen Swisshelm, Department of Pathology, University of Colorado, Denver. B) Morphologic heterogeneity. Hematoxylin and eosin section from a large cell, undifferentiated lung cancer demonstrating a highly pleiomorphic cellular population. Courtesy of Dr. Ray Monnat, Department of Pathology, University of Washington.
The primacy of DNA as the critical macromolecule involved in the etiology of cancer is strongly supported by the inherited human diseases that are associated with, and increase the incidence of, specific cancers (2). In addition, multiple genetic changes in cancer cells have been frequently documented by microscopic examination of chromosomes or by hybridization with specific probes. These changes include: deletions, insertions, amplifications and translocations and frequently involve millions of nucleotides. Chromosomal studies on adenocarcinoma of the breast (3) and ovary (4), as well as in leiomyosarcoma (5) documented tumors harboring more than 20 different chromosomal alterations. Measurements of loss-of-heterozygosity in tumors using PCR-amplified gene fragments reveals an even greater number of changes (6). Klein et al. used both of these techniques and demonstrated the presence of multiple alterations within single tumor cells (7). These chromosomal changes have often been considered to result from chromosomal instability and in some tumors may occur sequentially (8).
The technology for dissecting chromosomes into their finest nucleotide elements has exponentially improved in recent years, both in terms of throughput and cost-effectiveness. It is now possible to analyze the processes that generate mutations in normal and malignant cells and begin the ambitious task of cataloging cancer-associated nucleotide changes by DNA sequencing. For some, the surprise has been the unexpectedly large number and diversity of mutations present in human tumors. In light of this emerging mutational complexity, it seems timely to review mechanisms that guarantee the high fidelity of DNA replication in normal human cells, to consider how mechanisms for preventing mutations might be altered in tumors and to interpret the recently reported results on mutations identified in different human cancers.
The Accuracy of DNA Replication in Normal Human Cells
DNA is a living molecule; it continually breaths and is exposed to modifications. Yet, in normal cells it is faithfully copied during each division cycle. Each human cell contains more than 6 billion nucleotides that are replicated with exceptionally high accuracy. Approximately one mutation is introduced into DNA during each division cycle. Most mutation rate measurements have been carried out at the hgprt locus because it is present as a single copy on the X-chromosome. Spontaneous mutations in this gene render a cell dominantly resistant to the toxic effects the nucleoside analog 6-thioguanine and form countable colonies under appropriate culture conditions. A tabulation of data derived from hgprt studies indicates that the overall mutation frequency in mammalian cells varies between 10−5 to 10−7 mutations/gene, or approximately 10−9 to 10−10 substitutions/DNA-nucleotide (9–12). It should be noted that cancers arise in stem cells, and detailed studies of mouse embryonic stem cells indicate that mutation rates in these pluripotent progenitors are as much as 100-fold lower than those observed in cultured fibroblasts derived from adult tissues (13). Based on the conservative assumption that the accuracy of DNA replication in stem cells is similar to that in other cells, it can be estimated that each stem cell would amass, on average, only 1 to 2 mutant genes during 100 cell divisions in a normal human life span (14).
This remarkably high accuracy results from sequential processes, each contributing a 100- to 1000-fold increase in the fidelity of DNA replication. First, based on simple thermodynamics, the difference in free energy of hydrogen bonding between complementary and non-complementary base-pairings during DNA synthesis can provide base selection down to approximately one error per 102 nucleotides incorporated (15–16). Second, DNA polymerases are believed to undergo allosteric transformations with each nucleotide addition step that tightens the bonding of complementary nucleotides at the active site on the polymerase (17). Third, replicative DNA polymerases have an associated “proofreading” 3′->5′ exonuclease activity that preferentially excises non-complementary nucleotides prior to incorporation of the next nucleotide (18). Finally, remaining non-complementary nucleotides are removed by mismatch repair once the replication fork has passed (19). Together these processes have the potential to synthesize DNA in vitro with an accuracy that approximates the fidelity of DNA replication observed in vivo. However, it is of note that experimental values are based on reactions carried out under simplified, optimal conditions that may not exist in cells; other components of the replicative machinery are likely to play a role in replication fidelity. Also of note, the error rates of DNA polymerases are proportional to the concentrations of non-complementary to complementary nucleotides in the reaction (20). This infers that size of cellular nucleotide pools will have a significant impact on the accuracy of DNA replication. Alterations in the accuracy of DNA polymerases by mutation, damage, or imbalances in nucleotide pools could have profound effects of the overall fidelity of DNA replication (21).
DNA Damage by Endogenous and Environmental Agents
DNA is subject to attack by both endogenous and exogenous reactive molecules. A major source of DNA modifications in human cells are reactive oxygen species (ROS) arising as a byproduct of energy metabolism in mitochondria. (22). Based on the measurements of 8-oxo-dG and other oxidative modifications, it has been estimated that more than 10,000 nucleotide residues in DNA are altered by ROS per cell per day (23). Other modifications include: methylation, alkylation, inter/intrastrand cross-links and apurinic sites; in total there may be as many as 50,000 alterations per cell per day resulting solely from normal cellular metabolism (24). Many human cancers arise in the setting of chronic inflammation (25–28), where extracellularly derived ROS are likely contribute to the burden of DNA damage in affected tissues (29). Mutagens are ubiquitous in our environment and it is important to recognize their contribution to spontaneous human cancer. Exposure of individuals to high concentrations of mutagens has frequently been associated with an increased incidence of cancer (30–32) and recognition of tobacco smoke as a human carcinogen (33–34) has led to the most significant and successful effort at reducing cancer incidence in human history (35).
The Repair of DNA Damage in Human Cells
Against this onslaught of DNA damage is an armamentarium of repair systems with overlapping specificities (Figure 2). These systems continuously monitor the genome and correct many forms of DNA damage. So far, more than one hundred repair genes have been identified (36). Pathways of DNA repair include base excision repair, nucleotide excision repair, transcription coupled repair, mismatch repair, double strand break repair and even direct reversal of adduct-mediated lesions (37). DNA damage by environmental agents is predominantly a stochastic process. Recognition of damage is generally dependent on the nature of the lesion, and less a function of sequence context. Small adducts on bases are excised by both short patch and long patch pathways with re-synthesis of the excised segment carried out by DNA polymerase-β and presumably DNA polymerases-δ and -ε, respectively (17). Bulky adducts such as thymine dimers, resulting from UV-irradiation, or benzo[a]pyrene, resulting from tobacco products, are subject to nucleotide excision repair. In the presence of DNA damage, many sensing systems signal to cell cycle control genes, such as p53, to arrest the cycle and allow additional time for DNA repair (38). The high efficiency of these mechanisms for DNA repair guarantees that only a few of the tens of thousands of DNA lesions generated persist prior to DNA replication and have the potential to cause mutations.
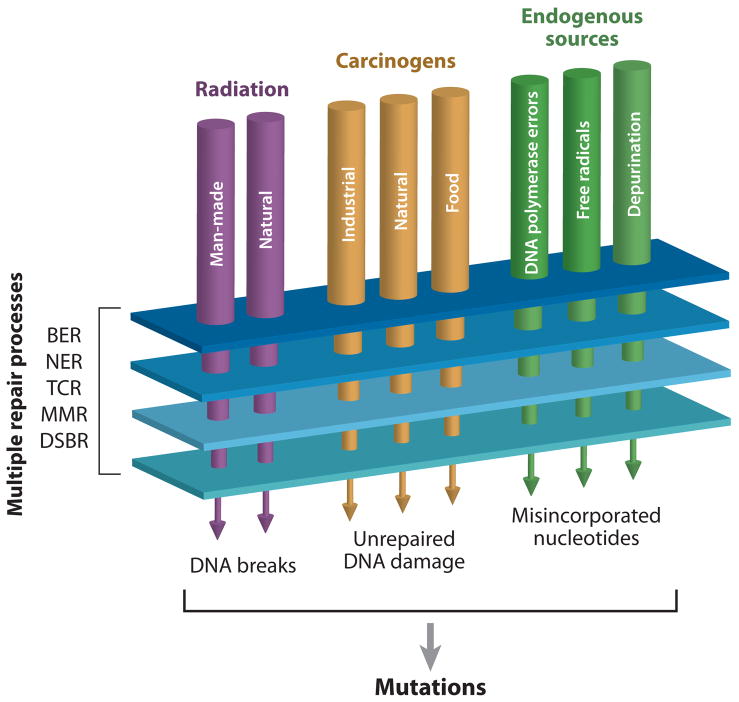
Figure 2. Mutational homeostasis.
In each human cell DNA is damaged thousands of times per day by both exogenous and endogenous sources. Most alterations are corrected by cellular mechanisms including: base excision repair (BER), nucleotide excision repair (NER), transcription coupled repair (TCR), mismatch repair (MMR) and double strand break repair (DSBR). Lesions that escape repair have the potential to cause mutations during DNA replication.
Inherited Cancers Frequently Involve Mutations in DNA Repair Genes
A number of rare inherited diseases, caused by germline mutations in genes involved in DNA repair, are associated with elevated risks of specific cancers. Investigation of many of these diseases has been instrumental in deciphering the different cellular mechanisms for DNA repair. The seminal findings on the defects of UV-induced DNA damage repair in patients with xeroderma pigmentosum highlighted the association of DNA repair with suppression of carcinogenesis and provided a powerful tool for identifying genes involved in nucleotide excision repair (2). Subsequently, inherited defects in members of several other DNA-repair pathways have been shown to underlie a variety of cancer syndromes including: mismatch repair (hereditary non-polyposis colorectal cancer (39)), base excision repair (MYH-associated polyposis (40)), homologous recombination (early onset breast cancer (41)), non-homologous recombination (Lig4 syndrome (42)) and translesion synthesis (xeroderma pigmentosum variant (43)).
