Abstract
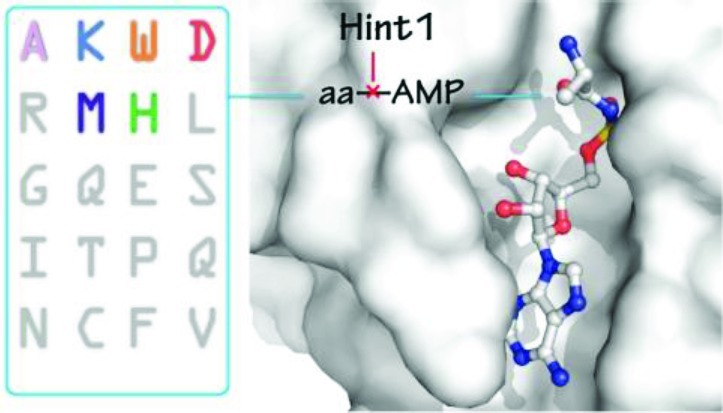
Human Hint1 suppresses specific gene transcription by interacting with the transcription factor MITF in mast cells. Hint1 activity is connected to lysyl-tRNA synthetase (LysRS), a member of the universal aminoacyl tRNA synthetase family that catalyzes specific aminoacylation of their cognate tRNAs, through an aminoacyl adenylate (aa-AMP) intermediate. During immune activation, LysRS produces a side-product diadenosine tetraphosphate (Ap4A) from the condensation of Lys-AMP with ATP. The pleiotropic signaling molecule Ap4A then binds Hint1 to promote activation of MITF-target gene transcription. Earlier work showed that Hint1 can also bind and hydrolyze Lys-AMP, possibly to constrain Ap4A production. Because Ap4A can result from condensation of other aa-AMP's with ATP, the specificity of the Hint1 aa-AMP–hydrolysis activity is of interest. Here we show that Hint1 has broad specificity for adenylate hydrolysis, whose structural basis we revealed through high-resolution structures of Hint1 in complex with three different aa-AMP analogues. Hint1 recognizes only the common main chain of the aminoacyl moiety, and has no contact with the aa side chain. The α-amino group is anchored by a cation-pi interaction with Trp123 at the C-terminus of Hint1. These results reveal the structural basis for the remarkable adenylate surveillance activity of Hint1, to potentially control Ap4A levels in the cell.
Introduction
Hint1 is a member of a histidine triad (HIT) protein family that is widespread in eukaryotes and bacteria.1 Hint1 acts as a haplo-insufficient tumor suppressor whose deficiency in mice results in increased susceptibility to both spontaneous and carcinogen-induced tumor formation.2,3 Hint1 deficient cells are resistant to ionizing radiation-induced apoptosis.4,5 Overexpression of Hint1 in nonsmall lung cancer cells led to cell growth inhibition in vitro and to a reduction of tumorigenicity in vivo.6 Additionally, Hint1 can bind to and has hydrolase activity toward phosphoramidates and acyladenylates.7,8 Hint1 also associates with and suppresses the microphthalmia transcription factor (MITF) and the upstream stimulatory factor 2 (USF2).9,10 How these activities are associated with Hint1 suppressor function remains unclear. Interestingly, regulation of Hint1 in mast cells is at least in part through collaboration with a member of the aminoacyl tRNA synthetase family.11−13
Aminoacyl-tRNA synthetases (aaRS's) are essential enzymes that catalyze the attachment of amino acids onto their cognate tRNAs in a two-step reaction.14,15 The amino acid (aa) is first condensed with ATP to form a tightly bound aminoacyl adenylate (aa-AMP), and inorganic pyrophosphate (PPi) is released. The activated aa-AMP is then transferred from the adenylate to the 3′ end of the tRNA to form aa-tRNA:
| 1 |
| 2 |
Some aaRS's also generate a signaling molecule diadenosine tetraphosphate (AppppA or Ap4A) by catalyzing a side reaction:16
| 3 |
Lysyl-tRNA synthetase (LysRS) is one of the best-known aaRS's that produces Ap4A.17−19 Higher eukaryotic LysRS also joins with eight other aaRS's and three auxiliary proteins (MSC p43, MSC p38, MSC p18, or named as AIMP1, AIMP2, AIMP3, respectively) to form a high molecular mass multisynthetase complex (MSC).20−23 Harboring almost half of the cellular tRNA synthetases, MSC is regarded as a reservoir that controls the flow of tRNA synthetases between translational and ex-translational functions.24,25 For example, in immunologically activated mast cells, LysRS is released from the MSC, produces Ap4A, and drives the expression of microphthalmia associated transcription factor (MITF)-downstream genes that regulate the immune response.11,12
Through binding to MITF, Hint1 suppresses MITF activity in quiescent mast cells.9 In stimulated mast cells, MITF activation is accompanied by an increase of intracellular Ap4A, which promotes dissociation of the Hint1-MITF complex.9,11It has been demonstrated that Ap4A binds to Hint1 and specifically dissociates the Hint1-MITF interaction.26 Additionally, directly adding Ap4A to the cell medium, or siRNA knockdown of the Ap4A hydrolase, caused dissociation of the Hint1-MITF complex and activated a subset of MITF-targeted gene transcription.11,13,26 It was suggested that LysRS-produced Ap4A activates MITF by binding and releasing Hint1.12
Hint1 proteins contain a conserved HxHxHxx motif (x is hydrophobic) that binds to and hydrolyzes purine-containing nucleotide derivatives such as adenosine monophosphoramidate (AMP-NH2), AMP-N-ε-(N-α-acetyl lysine methyl ester) (AMP-N-ε-lysine), and AMP-N-alanine methyl ester.27 On the other hand, Hint1 also hydrolyzes Lys-AMP, the canonical intermediate product of LysRS (eq 1).8 Therefore, Lys-AMP may play a role in the LysRS-Hint1-MITF pathway.28 For example, the destruction of Lys-AMP would limit the amount of Ap4A that can be produced (eq 3 above). In addition, because 5 mammalian aaRS's (LysRS, ThrRS, SerRS, PheRS, GlyRS) and more than 10 aaRS's from other species have been demonstrated to synthesize Ap4A,18,29−32 and because these synthetases also synthesize their respective aa-AMP intermediate, we considered the possibility that the hydrolytic activity of Hint1 may extend beyond Lys-AMP. Thus, if Ap4A was generated by various synthetases, then the control of its level would necessitate a broader specificity for aa-AMP hydrolysis, either by Hint1 or by one or more other enzymes.
