Abstract
Mass spectrometry (MS)-based proteomics is emerging as a broadly effective means for identification, characterization, and quantification of proteins that are integral components of the processes essential for life. Characterization of proteins at the proteome and sub-proteome (e.g., the phosphoproteome, proteoglycome, or degradome/peptidome) levels provides a foundation for understanding fundamental aspects of biology. Emerging technologies such as ion mobility separations coupled with MS and microchip-based-proteome measurements combined with MS instrumentation and chromatographic separation techniques, such as nanoscale reversed phase liquid chromatography and capillary electrophoresis, show great promise for both broad undirected and targeted highly sensitive measurements. MS-based proteomics is increasingly contribute to our understanding of the dynamics, interactions, and roles that proteins and peptides play, advancing our understanding of biology on a systems wide level for a wide range of applications including investigations of microbial communities, bioremediation, and human health.
Keywords: Proteomics, mass spectrometry, ion mobility, post translation modifications
Introduction
Understanding the biochemical processes that constitute life requires not only knowledge of the genetic instructions encoded in the genome but also a detailed comprehension of the participating proteins and metabolic substrates. Characterization of the proteins present in a biological system, or the proteome, provides a foundation for better understanding the complexities inherent in biology. The proteome is not only complex, it is spatially, temporally, and chemically dynamic. Mass spectrometry (MS)-based proteomics includes a growing set of ancillary technologies, that provide a means for high-throughput characterization and quantification of proteins in a biological sample or system.1
The genome sequence and identified genes provide an incomplete picture of the inherent systems-wide biological complexity of an organism. While the proteome is highly complementary to the genome, it differs from cell-to-cell and time-to-time and should be profoundly descriptive of biological phenotype. Post-translational modifications (PTMs) of proteins generally contribute to a much broader (but still often poorly understood) diversity of protein species than are predicted based on the a priori knowledge of an organism’s genome alone. A wide variety of MS-based proteome studies, with broad impact on biology, biomedical research, and systems biology have been reported. Applications of MS-based proteomics range from descriptive to quantitative, providing insight into emergent biological properties through systems biology initiatives2 and driving biomarker discovery efforts for the development of new diagnostics. Advancement in MS technologies combined with improvement in sample preparation have provided greater insight into the biological complexity of a wide variety of sample types including organelles, membranes, biofluids (e.g. blood, cerebrospinal fluid, saliva, urine, sweat), tissues, organs, and microbial communities.
The last decade of rapid developments in MS-based proteomics have included key efforts to increase the depth and breadth of proteome coverage, data quality, and identification confidence as well as increased sample throughput necessary for enabling population-scale proteome measurements. In this review, we provide an assessment of MS-based proteomics strategies and highlight recent developments and their potential impacts.
Mass spectrometry based proteomics
Detecting and quantifying the rich diversity of potentially hundreds of thousands of protein isoforms present in a biological sample, often spanning as much as 12 orders of magnitude in relative abundance, poses an enormous analytical challenge. Coupling liquid chromatography (LC) separations with MS (our definition of MS-based proteomics implicitly includes a range of ancillary fractionation, separation, and other analytical methods and technologies) allows for analysis of thousands of proteins per measurement and has addressed many of the analytical challenge inherent in proteomics.
Analysis of biomolecules, such as proteins and peptides, in the mass spectrometer requires the analyte form a charged ion in the gas phase. Development of efficient, nondestructive ionization methods enables analysis of intact biomolecules by MS without significant sample degradation and historically facilitated development of the field of proteomics. The most commonly applied of these soft ionization processes are electrospray ionization (ESI)3 and matrix assisted laser desorption ionization (MALDI).4 As illustrated in Figure 1, the identification of biomolecules by MS is a key component in the typical proteomics workflow.
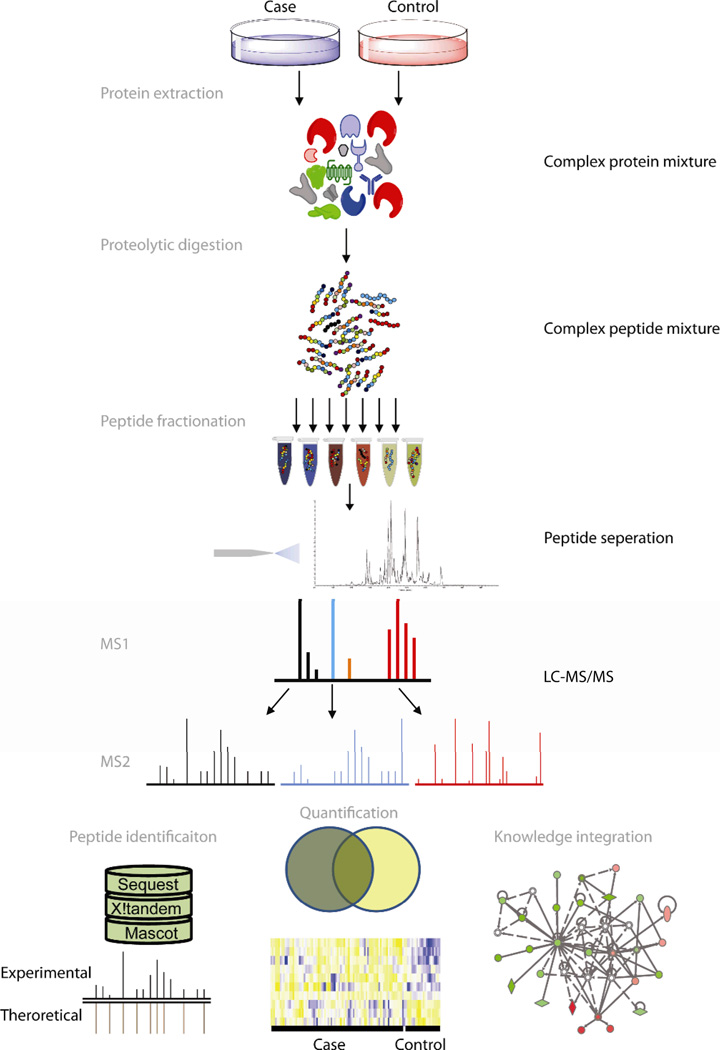
Figure 1.
Overview of bottom-up proteomics. In MS-based bottom-up ‘shotgun’ proteomics studies complex mixtures of proteins are isolated from the biological sample of interest and enzymatically or chemically cleaved into peptides. The peptide mixture is often fractionated and analyzed by tandem LC MS (MS/MS). Tandem LC-MS analysis involves the acquisition of a preliminary mass spectrum (MS1) of the intact (precursor) species, dissociation of selected ions of interest into smaller fragments, and then mass analysis of the fragments (MS2). Peptide identification of tandem MS spectra is often performed by matching mass measurements for intact peptides and MS/MS fragment ions to theoretical sequences derived from genome sequence information. Quantification and visualization tools facilitate interpretation of data.
Bottom-up or shotgun proteomics is the most common MS-based method for studying proteins. In bottom-up proteomics studies, a mixture of proteins is isolated and enzymatically or chemically cleaved into peptides (Figure 1). The resultant complex peptide mixture is fractionated using chromatography and other methods. Typically following reversed phase chromatographic separation, the peptides eluting from the chromatographic column are ionized by electrospray ionization (ESI) and analyzed by MS. The power of MS lies not only in its parts per million (ppm) mass measurement accuracy, but in the ability to perform tandem MS (MS/MS) measurements that provide additional information specific for the peptide amino acid sequence. Typical LC MS/MS involves the acquisition of a preliminary mass spectrum (MS1) of the intact (precursor) peptide, dissociation of the isolated precursor ion of interest into smaller fragments, and subsequent mass analysis of the fragments (MS2). The process is repeated for the duration of the LC separation of the peptide mixture. Peptide fragmentation typically results from collision-induced dissociation (CID), or alternative techniques such as electron capture dissociation (ECD) or electron transfer dissociation (ETD).1 Both electron-based fragmentation methods provide better sequence coverage of larger analytes that are highly charged and show great promise for improved characterization of labile PTMs such as phosphorylation.
Frequently, peptide identification using tandem MS data utilizes genomics data by matching mass measurements for intact peptides and MS/MS fragment ions to theoretical sequences derived from genome sequence data. Database matching strategies pairing MS data with genomic sequences are typically implemented using bioinformatics tools such as Mascot, Sequest, and X!tandem.5 Reverse or scrambled database searches are often used for false discovery rate (FDR) estimation, leading to empirical estimation of the relative FDR for the analysis of an entire dataset.5 These methods have been widely adopted and employed, but can significantly underestimate actual FDRs, and may not be compatible and appropriate with all database search tools.6 A relatively new alternative approach for assessment of data quality that provides statistical significance of spectrum-to-peptide matches following database searching is calculated using MS-GF.7 Validating MS/MS database matches can be performed with tools such as Peptide Prophet, and MS-GF. These tools are of value as they provide a means for estimating the relative quality of individual spectrum-to-peptide matches.
Relative quantification of identified proteins increases the available biological knowledge from a proteomics experiment. Relative peptide and protein abundance from LC-MS/MS measurements is often estimated by counting the number of times a peptide mass spectrum is measured and identified (spectral counting). The sum of the spectral counts for peptides derived from the same protein for samples measured on the same instrument provides a means for estimating relative protein abundance. Peptide quantification by integrated ion intensity or employing isotopic labeling is typically more precise compared to spectral counting and is covered in detail below. Interpretation of the biological significance of qualitative and quantitative data is often assisted by integrating publically available information, such as protein tissue origin, functionality, and reported role in biochemical processes. Integration of information from public repositories as well as assessment of functional enrichment can be accomplished utilizing publically available bioinformatics tools such as DAVID8 and a variety of others included in Bioconductor.9
Challenges and opportunities for LC-MS-based characterization of complex biological samples
There are many challenges for LC-MS-based characterization of complex biological samples since the typical dynamic range of detection for LC-MS is 4–6 orders of magnitude10, while in biological samples such as biofluids (e.g. plasma), the protein concentration range can span 12 orders of magnitude. For example, in human blood plasma individual protein concentrations range from high mg/mL for albumin and immunoglobulins down to pg/mL for cytokines.11 A dynamic range of measurement greater than 7 orders of magnitude is required for the detection of many clinically relevant proteins in plasma that are present at ng/mL levels or lower. Current one dimension (1D) LC-MS/MS analysis falls short of the required dynamic range of detection, highlighting a limitation with current MS technologies for comprehensive proteome coverage and biomarker discovery.12 In-depth characterization of the proteome of biofluids has been made possible through the development and application of analytical strategies such as immunoaffinity depletion chromatography.13 Specifically, the application of immunoaffinity multi-protein depletion using Agilent MARS or Seppro IgY-14 columns provides a means for reducing or eliminating the “masking” effects of highly abundant species. A further round of depletion using a Seppro Supermix affinity column results in the removal of moderately abundant proteins improving coverage of low abundant proteins.14
Increased depth of proteome coverage can be accomplished e.g., by sample fractionation at either the protein or the peptide level, or both, which leads to a reduction in sample complexity. Coupling fractionation with ultra-high resolution nanocapillary LC (UPLC) separations further reduces the number of co-eluting peptides by decreasing the complexity of analytes present in each peak and increasing the chromatographic peak capacity. This reduced complexity results in fewer analytes simultaneously in the mass spectrometer allowing for less instrumental undersampling in MS/MS measurements and subsequently broader proteome coverage. Increasing LC separation efficiency either by increasing the pressure used during the LC separation or reducing the LC column stationary phase particle size along with increasing separation time results in greater numbers of peptides and proteins identified using tandem MS. Ultra high pressure (20 kpsi) reverse phase separations have led to faster separations and allow for the use of longer columns which provide increased peak capacity, resulting in identification of several thousand proteins in a single LC-MS analysis.15
A wide variety of chromatographic fractionation approaches and their multi-dimensional combinations have been developed for improved proteome coverage. Coupling orthogonal fractionation methods such as strong cation exchange or high pH reversed-phase fractionation with 1D LC-MS leads to significant increases in peptide and protein identifications.16 Multidimensional separations have been widely adopted for analysis of complex protein mixtures. The Mudpit (multidimensional protein identification technology) approach coupling strong cation exchange with reversed phase chromatographic separations, initially developed by Link et al.,17 has proven to be a very effective means for in-depth proteome analysis. However, it should be noted that there is a significant trade off associated with sample fractionation as, sample throughput is reduced in proportion to the number of fractions generated for analysis.