Hereditary mutations in other DNA maintenance enzymes are also associated with cancer. Defects in genes encoding members of the RecQ helicases are found in Bloom and Werner syndromes–rare inherited diseases characterized by large-scale DNA rearrangements and a high incidence of specific cancers (44–45). Mutations in TP53 are found in Li-Fraumeni syndrome (46), a highly cancer-prone condition most frequently associated with sarcomas and breast adenocarcinomas. Additionally, polymorphisms in a wide variety of DNA repair genes, including OGG1 and XRCC1, are increasingly being considered as a risk factor for cancer (47). Why defects in particular DNA repair genes result in specific types of cancer remains largely unresolved.
Prevention of Mutations by Growth Limitation, Clonal Hierarchy and Programmed Cell Death
Despite these extensive DNA repair mechanisms, every time a cell divides there remains an opportunity for fixation of new mutations through miscopying across unrepaired damage, missegregation of replicated chromosomes and/or failure to recognize improperly repaired sequences. Hence, a key mechanism for preventing the accumulation of DNA mutations is limiting the number of cell divisions that occur. The importance of proliferation in oncogenesis has been appreciated from experiments demonstrating that liver regeneration is associated with an increased incidence of cancer (48). The initiation of skin cancer in mice by mutagens was shown to be markedly accelerated by the subsequent application of phorbol esters that promote cell proliferation (49). Many of the molecular mechanisms that control cellular growth control were first identified in the context of their disruption in cancer. The identification of replication-enhancing avian viral oncogenes (50), and the discovery that these represent constitutively active versions of endogenously present genes permanently activated through mutation in some cancers (51), lead to the characterization of one of the earliest known growth control pathways in human cells. Extensive work during the last three decades has revealed the cellular network of defenses preventing unregulated proliferation to be staggeringly complex, with many redundant protections (52).
The necessity of replacing worn out or damaged tissues must be carefully balanced against the risk of proliferation-induced mutations. To allow cellular repopulation with minimal risk of mutation, tissues in the body are frequently organized hierarchically, whereby the ability to continuously proliferate is relegated to a specialized subset of cells (53). Stem cells are believed to have an inherently lower rate of mutation than the majority of their daughter cells that have only limited replication potential (13). Among the most studied example of this hierarchical organization is colonic epithelium. Here, a small number of long-lived stem cells reside at the base of each crypt and produce progeny that migrate luminally to populate the upper levels of the crypt – first as transiently amplifying cells, then as terminally differentiated colonocytes destined to slough-off after several days (54). Because the majority of mutations that arise during division occur in short-lived daughter cells, most mutant cells are rapidly purged from the population. It has been hypothesized that the same “immortal” strand of DNA is maintained in the parental stem cell, while always transferring the (potentially imperfect) newly replicated strand to its daughter (55), though at least one recent study (21) suggests this not to be the case. Whether tumors derive from abnormally replicating stem cells, or from dedifferentiated progeny, remains an open question (56).
Cancer as a Somatic Evolutionary Process
This November marks the 150th anniversary of the publication of Darwin’s seminal work, On The Origin Of Species. It was therein first postulated that heritable phenotypic variation underlies natural selection and is responsible for adaptation as well as for the emergence of new species. Though initially used to describe how organisms evolve across generational time, the idea that evolutionary forces also drive intra-organismal neoplastic development has frequently been noted (57–59). In this model individual cancer cells, rather than complete metazoans, are considered the reproductive units within a population (Figure 3). New mutations are acquired somatically and genetic alterations that bestow the cancer cell with a growth advantage will enable them to clonally expand. Additional mutations that arise in the expanding population will generate further selectable phenotypes, such as the ability to invade adjacent tissues, recruit a blood and lymphatic supply, overcome nutritional deficiencies and resist immune attack. After bypassing all antineoplastic defense mechanisms, tumor growth may continue indefinitely until the death of the host.
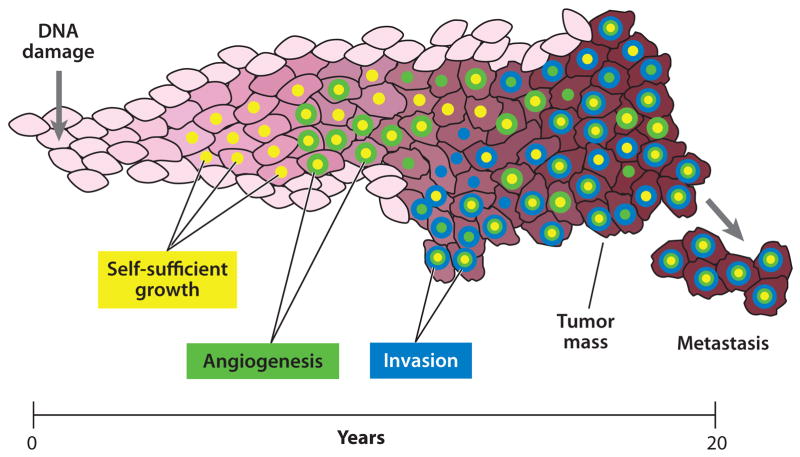
Figure 3. Cancer recapitulates evolution.
Within a developing tumor mutations accumulate over time as a result of unrepaired DNA damage. Most of these are either neutral or detrimental; only a small number bestow a cell with growth and survival benefits. These beneficial variants will preferentially multiply and produce additional mutations that may undergo further selection and expansion. Adventagous phenotypes for tumor growth include, among others, the ability to divide independently of extracellular signals (yellow), the ability to recruit a blood supply (green) and the ability to invade adjacent and distant tissues (blue). After overcoming all antineoplastic defense mechanisms, a tumor may proliferate indefinitely until the death of the host.
Evolutionary adaptation, somatic or otherwise, requires both genetic diversity and the ability to undergo selection in response to this diversity. In the absence of either of these key parameters, the process cannot proceed. If one or other of these features is limiting, any event that diminishes this limitation will accelerate adaptation. In multicellular organisms, the intact genome of normal tissues encodes strict defenses against the emergence of new mutations to suppress evolution which, on a cellular level, would be detrimental to the fitness of the individual as a whole. In the context of neoplastic evolution, genetic heterogeneity is not merely a feature of cancer, it is the fuel with which the selection process is driven.
The Number of Mutations to Cancer
Given that carcinogenesis may be viewed as an evolutionary process that sequentially increases neoplastic cell fitness through a series of (epi)genome modifying events, an important question arises: how many mutations are needed to produce a tumor? The increased incidence of most human cancers as the fifth or sixth power of age (60) has been taken to indicate that there are five or six events (presumably mutations) that drive the carcinogenic process, each event increasing the probability of the next. Of exception are certain pediatric tumors, such as retinoblastoma, in which significantly fewer mutations appear to be necessary (61) and some late onset adult tumors, such as those of the prostate, that may require as many as 10 to 12 events (62). Weinberg has demonstrated that at least three or four altered genes are required for the expression of the malignant phenotype in cultured cells (63). Passage in tissue culture or as implanted xenographs may, in itself, select for additional mutations as highlighted by the recent work of Mahale et. al. (64). Thus, if cancer requires as many as 12 different rate limiting mutations to arise, and the normal per-division mutation rate of human stem cells is as low as calculated, and the number of long-lived stem cell divisions limited, how can a cancer possibly occur within the human lifetime? It is hypothesized that early in the neoplastic process at least one, and likely several of the mechanisms for preventing mutations, must be reduced.
Antimutational Defenses: Primary Versus Secondary Mechanisms
From a simplified perspective, one may divide antimutational processes that suppress the emergence of new genetic diversity into two classes. Primary mechanisms, those that act at the level of DNA to prevent genetic and epigenetic mutations from occurring or persisting until cell division include: proofreading by DNA polymerases, DNA repair processes, ability to quench reactive oxygen species and other means of limiting the per-cell-division mutation rate. Secondary mechanisms, in contrast, prevent the accumulation of mutations in the population at large by limiting the total number of divisions in long-lived cells. These include means of controlling cell growth, confinement of reproductive abilities to stem cells in a well-protected niche and culling of irreversibly damaged cells through programmed cell death. Secondary mechanisms do not increase the per-division rate at which individual cells accumulate mutations; instead they limit the overall number of different mutations in the population as a whole.
For evolution to occur in a somatic setting there must be (epi)genetic variation within a population of cells and these cells must be able to undergo selection (i.e. through more efficient division and survival) in response to advantageous environments. If either of these features is rate-limiting to the process and the factor responsible for the limitation is genetically encoded within the evolving cells, disruption of the responsible genes through mutation will accelerate the adaptive process. The question of which parameter is most limiting in different stages of tumorigenesis is complex and, rightly, has been the subject of extensive debate (65–66). Given that both therapeutic interventions and preventative measures might be better directed if this was known, the question is not merely of academic interest.
Within the view that somatic evolution drives carcinogenesis, at least two schools of thought have emerged regarding the relative importance of overcoming primary and secondary anti-mutational defenses. One hypothesis asserts that the mutation frequency existing within non-malignant cells provides sufficient genetic diversity to fuel tumorigenesis (65, 67–68). Some of these variants in a normal population can confer a phenotype of increased proliferative abilities that enables subsequent clonal outgrowth and consequent generation of more mutations upon which further selection may act. Not only does escape from growth limitations facilitate a cell acting upon preexisting genetic diversity through selection, it also enhances the production of additional variants by increasing the number of fallible DNA replication cycles. This school of thought conceptually favors the importance of defects in secondary mechanisms but relies upon the fact that primary mechanisms are imperfect in non-neoplastic cells. Eventually, after enough successive rounds of mutation, selection and expansion, the threshold number of events required to drive carcinogenesis is reached.
