Abstract
Many of the known xylose-fermenting (X-F) yeasts are placed in the Scheffersomyces clade, a group of ascomycete yeasts that have been isolated from plant tissues and in association with lignicolous insects. We formally recognize fourteen species in this clade based on a maximum likelihood (ML) phylogenetic analysis using a multilocus dataset. This clade is divided into three subclades, each of which exhibits the biochemical ability to ferment cellobiose or xylose. New combinations are made for seven species of Candida in the clade, and three X-F taxa associated with rotted hardwood are described: Scheffersomyces illinoinensis (type strain NRRL Y-48827T = CBS 12624), Scheffersomyces quercinus (type strain NRRL Y-48825T = CBS 12625), and Scheffersomyces virginianus (type strain NRRL Y-48822T = CBS 12626). The new X-F species are distinctive based on their position in the multilocus phylogenetic analysis and biochemical and morphological characters. The molecular characterization of xylose reductase (XR) indicates that the regions surrounding the conserved domain contain mutations that may enhance the performance of the enzyme in X-F yeasts. The phylogenetic reconstruction using XYL1 or RPB1 was identical to the multilocus analysis, and these loci have potential for rapid identification of cryptic species in this clade.
Introduction
D-xylose is a five-carbon backbone molecule of the hemicellulose component of plant cell walls and is one of the most abundant renewable carbon resources on Earth. Some bacteria and certain fungi, including fewer than twenty-five species of the more than 1500 described ascomycete yeasts, share the ability to produce ethanol by the fermentation of D-xylose [1]. In order to ferment D-xylose, yeasts express xylose reductase (XR), xylitol dehydrogenase (XDH), and xylulose kinase (XK) to convert D-xylose to D-xylulose-5-phosphate; D-xylulose-5-phosphate is then incorporated into the pentose phosphate pathway to be catalyzed to ethanol [2]–[4].
Xylose fermentation has been the focus of several studies in order to identify differences in the catabolic rate between strains [2], [5]-[14]. Overexpression, homologous and heterologous expression, and direct mutagenesis of genes involved in D-xylose assimilation and fermentation have only modestly enhanced the quantity of ethanol production by yeasts due to several metabolic constraints; these include rate of regeneration of the cofactor NADP(H) required by XR and XDH, repression by glucose, and anaerobic respiration regulatory control [15]-[17]. These studies have resulted in the present understanding of the biochemical pathway, but the main goal of bioengineering yeasts capable of fermenting D-xylose at a high rate to be used at industrial scales has not yet been achieved. Consequently, recent research has been focused on the discovery of new X-F yeasts, e.g. Spathaspora passalidarum and Candida jeffriesii [18], and Spathaspora arborariae and other taxa [19], [20], from the guts of lignicolous beetles and rotted wood, niches from which a number of X-F yeasts have been isolated [21]-[23].
Although, X-F yeasts appear scattered throughout the Saccharomycotina, the yeasts that have been reported to exhibit the highest rate of xylose fermentation under certain conditions are members of the Scheffersomyces clade [9], [21], [24], and for this reason fermentative ability has been intensively studied in this clade [9], [25]-[51]. Few studies, however, have undertaken clarification of the phylogenetic relationships among the yeasts of the clade [1], [52]-[58]. Therefore, a robust, well-supported phylogeny including as many taxa as possible is necessary to clarify the phylogenetic relationships among the Scheffersomyces clade members along with a comparison of the nucleotide mutations and enzymatic activity of the XR to understand the importance of the biochemical ability in the speciation process of this yeast clade.
In order to distinguish the species in the Scheffersomyces clade we used BLAST searches, biochemical and morphological characterization, and a multilocus phylogenetic analysis that included the traditional SSU and LSU markers, the orthologous RPB1, and the recently proposed ITS barcoding region for fungi [59]. We present a taxonomic revision of the Scheffersomyces clade, trace the nucleotide differences in the XYL1, and propose three new species of X-F yeasts, Scheffersomyces illinoinensis, Scheffersomyces quercinus, and Scheffersomyces virginianus associated with rotted hardwoods, Carya illinoinensis (pecan), Quercus nigra (water oak), and Quercus virginiana (live oak). We performed a molecular study of the XYL1 that codifies XR in certain members of the Scheffersomyces clade.
Materials and Methods
Yeast Isolation and Culture
Partially decayed logs and fallen branches of Carya illinoinensis (pecan, 30 cm diam), Quercus nigra (water oak, approximately 10 cm diam), and Quercus virginiana (live oak, approximately 10 cm diam) were collected from Pecan Drive, Saint Gabriel, Ascension Parish, Louisiana, and LSU Burden Center and the corner of Highland Road and S. Stadium Drive on the LSU campus, Baton Rouge, East Baton Rouge, Louisiana, respectively, between Sep and Oct 2007. The wood samples were divided into approximately 1 cm2 samples, and each sample was placed in a 1.5 mL microcentrifuge tube with 1 mL of sterile water. Tubes were vortexed for 30 s and a 100 µL aliquot was plated on acidified yeast medium agar [60]. Plates were incubated for 3 d at 25 C, and single colonies were isolated and streaked 4 times to obtain pure cultures.