We thus investigated the ability of Hint1 to hydrolyze adenylates other than Lys-AMP. We showed that Hint1 has broad specificity for aa-AMP hydrolysis, and does so through a mechanism that exploits the histidine triad configuration of the protein. To understand the molecular basis of this broad specificity, we determined the crystal structures of Hint1 in complex with a stable Lys-AMP analogue and, separately, with two other stable aminoacyl adenylate analogues. The results reveal a new mechanism for binding of aminoacyl adenylates (aa-AMP's), which is distinct from that seen for the highly specific complexes of aa-AMP's with their cognate aminoacyl-tRNA synthetases. Therefore, Hint1 binds and hydrolyzes a broad array of aa-AMP's, suggesting that the levels of free adenylates and of Ap4A may be regulated by the Hint1 hydrolytic activity in the cell.
Results
Hint1 Reacts with Different aa-AMP's
The intermediate product of LysRS, Lys-AMP, contains a high-energy ester-phosphate bond that can be cleaved when dissociated from LysRS and bound to Hint1. The hydrolysis by Hint1 of LysRS-generated Lys-AMP can be monitored by the formation of the Hint1-AMP intermediate.28 Because Hint1-AMP has a half-life of ∼0.3 s and is less sensitive to base catalyzed hydrolysis,8,28 it represents a transiently modified form of Hint1 that could in principle also have a specific function distinct from Hint1 itself. When l-lysine and α-32P-radiolabeled ATP are incubated with an E. coli LysRS known as LysU, both human and E. coli Hint hydrolyze Lys-AMP and form the Hint1-AMP intermediate state that can be detected by SDS-PAGE.28,33,34 This method of detection of Hint1-AMP is a definitive way to measure the ability of Hint1 to react with various aminoacyl-tRNA synthetase-produced aa-AMP's.
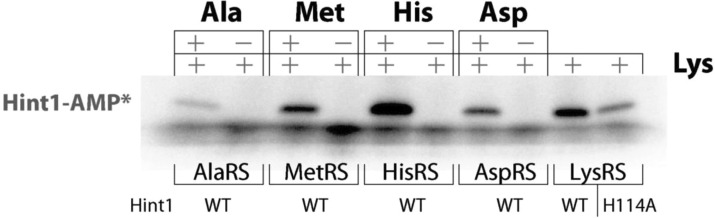
For this purpose, several amino acids (l-Ala, l-Asp, l-Met, l-His, and l-Lys) with distinct side chains were incubated with their cognate tRNA synthetase and with human Hint1. Formation of Hint1-AMP was detected for l-Lys/LysRS, indicating the Lys-AMP generated by LysRS reacts with Hint1 wild type (WT), and to a lesser extent with an active site mutant Hint1_H114A (see below) (Figure 1). In addition, formation of Hint1-AMP was also detected for each other amino acid/tRNA synthetase pair, with each yielding approximately the same amount of product within the same time period. Importantly, no Hint1-AMP was formed when Hint1 was incubated with the four noncognate tRNA synthetases and l-Lys. This observation is consistent with the expectation that Hint1 is labeled by capture of the aa-AMP generated from each tRNA synthetase (Figure 1). Thus, Hint1 binds and reacts with different aa-AMP's produced by other tRNA synthetases.
Figure 1.
aa-AMP hydrolysis by Hint1 is general. Hint1 was incubated with alpha-32P-ATP, l-lysine, and separately with five different tRNA synthetases, with/without the cognate amino acids. The reaction was quenched at 1 min and analyzed by SDS-PAGE and autoradiography. Although lysine is present in each reaction mixture, Hint1-AMP only forms when LysRS is present, or when there is another tRNA synthetase with its cognate amino acid. An active site mutant Hint1_H114A, which was characterized in this work, was also used to coincubate with LysRS and l-Lys.