A second school of thought emphasizes the importance of defects in the primary class of antimutational mechanisms for fueling neoplastic evolution (69–70). Deficits in these cellular components renders DNA replication more error-prone and increases the number of potentially advantageous variants produced per generation. It is argued that, even with additional mutations generated from increased cell division, the number of variants in the population will still be a limiting parameter for evolutionary adaptation. Proponents of this view suggest that mutation-prone variants would overcome this limitation, and therefore be likely to emerge during clonal selection. It is important to recognize that a genetically encoded mutator phenotype need not, in itself, be a driver that directly increases reproductive fitness. Instead, by virtue of increasing the probability of advantageous new mutations, it would be passively carried along on the resulting clonal expansion as a passenger. Because of this unique position, it can be argued that generation of a mutator phenotype is unlikely to be the very first “hit” on the path to cancer. Similar to inherited deficiencies of DNA repair, it is only when the phenotype is expressed in a plurality of cells that it can meaningfully increase the total number of genetic variants in the population. Thus, defects in secondary antimutational defenses leading to increased proliferation remain a critical component of this model of tumor development.
Those on both sides of the debate have made arguments for (69) and against (71–72) the necessity of a primary mutator phenotype in carcinogenesis based on calculations that assume the number of mutations needed for cancer, the mutation rate in normal cells and the estimated number of divisions that occur between conception and a late stage tumor. Given that these values are, themselves, not easily quantified, the disparate results are not surprising. An alternate approach that our group has more recently taken is to consider, instead, the relative efficiency of mutator and non-mutator pathways to cancer (73). Modeling of fitness landscape suggests that in spite of the “cost” of an extra step to produce the phenotype, a primary mutator pathway is generally a more efficient trajectory to cancer, as long as the total number of mutations required exceeds 3–5 (Beckman, submitted). Moreover, although mutator lineages are also more likely to suffer deleterious mutations that terminate their lineage through negative clonal selection (74), this negative effect does not predominate until mutation rates become very high (Beckman, submitted). One additional consideration that has been frequently overlooked is that newly arising mutations, including those with a fitness advantage, have a high probability of becoming extinct from random drift (58). Depending on the population size and precise fitness advantage, a given mutation may have to arise on multiple independent occasions before being able to expand to a clinically meaningful size. Hence, calculating the mutation rate required for a defined number of genetic events to occur once per tumor will underestimate the rate required for each to arise and expand.
Making a sharp distinction between primary and secondary mechanisms of mutation suppression is conceptually interesting, but ultimately artificial, given their intimate link within the cell. It is likely that both mechanisms are operative; the relative contribution of each may depend on the tumor. Cell-cycle progression, DNA repair and programmed cell death are, in fact, coordinately regulated. TP53, for example, the most commonly mutated gene in human cancer, encodes a multi-functional protein that acts as a network hub to integrate information about genome state from more than a dozen sources (75). Many types of DNA damage can trigger activation of p53; this may lead to cell cycle arrest, upregulation of DNA repair processes or activation of programmed cell death if damage is severe. Hence, TP53 exemplifies a gene directly involved in both primary and secondary antimutational pathways. It has been argued that for such dual-function tumor-suppressor genes, the pressure for loss in neoplastic cells stems from the immediate proliferative advantage gained, rather than from an increased mutation frequency (65). While this is very likely to be the case given that a primary mutator phenotype is not, itself, selectably advantageous, disruption of such genes will, nevertheless, increase the genetic diversity of a developing cancer through both primary and secondary mutational pathways and facilitate continued evolution.
Deterministic Versus Plastic Tumorigenesis
DNA damage by chemical agents and physical processes are predominantly stochastic events. For the most part, damage occurs randomly throughout the genome and mechanisms governing the correction of affected nucleotides are specified primarily by the nature of the chemical alterations and not by the surrounding nucleotide sequence. Of exception to this random hit model are nucleotide sequences that can form alternative DNA structures (76) that are resistant to DNA repair and highly repeated sequences of the genome (77)–both of which constitute mutational hot-spots. The specific role of histones and other DNA associated proteins in preventing DNA damage has not been delineated fully. Similarly, transcriptional status (78) as well as local replication timing (79), have general, yet incompletely predictive influences on local mutation rate. Only a small fraction of all mutations that occur will confer a selectable fitness advantage with the ability to initiate or promote tumor growth. Given that different individuals will acquire a unique, yet random, selection of mutations prior to and during tumorigenesis, the order and specific nature of advantageous variants will be determined stochastically. Just how similar are two tumors to each other with respect to specific genes mutated and the order in which the selected mutations arise? Do tumors evolve along a pathway characterized by sequential rounds of mutation and selection of a defined set of targets or is the process more variable, selecting instead for phenotypes that may be encoded by many different or multiple loci (Figure 4)?
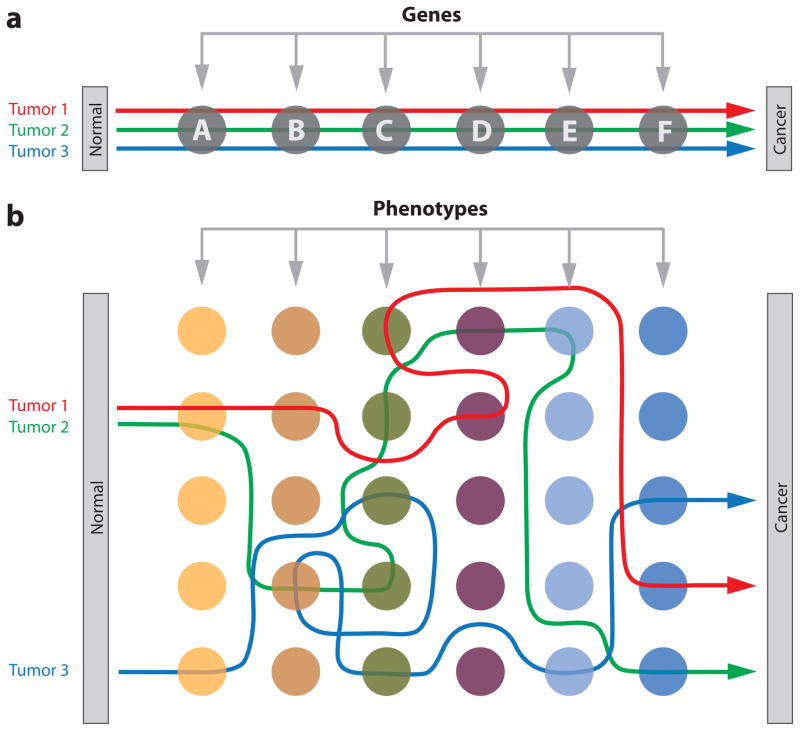
Figure 4. Pathways to cancer.
A) Deterministic. In this model, different tumors of the same cancer type occur reproducibly through sequential mutation of each gene within a defined series. Although mutation occurs randomly, the order of selection is fixed. B) Plastic. In this model, different tumors evolve along highly variable pathways, selecting for specific cancer phenotypes that may be achieved through (epi)mutation of many possible sites in the genome. Although some mutated loci may be shared by different tumors, most are not and the order of selection is predominantly stochastic.
There is considerable evidence for sequential mutations during tumor progression. In melanomas (80), colon (8) and esophageal (25) cancers, a series of chromosomal alterations frequently characterize different phases of neoplastic progression. These changes have been most extensively documented in adenocarcinomas of the large intestine where the order of DNA alterations has been correlated with tumor grade and stage (8). From a clinical perspective, a chronologically ordered series of mutations driving malignancy is particularly attractive, as it implies that the evolutionary process must bottleneck through a defined set of genes which might be therapeutically targeted. Unfortunately, even in the most studied model, early investigations indicated that only 7% of advanced colon cancers simultaneously bear mutations in the three most frequently mutated genes (81).
It has long been clear from traditional genetic and molecular methods, that, despite some commonalities, overall the profile of clonal somatic mutations occurring in different tumors is highly variable. The resolution of these early findings was inherently limited to a moderate number of genes by the biochemical tools available. The complete human genome, however, comprises more than six gigabases of sequence information and, until very recently, methods for high-throughput analysis were inadequate for the task of whole-genome exploration. Many long-standing questions remain to be answered: How frequently do different tumors overcome hard-wired barriers to neoplasia in the same way, both in terms of altering specific genes and specific pathways? Does this vary from one cancer type to another? What, if any, mutational changes differ between metastatic lesions and their primary tumor? What is the relative importance of different types of mutations such as single-base changes versus deletions, rearrangements or epigenetic phenomena? Can this mutational spectrum of a tumor inform us of likely environmental contributors or specific dysfunctional antimutational pathways? The emergence of high-throughput capillary sequencing robotics and more recent “next-generation” sequencing methods have provided an exciting opportunity to delve more deeply into these questions.