Molecular Identification
Genomic DNA was extracted using a Wizard® Genomic DNA purification kit (Promega). The concentration, integrity, and purity of total DNA extracted were confirmed by gel electrophoresis in 0.8% agarose in 0.5 × Tris-Borate-EDTA (TBE) buffer. Initial rapid identification was carried out by PCR amplification and sequencing of the LSU (D1/D2 region ∼600 bp) rRNA gene for use in BLAST searches [1], [54]. In order to increase the robustness of the phylogenetic analyses, PCR amplifications of the small subunit (SSU ∼1.6 Kbp) and internal transcribed spacers 1 and 2 (ITS ∼500 bp) of the rRNA marker were carried out in addition to the D1/D2 region [61], [62] and the SSU rRNA gene was amplified using the combination of primers NS1 (forward) (5′-GTAGTCATATGCTTGTCTC-3′) and NS8 (reverse) (5′-TCCGCAGGTTCACCTACGGA-3′); ITS1-LSU markers were amplified using the combination of primers ITS1 (forward) (5′-TCCGTAGGTGAACCTGCGG-3′) and LR3 (reverse) (5′-CCGTGTTTCAAGACGGG-3′) in a PCR reaction with 20 µg of total DNA, 0.5 mM DTPs, 2.5 mM MgSO4, 0.8 µM of each primer set, 1 × PCR buffer, and 1 U of Taq polymerase (Promega) in 25 µL of final volume. The PCR amplification protocol included 5 min of DNA pre-denaturation at 95 C followed by 35 cycles of 1 min of DNA denaturation at 95 C, 45 s of primer annealing at 55 C, and a 2 min extension at 72 C, and 10 min final PCR extension.
In addition, another nuclear locus, RNA polymerase II, was used in the phylogenetic analysis. A fragment of ∼700 bp of the subunit I (RPB1) gene was amplified by the primer pair RPB1-Af (forward) 5′-GARTGYCCDGGDCAYTTYGG-3′ and RPB1-Cr (reverse) 5′-CCNGCDATNTCRTTRTCCATRTA-3′ [63], [64]. The PCR reaction was performed using 100 µg of total DNA, 0.6 mM DTPs, 2.5 mM MgSO4, 1 µM of each primer, 1 × PCR buffer, and 1.5 U of Taq polymerase (Promega) in 35 µL total final volume of reaction. The PCR amplification program included 5 min of DNA pre-denaturation at 95 C followed by 35 cycles of 1 min of DNA denaturation at 95 C, 45 s of primer annealing at 55 C, and 2 min of extension at 72 C, and 10 min final PCR extension.
The purified PCR products were sequenced in both directions by Beckman Coulter Genomics (Danvers, MA). Each molecular marker was sequenced on three independent occasions in order to avoid nucleotide differences due to sequencing errors.
RAPD-PCR Fingerprinting
RAPD–PCR analysis was performed with the oligonucleotide primer CDU (5′-GCGATCCCCA-3′) [65]-[68]. Aliquots of 25 µL of amplified product were analyzed by electrophoresis on 1.8% agarose gel in 1× TBE buffer with 1 × SYBR® Safe DNA Gel Stain (Invitrogen, Grand Island, NY) at 70 V for 80 min. DNA fragments were visualized with a UV-light transilluminator and photographed using a Polaroid system.
Morphological, Biochemical and Physiological Characteristics
The yeast standard description based on phenotypic characters was executed following standardized protocols [1], [69], [70].
Xylose Reductase (XR) Molecular Studies
The ∼600 bp fragment of the XYL1 was amplified using the following degenerate primers: XYL1-forward (5′-GGTYTTYGGMTGYTGGAARSTC-3′) and XYL1-reverse (5′-AAWGATTGWGGWCCRAAWGAWGA-3′) designed in this study, in a PCR reaction with 100 µg of total DNA, 0.4 mM DTPs, 4 mM MgSO4, 1 µM of each primer, 1× PCR buffer, and 1U of Taq polymerase (Promega) in a final volume of 25 µL. The PCR amplification program included 5 min of DNA pre-denaturization at 95 C followed by 35 cycles of 1 min of denaturization at 95 C, 1 min primer annealing at 57 C, 1 min extension at 72 C, and 10 min final PCR extension. The purified PCR products were sequenced as described above.