Complex Structures of Hint1 with Lys-AMS
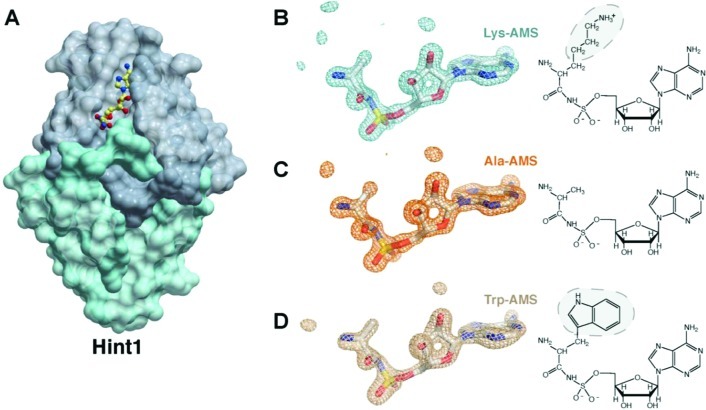
Although the hydrolysis of Lys-AMP by Hint1 was previously established,28,35 no detailed understanding of the interaction of Hint1 with adenylates has been reported. In order to obtain a structure of a complex, Lys-AMS ([N-(l-lysyl)sulfamoyl]adenosine), the nonhydrolyzable analogue of Lys-AMP, was cocrystallized at a concentration of 5 mM with human Hint1 (Figure 2A). The Hint1/Lys-AMS complex structure was solved at a resolution of 1.52 Å. Each asymmetric unit in the crystal contained two Hint1 molecules. As previously observed in other human Hint1 structures, the two Hint1’s form an extended β-sheet-like dimer that creates two purine nucleotide-binding sites.36,37 Interestingly, only one subunit of the Hint1 dimer bound Lys-AMS. The binding site in the second subunit of the homodimer was blocked by crystal packing interactions.
Figure 2.
Complex structure of human Hint1 with Lys-AMS, Ala-AMS, and Trp-AMS. (A) Dimeric structure of Hint1 in complex with Lys-AMS. One subunit of the Hint1 dimer contains the ligand. (B–D) Electron density maps of the bound aa-AMS analogues. The refined 2Fo-Fc density maps are shown at 1.0 σ. Chemical structures of the analogues are shown on the right, with electron densities missing from the side chains highlighted by shading on the chemical structures.
The electron density of the ligand could be clearly identified, including the adenosine moiety, the sulfate-substituted group, and the N-substituted covalent bond between the lysyl moiety’s backbone atoms C, Cα, and N and the sulfate. However, no density for the lysyl side chain could be seen beyond Cβ in the electron density map (shown at contour level of 1 σ of 2Fo-Fc map) (Figure 2B). This observation suggests that the side chain of Lys-AMS adopts a free conformation, such that its electron density is averaged out in the crystal.
Side Chain Independent Recognition of Hint1 with aa-AMP Analogues
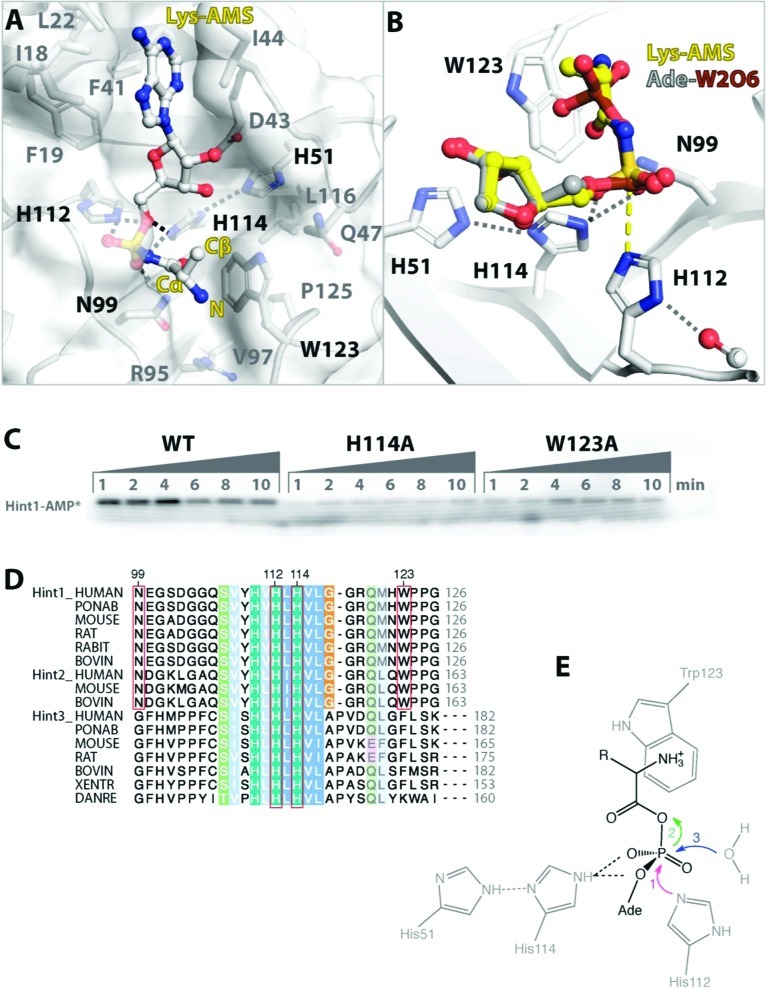
The Hint1/Lys-AMS structure showed that Hint1 only recognized the main chain of the lysyl group. The α-amino group of the lysyl backbone is placed on top of Trp123 of Hint1. Because the amino group is likely to be protonated under the conditions of crystallization (pH 7.5), at the close distance of 3.5 Å, it forms a cation-pi interaction with the aromatic side chain of Trp123.38 With lateral support from the side chains of Gln47, Arg95, Val97, and Leu116, and from Pro125 from the back, the indole side chain of Trp123 forms a rigid wall for the lysyl-binding pocket (rmsd of 0.3 Å between the structure of the apoprotein and of the Lys-AMS complex) (Figure 3A).37 Consistent with the important role of Trp123 for recognizing the lysyl backbone, mutation of Trp123 to Ala showed both a lag in the formation of Hint1-AMP and a decrease in the total amount of Hint1-AMP that was generated (Figure 3C). In addition, the carbonyl group of the lysyl moiety formed one hydrogen bond with the side chain of N99, together with two internal hydrogen bonds with the α-amino group of the lysyl and sulfate groups. The N-substituted oxygen of Lys-AMS also formed hydrogen bonds with Ser107, Gly105, and Asn99. Thus, Lys-AMS is bound at a defined position in Hint1.