The Human Cancer Genome Atlas
Within the last two years, a large number of DNA sequences from human cancers have been published. Included among these is the first complete genome of a human cancer and its paired normal (82). With the passing of this milestone, it is important to consider the likely implications of this data and how it might frame both basic and clinical research in the near future. Prevailing models of tumorigenesis stress tumor progression to be the result of sequential mutations in a few key cancer genes; each mutation driving a new round of clonal proliferation. In large part, the effort to systematically tabulate mutations found in different human cancers encompasses the expectation that a cancer’s most significant mutated genes will be potent targets for chemotherapy. This supposition has been reinforced by the success of targeted treatments in some hematological cancers (83) and the hope that, by identifying analogous key mutational events in solid tumors, specific therapeutic targets might similarly be identified and exploited. However, an increasingly complex picture has emerged from nearly twenty studies detailing the genome of many solid tumors; the findings suggest that the extent of prevalent, new targets may be far more limited than anticipated.
Initial Studies on Nucleotide Variation Within Human Cancers
The first large-scale efforts to systematically screen individual tumors for somatic mutations identified remarkably few previously unknown genes that were mutated in a significant proportion of specific cancers (84–86). The relatively limited sequence coverage of these initial studies prompted more comprehensive screens of larger portions of the genome. The first complete sequencing of all predicted coding exons, conducted in breast and colon cancer, concluded that these cancers, respectively, contain a median of 84 and 76 clonal mutations likely to alter protein function (87–88). Although nearly one tenth of the 18,197 genes sequenced were detectably mutated in at least one specimen, each tumor displayed a unique and diverse profile of mutated genes. Other than those previously known, such as TP53, APC and KRAS, no new prevalently mutated genes were identified. The authors of this study proposed that the cancer genome may be considered as a landscape composed of a handful of commonly mutated gene “mountains” and a larger number of less frequently mutated gene “hills”; a view consistent with multiple, pathways to cancer. The authors additionally affirmed that tumor-to-tumor heterogeneity of clonally present mutations may underlie the wide variation in tumor behavior and responsiveness to therapy.
The initial studies of Sjoblom et al. and Wood et al. served to highlight several important technical challenges faced by The Cancer Genome Project (87–88). First, with large amounts of tumor sequence data comes significant experimental noise that complicates detection of true clonal mutations. Such noise derives from PCR introduced mutations, automated base calling errors, mutations arising in the germline rather than somatically, and previously unknown SNPs. The most rigorous cancer genotyping approach would entail sequencing of a matched non-neoplastic sample for every tumor to rule out germline variation as well as automatic resequencing of every tumor-specific mutation identified. Unfortunately, even the highest-throughput capillary sequencing systems are cost- and time-limited when used on the scale of multiple human exomes. Thus, in these initial studies, as well as several that followed, compromises to preferred protocols have been made; for example, eliminating non-coding changes and known SNPs prior to confirming the small fraction of mutations remaining against a tumor’s corresponding control sample.
A second impediment to high-throughput capillary sequencing is the substantial amount of DNA required for the hundreds of thousands of PCRs. One way to overcome this limitation has been to expand tumor cells in culture or as mouse xenographs. Such ex vivo passaging adds the possibility of artifactually introducing new mutations as a result of artificial growth conditions (64). Direct biochemical methods of whole-genome amplification have also been used to extend DNA samples. It has been suggested that the differing mutational spectrums reported within the discovery and validation experiments of Sjoblom et al. (87) are likely reflective of such differences in ex vivo treatments between the two groups (Rubin and Green, submitted).
A third, and arguably most significant challenge to cancer genome sequencing, lies in the complex problem of determining which mutations in a tumor are causative and which are merely present by chance. Mutagenesis is largely a random process; only a small subset will confer a proliferative advantage to their host cell. Differentiating “drivers” from neutral “passengers” (or “hitchhikers”) that happen to co-occur and be swept along with the same expanding genome poses a formidable challenge to deciphering megabases of sequencing data (89). The conceptual approach taken by these, as well as later studies has been to sequence multiple tumors of the same variety and look for genes that are commonly mutated. The appropriate statistical methods to be used for determining which genes are found clonally mutated more frequently than would be expected by chance alone has been heavily debated (90–93). More importantly, implicit in this approach is the de facto assumption that a limited subset of genes will frequently drive tumorigenesis. The alternate hypothesis – that a large number of loci may combinatorally serve as weak drivers and any one may arise only infrequently – cannot easily be addressed by such methods, given that rare drivers are filtered out as probable passengers. The number of tumors needing to be sequenced to resolve minority drivers increases substantially as the prevalence of the driver declines. Attempts have also been made to identify likely driver mutations by bioinformatically predicting the probable functional impact of specific mutations (88). While such approaches are useful in a limited number of instances, the current technology for accurately modeling the resulting changes to protein activity remains limited. Despite these complexities, the demonstrated ability of random exon sequencing to re-identify the majority of previously characterized genes known to play a role in colorectal cancer was a noteworthy technical achievement that set the stage for cancer genome sequencing studies that followed.
Multiple Mutations in Diverse Tumors
While the initial studies of Sjoblom et al. and Woods et al. (87–88) focused on the exhaustive comparison of mutations in two tumor types, a second milestone project analyzed an extensive gene family in a wide variety of different cancers. Greenman et al. (94) sequenced 518 protein kinase genes in 210 tumors of diverse origin including: breast, colorectal, lung, brain and blood. They observed 1007 likely-driver mutations, of which 921 were single-base substitutions. Similar to previous studies (95–97), there was substantial variation in the number of genes mutated per tumor regardless of type; again, few commonly mutated genes were found in any of the cancer types examined. Although these studies generated an extensive catalog of somatic point mutations, only a small number of prevalently mutated genes were identified. The data reinforced the notion that mutational patterns of solid tumors evolve stochastically and are highly diverse, in contrast to the relatively predictable stepwise patterns of cytogenetic abnormalities in some hematological cancers.
Subsequent studies (Table 1) increasingly have relied on associating sequence data with other complementary genomic information. This has been paralleled by a shift to a more integrated interpretation of the significance of individual mutations: from one of specific genes into one of pathways and processes. One follow-up study focused on mutations arising during the progression of adenoma-to-carcinoma-to-metastasis (98). No metastasis-specific mutations were detected in the vast majority of specimens and, as expected, the number of mutations was markedly increased in the carcinoma compared with its matched precursor adenoma. Building on the observation that individual tumors express unique immune profiles (99), Segal et al. demonstrated that the diverse mutational pattern of breast and colorectal tumors likely underlies their immunological heterogeneity (100). Leary et al. examined homozygous deletions and focal amplifications in the breast and colorectal cancer genomes (101). Each of these studies further confirmed the heterogeneity and inter-tumor diversity between breast and colorectal cancer genomes.
Table 1.
Cancer Genome – Sequencing Studies
| Study type | Number of genes screened | Total number of mutations | Number of genes mutated | Average number of mutations per tumor | Estimated number of driver mutations | Reference(s) |
|---|---|---|---|---|---|---|
| Exomic | ||||||
| Breast (n = 11) | 18,191 | 1243 | 1137 | 84 | 140 | 87, 88 |
| Colorectal (n = 11) | 18,191 | 942 | 848 | 76 | 140 | 87, 88 |
| Diverse (n = 210) | 518 | 798 | 581 | – | 119 | 94 |
| Pancreatic (n = 24) | 20,661 | 1163 | 1007 | 48 | 160 | 98 |
| Glioblastoma (n = 21) | 20,661 | 748 | 685 | 47 | 155 | 102 |
| Glioblastoma (n = 91) | 601 | 453 | 223 | – | 8 | 103 |
| Lung (n = 188) | 623 | 1013 | 348 | – | 26 | 108 |
| Genomic | ||||||
| Acute myeloid leukemia (n = 1) | – | 500–1000 | 10 | Not applicable | 10 | 82 |
Paradigm Shift
This change of focus, away from the search for key, sequentially mutated genes that govern cancer progression towards a more systems-oriented description, is evident in recent studies on pancreatic cancer. Using a two part discovery and prevalence-determination strategy, along with copy number and transcriptomic analyses, Jones et al. concluded that pancreatic cancers contain an average of 63 clonal genetic alterations, the majority being point mutations (98). As with breast and colorectal cancer, there was considerable variation in both the number of mutations and in the specific genes mutated among different cancer specimens, again with no new prevalently mutated genes identified. Because nearly all of the predicted protein-coding genes in the human genome were evaluated, this data provided the opportunity to investigate groups of genes operating through specific signaling pathways and processes in a relatively unbiased manner. The authors concluded that pancreatic cancer results from genetic dysregulation of 12 core pathways and processes, including apoptosis, DNA damage control and regulation of invasion. Although these 12 processes are genetically altered in the majority of pancreatic cancers, the specific components mutated in individual tumors were largely different. It was proposed that agents be designed to target the physiological effects of the altered pathways and processes rather than individual genes. While this therapeutic logic is reasonable, and the analyses do demonstrate enrichment for specific cellular processes, the granularity of the results do not extend much beyond reinforcing the general hallmarks of cancer(52).
Glioblastoma multiforme (GBM) was the first cancer type to be screened systematically by two different groups (102–103). Both studies integrated gene sequencing, identification of focal amplifications and deletions through comparative hybridization arrays and expression analysis to comprehensively interrogate the GBM genome. One study, focusing on a group of 601 selected genes in 92 predominantly primary GBMs, found no novel commonly mutated genes among different tumors (103). Interestingly, the number of gene alterations in GBMs was smaller than that previously reported for colorectal and breast cancers. The second study focused on exhaustively sequencing all likely coding exons in a discovery screen and then determined the prevalence of any identified variants in a secondary screen (102). The discovery of one novel recurrent mutation (isocitrate dehydrogenase 1, IDH1) mutated in 12% of all GBMs and strongly associated with secondary GBMs in particular, was cited as a validation of the utility of genome-wide genetic analysis of tumors. Indeed, as of this writing, this finding is among the most significant yet unearthed by cancer genome sequencing. Two follow-up reports have indicated that active site mutations in IDH1 and, occasionally its homolog IDH2, are found in more than 70% of certain CNS tumors including grade II and III astrocytomas and oligodendrogliomas and secondary glioblastomas, though rarely in primary glioblastomas and in none of the tumors from outside the CNS that were tested (104–105). Reinforced by functional studies of these mutations in cultured cells indicating lowered enzymatic activity (105–106) this work has unequivocally identified an important new pathway for a specific subset of CNS tumors. Nevertheless, both initial sequencing studies independently concluded that for the bulk of GBMs, dysregulation of three core pathways, based around the previously well studied genes TP53, CDKN2A and EGFR, is central to tumor progression.