Phylogenetic Analyses
Contig sequences and sequencing manipulations were performed with Se-AL v2.01a11 (http://tree.bio.ed.ac.uk/software/seal/) and MESQUITE v2.74 [71]. The sequence alignments were carried out using the online interface MAFFT v6.859 (http://mafft.cbrc.jp/alignment/software/) with different advanced alignment strategies per locus: LSU and RPB1, global homology (G-INS-i); ITS, one conserved domain (L-INS-i); and SSU, secondary structure of RNA (Q-INS-i). In particular ITS loci were realigned using the software SATé v2.1.2 [72], and ambiguous sequence alignment ends were eliminated in all the alignments. Maximum likelihood (ML) phylogenic inference was performed in RAxML-VI-HPC [73] using a partitioned multilocus matrix (each partition for SSU, ITS1, 5.8 S, ITS2, LSU, and RPB1) under a general time reversible model with a gamma distribution of site rate variation (GTRGAMMA), and ML support was estimated using 1000 bootstrap replicates. The sequences for RPB1 and XR were obtained using SEQUIN v11.0 (http://www.ncbi.nlm.nih.gov/Sequin/) with the alternative yeast nuclear codon bias [45], [74]. Phylogenetic analysis using the amino acid matrix was performed in RAxML-VI-HPC under the VT AA selection model [75] and ML support was estimated using 1000 bootstrap replicates. Tree editing was done with FigTree v1.3.1 software (http://tree.bio.ed.ac.uk/software/figtree/).
Results
Diversity of Yeasts Isolated from Rotted Wood
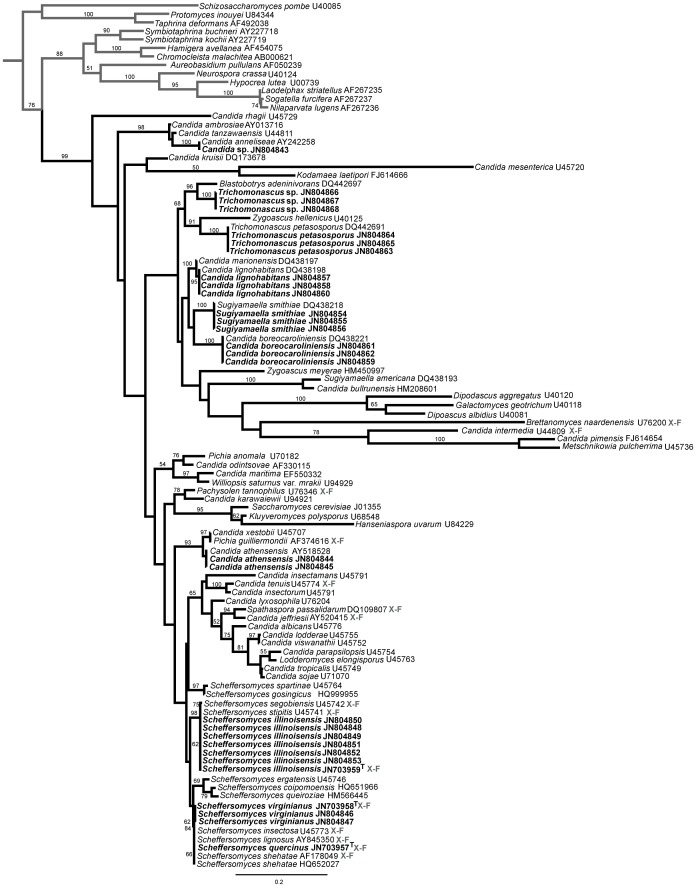
The initial rapid molecular identification of the 29 yeast strains isolated from the wood samples using the D1/D2 LSU region confirmed the presence of several closely related species classified in the Scheffersomyces (11 isolates), Sugiyamaella (10 isolates), Trichomonascus (5 isolates), Meyerozyma (2 isolates), and Candida tanzawaensis (1 isolate) clades (Fig. 1). Initial biochemical characterization of fermentation abilities performed on all the isolates showed that only strains classified in the Scheffersomyces clade had the ability to ferment xylose.
Figure 1. ML tree based of the D1/D2 LSU region using a 606-character matrix for yeast species isolated from the wood samples (in bold).
Schizosaccharomyces pombe was used as an outgroup taxon (in grey). X-F, xylose-fermenting yeasts. Numbers above each branch refer to bootstrap values out of 1000 repetitions. ML score -11353.90.