Proteome quantification strategies
Quantitative measurements of peptide abundance can be made with or without protein or peptide labeling (recently reviewed in 18). Several in vitro and in vivo labeling techniques have been developed for MS-based quantification by building on the basic bottom-up proteome profiling strategy outlined in Figure 1. Metabolic labeling of proteins can be accomplished by the addition of isotopically labeled amino acids to cell culture (referred to as SILAC) as part of normal protein biosynthesis. Metabolic labeling can be useful for monitoring proteome dynamics.10 After protein isolation, the labeled and unlabeled proteins from experimental and control samples are mixed and quantified either as intact proteins (i.e. without proteolysis) or following enzymatic (typically tryptic) digestion. The molecular weight difference between the light (natural version) and heavy (labeled) amino acids allows quantitative proteome comparisons.19 Methods employing global internal standards generated by SILAC labeling different cell lines in Super-SILAC20 or with trypsin-catalyzed O18 labeling of a pooled reference sample21 have enabled large scale quantitative studies. A popular alternative quantitative labeling strategy is isobaric tag for relative and absolute quantification (iTRAQ)10 in which primary amino groups (the N-terminus and lysine side chains) of peptides are labeled. However, unlike SILAC, iTRAQ is only apparent following MS/MS analysis and detection of reporter ions in the mass spectrometer. iTRAQ can be multiplexed using up to 8 different labels. However, it is important to note that some level of side reaction is unavoidable for most chemical derivatization procedures, and this may interfere with unbiased peptide and protein quantification.22 Label-free quantification is increasingly popular due to e.g., computational methods allowing sophisticated normalization of signals between LC-MS analyses.23 Label-free methods for proteome quantification have been shown to provide greater global proteome coverage than labeling strategies such as O18 labeling, whereas O18 labeling resulted in tighter standard deviations and better coefficient of variance (CV) values.24
The most common label-free means for peptide and protein quantification is to use data-dependent MS/MS analysis, where masses are chosen for MS/MS analysis based on an initial MS survey scan (as shown in Figure 1). One limitation of this approach is under-sampling, where peptide coelution and sample complexity results in an underestimation of abundance. To overcome the limitations associated with this standard tandem MS/MS measurement method and improve quantification, the accurate mass and time (AMT) tag strategy was developed (reviewed in 25). The AMT tag label-free quantification strategy couples traditional tandem MS bottom-up ‘shotgun’ proteomics with high-throughput LC-MS analysis of samples, which better utilizes instrument duty cycle (scan speed) and leads to reduced under-sampling of co-eluting peptides for improved quantitative fidelity. The premise of the AMT tag approach is that peptides can be identified based on the paired physiochemical and molecular mass uniqueness when measured with sufficient LC resolution and mass measurement accuracy. By combining high mass measurement accuracy with high efficiency capillary nanoLC separations, peptides are identified by accurate mass and LC retention time and quantified by integrated ion intensity for peptide species. Methods similar to the AMT tag approach such as PePPER have been shown to be effective for label-free peptide quantification.26
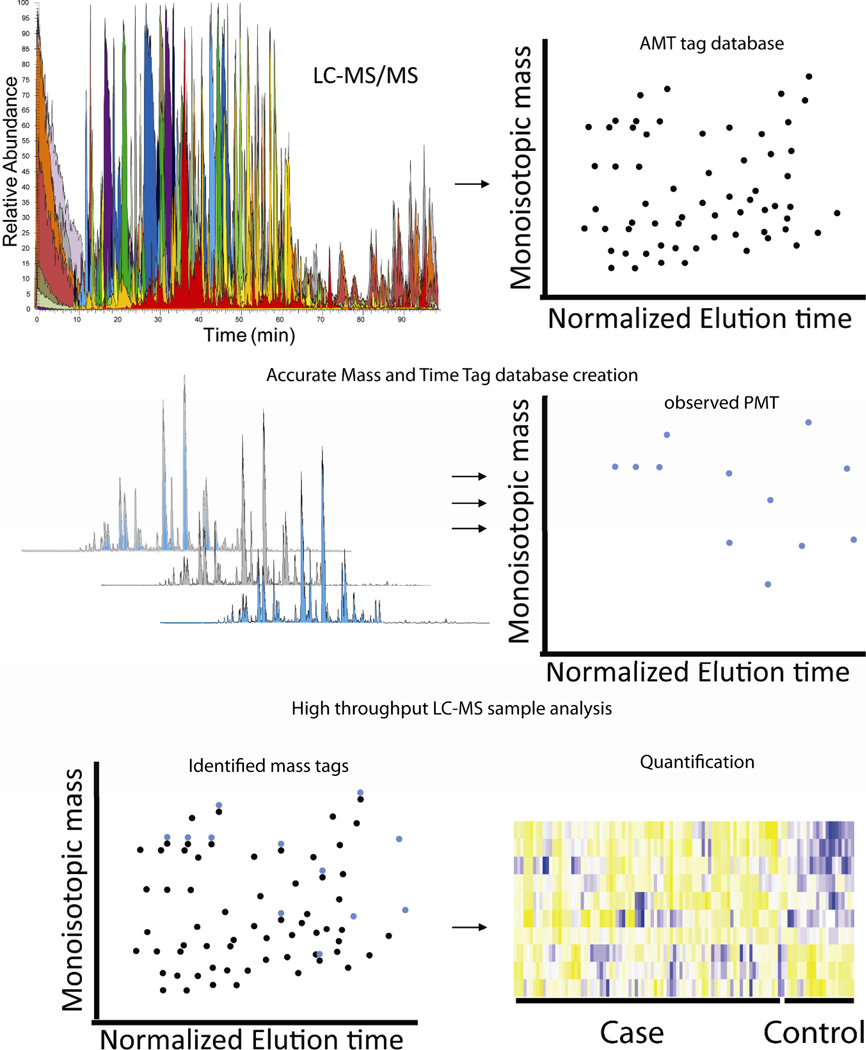
The typical AMT tag study involves two stages: 1) creation of a comprehensive AMT tag database from the MS/MS analysis of highly fractionated proteolytic peptides from representative sample pools of both experimental and control sample groups and 2) high-throughput, high mass accuracy LC-MS analysis of individual study samples. Peptides are identified by tandem LC MS/MS analysis of highly fractionated representative samples, as outlined in Figure 2. AMT tag databases are created from identified peptides by coupling accurate mass and normalized LC retention time assignments (NET), where identified peptides are assigned a potential mass and time (PMT) tag. Coupling normalized elution time and accurate mass measurements leads to an increase in the ability to distinguish similar peptides from one another compared to drastically increased measured mass accuracy (MMA) alone.25 The second stage of the AMT tag strategy involves analysis of a large number of unfractionated biological replicate samples, increasing the statistical power of the study. Following LC-MS analysis of individual samples, charge state, accurate mass, and normalized elution times are determined for the observed spectral features. These features are then aligned to an AMT tag database employing computational tools such as VIPER27 or Multialign28 (software available at omics.pnl.gov).
Figure 2.
Accurate Mass and Time (AMT) tag quantification. AMT tag quantification involves two stages: 1) representative samples are fractionated and analyzed in a typical shotgun proteomics approach, where identified peptides are assigned normalized elution times (NET) and accurate mass values, 2) biological replicates are analyzed by LC-MS in a high-throughput fashion and potential mass and time tag (PMT) are assigned for peptides in the sample. The PMT are then mapped onto the accurate mass and time tag database allowing for peptide identification and quantified by ion intensity.
AMT tag method allows tracking and comparison of a large numbers of analyte species that may be unidentified or ambiguous for numerous reasons (e.g., insufficient mass and time tag database coverage, poor peptide fragmentation or informatics limitations; presumably similar factors that leave a significant fraction of the species detected in LC-MS/MS analyses unidentified). However, the ability remains to quantify changes in such unassigned anonymous ‘features’ across datasets, allowing any features that change in a significant or ‘interesting’ fashion to be selected for subsequent targeted measurements that aim to either identify the features or to more sensitively and precisely measure its variation across a set of analyses. A limitation for label free quantification as compared to labeling approaches is that samples are analyzed individually and multiplexing of samples is not possible.