Analysis of the lung cancer genome identified 1,013 mutations in 188 cases of lung cancer (107–108). Twenty-five cases harbored no mutations in the 623 genes analyzed and only four genes had point mutations in more than 10% of tumors. Examining the distribution of genes across cellular pathways, the authors identified five key pathways in which components were frequently mutated. By far the most commonly affected of these was the MAPK pathway, with 70% of tumors sequenced having at least one mutation altering known MAPK proteins. This pathway, however, encompasses 56 genes, the majority of which, individually, are mutated in fewer than 1% of lung cancers. Again, the predominant results of this study were the mutational heterogeneity among tumors and an absence of prevalently mutated genes.
The First Complete Cancer Genome
The characterization of the first hematopoietic cancer genome represented an important methodological milestone in cancer genomics – truly whole genome sequencing of a tumor specimen (82). Prior efforts at re-sequencing tyrosine kinase genes in acute myeloid leukemia (AML) had yielded few mutations (109–110). By exhaustively screening the entire genome of a paired set of cancer and normal samples from a single AML patient using massively parallel sequencing, this group identified 500–1000 non-synonymous somatic changes uniquely present in the cancer. Of these, only ten mutations occurred in protein-coding genes, including two previously described indels (within FLT3 and NPM1) known to occur at high frequency in AML. Importantly, none of the eight newly identified genes were found to be mutated in a further 187 cases of AML.
This study marked a number of significant advances in large scale cancer genome sequencing. First, unbiased whole genome sequencing is inherently a more complete means of cataloging all the clonal alterations in a cancer genome. With the increasing recognition that so-called intergenic “junk” DNA and intronic sequences contain functional elements such as regulatory regions and non-coding RNAs, exon-centered genotyping may be missing important drivers occurring in regions of the genome not previously explored. Even if most of these turn out to be non-causative, the spectrum and total number of clonal mutations born by a tumor may provide important information about sources of mutations as well as tumor life-history.
Second, unlike previous studies where matched normal DNA was only sequenced to specifically validate suspicious candidate mutations, this study was the first to simultaneously apply an unbiased analysis to paired normal DNA, albeit with less sequence depth. One criticism of the earlier strategies (87) had been the exclusion of any sequence variant coincident with a previously described SNP, without considering it a possible de novo event. New mutations in known SNP sites, in fact, may represent some of the most likely selectable drivers given the strong familial component to many cancers for which no specific genes have yet been implicated. Only by sequencing of paired normal DNA can clonal mutations at polymorphic loci be scored.
Third, next-generation sequencing platforms have several advantages that will likely make them the preferred technologies for future studies. The most obvious is the significantly lower cost per base-pair sequenced. In addition, minimal input DNA requirement obviates the need for expanding tumor cells in culture or as xenographs. Three such devices are presently in commercial production with many more on the horizon. Genotyping is accomplished by randomly shearing the genome into many pieces, clonally amplifying individual fragments on a solid matrix and then sequencing these immobilized “clones” using one of a variety of chemistries. Although enriching for certain portions of the genome is possible, typically fragments to be sequenced are generated randomly rather than by user-selected sequencing primers. An important benefit to these random fragments is the greater ease with which breakpoints resulting from large idels or other rearrangements can be identified. Conventional targeted resequencing by standard capillary methods alone is likely to miss many such events because PCR amplification of a given region cannot occur if one or more primer sites is lost or distantly relocated.
Finally, the markedly lower cost of next-generation methods also means that automatic confirmation of mutant bases can be reasonably built into to the sequencing protocol. With such platforms, this is a simple matter of sequencing enough random fragments to have a high probability of genotyping every base several times. The lower-throughput of capillary methods has often necessitated the initial filtering of mutant calls such that only those deemed to have a high likelihood of being drivers are retested. For example, the first major study of Sjoblom et. al. triaged 260,000 non-coding changes without further confirmation (87). While unavoidable from the logistical standpoint of traditional sequencing, synonymous changes within tumors may be of importance, given that they can influence both transcription and translation (89).
Implications of the Cancer Genome Atlas: Inter-Tumor Heterogeneity
The primary goal of cancer genome sequencing studies has been the identification of genes and pathways that play a causal role in the neoplastic process (98). It was the expectation that sufficiently detailed genetic analysis would lead to the identification of a small set of commonly mutated genes that drive tumor progression and, thus, present new therapeutic targets. Collectively, the initial studies described above constitute the most systematic characterization of a disease genome ever undertaken and demonstrate the feasibility of producing a compendium of clonally altered somatic sequences. Additional analyses using complementary approaches, including those assessing rearrangements (111), deletions (112) and epimutations (113) as well as the impact of mutations in non-coding sequences is within the scope of our current technology and will soon provide an even more complete description of changes within the cancer genome.
The overarching conclusion to be drawn from completed cancer genome sequencing studies is that most cancer types display substantial inter-tumor mutational heterogeneity. Individual solid organ tumors harbor, on average, more than 50 non-silent clonal mutations (GBM < pancreatic ≪ colorectal ≈ breast) in the coding regions of different genes, yet only a small fraction of these genes are mutated in a high proportion of tumors. Although certain clonally disrupted genes are more prevalently represented within specific types of cancer, there remains a great deal of overlap. The large number and breadth of diversity in genes mutated among individual tumor specimens emphasizes the fundamentally stochastic nature of cancer evolution.
The therapeutic implications of these findings are considerable. Preliminary studies focusing on the most druggable portion of the genome, kinases, explored the possibility of identifying commonly mutated genes that might be exploited with targeted approaches (86). However, after increasing the number of samples profiled, it has become clear that any one of these is only mutated in a small fraction of tumors. To synthesize and test enough small molecule inhibitors to combat even half of only the kinase class of suspected tumor-drivers would be a daunting undertaking on a scale that is arguably beyond our current drug-developing and regulatory capacities. The alternative option of targeting general pathways rather than specific mutant proteins might be more achievable.
Non-Clonal Mutations and Intra-Tumor Heterogeneity
A significant limitation of these studies lies, not in the complexity of the clonal mutations they are attempting to annotate, but in the fact that, by design, they are unable to address deeper heterogeneity within individual tumors. Most investigations to date have been concerned only with identifying mutations in the dominant clone. A tumor is itself genomically heterogeneous, with each cell having a different mutational signature reflective of its unique lineage history within the evolving neoplasm. Analysis of disseminated single cells in minimal residual disease has demonstrated a high level of genomic heterogeneity within individual lesions as well as between primary tumors and metastatic cells (114). Irrespective of the predominant forces generating mutations during tumorigenesis, it is of critical importance to recognize that mutations occur randomly, and that only a tiny subset of these are likely to be selectably advantageous at a given stage of development. This leaves an exponentially larger number of unexpanded mutations to act as a dormant repository of genetic diversity. A tumor is a dynamic entity that never ceases to evolve. The specific fitness of a cancer cell depends on the context of its tumor environment. As this changes over time, new cellular stresses such as hypoxia, nutrient depletion and immune recognition arise and cancer cells with the requisite phenotypes are selected. Thus, subclonal heterogeneity is of paramount importance to tumor progression.
The clinical importance of subclonal mutations lies in the fact that genetic variants encoding resistance to all single-target drugs are likely to pre-exist in a tumor cell population (70). Imatinib, the prototypical targeted therapy for chronic myelogenous leukemias (CML) bearing activating mutations in the ABL gene, frequently loses clinical efficacy due to the emergence of resistant clones (83). The basis for this resistance is frequently attributable to new point mutations in the ABL kinase domain that decrease its affinity for the drug (115–116). It has been specifically demonstrated that resistance mutations can be found in CML prior to the initiation of therapy (117). A method for overcoming the problem of resistance, which has long been standard in the clinic with existing chemotherapy regimens, is to use multi-drug cocktails. A group of agents directed at multiple, unrelated tumor-relevant pathways is more likely to prevent or delay resistance given that the combinatorial probability of a single cell simultaneously bearing resistance mutations to each is very low. Although this is not conceptually new, the degree of inter-tumor heterogeneity revealed by cancer genome studies hints that an even larger cocktail of drugs than is presently used may be advantageous to combat resistance arising from intra-tumor genetic variants (118).