New Xylose-fermenting Yeasts
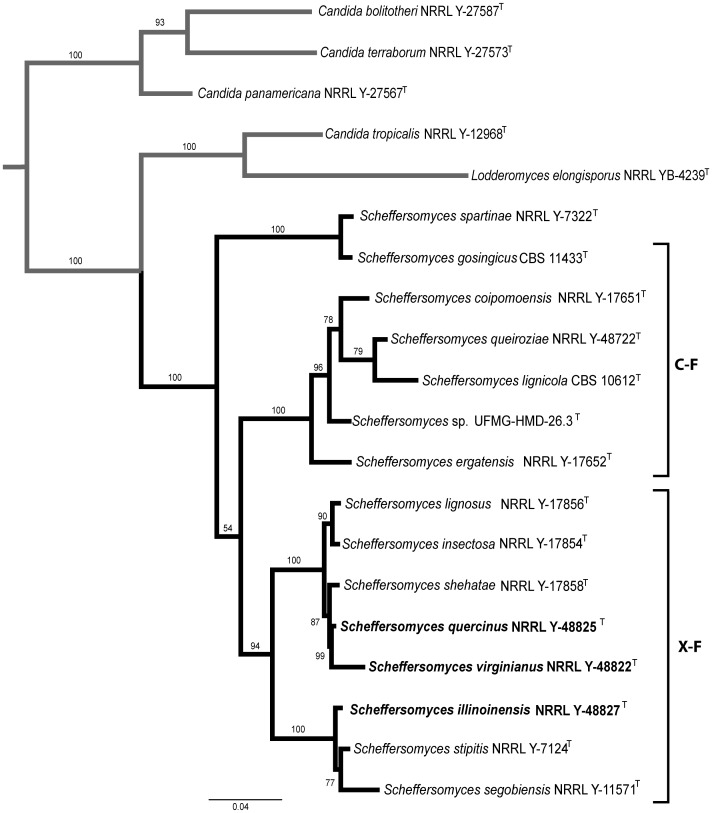
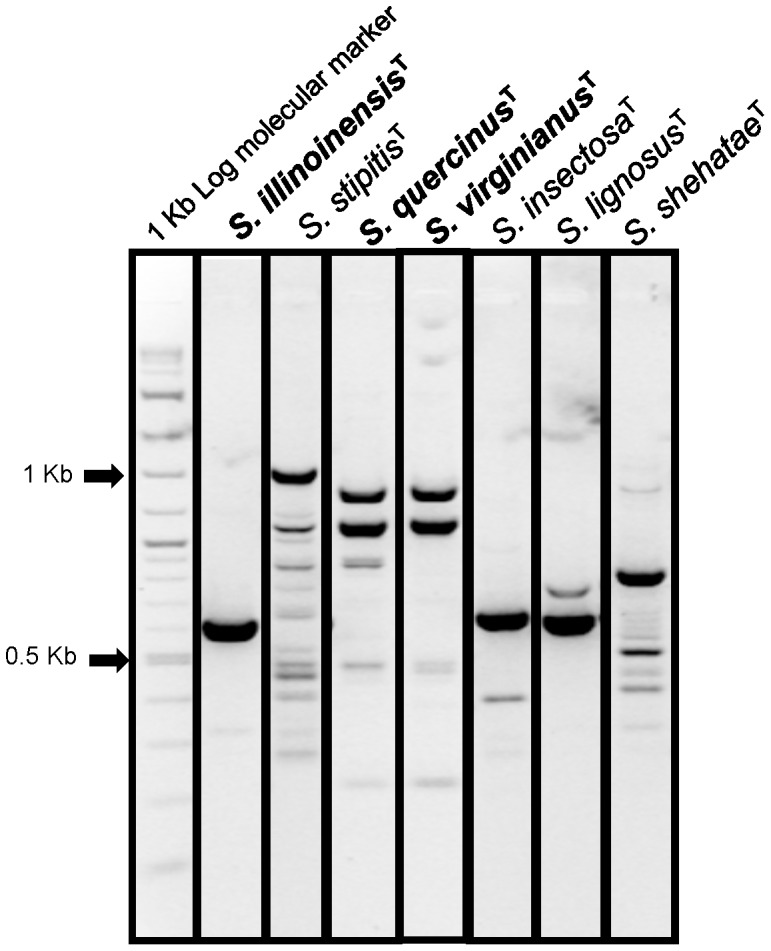
Species delimitation of S. quercinus, S. virginianus, and S. illinoinensis, was based on phylogenetic placement, nucleotide differences in the rRNA markers (SSU, ITS and LSU), RPB1 and XYL1, and biochemical assay differences compared with their closest relatives (Table 1, 2 and 3, Fig. 2). In addition, we obtained different banding patterns using CDU RAPID-PCR fingerprinting primers (Fig. 3). These fingerprinting primers have been used previously to discriminate among cryptic yeast species [66], [67], [76].
Table 1. GenBank accession numbers of the nucleotide sequences used in this study. Sequences generated in this work shown in bold.