Targeted proteomics measurements
The discovery and development of protein biomarkers, measurable indicators that correlate to specific biological or disease states, has become a key area of application for proteomics. Developments in MS-based proteomics methods have yielded a number of candidate biomarkers showing great promise for improvements in diagnosis, prognosis, and treatment of complex human diseases.12 A strategy that complements global proteome profiling is the targeted MS based quantification of a predefined list of proteotypic peptides, which are often discovered in a global proteome study. Targeted proteomics strategies have the potential for providing greater sensitivity and allowing for the detection of lower abundance candidate peptides and proteins. The difficult task of bridging the gap between biomarker discovery efforts and development of useful clinical assays requires the application of high-throughput analytical methods for verifying and prioritizing candidate biomarkers identified in discovery phase efforts. Currently, the most common approach for verification and validation of new biomarkers relies on development of immunoassays due to the high sensitivity and specificity that can be achieved with antibody-based affinity reagents. However, utilizing immunoassays for verification and validation of candidate biomarkers presents several limitations. Specifically, development of antibody-based assays is costly and requires extensive time, creating a significant bottleneck in the biomarker verification and validation pipeline.29
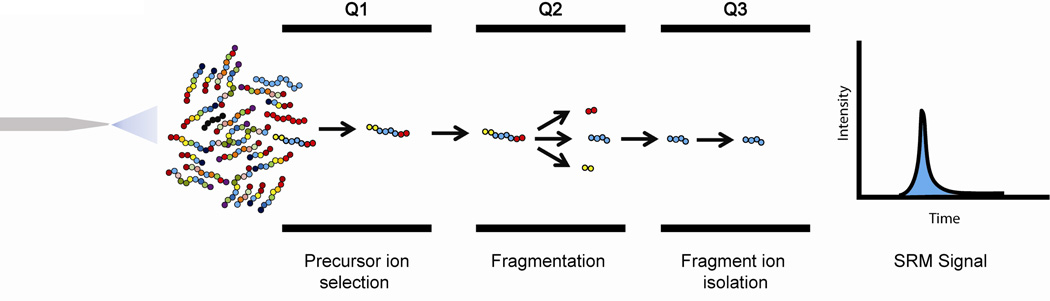
An alternative to the development of an antibody-based assay that avoids the need to develop paired affinity reagents for each candidate protein involves the application of targeted MS-based measurements such as selected reaction monitoring (SRM, also often referred to as multiple reaction monitoring; MRM). Targeted MS measurements with greatest sensitivity, specificity, and sampling rate (throughput) are measured in a triple quadrupole mass spectrometer. The triple quadrupole mass spectrometer provides a significant boost in sensitivity, dynamic range, and reduction in coefficient of variation (CV) for quantitative targeted MS-based assays (reviewed in 31). LC-SRM-MS has been demonstrated to have the potential for allowing the detection and quantification of proteins over the whole range of cellular concentrations.30 The SRM approach measures pre-selected analyte ions following two stages of mass selection. In the first stage of selection the m/z of an intact analyte (precursor ion) is isolated in the first quadrupole (Q1). After fragmentation of the parent ion, typically by CID in Q2, the resulting product ions (i.e., the fragmentation products of the selected precursor ion) are isolated in Q3 and the m/z for their corresponding fragment ions are recorded (Figure 3). Targeted MS analysis of immunodepleted and fractionated blood plasma using SRM was reported to enhance the lower detection limit for peptides by up to 1000-fold as compared to LC-MS/MS analysis32, thus providing increased sensitivity of measurements. The discriminating power of mass analyzers can provide high specificity for analyte identification and the ion current can provide accurate quantification of analyte concentration with addition of appropriate stable isotope-labeled internal standards. With modern triple quadrupole mass spectrometers, a large number of precursor-product transitions (1000’s) can be monitored during a single LC-MS/MS run. Identification of proteotypic peptide targets useful for quantification can be made using results from discovery data or public repositories such as Peptide Atlas and the Global Proteome Machine.31 Additionally, computational methods have been developed for prediction of proteotypic peptides given a protein amino acid sequence (reviewed in 31).
Figure 3.
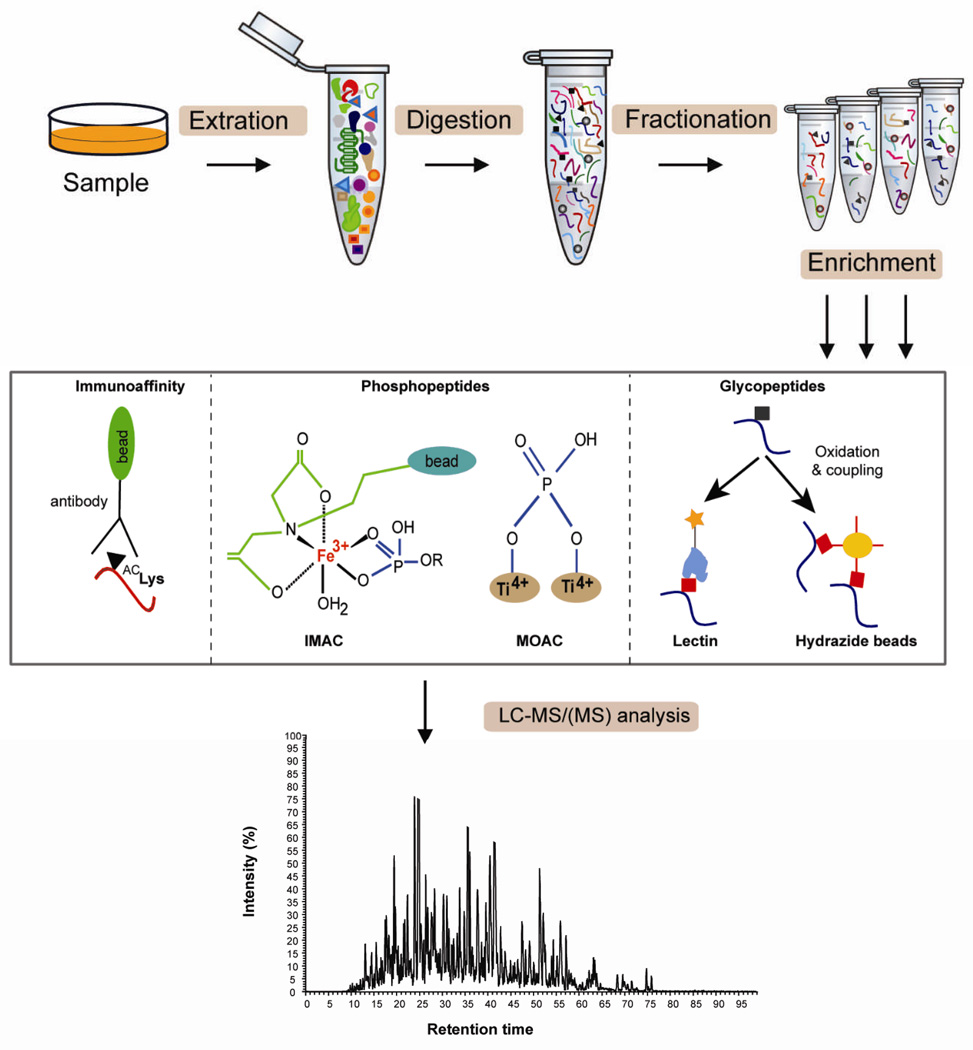
Experimental strategies to study protein post-translational modification. Enrichment of lysine acetylated (▲), phosphorylated ( ) and glycosylated peptides (■) using different affinity media is shown as examples. A typical workflow for PTM analysis involves extraction of proteins from cells or tissues followed by proteolysis of extracted proteins into peptides, reduction of sample complexity by fractionation (if required), enrichment of PTM peptides by using appropriate methods, LC-MS/(MS) and database searches for identification, quantification and PTM site matching. Different methods can be selected for protein extraction, fractionation, and enrichment of PTM peptides depending on sample type, complexity and targeted analysis.
) and glycosylated peptides (■) using different affinity media is shown as examples. A typical workflow for PTM analysis involves extraction of proteins from cells or tissues followed by proteolysis of extracted proteins into peptides, reduction of sample complexity by fractionation (if required), enrichment of PTM peptides by using appropriate methods, LC-MS/(MS) and database searches for identification, quantification and PTM site matching. Different methods can be selected for protein extraction, fractionation, and enrichment of PTM peptides depending on sample type, complexity and targeted analysis.
SRM (or MRM) technologies offer the capability for development of biomarker verification and validation assays in high-throughput manner given the multiplexing capabilities (e.g. analyzing hundreds of biomarkers in a single experiment) of this analytical platform. However, for SRM methods to approach the performance of immunoassays for measurement of protein biomarkers in biofluids, both the sensitivity and dynamic range must be significantly increased. The utility of current SRM-based assays is limited by the achievable sensitivity and dynamic range to quantitatively measure many “analytes of interest” in a complex matrix (e.g., blood plasma/serum). Without additional sample enrichment or fractionation, the demonstrated limit of detection (LOD) of SRM is typically in the tens of ng/mL range when candidate biomarkers are measured in blood plasma33; however, enhancements have recently been made to robustly measure peptide analytes in the single digit ng/mL range. While MS sensitivity continues to be improved, this LOD falls short of that provided by current antibody-based immunoassays which is usually at the low ng/mL to pg/mL detection limit, a level expected to be necessary for disease-specific biomarker validation applications. The accessible dynamic range of SRM measurement has also been limited to a maximum of five orders of magnitude in analyte concentrations34, which is insufficient for detecting low abundance protein biomarkers (e.g., <1 ng/mL concentration) in blood plasma. The insufficient sensitivity and measurement dynamic range is also reflected in inadequate selectivity of SRM measurements when low abundance protein biomarkers are either buried in the background ‘chemical noise’ or overlap (i.e. coelute) with high abundance matrix peptides/proteins.35
The key to realizing the full potential of SRM-based assays is to dramatically enhance the sensitivity, dynamic range and selectivity of the MS measurement while maintaining high-throughput capability. For example, SISCAPA35 (stable isotope standards and capture by anti-peptide antibodies) has been shown to enable increased sensitivity of analytical measurements and lead to improved detection and quantification of low abundant peptides in samples such as blood plasma with high concentrations of interfering species. To address these limitations, advances in high performance capillary LC systems, high sensitivity electrospray ionization (ESI) sources, efficient ESI-MS interfaces, and MS instrumentation are also being developed.33 A new instrument platform incorporating improvements addressing the aforementioned limitations was designed with an automated multi-column high pressure capillary LC system optimized for high-throughput separations, a more sensitive (e.g. multi-emitter ESI and multi-inlet) ionization source, and a tandem electrodynamic ion funnel MS interface to effectively focus and transmit the ion beam from the high intensity ion source to e.g. a triple quadrupole MS.36–38 While the full integration of improved LC-SRM-MS platforms that provide much higher sensitivity and selectivity than any existing platform is a work in progress, recently it has been shown that significantly lower LOD can be readily achieved by implementation of the multicapillary MS inlet and dual ion funnel technologies.33 A series of measurements were made using mouse plasma samples spiked with varied concentrations of proteolytic peptides to evaluate the detection sensitivity and reproducibility attained by the dual ion funnel and multi-capillary inlet interface relative to a standard instrumentation (single capillary inlet/skimmer) interface. Peak areas were enhanced approximately 70-fold, with peak area improvements ranging from ~20 to ~150-fold for individual peptides.33
Another LC-SRM-MS strategy termed PRISM (high-pressure high-resolution separations with intelligent selection and multiplexing) was also recently developed to further extend LODs.39 PRISM is an antibody free strategy for peptide enrichment allowing quantification of proteins at the low pg/mL range in blood plasma/serum. This method starts with a dimension of high-resolution reversed phase capillary separation using reverse phase high pH fractionation (pH 10) to allow for peptide enrichment that is highly compatible with the downstream LC-SRM-MS analysis. Then the fractions containing peptides of interest are targeted in an LC-SRM-MS analysis, increasing the overall throughput, and extending LOD 10 to 100-fold lower, e.g., to low pg/mL protein levels in plasma.