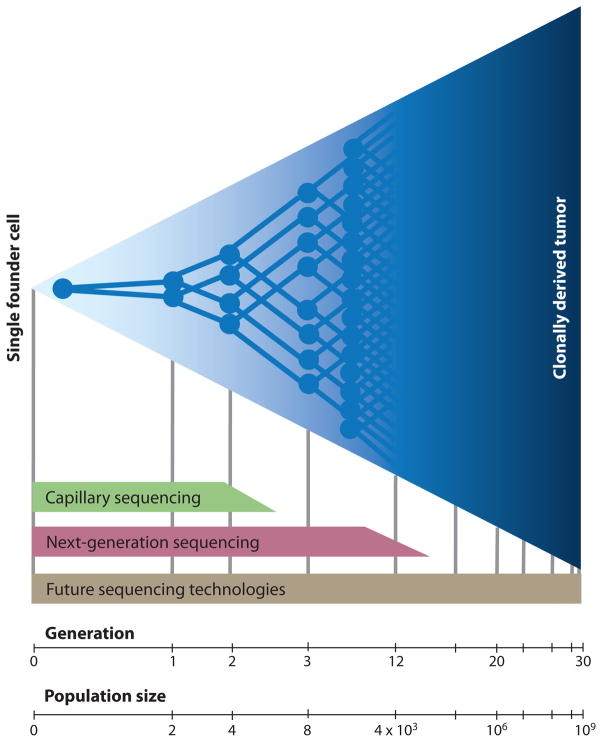
The ability to quantify subclonal genetic diversity might provide important clinical information about the likelihood of a tumor becoming resistant to specific therapies. Just how many unexpanded random mutations are there in a tumor? This has historically been a difficult question to answer because of the technical challenges facing low frequency measurements (Figure 5). Standard capillary sequencing technology measures average population genotype and will only detect minority clones down to approximately 25% with routine, automated use. Next generation sequencing methods are remarkably more sensitive, given that they genotype the amplified product of individual molecules. Sequencing of many fragments from a given region (“deep sequencing”) produces a digital histogram representing the frequency of different genotypes in a population of molecules. However, because of imperfections in detection hardware and chemistry, as well as the need for amplification steps by fallible polymerases, sensitivity is currently limited to about 1/5000 (119). Exquisitely sensitive methods of mutation detection do exist, including cell culture-based fluctuation assays (10) as well as systems involving transgenic animals bearing reporter genes, yet neither of these is amenable to the direct examination of human tumors.
Figure 5. Limit of subclonal detection.
Depicted is the clonal expansion of a single cell into a population of one billion cells. In a hypothetical scenario where no cell death occurs, this requires approximately 30 generations of division. Current capillary methods of DNA sequencing only detect mutations that have clonally expanded to represent 25% or more of a tumor. Only mutations that are present in the founding cell or that arise within the first two generations of division can be identified. Deep sequencing on current “next-generation” sequencing platforms are reported to detect subclonal mutations down to a frequency of 1/5000 (119) (thus, those occurring within the first twelve generations after founding). Sensitivity is limited by the error rate of PCR amplification steps and that of the sequencing chemistry itself. Future technologies may eventually enable ultra-accurate, high-throughput detection of mutations that arise during any stage of clonal expansion.
Our group has recently developed a method for the detection of random mutations that offers unprecedented sensitivity; one mutation can be identified among 108 wild-type nucleotides in nuclear DNA (120). The system is based on the concept that spontaneous mutations occurring in a non-coding, Taq1 restriction endonuclease recognition site will render it non-cleavable by this enzyme. After multiple rounds of enzymatic digestion, only the mutant sequences from a larger population will remain intact and be amplifiable by PCR primers flanking the restriction site. We (121) and others (122) have used this approach to demonstrate a markedly elevated frequency of random, unexpanded mutations in several types of cancer. Although highly sensitive, this approach can interrogate only four bases out of the entire genome at once. Hopefully the future will bring even more sophisticated methods that combine the throughput of present next-generation sequencing platforms with the ultra-high sensitivity needed to accurately identify single mutants.
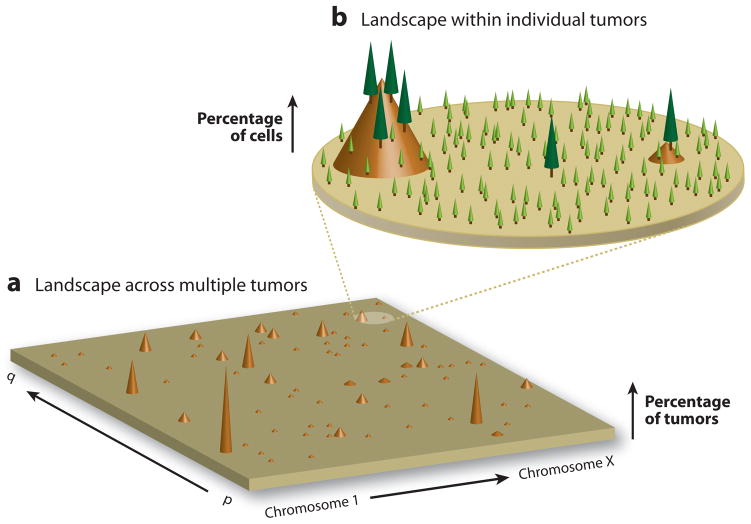
The cancer genomic landscape has been previously described from a multiple tumor perspective where “mountains” and “hills” represent, respectively, frequently and infrequently mutated genes (Figure 6a) (87). We posit that a complete description of the cancer genome must necessarily include a provision for the intra-tumor heterogeneity of individual neoplasms. To this landscape we add a small number of “trees” to represent clonally mutated genes present in a large number of cells and, surrounding these, a much larger number of “seedlings” representing mutations present in only one or a few cells (Figure 6b). We argue that it is this forest, undetected by the cancer genome sequencing studies above, that provides much of the basis for the wide variations in tumor behavior and responsiveness to therapy, and represents one of the clinically most important features of the cancer genome; when an old tree falls or is logged, many seedlings are poised to grow and take its place.
Figure 6. The mutational landscape of the cancer genome.
A) The cancer genome landscape proposed by Wood et. al. (88) graphically represents the mutational heterogeneity among different tumors of a single cancer type. The height of each brown peak indicates the percentage of tumors found to carry a clonal mutation in a particular gene. The landscape comprises a small number of “mountains”–genes which are clonally mutated in a large fraction of individual cancers–and a significantly greater number of “hills” – genes clonally mutated in only one or a few tumors. While there may be 50 or more genes clonally mutated within the genome of an individual tumor, most genes are rarely mutated in more than a few tumors. B) An additional level of the mutational landscape exists within individual tumors due to differences among the genomes of single tumor cells. Although a small number of mutations are clonally present in the majority of cells in an individual tumor (“trees”), an exponentially larger number exist subclonally in only one or a few cells (“seedlings”). Among this vast reservoir of non-clonal mutations exists many therapy-resistant variants.
Final Thoughts and Future Directions
Today we recognize the unidirectional fate of neoplastically transformed cells to be the result of unrepaired mutational events that become permanently fixed in the genome and epigenome of subsequent generations of progeny. The path that has brought us to our current understanding of the genetic basis for cancer has been a long and remarkable one punctuated by a wide breadth of discoveries. The ability of most cells in the human body to prevent the build-up of cancer-causing mutations is an impressive tribute to the billions of years of evolution leading up to the emergence of multicellular organisms. Mechanisms for mutation prevention and suppression are, nevertheless, imperfect; progressive accumulation of new genetic variants provides the fuel for evolution on a cellular level which forms the ultimate basis of tumorigenesis.
New technology has allowed us to begin to tabulate the mutations of cancer. First generation cancer genome sequencing studies were driven by the expectation that clonal mutations in a limited set of key genes would be commonly found in different tumors and might provide new druggable targets. However, the results so far indicate a more complex picture than initially hoped. Very few genes that have not been previously identified by other means are prevalently mutated in specific cancers. Many genes likely to be involved in driving tumorigenesis are altered in only a small fraction of tumors. The presence of many thousands of clonally expanded passengers, although playing no causal role in the cancer, serve as a reminder of the invisible legions formed by the exponentially larger number of unexpanded variants, many of which are drug-resistant and awaiting the opportunity to selectively proliferate upon induction of new treatments. In light of this emerging complexity, it is becoming increasingly difficult to envision how it will be possible to develop a realistic number of targeted chemotherapies to be directed against a discrete panel of commonly mutated cancer genes. These findings substantiate the concept that simultaneous use of multiple agents against different general pathways may be most efficacious.
Though sobering, the cancer genome studies so far have established an important baseline of information from which to advance. As technology improves and comes down in cost, large-scale genome analysis methods will become tractable to smaller groups who will be able to explore innovative and higher risk approaches. As we move forward in these endeavors, it will be critical not to lose sight of the need to confirm functional status of mutations identified. While it remains a powerful tool, genome sequencing cannot address all the questions of cancer research so we cannot neglect to spread our resources among many complementary means of identifying novel features of the disease and ways to prevent and target these. Most importantly, we need to recognize the many levels of heterogeneity inherent to cancer and ensure that this reality be integrated into future studies.
Summary points.
Cancer is a disease of somatic evolution occurring on a cellular level. Random mutations occur throughout an organism’s life and the small subset of these that bestow a cell with growth and survival benefits will clonally expand to form a tumor.
Mutations result from failure of DNA repair. Within a population of cells, mutations accumulate more rapidly during tumorigenesis from increased cell proliferation as well as from defects in DNA maintenance pathways. The relative importance of these two mechanisms is likely to vary from one tumor to another and remains the subject of debate.
Identifying causative mutations within a tumor’s genome by DNA sequencing is a complex problem from both a technical and analysis standpoint. Many mutations are clonally present by chance alone and differentiating these neutral passengers from causal drivers presents a significant challenge.
Recent large-scale cancer genome sequencing studies have indicated a great deal of variation in the clonal mutations found among different tumors. Very few genes are commonly mutated in any type of cancer and this finding suggests that it will be difficult to design a limited number of widely-usable targeted therapies that center on specific genes.