| Species | Codes | SSU | ITS | LSU | RPB1 | XYL1 |
| C. bolitotheri T | NRRL Y-27587, CBS 9832, BG 00-8-15-1-1 | AY242142 | FJ623599 | AY242249 | JN804828 | - |
| C. terraborum T | NRRL Y-27573, CBS 9826, BG 02-7-15-019A-2-1 | AY426956 | FJ623596 | AY309810 | JN804831 | - |
| C. panamericana T | NRRL Y-27567, CBS 9834, BG 01-7-26-006B-2-1 | AY426960 | FJ623601 | AY309872 | JN804835 | - |
| S. coipomoensis T | NRRL Y-17651, ATCC 58904, CBS 8178 | HQ651931 | HQ652070 | HQ651966 | EU344070 | - |
| S. lignicola T | ATCC MYA-4674, CBS 10610, BCC 7733 | AY845351 | HQ652074 | AY845350 | - | - |
| S. ergatensis T | NRRL Y-17652, ATCC 22589, CBS 6248 | AB013524 | EU343826 | U45746 | EU344098 | JQ436926 |
| S. insectosa T | NRRL Y-12854, ATCC 66611, CBS 4286 | AB013583 | HQ652064 | FM200041 | JN804842 | JQ235697 |
| S. lignosus T | NRRL Y-12856, ATCC 58779, CBS 4705 | HQ651941 | JN943262 | U45772 | JN804837 | JQ235693 |
| S. segobiensis T | NRRL Y-11571, ATCC 58375, CBS 6857 | AB054288 | DQ409166 | U45742 | EF599429 | JQ436925 |
| L. elongisporus T | NRRL YB-4239, ATCC 11503, CBS 2605 | HQ876033 | HQ876042 | HQ876050 | AY653537 | - |
| C. tropicalis T | NRRL Y-12968, ATCC 4563, CBS 616 | EU348785 | AB437068 | U45749 | - | - |
| S. queiroziae T | NRRL Y-48722, UFMG-CLM 5.1, CBS 11853 | - | HM566445 | HM566445 | - | - |
| S. gosingicus T | CBS-11433, BCRC 23194, SJ7S11 | HQ876040 | HQ999978 | HQ999955 | - | - |
| S. spartinae T | NRRL Y-7322, ATCC 18866, CBS 6059 | FJ153139 | HQ876044 | U45764 | - | - |
| S. stipitis T | NRRL Y-7124, ATCC 58376, CBS 5773 | AB054280 | JN943257 | U45741 | JN804841 | JQ235696 |
| Scheffersomyces sp. | NRRL Y-48762, CBS 12363, UFMG HMD-26.3 | - | JF826438 | JF826438 | - | - |
| S. shehatae T | NRRL Y-12858, CBS 5813, ATCC 34887 | AB013582 | JN943264 | AF178049 | JQ436927 | JQ235691 |
| S. quercinus T | NRRL Y-48825T, CBS 12625, W07-09-15-1-3-2 | JN940981 | JN943260 | JN703957 | JN804838 | JQ008829 |
| S. virginianus T | NRRL Y-48822T, CBS 12626, W07-10-04-4-6-2 | JN940969 | JN943259 | JN703958 | JN804839 | JQ235695 |
| S. illinoinensis T | NRRL Y-48827T, CBS 12624, W07-11-15-9-2-1 | JN940968 | JN943261 | JN703959 | JN804840 | JQ235694 |
Table 2. Nucleotide differences and percentages of homology between the new xylose-fermenting yeasts and the type cultures of closest relatives, S. shehatae or S. stipitis.
| Species | SSU | ITS | D1/D2 LSU | RPB1 | XYL1 |
| S. insectosa T | 100% | 98% (5 n) | 100% | 94% (34 n) | 92% (44 n) |
| S. lignosus T | 100% | 98% (5 n) | 100% | 94% (37 n) | 91% (46 n) |
| S. quercinus T | 98% (27n) | 99% (1 n) | 98% (7 n) | 98% (10 n) | 96% (11 n) |
| S. virginianus T | 97% (54n) | 99% (1 n) | 99% (4 n) | 98% (10 n) | 97% (8 n) |
| S. illinoinensis T | 99% (3n) | 99% (3 n) | 100% | 95% (28 n) | 97% (15 n) |
Table 3. Differences in physiological reactions of S. quercinus, S. illinoinensis, and S. virginianus and their closest relatives*.
| Species | Rhammose | Galactitol | Lactate | Cadaverine | Glucosamine | Tryptophan | Melibiose | Inulin |
| S. insectosa | - | - | - | + | - | - | - | - |
| S. lignosus | - | + | w | + | - | - | - | - |
| S. quercinus | - | +,f | + | + | + | + | - | + |
| S. shehatae | - | - | - | + | - | - | - | - |
| S. virginianus | w | w | - | - | + | + | + | + |
| S. stipitis | + | - | +/− | + | - | - | - | - |
| S. illinoinensis | + | +/d | +/d | w | + | + | w | w |
Biochemical assay results of previously described species were complied from Kurtzman (1990) and Barnett et al. (2000). Abbreviations for reaction results: +, positive; -, negative; d, delayed positive; w, weak positive.
Figure 2. ML tree based on a multilocus dataset using a 3488-character matrix for Scheffersomyces clade.
Candida tropicalis was used as an outgroup taxon (in grey). C-F and X-F, cellobiose- and xylose-fermenting yeasts respectively. Numbers above each branch refer to bootstrap values out of 1000 repetitions. ML score is -13300.52.
Figure 3. Characterization of S. quercinus, S. illinoinensis, and S. virginianus using RAPID-PCR CDU fingerprinting primers.