Proteome wide analysis of post-translational modifications
Biological functions of cells are mediated in part by protein PTMs. Genomic and transcriptomic approaches are blind to PTMs, making proteomics the only way to study these on a large scale. PTMs can be static or dynamic, altering the chemical state of a protein in subtle ways that are not easily detected by standard protein profiling techniques.40 They increase chemical diversity and complexity of the proteome, and comprehensive understanding requires the ability to perform dynamic systems-level analyses.40 Recent advances in the development of efficient large-scale PTM profiling technologies have resulted in many important insights into how cells process information41, however, there are still significant challenges. Thus far, most PTM studies have focused on common modifications, such as phosphorylation, glycosylation, ubiquitination, sumoylation, and acetylation.
PTMs determine protein function by altering activity, cellular location, turnover, and interactions with other proteins.41 Protein modifications are involved in most signaling events driving communication from the cell membrane to the nucleus in response to external stimuli. The organization and re-arrangement of modular domains in different signaling proteins due to PTMs creates complexity in signaling networks and pathways. In addition to modifying catalytic functions, PTMs direct the assembly of multi-protein complexes, and provide a means for crosstalk between convergent pathways in a transient and reversible fashion. Proteins are not only modified by small chemical moieties, but also through conjugation with other proteins. For example, ubiquitin, a small protein of 76 amino acids, is covalently attached to a specific lysine residue in substrate proteins by ubiquitin-ligase enzymes.42 Polyubiquitylation can target proteins for proteasome-mediated degradation providing an important function in the regulation of protein abundance and turnover in cells. Ubiquitin-like modifiers such as SUMO (small ubiquitin-related modifiers), NEDD8 (neuronal-precursor-cell-expressed developmentally down-regulated protein-8) and ISG15 (interferon-stimulated gene-15) are polypeptides that are conjugated to proteins and are involved in the regulation of a range of cellular processes and pathways.42 Because PTMs are a central mechanism for signal transduction and regulation, knowledge of the protein targets and modified amino acid sites shed light on important aspects of signaling and regulation.
A challenge inherent for PTM analysis is the sub-stoichiometric level, chemical diversity, and labile nature of modifications. More than 300 chemical PTMs have been identified making comprehensive PTM analysis impractical at any significant depth of coverage.43 Reduction in sample complexity by upstream sample processing such as organelle isolation, subcellular fractions or plasma membrane preparations can also enhance detection of sub-stoichiometric PTMs. Capture and enrichment have been the key to PTM analysis. Specific modifications, such as phosphorylation, occur on a minority of all proteins in a sample at any give time, often require selective enrichment prior to MS detection.
As with conventional bottom-up proteome analyses, targeted PTM analytical methods aim to achieve higher sensitivity and a wider range of proteome coverage. Efficient cellular preparations and protein-extraction protocols are keys to successful PTM analysis. Recently, there has been significant improvement in sample preparation and analytical techniques for large scale PTM analysis (Figure 4). Chemical and affinity based methods are commonly used to enrich modified proteins or peptides.22 Chemical approaches include the introduction of affinity tags onto modified proteins/peptides, such as with chemically engineered kinases and glycosyltransferases where affinity tags are introduced onto phosphorylated and glycosylated proteins allowing selective capture.44 One chemical method involves β-elimination of the phosphate group from phosphoserine (pSer) and phosphothreonine (pThr) converting these residues to dehydroalanine and dehydroaminobutyric acid allowing for direct detection. An alternative chemical strategy involves the selection of phosphorylated residues by coupling a solid phase support using phosphoramidate chemistry (PAC). However, chemical approaches can lead to unwanted side reactions, which results in increased sample complexity and complicates distinction of in vivo modifications.22
Figure 4.
Overview of selected reaction monitoring mass spectrometry (LC-SRM-MS). Proteotypic peptides are separated from complex biological samples by reversed phase liquid chromatography. The selected proteotypic peptides are isolated in Q1 reducing interfering background signal, and subsequently fragmented in Q2 and specific transition ions are isolated in Q3 prior to detection. Multiple rounds of isolation greatly reduce the background signal resulting in greatly improved signal to noise in typical SRM-MS quantification.
Advanced MS technologies allow identification of both known as well as novel PTMs, and thus offer a significant advantage over and complement antibody-based approaches. Other affinity based enrichment methods for phosphorylated (IMAC, MOAC) and glycosylated (lectin affinity chromatography, hydrazide capture) proteins/peptides coupled with MS have been applied with significant success, and are discussed below in detail. PTMs can either increase or decrease the molecular weight of peptides and result in modification-specific signals in MS/MS. Examples include the deamidation of asparagine and glutamine to aspartate and glutamate (+1 Da), the formation of Cys-Cys disulphide bonds (−2 Da), and the addition or removal of phosphate groups (+/− 80 Da) or aceytyl groups (+/− 42 Da). Although now routinely used in biological research, detecting and differentiating PTMs still poses a challenge for conventional bottom-up proteomics approaches, due to low PTM stoichiometry, more complicated MS/MS spectra, and informatics challenges associated with the increased database search space resulting in higher false discovery rates.41
Protein phosphorylation
Since its first detection on glycogen phosphorylase in 1955, protein phosphorylation has become a central focus in signaling studies. It is estimated that 30% of proteins in the human genome can be phosphorylated, and abnormal phosphorylation is now recognized as a cause of human disease.45 Phosphopeptide enrichment using immobilized metal ion (Fe3+ or Ga3+) affinity chromatography (IMAC) and metal oxide affinity chromatography (MOAC) using TiO2 or ZrO2 are widely used for phosphoproteomic studies.46 Other emerging techniques include protein and antibody-based arrays and fluorescence-based single cell analyses, which have the potential for high sensitivity and throughput, but require prior knowledge of targets. Phosphorylation increases molecular mass by 80 Da in phosphotyrosine (pY) due to the addition of HPO3 and assignment of the amino acid phosphorylation site can be performed through the observation of discrete mass increment of 80 Da (or 98 Da for H3PO4) on peptide fragment ions. Peptides containing phosphorylated serine and threonine undergo cleavage of the phosphoester bond resulting in the loss of H3PO4, termed a ‘neutral loss’ in the MS/MS spectrum resulting in a product with a mass decrease of 98 Da.
Protein glycosylation
Glycosylation plays an important role in protein secretion, stability, function, localization, and turnover.47 Two major types of protein glycosylation; N-linked and O-linked have been identified. N-linked glycans are attached to asparagine residues and O-linked glycans are most commonly connected to the -OH side chain of serine and threonine residues. N-linked glycosylation sites can be localized and predicted to be present at the amino acid motif N-X-S/T where X denotes any amino acid except proline. Glycans are synthesized by the coordinated expression of numerous genes that encode glycosyltransferases, glycosidases, and other enzymes that synthesize and remodel glycan chains. Glycosylation studies are typically performed using three different approaches: (i) characterization of the glycan in the intact glycoproteins; (ii) characterization of glycopeptides; and (iii) structural analysis of chemically or enzymatically released glycans. Intact glycoproteins can be purified using lectins such as Concanavalin A (ConA) or Wheat-Germ Agglutinin (WGA). Alternatively glycoproteins and peptides can be captured using hydrizide chemistry (reviewed in 48). Intact N-linked glycans are enzymatically released with an amidase (peptide N-glycosidase F, PNGase), which cleaves the linkage between the core GlcNAc and the asparagine residue. Two other enzymes, endoglycosidase D (endo D) (which release all classes of N-linked sugars) and endo H (which release oligomanose and hybrid type sugars), are also useful. There are fewer options for enzymatic cleavage of O-linked glycans, presently O-glycanase, which cleaves at core 1 structures of an O-linked glycans, is one of the only enzymatic methods available. Because glycoproteomics is still an emerging field, MS-based analysis is just beginning to provide a means for characterizing this heterogeneous family of PTMs.
Proteolytic PTMs
Proteolytic PTMs are irreversible ubiquitous protein modifications that affect the structure and function of proteins.49 Although, the importance in biological systems is admittedly largely unknown, emerging MS-based proteomics technologies to characterize degradomes50 have started to unfold the significant role proteolytic processing plays in development and progression of various diseases including neurodegeneration, heart disease, and cancer. Indeed many biomarkers of disease are stable protein fragments generated by proteolysis49, and systems-wide analysis of such proteolytic processing is expected provide better understanding of numerous human diseases. In vivo proteolysis of proteins by proteases modifying the structure and function of proteins in the cell as a part of normal physiological function such as immunity, development and repairs (e.g. blood clotting and wound healing) represents an important category of PTMs.49 The human genome encodes over 569 proteases, which constitute 5–10% of all drug targets.51 Proteases function by cleaving the covalent peptide backbone and many proteins undergo limited proteolytic processing as part of the normal temporal and spatial maturation process. These controlled proteolytic events do not result in the total degradation of proteins but rather are involved in protein-substrate maturation and tailoring processes. However, dysregulation of proteolysis is a feature of many diseases such as cancer and currently more than 53 specific hereditary diseases with effected proteolysis have been recognized.52 Many potential biomarkers of diseases are stable proteolytic fragments and identifying proteases and processing events that generate these fragments is crucially important for the development of targeted treatments. Unfortunately, considerably little is known about the relevant biological functions of proteases, and thus far the importance and ubiquity of proteolysis is underappreciated.
Due to recent advances in MS-based proteomics techniques, the significance of proteolytic PTMs is gradually emerging and systems-wide analysis of proteases and their substrates are being enabled.51 By combining CID, high energy C-trap dissociation (HCD) and ECD fragmentation techniques for peptidomic/degradomic analysis of human blood plasma50, improving strategies for peptidome isolation from human blood, and utilizing iTRAQ for quantitative N-terminome analysis51, the challenges for proteolytic PTM analyses are being met. Peptidomics/degradomics studies are expected to provide information critical for understanding the biology of health and disease. By taking advantage of technological and methodological advances including application of activity-based protein probes40, improved sample preparation and labeling techniques as well as better informatics tools, the biological significance of these protein species will become ever clearer.