Current methods of DNA sequencing cannot accurately portray the many mutations in a developing cancer that are present in only a minority of tumor cells. These sub-clonal mutations comprise a tumor’s pre-existing evolutionary potential for overcoming theraputic interventions. Characterization of this intra-tumor heterogeneity will be of clinical importance as new, more sensitive sequencing technologies are developed.
Acknowledgments
Grant Support: National Cancer Institute: CA102029, CA115802 (L.A. Loeb); DOD: W81XWH-07-0029 (L.A. Loeb); National Institute of Aging: AG033485 (J.J. Salk); NCI/Health Research Board of Ireland Joint Research Fellowship (E.J. Fox).
Mini-glossary
- Mutator phenotype
an increased (per-cell division) mutation rate resulting from heritable cellular defects, generally in DNA maintenance machinery
- Clonal expansion
the multiplication of a single cell to produce a population of genetically related progeny
- Clonal mutation
a mutation present in the majority of cells in a tumor that is detectable by conventional sequencing
- Subclonal mutation
a mutation present in a single cell or a minority of cells in a tumor and not detectible by conventional sequencing
- Driver mutation
a mutation that provides a selectable fitness advantage to the cell and facilitates its clonal expansion in the population
- Passenger mutation
a mutation having no effect on a cell’s fitness that clonally expands in the population as a result of a different driver mutation
- Cancer Genome Atlas
the complete catalog of genetic and epigenetic alterations found in cancers of all types
- Next-generation sequencing
new methods of high-throughput DNA sequencing carried out on amplified single DNA molecules affixed to a solid matrix
- Deep Sequencing
sequencing of many individual DNA fragments from an identical portion of the genome to identify subclonal mutations
References
- 1.Boveri T. Veh Dtsch Zool Ges. Wurzburg: 1902. Uber mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. [Google Scholar]
- 2.Cleaver JE, Kraemer KH. Xeroderma pigmentosum. In: Scriver CR, Beudet AL, Sktm WS, Valle D, editors. Metabolic Basis of Inherited Disease. New York, NY: McGraw-Hill; 1989. pp. 2949–71. Number of 2949–71 pp. [Google Scholar]
- 3.Kallioniemi A, Kallioniemi O-P, Piper J, Tanner M, Stokke T, et al. Detecton and mapping of amplified DNA sequences in breast cancer by comparative gnomic hybridization. Proc Natl Acad Sci USA. 1994;91:2156–60. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riopel MA, Spellerberg A, Griffin CA, Perlman EJ. Genetic analysis of ovarian germ cell tumors by comparative genomic hybridization. Cancer Res. 1998;58:3105–10. [PubMed] [Google Scholar]
- 5.El-Rifai W, Sarlomo-Rikala M, Knuutila S, Miettinen M. DNA copy number changes in development and progression in leiomyosarcomas of soft tissues. Am J Pathol. 1998;153:985–90. doi: 10.1016/S0002-9440(10)65640-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerangueven F, Noguchi T, Coulier F, Allione F, Wargniez V, et al. Genome-wide search for loss of heterozygosity shows extensive genetic diversity of human breast carnomas. Cancer Res. 1997;57:5469–74. [PubMed] [Google Scholar]
- 7.Klein CA, Schmidt-Kittler O, Schardt JA, Pantel K, Speicher MR, Riethmuller G. Comparative gnomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc Natl Acad Sci USA. 1999;96:4494–9. doi: 10.1073/pnas.96.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 9.Albertini RJ, Nicklas JA, O’Neill JP, Robison SH. In vivo somatic mutations in humans: measurement and analysis. Annu Rev Genet. 1990;24:305–26. doi: 10.1146/annurev.ge.24.120190.001513. [DOI] [PubMed] [Google Scholar]
- 10.Bielas JH, Heddle JA. Elevated mutagenesis and decreased DNA repair at a transgene are associated with prolifration but not apoptosis in p53-deficient cells. Proc Natl Acad Sci USA. 2003;100:12853–8. doi: 10.1073/pnas.2235595100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu EHY, Boehnke M, Hanash SM, Kuick RD, Lamb BJ, et al. Estimation of mutation rates based on the analysis of polypeptide constituents of cultured human lymphoblastoid cells. Genetics. 1988;119:693–703. doi: 10.1093/genetics/119.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMars R, Held KR. The spontaneous azaguanine-resistant mutants of diploid human fibroblasts. Humangenetik. 1972;16:87–110. doi: 10.1007/BF00393992. [DOI] [PubMed] [Google Scholar]
- 13.Cervantes RB, Stringer JR, Shao C, Tischfield JA, Stambrook PJ. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc Natl Acad Sci USA. 2002;99:3586–90. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson AL, Loeb LA. The mutation rate and cancer. Genetics. 1998;148:1483–90. doi: 10.1093/genetics/148.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrin LJ, Beckman RA, Loeb LA, Mildvan AS. Manganese in Metabolism and Enzyme Function. New York: Academic Press; 1986. Kinetic and magnetic resonance studies of the interaction of Mn2+, substrates and templates with DNA polymerases; pp. 259–73. [Google Scholar]
- 16.Petruska J, Sowers LC, Goodman MF. Comparison of nucleotide interactions in water, proteins and vacuum: model for DNA polymerase fidelity. Proc Natl Acad Sci USA. 1986;83:1559–62. doi: 10.1073/pnas.83.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loeb L, Monnat R. DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 18.Uyemura D, Lehman IR. Biochemical characterization of mutant forms of DNA polymerase I from Escherichia coli. I The polA12 mutation. J Biol Chem. 1976;251:4078–84. [PubMed] [Google Scholar]
- 19.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu Rev Biochem. 1996;65:101–33. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel TA, Schaaper RM, Loeb LA. Depurination-induced infidelity of DNA synthesis with purified DNA replication proteins in vitro. Biochemistry. 1983;22:2378–84. doi: 10.1021/bi00279a012. [DOI] [PubMed] [Google Scholar]
- 21.Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, et al. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–42. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishimura S. 8-Hydroxyguanine: from its discovery in 1983 to the present status. Proc Jpn Acad. 2006;82:127–41. doi: 10.2183/pjab.82.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Floyd RA. Measurement of oxidative stress in vivo. In: Daview KJA, Ursini F, editors. The Oxygen Paradox. Cleup University Press; 1995. pp. 89–103. [Google Scholar]
- 24.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 25.Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bronner MP, O’Sullivan JN, Rabinovitch PS, Crispin DA, Chen L, et al. Genomic biomarkers to improve ulcerative colitis neoplasia surveillance. Am J Pathol. 2008;173:1853–60. doi: 10.2353/ajpath.2008.080250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baik SC, Youn HS, Chung MH, Lee WK, Cho MJ, et al. Increased oxidative DNA damage in Heliocobacter pylori-infected human gastric mucosa. Cancer Res. 1996;56:1279–82. [PubMed] [Google Scholar]
- 28.Fausto N, Webber EM. Mechanisms of growth regulation in liver regeneration and hepatic carcinogenesis. In: Boyer JL, Okner RK, editors. Progress in Liver Diseases. Vol. 11. Philadelphia: W.B. Saunders Co; 1993. pp. 115–37. [PubMed] [Google Scholar]
- 29.Feig DI, Loeb LA. Oxygen radical induced mutagenesis is DNA polymerase specific. J Mol Biol. 1994;235:33–41. doi: 10.1016/s0022-2836(05)80009-9. [DOI] [PubMed] [Google Scholar]
- 30.Ames BN, Durston WE, Yamasaki E, Lee FD. Carcinogens are mutagens: a simple test system combining liver homogenates for activation and bacteria for detection. Proc Natl Acad Sci USA. 1973;70:2281–5. doi: 10.1073/pnas.70.8.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auerbach C, Robson JM. Chemical production of mutations. Nature. 1946;154:302. doi: 10.1038/157302a0. [DOI] [PubMed] [Google Scholar]
- 32.Loeb L, Harris CC. Advances in chemical carcinogenesis: a historical review and prospective. Cancer Research. 2008;68:6863–72. doi: 10.1158/0008-5472.CAN-08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 34.Wynder EL, Graham EA. Tobacco smoking as a possible etiologic factor in bronchogenic carcinoma. JAMA. 1950;143:329–36. doi: 10.1001/jama.1950.02910390001001. [DOI] [PubMed] [Google Scholar]
- 35.Loeb LA, Ernster VL, Warner KE, Abbotts J, Laszlo J. Smoking and lung cancer: an overview. Cancer Res. 1984;44:5940–58. [PubMed] [Google Scholar]
- 36.Friedberg EC, Aguilera A, Gellert M, Hanawalt PC, Hays JB, et al. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006;5:986–96. doi: 10.1016/j.dnarep.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 37.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 38.Harris CC. p53: at the crossroads of molecular carcinogenesis and cancer risk assessment. Science. 1993;262:1980–1. doi: 10.1126/science.8266092. [DOI] [PubMed] [Google Scholar]
- 39.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 40.Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, et al. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet. 2002;30:227–32. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 41.King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–6. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 42.Spivak G, Itoh T, Matsunaga T, Nikaido O, Hanawalt P, Yamaizumi M. Ultraviolet-sensitive syndrome cells are defective in transcription-coupled repair of cyclobutane pyrimidine dimers. DNA Repair (Amst) 2002;1:629–43. doi: 10.1016/s1568-7864(02)00056-3. [DOI] [PubMed] [Google Scholar]
- 43.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, et al. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis NA, Groden J, Ye T-Z, Straughen J, Lennon DJ, et al. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–66. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 45.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. The Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci USA. 2007;104:10394–9. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 47.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–30. [PubMed] [Google Scholar]
- 48.Webber EM, Wu JC, Wang L, Merlino G, Fausto N. Overexpression of transforming growth factor-alpha causes liver enlargement and increased hepatocyte proliferation in transgenic mice. Am J Pathol. 1994;145:398–408. [PMC free article] [PubMed] [Google Scholar]
- 49.Hecker E. Phorbol esters from croton oil. Chemical nature and biological activities. Naturwissenschaften. 1967;54:282–4. doi: 10.1007/BF00620887. [DOI] [PubMed] [Google Scholar]