Species Description of Scheffersomyces quercinus H. Urbina & M. Blackw. sp. nov
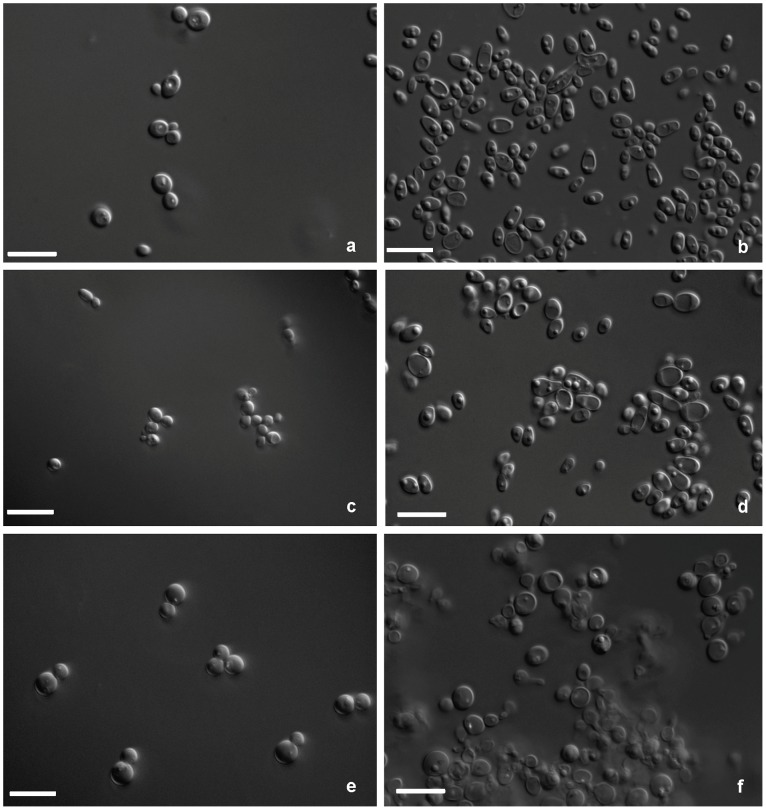
MycoBank accession number MB563719, (Fig. 4, a-b).
Figure 4. Morphological characterization of S. quercinus, budding cells (a-b); S. virginianus, budding cells (c-d); and S. illinoinensis, budding cells (e-f); grown at 25 C in YM broth and V8 agar at 7 d days respectively, bar 10 µm.
After 7 d growth in YM broth at 25 C, cells are subglobose (5–8×5–7.5 µm), and occur singly, in pairs, or in chains. Pseudohyphae are present; true hyphae are absent. After 7 days on YM agar at 25 C, colonies are cream-colored with pale-pinkish perimeter on some older colonies, smooth, flat, and/or with scattered filaments at the margin. After 10 d of Dalmau plate culture on corn meal agar at 25 C, true hyphae are present. Aerobic growth is white, shiny, and smooth with filamentous margin. Asci and ascospores are not observed on YM or V8 agar. Diazonium blue B reaction is negative. See Table 4 for physiological characterization.
Table 4. Biochemical characterization of S. quercinus, S. illinoinensis, and S. virginianus.
| S. quercinus | S. illinoinensis | S. virginianus | ||
| Fermentation | ||||
| F1 | D-Glucose | + | + | +, f |
| F2 | D-Galactose | + | + | + |
| F3 | Maltose | w | + | + |
| F4 | α-Methyl-D-glucoside | - | - | - |
| F5 | Sucrose | - | - | - |
| F6 | α,α- Trehalose | + | + | + |
| F7 | Melibose | - | w | - |
| F8 | Lactose | - | - | - |
| F9 | Cellobiose | - | - | - |
| F10 | Melezitose | - | - | - |
| F11 | Raffinose | - | - | - |
| F12 | Inulin | - | - | - |
| F13 | Starch | - | - | - |
| F14 | D-Xylose | + | + | + |
| Assimilation | ||||
| C1 | D-Glucose | +, f | +, f | + |
| C2 | D-Galactose | +, f | +, f | +, f |
| C3 | L-Sorbose | +,d | - | w |
| C4 | D-Glucosamine | w | - | + |
| C5 | D-Ribose | +,d | +,d | +, d |
| C6 | D-Xylose | + | + | + |
| C7 | L-Arabinose | +,d | + | + |
| C8 | D-Arabinose | w | + | w |
| C9 | L-Rhamnose | - | + | w |
| C10 | Sucrose | + | + | + |
| C11 | Maltose | + | +, f | + |
| C12 | Trehalose | + | +, f | + |
| C13 | α-Methyl-D-glucoside | + | + | + |
| C14 | Cellobiose | + | + | + |
| C15 | Salicin | + | + | + |
| C16 | Arbutin | + | + | + |
| C17 | Melibiose | - | + | + |
| C18 | Lactose | - | - | + |
| C19 | Raffinose | - | - | w |
| C20 | Melezitose | - | + | +, d |
| C21 | Inulin | w | w | + |
| C22 | Soluble Starch | + | + | + |
| C23 | Glycerol | + | + | + |
| C24 | Erythritol | + | + | + |
| C25 | Ribitol | + | + | + |
| C26 | Xylitol | +, d | + | w |
| C27 | L-Arabinitol | +, d | +,d | - |
| C28 | D-Glucitol | + | + | + |
| C29 | D-Mannitol | + | + | + |
| C30 | Galactitiol | +, f | +, f | w |
| C31 | myo-Inositol | - | - | w |
| C32 | D-Glucono-1,5-lactone | + | + | + |
| C33 | 2-Keto-D-gluconate | + | +,f | +, f |
| C34 | 5-Keto-D-gluconate | ? | ? | ? |
| C35 | D-Gluconate | + | +,d | + |
| C36 | D-Glucuronate | - | - | - |
| C38 | DL-Lactate | + | +,d | - |
| C39 | Succinate | + | + | + |
| C40 | Citrate | +, d | + | + |
| C41 | Methanol | - | - | - |
| C42 | Ethanol | +, f | +,f | +, f |
| C43 | Propane 1,2 diol | +, d | w | w |
| C44 | Butane 2,3 diol | w | - | w |