Top-down Proteomics
Top-down MS-based proteomics refers to the analysis of intact proteins, in contrast to bottom-up proteomics methods, which are not enzymatically digested prior to MS analysis. Characterization of biological systems through the analysis of intact proteins and protein complexes using MS provides information on post-translational processing through the identification of the intact mass of a given protein. Applications of top-down proteomics include identification of protein isoforms arising from amino acid modifications, gene variants, transcript variation, and PTMs as well as proteolytic processing of proteins. Although less time is required for sample preparation, MS analyses of intact proteins can be challenging with reduced ionization, and detection for proteins with increasing molecular weight.53 Longer MS acquisition times required for top-down analysis creates challenges when attempting to couple online chromatography with top-down analyses of protein mixtures. Furthermore, sample purity can have a large effect on distinguishing signal in the mass spectrum. Efforts to address some of these challenges have been made by coupling top-down and bottom-up approaches with online digestion.54
While peptides mixtures can be studied in a range of mass analyzers, the analysis of intact proteins is more restrictive due to requirements of mass accuracy and resolution. High resolving power is required to ‘deisotope’ charge state envelopes, generated by ESI, of high MW species to accurately determine the monoisotopic mass of intact proteins. The majority of top-down studies of larger proteins have used Fourier transform ion cyclotron resonance cell (FT-ICR) instrumentation because of the combined benefits of high resolution and mass accuracy when compared to other mass analyzers. Several reports of measured peak-to-peak resolution exceeding 106 have demonstrated the resolving power of FT-ICR instrumentation, providing measured mass accuracies of less than one ppm for intact proteins55 and making them the instruments of choice for intact protein molecular weight determination. Further improvements on ICR-cells have provided increased resolution and capabilities56 for accurate mass measurements of large proteins (>100 KDa), increasing the potential applications for top-down MS. Additionally, improved vacuum chambers and optimization of front end ion focusing continue to improve the sensitivity of such instruments. However, with further technological advances of non-FT-ICR mass analyzers, applications of top-down proteomics are increasingly feasible with a larger range of instrumentation (e.g. time of flight (TOF) and Orbitrap). Several recent reports using the hybrid Velos LTQ-Orbitrap and the new Orbitrap Elite have demonstrated sufficient resolution for intact protein analysis and high scan speeds compatible with LC time scales.57 For example, top-down histone analyses (11 – 15 kDa proteins) utilize fast scan speeds and efficient fragmentation of the Velos LTQ-Orbitrap to assign PTM locations on the highly modified N-terminal histone tails that are associated with chromatin folding and epigenetics.58 Alternatively, top-down experiments using MALDI TOF-TOF mass spectrometry, which predominantly produces singly charged ions, has been successful for intact protein analysis of species ≤ 12 kDa.59 Recently top-down TOF-TOF experimental data was used to identify biomarkers of strain specific bacteria.60
Accurate mass measurements greatly facilitate detailed analysis of intact proteins (and top-down proteomics), as a small mass difference can distinguish between different molecular species. For example, the mass difference of trimethylation versus acetylation is 0.03639 Da, corresponding to 3 ppm for a 12,000 Da protein, both of which are commonly observed on core histones. The mass difference between lysine and methionine residues is 2.94553 Da; for 12,000, 100,000, and 200,000 Da proteins, this corresponds to 245, 29 and 15 ppm respectively. For peptides, these differences in mass are detected with low-resolution instrumentation; however, in the case of intact large molecular weight proteins, high mass accuracy is required to distinguish between these potential isoforms. In addition to accurate mass measurements, high resolving power is crucial for intact protein analysis. Often the monoisotopic peak of a multiply charged protein is below the signal to noise level, and the ability to distinguish between isotopic peaks for a single charge state directly influences the assigned molecular weight and measured mass accuracy. Increased resolution reduces ambiguity in peak picking/deisotoping and isotopic alignment between multiple charge state distributions, ultimately leading to a more accurate determination of the protein molecular weight.
The ability to decipher the accurate mass of an intact protein provides invaluable information when identifying PTMs. Specifically, the mass provides information regarding modification stoichiometry. While bottom-up studies can reveal which amino acids are modified, once the original intact protein undergoes proteolytic digestion the link to the original protein isoform is lost. Further challenges arise if multiple peptide sequences belonging to the same protein are identified as modified, making several isoform/stoichiometry combinations possible. However, if the intact mass were known, this would provide information on how many modifications were present per protein molecule, greatly simplifying the analysis. In the case of proteins that undergo extensive modification (such as core histones and P5361, 62), without knowledge gained from analysis of the intact precursor mass, the number of possible isoforms from a bottom-up analysis becomes a great computational problem (e.g. in the case of histone H3, all previously cataloged modification site combinations is >109 isoforms). While performing bottom-up analyses of these proteins is informative, linking the identified peptides back to the original intact protein molecular weight is the next step in the characterization of the biologically relevant isoforms. Such experiments are the basis for an integrated top-down / bottom-up workflow, combining the sensitivity and high-throughput elements of bottom-up methods with the informative top-down studies that require high resolution instrumentation.63
In addition to detection of PTMs and sequence variants, intact accurate mass measurement is useful for uncovering signal peptide cleavages, splice variants, and protein processing required for biological processes such as translocation and protein recruitment.64, 65 The presence or absence of signal peptides can indicate compartmentalization and transport within a cell. Bottom-up analysis cannot distinguish between lack of sequence coverage due to sample handling or instrumental duty cycle and protein truncations of biological interest such as signal peptide cleavage.
As an example, in a normal system the N-terminal domain of filaggrin is translocated to the nucleus after phosphorylation signaling events and N terminal truncation. It has been demonstrated that after exposure to ionizing radiation, the filaggrin N-terminal domain is not cleaved and subsequent translocation to the nucleus is blocked, leading to a decrease in the protective nature of skin.66 Analysis of filaggrin, a 44 kDa protein, using a top-down approach would be informative on both the presence of the N-terminal domain and phosphorylation states.
While the popular bottom-up proteomics approach has been implemented across several research fields, high-throughput top-down approaches are only recently becoming a reality. Deisotoping, search algorithms, and databases have been tailored to bottom-up peptide analysis, and altering/adapting these tools for efficient top-down analysis is a challenge for the field. Several search algorithms have been introduced for top-down MS/MS acquisitions and show promise for increased throughput.67–69 In the case of a large number of co-occuring PTMs and other protein modifications, complications at both the experimental and data-analyses levels remain, demonstrating a need for further improvement in the top-down proteomics field. The potential information gleaned from top-down proteomics studies or by integrating both top-down and bottom-up approaches, is vast and combined with recent improvements in MS instrumentation will become an important avenue for proteomics studies.
Top-down approaches can be broadly applied when analyzing the degradome/peptidome of a sample. Application of technologies developed for top-down MS to the degradome/peptidome analysis, which has a more tractable molecular weight range for MS analysis, has been shown to be of great utility. Degradomics/peptidomics profiling of blood plasma from breast cancer patients proved to be highly sensitive to changes present but not detectable by traditional bottom-up analysis. The degradome was altered between cancer and control samples and exhibited disease associated signatures with possible diagnostic utility.70
Emerging Technologies
Ion Mobility Separations Coupled to Mass Spectrometry
In spite of the significant advances to the MS technologies described above, several performance metrics including measurement throughput and detection sensitivity are still far from ideal for effective biomarker discovery. These deficiencies result in low sampling numbers and measurement quality incapable of confidently detecting proteins present at low concentrations. Coupling fast LC separations, high sensitivity ion mobility separations (IMS) and MS offers a promising direction to address these shortcomings. IMS is a gas-phase separation technique based on the fact that isomers with differing shapes travel with different velocities when they are pulled by a weak electric field through a drift cell filled with buffer gas.71 Due to the shape of the isomers and the number of collisions they encounter with the buffer gas, compact ions with small collisional cross sections drift more quickly than extended ions with large collisional cross sections. IMS effectively allows ions to be separated by shape, and in combination with TOF-MS that separates ions based on mass, result in multidimensional separation. IMS-MS also offers a tremendous increase in ion utilization efficiency and a high-throughput advantage because both IMS and TOF analyzes can be performed quickly as IMS separations typically require only 10–60 ms, while a TOF-MS spectrum takes ~100 µs (allowing numerous mass spectra to be obtained during each IMS separation).
LC is generally coupled with IMS-MS platforms to analyze complex samples. Two advantages of coupling LC and IMS are that LC reduces the complexity of the samples by separating relatively slowly, while IMS provides much faster separation that are significantly different in character. The ability to ‘accumulate’ ions before the IMS step, allows nested LC-IMS-MS measurements that are effectively lossless in terms of the peptide ion signals (through the use of ion funnels), preserving and improving upon the sensitivity of LC-MS72 due to the concentrating effect of the ion accumulation step and the reduced background from the IMS separation. To understand this effect, the sensitivity of IMS-MS alone and LC-IMS-MS measurements were evaluated by determining the LOD for both. The LOD for IMS-MS was determined to be 40 attomoles while LC-IMS-MS was ~1 attomole, a factor of 40 lower.72 Another advantage of coupling LC to IMS is that they are significantly orthogonal methods and IMS is able to further separate features that are unresolvable with LC-MS alone. This orthogonality has allowed the use of shorter LC gradients to perform high-throughput analyses of complex samples while identifying a similar number or additional features to that of longer gradient LC-MS runs (Figure 5). Experiments have been performed to compare 15-min LC-IMS-MS analysis with 100-min LC-Linear Ion Trap Fourier Transform (FT) MS analysis of a tryptic digest of mouse blood plasma sample spiked with twenty reference peptides at varying concentrations from 1 ng/mL to 10 µg/mL.73 The LC-FT MS detected thirteen out of the twenty spiked peptides, all having concentrations ≥100 ng/mL. In contrast, the drift time selected mass spectra from the LC-IMS-TOF MS analyses yielded identifications for nineteen of the twenty peptides with all spiking levels present. Since potential candidate biomarkers are expected at low concentrations and require the examination of many samples, these results show LC-IMS-TOF MS has enormous potential for improving biomarker discovery and verification, and also suggests applications for targeted measurements.
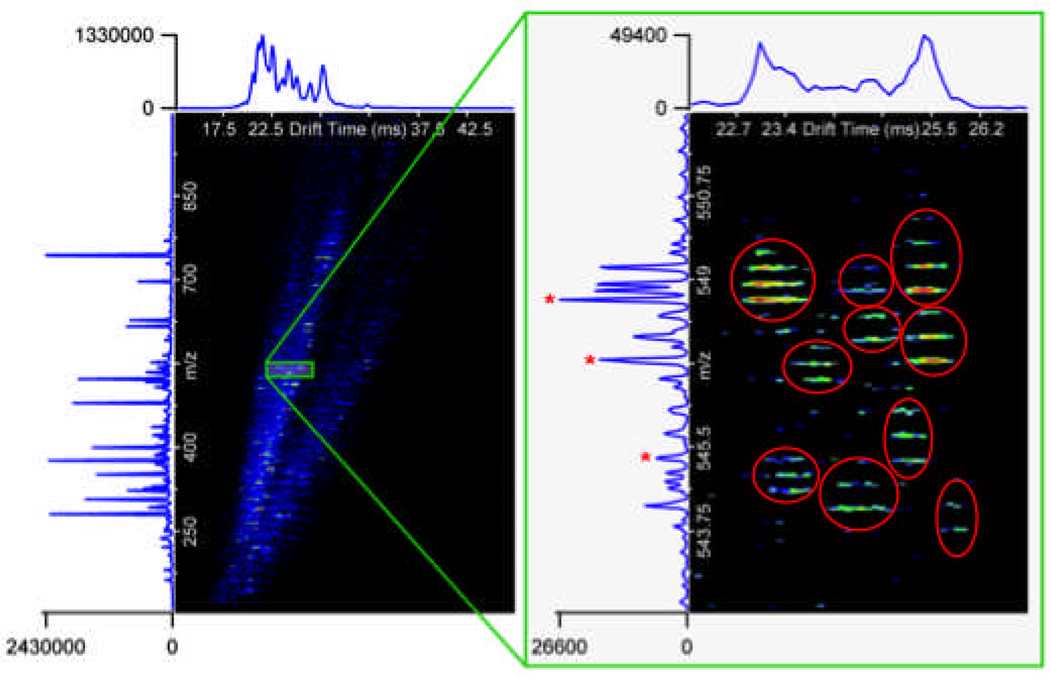
Figure 5.