- 50.Rous P. Transmission of a malignant new growth by means of a cell-free filtrate. JAMA. 1911:56. [PubMed] [Google Scholar]
- 51.Varmus HE, Shank PR, Hughes SE, Kung HJ, Heasley S, et al. Synthesis, structure, and integration of the DNA of RNA tumor viruses. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):851–64. doi: 10.1101/sqb.1979.043.01.091. [DOI] [PubMed] [Google Scholar]
- 52.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. This landmark paper presents a detailed analysis the required phenotyes for tumorigenesis. [DOI] [PubMed] [Google Scholar]
- 53.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–11. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 54.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nat Rev Cancer. 2008;8:415–24. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 55.Cairns J. Cancer and the immortal strand hypothesis. Genetics. 2006;174:1069–72. doi: 10.1534/genetics.104.66886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prindull GA, Fibach E. Are postnatal hemangioblasts generated by dedifferentiation from committed hematopoietic stem cells? Exp Hematol. 2007;35:691–701. doi: 10.1016/j.exphem.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 57.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. The seminal paper describing cancer an as evolutionary process. [DOI] [PubMed] [Google Scholar]
- 58.Merlo LM, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006;6:924–35. doi: 10.1038/nrc2013. [DOI] [PubMed] [Google Scholar]
- 59.Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 60.Armitage P, Doll R. The age distribution of cancer and a multistage theory of carcinogenesis. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knudson AG. Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol. 1996;122:135–40. doi: 10.1007/BF01366952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renan MJ. How many mutations are required for tumorigenesis? Implications from human cancer data. Mol Carc. 1993;7:139–46. doi: 10.1002/mc.2940070303. [DOI] [PubMed] [Google Scholar]
- 63.Rangarajan A, Hong SJ, Gifford A, Weinberg RA. Species- and cell type-specific requirements for cellular transformation. Cancer Cell. 2004;6:171–83. doi: 10.1016/j.ccr.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 64.Mahale AM, Khan ZA, Igarashi M, Nanjangud GJ, Qiao RF, et al. Clonal selection in malignant transformation of human fibroblasts transduced with defined cellular oncogenes. Cancer Res. 2008;68:1417–26. doi: 10.1158/0008-5472.CAN-07-3021. [DOI] [PubMed] [Google Scholar]
- 65.Bodmer W, Bielas JH, Beckman RA, Loeb LA. Genetic instability is not a requirement for tumor development. Cancer Res. 2008;68:3558–60. doi: 10.1158/0008-5472.CAN-07-6544. discussion 60–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–7. doi: 10.1158/0008-5472.CAN-07-5835. discussion 7. [DOI] [PubMed] [Google Scholar]
- 67.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 68.Moolgavkar SH, Luebeck EG. Multistage carcinogenesis and the incidence of human cancer. Genes Chromosomes Cancer. 2003;38:302–6. doi: 10.1002/gcc.10264. [DOI] [PubMed] [Google Scholar]
- 69.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant change. Cancer Res. 1974;34:2311–21. [PubMed] [Google Scholar]
- 70.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci USA. 2003;100:776–81. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tomlinson IP, Novelli MR, Bodmer WF. The mutation rate and cancer. Proc Natl Acad Sci U S A. 1996;93:14800–3. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beckman RA, Loeb LA. Efficiency of carcinogenesis with and without a mutator mutation. Proc Natl Acad Sci USA. 2006;103:14140–5. doi: 10.1073/pnas.0606271103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beckman RA, Loeb LA. Negative clonal selection in tumor evolution. Genetics. 2005;171:2123–31. doi: 10.1534/genetics.105.040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robins HGA, Harris SL, Levine AJ. The first twenty-five years of P53 research. In: Hainaut P, Wiman KG, editors. 25 Years of p53 Research. Dordrecht: Springer; 2005. pp. 1–26. [Google Scholar]
- 76.Wells RD, Sinden RR. Genome Analysis, Genome Rearrangement and Stability. Vol. 7. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 107–38. [Google Scholar]
- 77.Farber RA, Petes TD, Dominska M, Hudgens SS, Liskay RM. Instability of simple sequence repeats in a mammalian cell line. Hum Mol Genet. 1994;3:253–6. doi: 10.1093/hmg/3.2.253. [DOI] [PubMed] [Google Scholar]
- 78.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 79.Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–5. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Balaban GB, Herlyn M, Clark WH, Jr, Nowell PC. Karyotypic evolution in human malignant melanoma. Cancer Genet Cytogenet. 1986;19:113–22. doi: 10.1016/0165-4608(86)90378-x. [DOI] [PubMed] [Google Scholar]
- 81.Smith G, Carey FA, Beattie J, Wilkie MJV, Lightfoot TJ, et al. Mutations in APC, Kirsten-ras, and p53 -- alternative genetic pathways to colorectal cancer. Proc Natl Acad Sci USA. 2002;99:9433–8. doi: 10.1073/pnas.122612899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ley TJ, Mardis E, Ding L, Fulton B, Mclellan M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Druker BJ. Imatinib: paradigm or anomaly? Cell Cycle. 2004;3:833–5. [PubMed] [Google Scholar]
- 84.Wang TL, Rago C, Silliman N, Ptak J, Markowitz S, et al. Prevalence of somatic alterations in the colorectal cancer cell genome. Proc Natl Acad Sci USA. 2002;99:3076–80. doi: 10.1073/pnas.261714699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164–6. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 86.Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, et al. Mutational analysis of the tyrosine kinome in colorectal cancers. Science. 2003;300:949. doi: 10.1126/science.1082596. [DOI] [PubMed] [Google Scholar]
- 87.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. The concensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 88.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 89.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–8. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Getz G, Hofling H, Mesirov JP, Golub TR, Meyerson M, et al. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 91.Rubin AF, Green P. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138956. [DOI] [PubMed] [Google Scholar]
- 92.Forrest WF, Cavet G. Comment on “The consensus coding sequences of human breast and colorectal cancers“. Science. 2007;317:1500. doi: 10.1126/science.1138179. [DOI] [PubMed] [Google Scholar]
- 93.Parmigiani G, Lin J, Boca S, Sjoblom T, Jones S, et al. Response to Comments on “The Consensus Coding Sequences of Human Breast and Colorectal Cancers“. Science. 2007;317:1500. [Google Scholar]
- 94.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stephens P, Edkins S, Davies H, Greenman C, Cox C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–2. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 96.Davies H, Hunter C, Smith R, Stephens P, Greenman C, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65:7591–5. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 97.Bignell G, Smith R, Hunter C, Stephens P, Davies H, et al. Sequence analysis of the protein kinase gene family in human testicular germ-cell tumors of adolescents and adults. Genes Chromosomes Cancer. 2006;45:42–6. doi: 10.1002/gcc.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jones S, Zhang X, Parsons D, Lin J, Leary R, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prehn RT, Main JM. Immunity to methylcholanthrene-induced sarcomas. J Natl Cancer Inst. 1957;18:769–78. [PubMed] [Google Scholar]
- 100.Segal N, Parsons D, Peggs K, Velculescu V, Kinzler K, et al. Epitope Landscape in Breast and Colorectal Cancer. Cancer Research. 2008;68:889–92. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 101.Leary RJ, Lin JC, Cummins J, Boca S, Wood LD, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224–9. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.The Cancer Genome Atlas Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 105.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–5. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Weir BA, Woo MS, Getz G, Perner S, Ding L, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Loriaux MM, Levine RL, Tyner JW, Frohling S, Scholl C, et al. High-throughput sequence analysis of the tyrosine kinome in acute myeloid leukemia. Blood. 2008;111:4788–96. doi: 10.1182/blood-2007-07-101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lai LA, Paulson TG, Li X, Sanchez CA, Maley C, et al. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer. 2007;46:532–42. doi: 10.1002/gcc.20435. [DOI] [PubMed] [Google Scholar]
- 113.Shames DS, Girard L, Gao B, Sato M, Lewis CM, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. PLoS Med. 2006;3:e486. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stoecklein NH, Hosch SB, Bezler M, Stern F, Hartmann CH, et al. Direct genetic analysis of single disseminated cancer cells for prediction of outcome and therapy selection in esophageal cancer. Cancer Cell. 2008;13:441–53. doi: 10.1016/j.ccr.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 115.Michor F, Hughes TP, Iwasa Y, Branford S, Shah NP, et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–70. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 116.Radich JP, Dai H, Mao M, Oehler V, Schelter J, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci, USA. 2006;103:2794–9. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354:2531–41. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 118.Golub TR. Mining the genome for combination therapies. Nat Med. 2003;9:510–1. doi: 10.1038/nm0503-510. [DOI] [PubMed] [Google Scholar]
- 119.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–6. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bielas J, Loeb L. Quantification of random genomic mutations. Nat Meth. 2005;2:285–90. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 121.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–42. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng L, Dai H, Zhou M, Li M, Singh P, et al. Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nature Med. 2007;13:812–9. doi: 10.1038/nm1599. [DOI] [PubMed] [Google Scholar]