Figure 3.
Hydrolysis of Lys-AMP by Hint1. (A) Detailed interaction of Lys-AMS with human Hint1. The α-amino group of the lysyl group in Lys-AMS forms a cation-π interaction with the aromatic ring of Trp123. (B) Structural superimposition of Hint1/Lys-AMS with the transition state complex of Hint1/adenosine-5′-ditungstate (Ade-W2O6, PDB 6FIT). The sulfate group of Lys-AMS is located close to the α tungstate group of Ade-W2O6. A yellow dashed line indicates the covalent bond formed between the α tungsten (brown) and Nε of the H112 side chain. The overlapped adenosine moieties of these two ligands are not shown. (C) Formation of the covalent Hint1-AMP intermediate shows the important role of H114 and W123 for hydrolysis of Lys-AMP. The reaction conditions are as described in the legend to Figure 1. (D) Sequence alignment of vertebrate HINT proteins. Critical residues involved in the binding and hydrolysis of aa-AMP's are boxed in red, and are conserved in Hint1’s and its Hint2 paralogs (which function in mitochondria). Different residues are found in Hint3's, whose function is less characterized.52 (E) Catalytic mechanism proposed for the hydrolysis of aa-AMP's by human Hint1.
Because of the confined binding pocket shaped by Trp123, the side chain of Lys-AMS is oriented toward the solvent (Figure 3A). Having no interaction with Hint1, the side chain of the lysyl moiety is free to rotate and therefore is completely disordered in the crystal. For this reason, we speculated that Hint1 would not be specific to Lys-AMP and would likely bind different aa-AMP's from other tRNA synthetases.
To test this hypothesis, we crystallized Hint1 in complex with two other aminoacyl-AMP analogues, i.e., Ala-AMS and Trp-AMS. Consistent with expectations, the electron density of the tryptophanyl side chain was not seen in the Trp-AMS complex with Hint1 (Figure 2D). Only the adenylate moiety, the main chain, and the Cβ of the side chain of Trp-AMS had clear electron densities. These densities resembled the electron densities of Ala-AMS within its Hint1 complex (Figure 2C). Thus, the three crystal structures of the Hint1/aa-AMS complexes suggest that the side chain of aa-AMP is not recognized by Hint1.
Mechanism of Lys-AMP Hydrolysis by Hint1
Several structures of mammalian Hint1 in apo-form, in complex with AMP (one of the products of adenylate hydrolysis), GMP, the substrate analogue AMPCP, and the transition state analogue adenosine-5′-ditungstate have been reported.36,37,39,40 Structural comparisons showed that the AMS moiety of Lys-AMS (with sulfur substitution of the phosphate) occupied the same position as AMP in the Hint1/AMP complex. The sulfate group of Lys-AMS binds to two (His112, His114) of the three characteristic histidines in Hint1 (Figure 3A). Previous results showed that adenosine and sodium tungstate formed a covalently linked complex (adenosine-5′-ditungstate) with Hint1.36 The tungstate ion reacted with the imidazole side chain of His112 of Hint1 through a pentacovalent complex with the Nε of His112, to thus mimic the transient Hint1-AMP-P complex. Superimpositions of the different Hint1 complexes showed that the sulfur atom of Lys-AMS is at the position equivalent to the α-phosphorus atom of AMPCP in the substrate analogue complex (0.4 Å distance). This sulfur is also close to the tungsten atom in the adenosine-5′-ditungstate complex (distance of 0.4 Å, Figure 3B). In addition, the sulfate of Lys-AMS interacts with His114 and Asn99, both of which are conserved in Hint1 proteins (Figure 3B,D). Thus, Lys-AMS binds to Hint1 in a productive manner for hydrolysis, and the Hint1/Lys-AMS structure appears to recapitulate the bound conformation for native Lys-AMP.
Hydrolysis of Lys-AMP is likely through the same mechanism that Hint1 hydrolyzes other substrates, such as ADP or AMP-NH2.27,28,36 Binding of Lys-AMP initiates an in-line attack on the α phosphate by histidine (His112) as the nucleophile (Figure 3E). A pentacovalent transition state inverts the α-phosphate position (as also seen in the amino acid activation step by tRNA synthetases).36,41 Next, AMP is covalently linked to His112 of Hint1 to form a nucleotidyl phosphoprotein (Hint1-AMP) intermediate, and lysine is subsequently released. Finally, AMP is hydrolyzed from Hint1 by a water molecule probably facilitated by Ser107.40 The other conserved His residue (His114) forms an electron transfer chain between His51 and the sulfate group of Lys-AMS (Figure 3A,B,E), and may thereby help to stabilize the transition state. Mutating this His residue (His114) to Ala dramatically decreased the amount of Hint1-AMP that was formed, thus supporting the idea that His114 is also a critical residue for the hydrolysis of Lys-AMP (Figure 3C). Importantly, binding and hydrolysis of Lys-AMP involves only the recognition of AMP and the lysyl backbone. Therefore, this hydrolysis mechanism would be applicable to aa-AMP's generated from other aaRS's (Figure 3E).