A nested IMS-MS spectrum for a selected LC elution time region from a complex sample of 20 proteins covering 108 dynamic range. Results show IMS-TOF MS can detect more proteins in the sample than FT MS. Ten peptides are easily detected in the zoomed region of the IMS spectra, but only 3 can be deisotoped and effectively identified by MS only (shown by *).
Because the high-throughput LC-IMS-TOF MS platform evaluates samples in 4 different dimensions (mass, elution time, drift time and intensity), and when LC-IMS-CID-MS is performed deconvolution is also necessary, the large quantities of multidimensional data represent a major computational challenge. Considerable efforts have been placed on advancing the available MS informatics tools to rapidly perform multidimensional analyses of the raw data. The most reasonable way to quickly identify peptides based on mass, elution time, and drift time is by adapting the AMT tag approach. Since current AMT tag databases only have information for peptide mass and elution time, the drift time dimension has been populated with experimental values to create extended AMT (xAMT) tag databases. The development of xAMT tag databases by adding the experimental IMS drift times to existing NET and MS information has been reported to drop the FDR by nearly 50% when drift time tolerance was used as a filter, indicating that the additional dimension is very important for high confidence IMS-TOF MS identifications.74 While experimental drift time population of the xAMT tag databases is viable for development and demonstration, a high-throughput method is necessary to populate future xAMT databases. Utilizing the known experimental drift times from the xAMT tag databases, a support vector regression-based generic model has been used to accurately predict drift times to within 3–4% error.75 LC-IMS-(CID)-MS techniques may also be used to identify ions while simultaneously populating drift times to xAMT tag databases. These database populating capabilities should allow significant improvement in measurement throughput and identification accuracy addressing existing needs of biomarker discovery and studies with samples of limited quantity.
To further increase throughput and peak capacity, field asymmetric IMS (FAIMS) has also been coupled to IMS-MS analyses. FAIMS-IMS-MS peptide analyses show substantial orthogonality between FAIMS and IMS separations, since IMS measures mobility while FAIMS measures its derivative with respect to electric field, which are not a priori correlated. The 2-D FAIMS-IMS peak capacity for tryptic peptides has been measured at ~500, which is comparable with high-quality condensed-phase separations that are orders of magnitude slower. Additional value of this technique is also expected for more complex samples. Tryptic peptide ions constitute a set of low chemical and structural diversity analytes spanning a limited m/z range, while more diverse mixtures (e.g., including metabolites, nucleotides, lipids, or whole proteins) inhabit a broader separation space in both their FAIMS and IMS dimensions, resulting in much higher peak capacities. Thus, indicating that the utility of the FAIMS-IMS-MS platform extends to applications well beyond bottom-up proteomics.
Moving towards single cell proteomics
The dramatic and ongoing improvements to MS sensitivity resulting from enhanced ion utilization efficiency open the door to what was previously considered unattainable: direct proteome analysis at the single cell level. Successful implementation of single cell proteomics will have a major impact on biological research, as existing proteomics approaches aimed at better understanding the cellular processes, interactions, and dynamics that are fundamental to all biological systems require large populations of cells. Such bulk measurements average over and obscure important cell-to-cell heterogeneity that is present even in clonal populations.76 As a consequence, stochastic gene and protein expression, when measured at the population level, leads to an averaged result that is not representative of any individual cell.77 Limitations that arise from performing bulk measurements become even more severe when the sample of interest comprises a small portion of the total population analyzed. For example, cancer stem cells, which represent just a fraction of the cells present in a tumor, may be responsible for a tumor’s adaptive behavior during treatment.78 Analyzing these rare cells separately from other tumor cells would undoubtedly lead to important insights into these potential therapeutic targets, whereas the biological noise imparted by other cells when measured in bulk obscures our understanding of these species.
The historical impediment precluding the application of MS to single cell analyses has been its insufficient sensitivity for such small amounts of material. These limitations are primarily due to a combination of inefficient ion production, ion losses within the instrument, and chemical noise sources including solvent clusters and contaminants. The technological advances described above have now produced ion source efficiencies that are within a factor of two of the theoretical limit (i.e., where every solution phase species can be ionized and transmitted to high vacuum 79). In early explorations aiming at analyzing small samples, ~50 proteins were identified in our laboratory from 50 pg samples80, corresponding to the amount of protein present in an average sized mammalian cell. While this suggests the possibility of single-cell proteomics, dramatic improvements in detection resulting from further increases in instrument sensitivity (e.g., more efficient ion utilization at the mass analyzer) as well as reduced chemical noise will be necessary to achieve broad coverage of such trace samples.
Besides enhancing MS sensitivity, new tools for individual cell selection, lysis, and preparation need to be developed, as conventional methods that rely on pipetting, centrifugation, and transferring of samples between multiple reaction vessels are incompatible with single cell analysis. To date, efforts to simplify sample preparation and minimize transfer events while still using conventional sample vials have yielded limited proteome coverage from 500–5000 cells81, 82, indicating that a dramatically different approach will be needed to analyze individual cells. One such strategy is to load a cell at the beginning of a capillary-based separation column, lyse it within the capillary, and then separate and detect the contents, thus greatly reducing any surface-related losses. This method was demonstrated 15 years ago for detecting hemoglobin from individual red blood cells using capillary electrophoresis (CE) with FTICR MS.83 However, the absence of sample processing other than lysis and the painstaking manual nature of sample loading limit the applicability of the approach for real-world applications where many cells will be needed to e.g. rapidly ascertain intercellular heterogeneity.
Microfluidic devices, with their ability to manipulate and dispense ultra-small volumes (femto- to nanoliters)84, 85 and combine multiple sample preparation steps in a single integrated device, provide a more promising platform for preparing single cells for analysis. In one recent example, Ramsey and coworkers86 used a microchip CE platform with an integrated electrospray source to analyze individual cells. Cells passing through an intersection were subjected to an abrupt change in solvent environment and electric field, which led to rapid lysis. Following a brief CE separation, the contents were ionized on chip and detected by MS. Again, the sample used for this demonstration was red blood cells, which are uncharacteristically easy to lyse and contain very large (femtomole) quantities of the detected hemoglobin subunits. Still, the platform was capable of detecting ~12 cells/min, a notable improvement in throughput over capillary-based approaches.
An emerging technology in the microfluidics field that shows perhaps the most promise for single cell analysis involves the use of picoliter-sized aqueous droplets surrounded by an immiscible oil phase such that each droplet constitutes an individual sample vessel.85 Such droplet-based approaches have already found commercial application in next-generation genomic sequencing platforms due to their amenability to massively parallel analyses. Reagents can be combined and rapidly mixed by merging droplets of differing composition, suggesting the use of the platforms for single cell sample preparation. Importantly, by shrinking the size of the vessel to the same scale as that of a cell, dilution can be controlled and sample losses minimized even for reactions that require extended times (e.g., proteolytic digestion).
Several microfluidic technologies have recently been developed with the aim of creating a platform for MS-based single cell analysis. First, a microfluidic nanoESI interface was created that provides the highest sensitivity for chip-based ESI-MS reported to date (~80 zmol mass detection limits, sufficient for many cellular species).87, 88 An illustration of the membrane-based electrospray source, along with resulting high-sensitivity mass spectra, is shown in Figure 6A. A method for efficiently transferring aqueous droplets or plugs from an oil stream to an aqueous stream was also recently reported, which enabled the contents of picoliter-sized droplets to be analyzed by nanoESI-MS in a nearly dilution free fashion89 (see Figure 6B). In addition, pneumatic valves have been developed to create droplets with a high degree of control over droplet size, generation frequency and composition. Figure 6C shows the detection of droplets containing varying ratios of two components created at a multi-analyte droplet generator. This controlled mixing strategy will be used for in-droplet sample preparation of single cells, where agents for lysis and proteolytic digestions can be added to cell-containing droplets and incubated before separation and detection. While the above technologies are at an early stage of development, their combination into an integrated platform, used in conjunction with the next-generation ion mobility platforms and electrospray sources is expected to enable high-throughput proteomics analysis at the single cell level.
Figure 6.
Microfluidic technologies underdevelopment for MS-based single cell analysis. A. Mass spectra of 1 nM leucine enkephalin directly infused from a microchip (schematic shown in inset) at 50 nL/min. Signal was averaged for 30 s (top) and 0.2 s (bottom). Figure 6B. Nearly dilution-free analysis of subnanoliter aqueous plugs by nanoESI-MS. Figure 6C. Valve-based droplet generation enabling controlled reagent mixing for in-droplet sample preparation. Droplets 1 – 6 contain different ratios of dye molecules for demonstration. (6B adapted from 89)
Outlook
MS-based proteome analysis is accomplished employing a number of approaches utilizing many technologies; which together show great promise for providing insight into the complexities of biological systems. Coupling MS with a variety of sample processing methods allows for the characterization and quantification of the global protein complement of a biological sample as well as the PTMs on a global scale. Advances in sample processing with microfluidic devices, ion utilization with the ion funnel, and instrumental design with the LC-IMS-TOF platform development are addressing current limitations in mass spectrometry of biomolecules and MS-based proteomics. As analytical throughput increases and sample size requirements decrease, due to advances in instrument design, the parallel measurement of many ‘omics can be made leading to what is collectively referred to as pan-omics. The power of pan-omics will be increasingly realized by the integration of information from a range of measurements, enabling modeling and predicting biological processes and response to external stimuli, which collectively constitutes a systems biology approach to biological sciences.