| C45 | Quinic acid | - | - | w |
| C46 | D-Glucarate | - | - | - |
| Temperature | ||||
| T1 | 30°C | + | + | + |
| T2 | 35°C | +, f | +,f | + |
| T3 | 37°C | - | +,d | - |
| Osmotic pressure | ||||
| O1 | 0.01% Cycloheximide | +, f | +, f | +, f |
| O2 | 0.1% Cycloheximide | +, f | +, f | +,f |
| O3 | 1% Acetic Acid | - | - | - |
| O4 | 50% D-Glucose | + | + | + |
| O5 | 60% D-Glucose | w | + | w |
| O6 | 10% NaCl | + | + | w |
| O7 | 16% NaCl | - | - | - |
| Nitrogen assimilation | ||||
| N1 | Nitrate | - | +, d | + |
| N2 | Nitrite | - | +, d | - |
| N3 | Ethylamine | + | + | + |
| N4 | L-Lysine | +, d | - | + |
| N5 | Cadaverine | + | w | - |
| N6 | Creatine | - | - | - |
| N7 | Creatinine | - | - | - |
| N8 | D-Glucosamine | + | + | + |
| N9 | Imidazole | - | - | - |
| N10 | D-Tryptophan | + | + | + |
| Vitamins | ||||
| V1 | w/o Vitamins | - | - | - |
| V2 | w/o myo-Inositiol | +, d | w | + |
| V3 | w/o Pantothenate | + | + | + |
| V4 | w/o Biotin | + | - | - |
| V5 | w/o Thiamin | + | + | + |
| V6 | w/o Biotin & Thiamin | - | - | w |
| V7 | w/o Pyridoxine | + | + | + |
| V8 | w/o Pyrid. &Thiam | + | + | + |
| V9 | w/o Niacin | + | + | + |
| V10 | w/o PABA | + | + | + |
Abbreviations for reaction results: +, positive; -, negative; d, delayed positive; w, weak positive.
Type strain
NRRL Y-48825T ( = CBS 12625; W07-09-15-1-3-2) is preserved as a lyophilized preparation in the Agricultural Research Service Culture Collection (NRRL), Peoria, Illinois, USA. The strain was isolated from rotted wood (Quercus niger), collected on 15 Nov 2007 at Louisiana State University, Burden Research Plantation, Baton Rouge, East Baton Rouge Parish, Louisiana, USA, N30°40′59″-W91°10′29″.
Etymology
The species name quercinus (N.L. gen. n.) refers to the genus of the substrate, Quercus niger, from which this species was isolated.
Species Description of Scheffersomyces virginianus H. Urbina & M. Blackw. sp. nov
MycoBank accession number MB 563720 (Fig. 4, c-d).
After 7 d growth in YM broth at 25 C, cells are globose to ellipsoidal (5.5–10×4–6.5 µm), and occur singly, in pairs, in short chains, or in small clusters. Pseudohyphae are present; true hyphae are absent. After 7 d on YM agar at 25 C, cream-colored to light pink with abundant filaments at margin. After 10 d Dalmau plate culture on corn meal agar at 25 C, pseudohyphae are present; septate hyphae are absent. Aerobic growth is white, shiny, and smooth with filaments at margin. Asci and ascospores are not observed on YM or V8 agar. Diazonium blue B reaction is negative. See Table 4 for physiological characterization.
Type Strain
NRRL Y-48822T ( = CBS 12626; W07-10-04-4-6-2) is preserved as a lyophilized preparation in the Agricultural Research Service Culture Collection (NRRL), Peoria, Illinois, USA. The strain was isolated from rotted wood (Quercus virginiana) collected on 15 Sep 2007, Highland Rd. and S. Stadium Dr., Louisiana State University, Baton Rouge, East Baton Rouge Parish, Louisiana, USA, N30°40′90″-W91°17′60″.
Etymology
The species name virginianus (N.L. gen. n.) refers to the species of the substrate, Quercus virginiana, from which this species was isolated.