Discussion
Trp123 Has an Important Role for Substrate Recognition by Hint1
Hint1 is a member of the HIT superfamily that is divided into many branches including the Hint branch, the fragile histidine triad (Fhit) branch, and the galactose-1-phosphate uridyltransferase (GalT) branch.42 Each branch has a different substrate specificity.27 In contrast to Fhit, which hydrolyzes diadenosine-polyphosphates (such as Ap3A), Hint1 shows weak activity toward ADP (kcat of 10–3 s–1 and Km of 800 μM) and no activity toward other nucleotides with multiple phosphates such as ATP and ApnA (n = 3–5).36 On the other hand, Hint1 preferentially binds and hydrolyzes adenosine derivatives with a single phosphate, including phosphoramidates (e.g., AMP-NH2, AMP-N-Ala, AMP-N-ε-Lys,) and acyl adenylates (e.g., Lys-AMP), and has turnover rates kcat as high as ∼2 s–1 and Km values as low as ∼0.1 μM.34,39 Except for the characteristic AMP-binding pocket in Hint1, the structures of the Hint1/aa-AMS complexes showed that the main chain of the aminoacyl group was placed into a groove formed by Trp123. The α-amino group of the aminoacyl moiety stacked on top of the imidazole side chain of Trp123, by forming a cation-pi interaction (Figure 2). Similar interactions are also found in the amino acid binding pocket of AlaRS and SerRS, where a strictly conserved Trp residue supports the backbone of the bound amino acid.41,43 (Presumably, a phosphate group in ADP or ApnA at the position of the α-amino group will form a less favorable interaction with Trp123, and this circumstance would give a lower affinity for Hint1.) However, for AlaRS and SerRS, and unlike Hint1, there are also specific interactions and pockets for the alanyl and seryl side chains.
Interestingly, Trp123 is located at the C-terminus of human Hint1 (126 residues) and is one of the 15 residues that are sexually dimorphic in avian HINT-related genes, which were suggested to be responsible for feminization.44 Recent biochemical studies established an important role for the C-terminus of Hint1. Swapping of the C-terminal seven residues of human Hint1 to E. coli Hint changed the substrate specificity.33 In addition, deletion of three C-terminal residues abolished the ability of E. coli Hint to hydrolyze Lys-AMP. However, the deletion only had a modest effect on acyl-adenylate substrates that do not contain the aminoacyl backbone.35 In the structure of rabbit Hint1 in complex with an AMP-N-ε-Lys analogue, Trp123 interacts with the alkyl portion of the lysyl leaving group.39 Substitution of Trp123 to Gln, which is present in female-specific avian HINT-related genes, reduced kcat/Km for AMP-N-ε-lysine hydrolysis by 17-fold.44 Consistent with these results, our work showed that the Trp123Ala mutation at the C-terminus of Hint1 caused a dramatic loss of activity in forming the human Hint1-AMP intermediate, and thereby emphasized the importance of Trp123 in the hydrolysis of Lys-AMP.
General Specificity of Hint1 for aa-AMP's
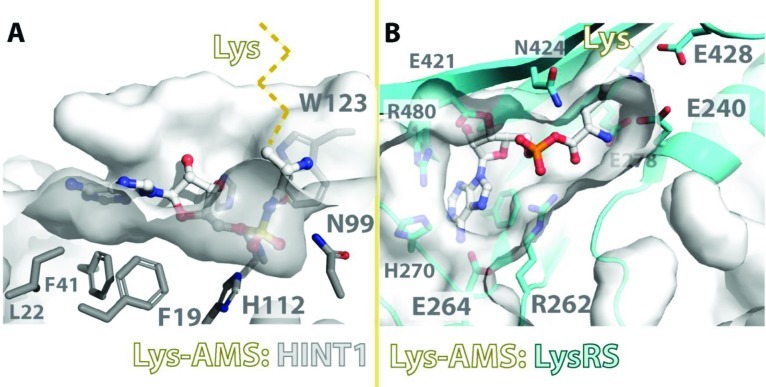
In the structures of Hint1/aa-AMS complexes, the aminoacyl backbone of Lys-AMS (or Ala-AMS, Trp-AMS) was oriented toward the outside of Hint1 (Figure 4). The interaction mode between Hint1 and Lys-AMP contrasts with that of the LysRS/Lys-AMP complex, as seen in structures of LysRS in complex with Lys-AMP, or with l-lysine and ATP.45,46 In order to recognize lysine and attach it to the cognate tRNALys, LysRS forms a deep pocket that specifically accommodates only lysine. Two strictly conserved glutamic acid side chains (Glu240 and Glu428 in E. coli LysU and Glu301 and Glu501 in human LysRS) are located at the end of the pocket, where they recognize the ε-amino of the lysine side chain (Figure 4B). Also different from the cation-pi interaction in Hint1, the α-amino group of the lysyl moiety is anchored by hydrogen bonds with two Glu’s in LysRS (Glu240 and Glu278 in LysU and Glu301 and Glu339 in human LysRS). The carboxyl group of the lysyl moiety interacts with the characteristic Arg262 in LysU and is positioned to react with the α-phosphate group of ATP. Comparison of the Hint1/Lys-AMS and LysRS/Lys-AMS cocrystal structures shows that the Lys-AMS pockets in these two proteins are in opposite directions. While in LysRS the lysyl moiety is facing inside and AMP/ATP faces outside, AMP is inside and the lysyl group is facing outside in Hint1 (Figure 4). Similarly, the ligand electron densities in the structures of Hint1 with Ala-AMS and Trp-AMS also show that no side chain of any aminoacyl group is involved in an interaction with Hint1 (Figure 2).