Only in the last ten years, can it be said that mass spectrometry has had a small but increasingly significant role in biological research. As its roles in proteomics have increased, so too have its applications e.g., to the discovery and development of new biomarkers. While successes to date have been quite modest, and essentially confined to preclinical stages, it is appropriate to ask, if and when mass spectrometry may move into broader or even routine clinical applications. Biomarker development, as an example, can be described in terms of a multi-stage process that consists of discovery, qualification, verification, research assay optimization, validation, and commercialization.90 From a mass spectrometric perspective, it is possible to ‘bin’ measurements into one of two categories – those aimed at discovering potential protein biomarkers and those seeking to verify and validate these biomarkers. Approaches in both categories generally involve digesting proteins (e.g., with trypsin) as a first step to yield peptides that can be effectively detected and identified using MS. Discovery-based approaches employ broad (i.e., ‘unbiased’ or ‘undirected’) measurements attempt to identify as many proteins as possible in the hope of revealing promising biomarker candidates. A key challenge here stems from the extremely large dynamic range (i.e., relative stoichiometry) of proteins in biofluids such as plasma. Protein concentrations in plasma extend from ~1010 pg/mL for albumin to ~10 pg/mL and below for interleukins and other cytokines; proteins secreted or leaking into blood from specific early stage tumors could be even lower in concentration. Currently, most FDA approved protein biomarkers fall in the ~102 to 105 pg/mL range, a challenge for detection by broad discovery-oriented proteomics measurements that are still largely confined to proteins at the upper end of this concentration range (above ~103 pg/mL). Because of the constrained dynamic range of present mass spectrometers (~103 for a single spectrum from Orbitrap MS platforms currently popular for discovery efforts), broad coverage of lower abundance proteins typically requires larger starting amounts of sample and extensive fractionation and/or separations that limit measurement throughput. An on-line high resolution LC separation requires on the order of an hour, and the resulting ~104 spectra provide information on typically hundreds of proteins. A proteomics ‘deep dive’ through the use of extensive fractionation (e.g., ~100 fractions using strong cation exchange chromatography prior to LC-MS) to expand coverage to thousands of proteins further exasperates throughput, requiring days or weeks of measurement time. The latter is highly attractive for detecting potential biomarkers, but the inherently low throughput largely precludes studies of populations that can effectively account for both human and disease diversity. While the minimum useful study size for biomarker discovery remains an open question, it is expected to be much larger than generally practical at present, and likely is numbered in the thousands. For these reasons MS-based blood protein biomarkers development efforts have been increasingly focused on verification and validation. These applications typically make use of targeted measurements that provide greater sensitivity, throughput, and more accurate quantification than broad discovery-based measurements and without the need for extensive pre-fractionation, but only for a limited number of ‘targeted’ peptides/proteins. In particular, widely used triple quadrupole SRM or MRM measurement platforms afford targeted multiplexed MS/MS detection of up to hundreds of peptides during an LC separation, providing limits of detection and quantification (LOD and LOQ) several orders of magnitude lower than presently feasible with discovery-based platforms. The larger signals (i.e., peptide ion currents) associated with such measurements and the general application of stable isotope-labeled internal standards are primary reasons for the improvements in sensitivity and data quality (e.g., lower CVs). Moreover, recent platform advances such as improved ion sources and interfaces (e.g., incorporating dual-stage ion funnels33) and the use of immunoaffinity major protein depletion or targeted peptide enrichment methodologies35 extend practical LOQ values to ng/mL for plasma levels, and low pg/mL levels have recently been demonstrated.35, 39
Most exciting are the developments and incipient trends pointing towards a convergence of the two major ‘untargeted/discovery’ and ‘targeted/verification’ bins in terms of their MS measurement platforms. One key driver for this is the dramatic increase in effectiveness of ion sources; e.g. it has been shown that as much as 50% of peptides in solution can be ionized and effectively transported through the MS interface to the m/z analyzer stage (whereas effectiveness of analysis and detection can be very high). It has also been shown that >1010 ions per second can be introduced into the mass analyzer (with an upper limit of ~1012 resulting from the disruptive effects of space charge on e.g. ion focusing).79, 91 Such developments are just beginning to be felt as they have required considerable redesign to MS platforms to deal with larger ion currents and incorporate other developments needed to exploit them (e.g. greater dynamic range for detection). One implication of these developments is that they favor platforms capable of utilizing the greater ion currents (e.g. triple quadrupole or TOF analyzers) as opposed to ion trap based platforms due to the finite charge capacity of trap based analyzers (about 104 ions in a linear ion trap MS, and 106 for the Orbitrap MS, a ceiling set by well understood space charge effects). A second implication is the increased use of targeted analyses with such platforms in which specific sets of ions are selected e.g., for detailed MS/MS analysis using hybrid quadrupole-orbitrap or quadrupole-TOF MS platforms. A related trend is multiplexing of MS/MS measurements. In this case, multiplexing is qualitatively different from that used for targeted SRM measurements, and involves the simultaneous dissociated of multiple peptides following one of any number of first stage selection events, e.g. limited m/z ranges, combined with the use of more sophisticated informatics tools for data analysis. These developments also greatly benefit from improvements of MS/MS resolution and mass measurement accuracy to enable effective deconvolution of the multiplexed peptide fragmentation spectra. The ability to correctly discern contributions from low-level species in the presence of much more abundant species in such measurements depends not only on having sufficient peptide signal, but also on sufficient analyzer specificity and detector dynamic range. These is also the basis for much more powerful targeted measurements, where the increased ion signals now achievable cannot only exceed detector capabilities, but are more significantly limited by the analyzer specificity. For example in targeted SRM measurements the LOD or LOQ is limited either by signal intensity or by the presence of interfering signals (e.g. from co-eluting species, or ‘chemical noise’). The sensitivity limitation is being increasingly addressed by the developments noted above (which suggest approximately 103 to 104 gains over the best present performance should be achievable). However, realizing this potential will more generally also require concomitant advances in analyzer specificity; gains that can potentially be achieved by higher separation power (either increased resolution or additional analyzer stages, e.g., MS3), or to a more limited extent at present by broad detection of MS2 fragment transitions (where again MS resolution and detector performance are presently limiting).
A key trend to be discerned here is that we can expect technological developments such as faster separations, more effective ion sources, higher MS resolution, and higher dynamic range detectors to increasingly allow broad untargeted measurements that retain the benefits of targeted measurements. An initial step in this direction exploits very fast gas phase IMS, which take place on the time scale of tens of milliseconds and can provide peak widths of less than a millisecond in combination with a TOF-MS that can acquire ~10 spectra every millisecond. This allows placement of IMS between the LC and TOF-MS stages, while the use of ion funnels makes operation essentially lossless, thereby making two-dimensional separations possible without the need for LC prefractionation, as well as any loss of sensitivity or throughput. As an example, an early LC-IMS-MS platform implemented in our laboratory73 consistently reached detection levels of 1 to 10 ng/ml for 20 peptides spiked into mouse blood plasma, an order of magnitude better than achieved with ion trapping-based FTMS platforms. LC-IMS-MS platforms also allow highly multiplexed peptide dissociation (e.g., between the IMS and MS stages)92, which translates to more effective information on ‘all the ions, all the time’.
Such developments are just a first step in the coming convergence of untargeted and targeted platforms, which will be accelerated by the emerging capabilities for faster and higher resolution separations, improved MS resolution, and extended detector dynamic range. The potential for higher throughput measurements with such platforms also presents an opportunity for considerably more effective discovery efforts, and ultimately, a ‘grand convergence’ of discovery and verification measurements. In the shorter term, more sensitive and increasingly multiplexed SRM measurements will lead this advance.
Supplementary Material
ACKNOWLEDGEMENTS
Portions of this research were supported by the National Center for Research Resources (5P41RR018522-10) and the National Institute of General Medical Sciences (8 P41 GM103493-10) from the National Institutes of Health and Laboratory Directed Research and Development program at Pacific Northwest National Laboratory. All PNNL proteomics research described was performed in the Environmental Molecular Sciences Laboratory, a U. S. Department of Energy/BER national scientific user facility at Pacific Northwest National Laboratory.
References
- 1.Domon B, Aebersold R. Science. 2006;312:212–217. doi: 10.1126/science.1124619. and references cited therein. [DOI] [PubMed] [Google Scholar]
- 2.Ideker T, Galitski T, Hood L. Annu Rev Genomics Hum Genet. 2001;2:343–372. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 3.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 4.Karas M, Hillenkamp F. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 5.Nesvizhskii AI. J Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. and references cited therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta N, Bandeira N, Keich U, Pevzner PA. J Am Soc Mass Spectrom. 2011;22:1111–1120. doi: 10.1007/s13361-011-0139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Gupta N, Pevzner PA. J Proteome Res. 2008;7:3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang DW, Sherman BT, Lempicki RA. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 9.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge YC, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang JH. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yates JR, Ruse CI, Nakorchevsky A. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. and references cited therein. [DOI] [PubMed] [Google Scholar]
- 11.Anderson NL, Anderson NG. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 12.Surinova S, Schiess R, Huttenhain R, Cerciello F, Wollscheid B, Aebersold R. J Proteome Res. 2011;10:5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 13.Schutzer SE, Liu T, Natelson BH, Angel TE, Schepmoes AA, Purvine SO, Hixson KK, Lipton MS, Camp DG, Coyle PK, Smith RD, Bergquist J. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010980. and references cited therein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou JY, Petritis BO, Petritis K, Norbeck AD, Weitz KK, Moore RJ, Camp DG, Kulkarni RN, Smith RD, Qian WJ. J Proteome Res. 2009;8:5387–5395. doi: 10.1021/pr900564f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakur SS, Geiger T, Chatterjee B, Bandilla P, Frohlich F, Cox J, Mann M. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003699. M110 003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Yang F, Gritsenko MA, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG, 2nd, Smith RD. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolters DA, Washburn MP, Yates JR., 3rd Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 18.Xie F, Liu T, Qian WJ, Petyuk VA, Smith RD. J Biol Chem. 2011;286:25443–25449. doi: 10.1074/jbc.R110.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goshe MB, Smith RD. Curr Opin Biotechnol. 2003;14:101–109. doi: 10.1016/s0958-1669(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 20.Geiger T, Cox J, Ostasiewicz P, Wisniewski JR, Mann M. Nat Methods. 2010;7:383–385. doi: 10.1038/nmeth.1446. [DOI] [PubMed] [Google Scholar]
- 21.Qian WJ, Petritis BO, Kaushal A, Finnerty CC, Jeschke MG, Monroe ME, Moore RJ, Schepmoes AA, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Herndon DN, Camp DG, 2nd, Smith RD. J Proteome Res. 2010;9:4779–4789. doi: 10.1021/pr1005026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen ML, Vermeulen M, Bonaldi T, Cox J, Moroder L, Mann M. Nat Methods. 2008;5:459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 23.Petyuk VA, Mayampurath AM, Monroe ME, Polpitiya AD, Purvine SO, Anderson GA, Camp DG, 2nd, Smith RD. Mol Cell Proteomics. 2010;9:486–496. doi: 10.1074/mcp.M900217-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian WJ, Liu T, Petyuk VA, Gritsenko MA, Petritis BO, Polpitiya AD, Kaushal A, Xiao W, Finnerty CC, Jeschke MG, Jaitly N, Monroe ME, Moore RJ, Moldawer LL, Davis RW, Tompkins RG, Herndon DN, Camp DG, Smith RD. J Proteome Res. 2009;8:290–299. doi: 10.1021/pr800467r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffe JD, Mani DR, Leptos KC, Church GM, Gillette MA, Carr SA. Mol Cell Proteomics. 2006;5:1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashworth M, Lloyd D, Smith RS, Wagner A, Rowlands G. J Public Health (Oxf) 2007;29:40–47. doi: 10.1093/pubmed/fdl068. [DOI] [PubMed] [Google Scholar]
- 28.Manes NP, Dong L, Zhou W, Du X, Reghu N, Kool AC, Choi D, Bailey CL, Petricoin EF, 3rd, Liotta LA, Popov SG. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.000927. M110 000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson NL. Clin Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 30.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallien S, Duriez E, Domon B. Journal of mass spectrometry : JMS. 2011;46:298–312. doi: 10.1002/jms.1895. and references cited therein. [DOI] [PubMed] [Google Scholar]
- 32.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hossain M, Kaleta DT, Robinson EW, Liu T, Zhao R, Page JS, Kelly RT, Moore RJ, Tang K, Camp DG, 2nd, Qian WJ, Smith RD. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M000062-MCP201. M000062-MCP000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. Nat Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 35.Whiteaker JR, Zhao L, Anderson L, Paulovich AG. Mol Cell Proteomics. 2010;9:184–196. doi: 10.1074/mcp.M900254-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livesay EA, Tang K, Taylor BK, Buschbach MA, Hopkins DF, LaMarche BL, Zhao R, Shen Y, Orton DJ, Moore RJ, Kelly RT, Udseth HR, Smith RD. Anal Chem. 2008;80:294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang K, Page JS, Marginean I, Kelly RT, Smith RD. J Am Soc Mass Spectrom. 2011;22:1318–1325. doi: 10.1007/s13361-011-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly RT, Tolmachev AV, Page JS, Tang K, Smith RD. Mass Spectrom Rev. 2010;29:294–312. doi: 10.1002/mas.20232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi TLFT, Sun X, Zhao Rui, Schepmoes AA, Hossain M, Wu S, Kim J-S, Jones N, Moore RJ, Paša-Tolić L, Kagan J, Rodland KD, Liu T, Tang K, Camp DG, II, Smith RD, Qian WJ. Unpublished. 2011 [Google Scholar]
- 40.Simon GM, Cravatt BF. Nat Chem Biol. 2008;4:639–642. doi: 10.1038/nchembio1108-639. [DOI] [PubMed] [Google Scholar]
- 41.Choudhary C, Mann M. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 42.Welchman RL, Gordon C, Mayer RJ. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 43.Armah HB, Wang G, Omalu BI, Tesh RB, Gyure KA, Chute DJ, Smith RD, Dulai P, Vinters HV, Kleinschmidt-DeMasters BK, Wiley CA. Brain Pathol. 2007;17:354–362. doi: 10.1111/j.1750-3639.2007.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. Nucleic acids research. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen P. Nat Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 46.Aryal UK, Ross AR. Rapid Commun Mass Spectrom. 2010;24:219–231. doi: 10.1002/rcm.4377. [DOI] [PubMed] [Google Scholar]
- 47.Marino K, Bones J, Kattla JJ, Rudd PM. Nat Chem Biol. 2010;6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- 48.Bond MR, Kohler JJ. Curr Opin Chem Biol. 2007;11:52–58. doi: 10.1016/j.cbpa.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 49.Doucet A, Butler GS, Rodriguez D, Prudova A, Overall CM. Mol Cell Proteomics. 2008;7:1925–1951. doi: 10.1074/mcp.R800012-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Shen Y, Tolic N, Xie F, Zhao R, Purvine SO, Schepmoes AA, Moore RJ, Anderson GA, Smith RD. J Proteome Res. 2011;10:3929–3943. doi: 10.1021/pr200052c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prudova A, auf dem Keller U, Butler GS, Overall CM. Mol Cell Proteomics. 2010;9:894–911. doi: 10.1074/mcp.M000050-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 53.Compton PD, Zamdborg L, Thomas PM, Kelleher NL. Anal Chem. 2011;83:6868–6874. doi: 10.1021/ac2010795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez-Ferrer D, Petritis K, Robinson EW, Hixson KK, Tian Z, Lee JH, Lee SW, Tolic N, Weitz KK, Belov ME, Smith RD, Pasa-Tolic L. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.001479. M110 001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tolmachev AV, Robinson EW, Wu S, Pasa-Tolic L, Smith RD. Int J Mass Spectrom. 2009;281:32–38. doi: 10.1016/j.ijms.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikolaev EN, Boldin IA, Jertz R, Baykut G. J Am Soc Mass Spectrom. 2011 doi: 10.1007/s13361-011-0125-9. in press. [DOI] [PubMed] [Google Scholar]
- 57.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian Z, Zhao R, Tolic N, Moore RJ, Stenoien DL, Robinson EW, Smith RD, Pasa-Tolic L. Proteomics. 2010;10:3610–3620. doi: 10.1002/pmic.201000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin M, Campbell JM, Mueller DR, Wirth U. Rapid Commun Mass Spectrom. 2003;17:1809–1814. doi: 10.1002/rcm.1102. [DOI] [PubMed] [Google Scholar]
- 60.Fagerquist CK, Garbus BR, Miller WG, Williams KE, Yee E, Bates AH, Boyle S, Harden LA, Cooley MB, Mandrell RE. Anal Chem. 2010;82:2717–2725. doi: 10.1021/ac902455d. [DOI] [PubMed] [Google Scholar]
- 61.Young NL, Dimaggio PA, Garcia BA. Cell Mol Life Sci. 2010;67:3983–4000. doi: 10.1007/s00018-010-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hock A, Vousden KH. Int J Biochem Cell Biol. 2010;42:1618–1621. doi: 10.1016/j.biocel.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 63.Wu S, Lourette NM, Tolic N, Zhao R, Robinson EW, Tolmachev AV, Smith RD, Pasa-Tolic L. J Proteome Res. 2009;8:1347–1357. doi: 10.1021/pr800720d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.von Heijne G. Nature. 1998;396:111–113. doi: 10.1038/24036. [DOI] [PubMed] [Google Scholar]
- 65.Shen F, Kirmani KZ, Xiao Z, Thirlby BH, Hickey RJ, Malkas LH. J Cell Biochem. 2011;112:756–760. doi: 10.1002/jcb.23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sandilands A, Sutherland C, Irvine AD, McLean WH. J Cell Sci. 2009;122:1285–1294. doi: 10.1242/jcs.033969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai YS, Scherl A, Shaw JL, MacKay CL, Shaffer SA, Langridge-Smith PR, Goodlett DR. J Am Soc Mass Spectrom. 2009;20:2154–2166. doi: 10.1016/j.jasms.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 68.Taylor GK, Kim YB, Forbes AJ, Meng F, McCarthy R, Kelleher NL. Anal Chem. 2003;75:4081–4086. doi: 10.1021/ac0341721. [DOI] [PubMed] [Google Scholar]
- 69.Frank AM, Pesavento JJ, Mizzen CA, Kelleher NL, Pevzner PA. Anal Chem. 2008;80:2499–2505. doi: 10.1021/ac702324u. [DOI] [PubMed] [Google Scholar]
- 70.Shen Y, Tolic N, Liu T, Zhao R, Petritis BO, Gritsenko MA, Camp DG, Moore RJ, Purvine SO, Esteva FJ, Smith RD. PLoS One. 2010;5:e13133. doi: 10.1371/journal.pone.0013133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mason, EW EAM. Transport Protperties of Ion Gases. New York: Wiley; 1988. [Google Scholar]
- 72.Baker ES, Clowers BH, Li F, Tang K, Tolmachev AV, Prior DC, Belov ME, Smith RD. J Am Soc Mass Spectrom. 2007;18:1176–1187. doi: 10.1016/j.jasms.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baker ES, Livesay EA, Orton DJ, Moore RJ, Danielson WF, 3rd, Prior DC, Ibrahim YM, LaMarche BL, Mayampurath AM, Schepmoes AA, Hopkins DF, Tang K, Smith RD, Belov ME. J Proteome Res. 2010;9:997–1006. doi: 10.1021/pr900888b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Belov ME, Clowers BH, Prior DC, Danielson WF, 3rd, Liyu AV, Petritis BO, Smith RD. Anal Chem. 2008;80:5873–5883. doi: 10.1021/ac8003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shah AR, Agarwal K, Baker ES, Singhal M, Mayampurath AM, Ibrahim YM, Kangas LJ, Monroe ME, Zhao R, Belov ME, Anderson GA, Smith RD. Bioinformatics. 2010;26:1601–1607. doi: 10.1093/bioinformatics/btq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spiller DG, Wood CD, Rand DA, White MR. Nature. 2010;465:736–745. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 77.Toriello NM, Douglas ES, Thaitrong N, Hsiao SC, Francis MB, Bertozzi CR, Mathies RA. P Natl Acad Sci USA. 2008;105:20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bao SD, Wu QL, McLendon RE, Hao YL, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 79.Marginean I, Page JS, Tolmachev AV, Tang K, Smith RD. Anal Chem. 2010;82:9344–9349. doi: 10.1021/ac1019123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shen Y, Tolić N, Masselon C, Paša-Tolić L, Camp DG, Hixson KK, Zhao R, Anderson GA, Smith RD. Anal Chem. 2004;76:144–154. doi: 10.1021/ac030096q. [DOI] [PubMed] [Google Scholar]
- 81.Wang H, Qian WJ, Mottaz HM, Clauss TR, Anderson DJ, Moore RJ, Camp DG, 2nd, Khan AH, Sforza DM, Pallavicini M, Smith DJ, Smith RD. J Proteome Res. 2005;4:2397–2403. doi: 10.1021/pr050160f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang N, Xu M, Wang P, Li L. Anal Chem. 2010;82:2262–2271. doi: 10.1021/ac9023022. [DOI] [PubMed] [Google Scholar]
- 83.Hofstadler SA, Severs JC, Smith RD, Swanek FD, Ewing AG. Rapid Commun Mass Spectrom. 1996;10:919–922. doi: 10.1002/(SICI)1097-0231(19960610)10:8<919::AID-RCM597>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 84.Zhong JF, Chen Y, Marcus JS, Scherer A, Quake SR, Taylor CR, Weiner LP. Lab Chip. 2008;8:68–74. doi: 10.1039/b712116d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chiu DT, Lorenz RM, Jeffries GDM. Anal Chem. 2009;81:5111–5118. doi: 10.1021/ac900306q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mellors JS, Jorabchi K, Smith LM, Ramsey JM. Anal Chem. 2010;82:967–973. doi: 10.1021/ac902218y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kelly RT, Tang K, Irimia D, Toner M, Smith RD. Anal Chem. 2008;80:3824–3831. doi: 10.1021/ac8000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sun X, Kelly RT, Tang K, Smith RD. Analyst. 2010 doi: 10.1039/c0an00253d. Available on the Web. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelly RT, Page JS, Marginean I, Tang K, Smith RD. Angew Chem Int Ed Engl. 2009;48:6832–6835. doi: 10.1002/anie.200902501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rifai N, Gillette MA, Carr SA. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 91.Page JS, Kelly RT, Tang K, Smith RD. J Am Soc Mass Spectrom. 2007;18:1582–1590. doi: 10.1016/j.jasms.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 92.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth HR, Smith RD, Clemmer DE. Anal Chem. 2006;78:4161–4174. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.