Figure 4.
Recognition of Lys-AMP by Hint1 vs LysRS. The Lys-AMS binding pocket of human Hint1 in comparison with the pocket of LysRS (PDB 1E1T) shows side chain independent recognition of Lys-AMP by Hint1. Residues responsible for recognition of the lysyl-AMS in Hint1 and LysRS are depicted. The potential direction of the Lys side chain in the Hint1/Lys-AMS structure is shown as a dashed line.
An important link between Hint1 and aaRS's was found during the immune activation of mast cells, where LysRS contributed to an ∼3-fold increase of intracellular Ap4A.12 Earlier work showed that Ap4A binding to Hint1 could dissociate the Hint1-MITF complex. This effect appears to be specific because other ApnA (n = 3, 5) did not cause dissociation.11 Lys-AMP, with a size between Ap3A and AMP, is less likely to dissociate the Hint1-MITF interaction. In addition, because the hydrolysis product AMP could not dissociate the Hint1-MITF interaction, the hydrolysis of Lys-AMP by Hint1 may also alleviate the effect of Lys-AMP.11 On the other hand, the adenylation of Hint1 by AlaRS, MetRS, HisRS, and AspRS shows that Hint1 hydrolyzes various aa-AMP's (Figure 1). Binding and hydrolysis of aa-AMP's by Hint1 is through a common mechanism, as evidenced by the formation of the Hint1-AMP intermediate with all tested aaRS's. This broad activity of human Hint1 to bind and hydrolyze aa-AMP's is caused by the unique binding pocket involving Trp123, which is conserved in Hint1 proteins from C. elegans to human.1 With more than half of the aaRS members including LysRS, AlaRS, and MetRS known to synthesize Ap4A in vitro and/or in cells,18,30,47 Hint1 may have a general ability to constrain the formation of Ap4A by the aaRS family. In addition, we cannot rule out the possibility that the transient Hint1-AMP complex, and complexes of Hint1 with other aminoacyl adenylates, also play a role in regulating the functions of Hint1.
Methods
Protein Preparation
The gene encoding human Hint1 was synthesized by codon optimization. Human Hint1 was constructed with a N-terminal 6xHis-tag with a TEV protease cleavage site in a pHisTEV vector. Human Hint1 was expressed in the bacterial strain BL21(DE3) at 16 °C for 20 h and purified to homogeneity with a Ni-HiTrap affinity column (GE Healthcare, Piscataway, NJ). The 6xHis-TEV tag was cleaved by TEV protease, and the nontagged Hint1 was further purified by a HiTrap SP Sepharose column (GE Healthcare, Piscataway, NJ). It was concentrated to 50 mg/mL in a final buffer (10 mM Tris–HCl, pH 8.0, 50 mM NaCl) for crystallization. Full-length AlaRS, AspRS, HisRS, and MetRS were cloned from the E. coli genome K12 and were inserted into pET vectors (Novagen, Darmstadt, Germany) with the 6xHis-tag. These proteins were expressed in the bacterial strain BL21(DE3) (NEB, Ipswich, MA) at 37 °C for 6 h and purified to homogeneity with the Ni-HiTrap affinity column.
Crystallization, Data Collection, and Structure Refinement
Crystallization was done by the sitting drop method. Protein solution (nontagged recombinant human Hint1 (see above)) was preincubated with 5 mM Lys-AMS, Ala-AMS, or Trp-AMS for 10 min at 22 °C. A drop was prepared by mixing 0.5 μL of protein solution with 0.5 μL of precipitant solution, containing 25–28% PEG3350, 100 mM HEPES pH 7.5, and was equilibrated against 90 μL of precipitant solution. Crystals were collected after incubation at 18 °C for 3–7 days and were flash-frozen in liquid nitrogen for data collection.
Three data sets were obtained from Beamline 11-1 at the Stanford Synchrotron Radiation Laboratory (Menlo Park, CA). The crystals belong to space group C2 with two molecules of Hint1 per asymmetric unit. The structures were solved by molecular replacement using the Hint1 apo structure (PDB 6RHN) with the program Molrep.48 Iterative model building and refinement were performed by using Coot and Phenix.49,50 Data collection and refinement statistics are given in Table 1.
Table 1. Data Collection and Refinement Statistics.
| Hint1/Ala-AMS | Hint1/Lys-AMS | Hint1/Trp-AMS | |
|---|---|---|---|
| Data Collection | |||
| space group | C2 | C2 | C2 |
| cell dimensions | |||
| a, b, c (Å) | 78.31, 46.37, 64.11 | 79.00, 46.41, 64.23 | 79.00, 46.43, 63.88 |
| α, β, γ (deg) | 90.00, 95.02, 90.00 | 90.00, 95.46, 90.00 | 90.00, 95.24, 90.00 |
| resolution (Å) | 50.00–1.52(1.57–1.52)a | 25.00–1.52(1.57–1.52) | 25.00–1.67(1.73–1.67) |
| Rsym or Rmerge (%) | 3.9 (11.5) | 4.8 (29.9) | 5.3 (48.9) |
| I/sI | 42.5 (16.8) | 38.96 (6.4) | 35.2 (3.4) |
| completeness (%) | 98.4 (97.1) | 99.3 (98.2) | 98.4 (96.7) |
| redundancy | 7.5 (6.7) | 7.5 (6.5) | 7.5 (7.3) |
| Refinement | |||
| resolution (Å) | 50.00–1.52(1.56–1.52) | 25.00–1.52(1.56–1.52) | 25.00–1.67(1.74–1.67) |
| no. reflections | 34772 | 34771 | 25244 |
| Rwork/Rfree (%) | 14.1/16.4 | 14.2/16.2 | 14.4/17.3 |
| no. atoms | |||
| protein | 1826 | 1805 | 1788 |
| ligand | 28 | 28 | 28 |
| solvent | 306 | 300 | 302 |
| B-factors (Å2) | |||
| protein | 12.11 | 13.98 | 13.95 |
| ligand | 16.07 | 19.25 | 26.33 |
| solvent | 28.20 | 31.48 | 31.42 |
| rms deviations | |||
| bond lengths (Å) | 0.007 | 0.008 | 0.009 |
| bond angles (deg) | 1.242 | 1.189 | 1.250 |
| Ramachandran plot | |||
| most favored (%) | 98.8% | 98.7% | 98.7% |
| additional allowed (%) | 1.2% | 1.3% | 1.3% |
Values in parentheses are for the highest-resolution shell.
Enzymatic Assays
All assays for Hint1-AMP formation were carried out in a standard buffer (50 mM Hepes-Na, pH 7.5, 20 mM KCl, 10 mM MgCl2, 2 mM DTT, 0.16 μM α-32P-ATP, 3 μg/mL inorganic pyrophosphatase, and 100 μM amino acid) at 22 °C. Wild-type or mutant Hint1 (0.5 μM) was coincubated with human LysRS,51 or E. coli AlaRS, AspRS, HisRS, or MetRS (0.5 μM). The reaction was quenched at each time point by adding equal amounts of 2× SDS-PAGE sample buffer and boiled for 5 min at 95 °C. Equal amounts of each sample were load onto a 4–20% SDS-PAGE gel, which ran to the point that most of the free ATP was eluted from the gel. Radioactive bands of Hint1 and AMP were detected by phosphoimaging (Molecular Imager FX, Biorad, Hercules, CA) over a time period of 12–30 h.
Acknowledgments
Crystal data collection was carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. This work was supported in part by GM15539 from the NIH, by a fellowship from the National Foundation for Cancer Research, by a Kimmel Scholar Award for Cancer Research, and by funding from The State of Florida to Scripps Florida.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Brenner C.; Bieganowski P.; Pace H. C.; Huebner K. J. Cell. Physiol. 1999, 181, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T.; Suzui M.; Wang L.; Lin C. S.; Xing W. Q.; Weinstein I. B. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 7824–7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Zhang Y.; Su T.; Santella R. M.; Weinstein I. B. Oncogene 2006, 25, 713–721. [DOI] [PubMed] [Google Scholar]
- Li H.; Balajee A. S.; Su T.; Cen B.; Hei T. K.; Weinstein I. B. J. Cell Biol. 2008, 183, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiske J.; Huber O. J. Biol. Chem. 2006, 281, 27356–27366. [DOI] [PubMed] [Google Scholar]
- Yuan B. Z.; Jefferson A. M.; Popescu N. C.; Reynolds S. H. Neoplasia 2004, 6, 412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieganowski P.; Garrison P. N.; Hodawadekar S. C.; Faye G.; Barnes L. D.; Brenner C. J. Biol. Chem. 2002, 277, 10852–10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. F.; Baraniak J.; Kaczmarek R.; Zhou X.; Cheng J.; Ghosh B.; Wagner C. R. Mol. Pharm. 2007, 4, 208–217. [DOI] [PubMed] [Google Scholar]
- Razin E.; Zhang Z. C.; Nechushtan H.; Frenkel S.; Lee Y. N.; Arudchandran R.; Rivera J. J. Biol. Chem. 1999, 274, 34272–34276. [DOI] [PubMed] [Google Scholar]
- Lee Y. N.; Razin E. Mol. Cell. Biol. 2005, 25, 8904–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. N.; Nechushtan H.; Figov N.; Razin E. Immunity 2004, 20, 145–151. [DOI] [PubMed] [Google Scholar]
- Yannay-Cohen N.; Carmi-Levy I.; Kay G.; Yang C. M.; Han J. M.; Kemeny D. M.; Kim S.; Nechushtan H.; Razin E. Mol. Cell 2009, 34, 603–611. [DOI] [PubMed] [Google Scholar]
- Carmi-Levy I.; Motzik A.; Ofir-Birin Y.; Yagil Z.; Yang C. M.; Kemeny D. M.; Han J. M.; Kim S.; Kay G.; Nechushtan H.; Suzuki R.; Rivera J.; Razin E. Mol. Cell. Biol. 2011, 31, 2111–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. W. Jr. Annu. Rev. Biochem. 1993, 62, 715–748. [DOI] [PubMed] [Google Scholar]
- Ibba M.; Soll D. Annu. Rev. Biochem. 2000, 69, 617–650. [DOI] [PubMed] [Google Scholar]
- Goerlich O.; Foeckler R.; Holler E. Eur. J. Biochem. 1982, 126, 135–142. [DOI] [PubMed] [Google Scholar]
- Wahab S. Z.; Yang D. C. J. Biol. Chem. 1985, 260, 5286–5289. [PubMed] [Google Scholar]
- Brevet A.; Chen J.; Leveque F.; Plateau P.; Blanquet S. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 8275–8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright M.; Boonyalai N.; Tanner J. A.; Hindley A. D.; Miller A. D. FEBS J. 2006, 273, 3534–3544. [DOI] [PubMed] [Google Scholar]
- Kellermann O.; Tonetti H.; Brevet A.; Mirande M.; Pailliez J. P.; Waller J. P. J. Biol. Chem. 1982, 257, 11041–11048. [PubMed] [Google Scholar]
- Kerjan P.; Cerini C.; Semeriva M.; Mirande M. Biochim. Biophys. Acta 1994, 1199, 293–297. [DOI] [PubMed] [Google Scholar]
- Rho S. B.; Kim M. J.; Lee J. S.; Seol W.; Motegi H.; Kim S.; Shiba K. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 4488–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminska M.; Havrylenko S.; Decottignies P.; Gillet S.; Le Marechal P.; Negrutskii B.; Mirande M. J. Biol. Chem. 2009, 284, 6053–6060. [DOI] [PubMed] [Google Scholar]
- Park S. G.; Ewalt K. L.; Kim S. Trends Biochem. Sci. 2005, 30, 569–574. [DOI] [PubMed] [Google Scholar]
- Ray P. S.; Arif A.; Fox P. L. Trends Biochem. Sci. 2007, 32, 158–164. [DOI] [PubMed] [Google Scholar]
- Carmi-Levy I.; Yannay-Cohen N.; Kay G.; Razin E.; Nechushtan H. Mol. Cell. Biol. 2008, 28, 5777–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C. Biochemistry 2002, 41, 9003–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. F.; Wagner C. R. J. Biol. Chem. 2007, 282, 4719–4727. [DOI] [PubMed] [Google Scholar]
- Blanquet S.; Plateau P.; Brevet A. Mol. Cell. Biochem. 1983, 52, 3–11. [DOI] [PubMed] [Google Scholar]
- Brevet A.; Plateau P.; Cirakoglu B.; Pailliez J. P.; Blanquet S. J. Biol. Chem. 1982, 257, 14613–14615. [PubMed] [Google Scholar]
- Guo R. T.; Chong Y. E.; Guo M.; Yang X. L. J. Biol. Chem. 2009, 284, 28968–28976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plateau P.; Blanquet S.. Synthesis of NpnN′ (n = 3 or 4) in vitro and in in vivo. In Ap4A and Other Dinucleoside Polyphosphates; McLennan A. G., Ed.; CRC Press, Inc.: Boca Raton, FL, 1992; pp 63–80. [Google Scholar]
- Chou T. F.; Sham Y. Y.; Wagner C. R. Biochemistry 2007, 46, 13074–13079. [DOI] [PubMed] [Google Scholar]
- Chou T. F.; Tikh I. B.; Horta B. A.; Ghosh B.; De Alencastro R. B.; Wagner C. R. J. Biol. Chem. 2007, 282, 15137–15147. [DOI] [PubMed] [Google Scholar]
- Bardaweel S.; Pace J.; Chou T. F.; Cody V.; Wagner C. R. J. Mol. Biol. 2010, 404, 627–638. [DOI] [PubMed] [Google Scholar]
- Lima C. D.; Klein M. G.; Hendrickson W. A. Science 1997, 278, 286–290. [DOI] [PubMed] [Google Scholar]
- Brenner C.; Garrison P.; Gilmour J.; Peisach D.; Ringe D.; Petsko G. A.; Lowenstein J. M. Nat. Struct. Biol. 1997, 4, 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallivan J. P.; Dougherty D. A. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakowiak A.; Pace H. C.; Blackburn G. M.; Adams M.; Mekhalfia A.; Kaczmarek R.; Baraniak J.; Stec W. J.; Brenner C. J. Biol. Chem. 2004, 279, 18711–18716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozga M.; Dolot R.; Janicka M.; Kaczmarek R.; Krakowiak A. J. Biol. Chem. 2010, 285, 40809–40818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M.; Chong Y. E.; Shapiro R.; Beebe K.; Yang X. L.; Schimmel P. Nature 2009, 462, 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner K.; Saldivar J. C.; Sun J.; Shibata H.; Druck T. Adv. Enzyme Regul. 2011, 51, 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S.; Maier T.; Ahel D.; Gruic-Sovulj I.; Soll D.; Weygand-Durasevic I.; Ban N. EMBO J. 2006, 25, 2498–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks K. P.; Seidle H.; Wright N.; Sperry J. B.; Bieganowski P.; Howitz K.; Wright D. L.; Brenner C. Physiol. Genomics 2004, 20, 12–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desogus G.; Todone F.; Brick P.; Onesti S. Biochemistry 2000, 39, 8418–8425. [DOI] [PubMed] [Google Scholar]
- Guo M.; Ignatov M.; Musier-Forsyth K.; Schimmel P.; Yang X. L. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 2331–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquet S.; Plateau P.; Onesti S.. Class II Lysyl-tRNA Synthetases. In The Aminoacyl-tRNA Synthetases; Ibba M., Francklyn C., Cusack S., Eds.; Eurekah: Georgetown, TX, 2005; pp 227–240. [Google Scholar]
- Winn, M. D.; et al. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Mustyakimov M.; Afonine P. V.; Langan P. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009, 65, 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P.; Zhang H. M.; Shapiro R.; Marshall A. G.; Schimmel P.; Yang X. L.; Guo M. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 8239–8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.; St-Pierre M. V.; Dufour J. F. Biochim. Biophys. Acta 2011, 1807, 626–632. [DOI] [PubMed] [Google Scholar]