Species Description of Scheffersomyces illinoinensis H. Urbina & M. Blackw. sp. nov
MycoBank accession number MB563457 (Fig. 4, e-f).
After 7 d growth in YM broth at 25 C, cells are globose (5−7.5 µm), and occur singly, in pairs, or in short chains. Pseudohyphae and hyphae are not present. After 7 d on YM agar at 25 C, colonies are cream-colored, smooth, and flat with a smooth margin. After 10 d Dalmau plate culture on corn meal agar at 25 C, pseudohyphae are present; septate hyphae are absent. Asci and ascospores are not observed on YM and V8 agar. Diazonium blue B reaction is negative. See Table 4 for physiological characterization.
Type strain
NRRL Y-48827T ( = CBS 12624; W07-11-15-9-2-1) is preserved as a lyophilized preparation in the Agricultural Research Service Culture Collection (NRRL), Peoria, Illinois, USA. The strain was isolated from rotted wood (Carya illinoinensis) collected on 15 Nov 2007 at 810 Pecan Dr., St. Gabriel, Ascension Parish, Louisiana, USA, N30°30′56″-W91°10′30″.
Etymology
The species name illinoinensis (N.L. gen. n.) refers to the species of the substrate, Carya illinoinensis, from which this species was isolated.
Multilocus Phylogenetic Study
The phylogenetic placement of S. quercinus, S. virginianus, and S. illinoinensis was based on ML analysis results of a concatenated nucleotide dataset containing 3488 characters (Fig. 2). The Scheffersomyces clade was divided into three subclades: 1) the early diverging S. spartinae and S. gosingicus subclade, 2) the cellobiose-fermenting S. ergatensis subclade, and 3) the largest, xylose-fermenting S. stipitis subclade to which the three new species belong.
Addition of more taxa and more molecular data in phylogenetic analyses has helped to define monophyletic clades, among a number of genera now recognized as polyphyletic. One problematic taxon, Pichia, was based primarily on ascospore shape. Hat-shaped ascospores, however, are found among several distant clades of yeasts as well as distantly related members of the Pezizomycotina. Scheffersomyces was proposed recently for species in the Pichia stipitis clade [55]. The genus included the type species, S. stipitis, and S. ergatensis and S. spartinae [55]. We propose additional new combinations in the genus Scheffersomyces by including clade members that previously were described as asexual species of the polyphyletic genus Candida [77].
Scheffersomyces coipomoensis (C. Ramírez & A. González) H. Urbina & M. Blackw. comb. nov.
MycoBank accession number: MB563714.
Basionym: Candida coipomoensis C. Ramírez & A. González, Mycopathologia 88:84, 1984.
Scheffersomyces gosingicus (C.F. Lee) H. Urbina & M. Blackw. comb. nov.
MycoBank accession number: MB563799.
Basionym: Candida gosingica C.F. Lee, Int J Syst Evol Microbiol 61:670, 2011.
Scheffersomyces insectosa (Kurtzman) H. Urbina & M. Blackw. comb. nov.
MycoBank accession number: MB 564799.
Basionym: Candida shehatae var. insectosa Kurtzman, Antonie van Leeuwenhoek 57:218, 1990. Synonym: Candida insectosa (Kurtzman) Kurtzman, in Kurtzman & M. Suzuki, Mycoscience 51:10, 2010.
Scheffersomyces lignicola (Jindam., Limtong, Yongman., Tuntir., Potach., H. Kawas & Nakase) H. Urbina & M. Blackw. comb. nov.
MycoBank accession number: MB 563803.
Basionym: Candida lignicola Jindam., Limtong, Yongman., Tuntir., Potach., H. Kawas & Nakase, FEMS Yeast Res 7:1412, 2007.
Scheffersomyces lignosus (Kurtzman) H. Urbina & M. Blackw. comb. nov.
MycoBank accession number: MB 563801.
Basionym: Candida shehatae var. lignosa Kurtzman, Antonie van Leeuwenhoek 57:218, 1990. Synonym: Candida lignosa (Kurtzman) Kurtzman, in Kurtzman & M. Suzuki, Mycoscience 51:10, 2010.
Scheffersomyces queiroziae (R.O. Santos, R.M. Cadete, Badotti, A. Mouro, Wallheim, F.C.O. Gomes, Stambuk, Lachance & C.A. Rosa) H. Urbina & M. Blackw. comb. nov.
MycoBank accession number: MB563717.
Basionym: Candida queiroziae R.O. Santos, R.M. Cadete, Badotti, A. Mouro, Wallheim, F.C.O. Gomes, Stambuk, Lachance & C.A. Rosa, Antonie van Leeuwenhoek 99:639. 2011.
Scheffersomyces shehatae (H.R. Buckley & van Uden) H. Urbina & M. Blackw. comb. nov.
MycoBank accession number: MB563716.
Basionym: Candida shehatae H.R. Buckley & van Uden, Mycopathologia Mycol. appl. 34:297, 1966.
Xylose reductase among X-F yeast members of the S. stipitis subclade
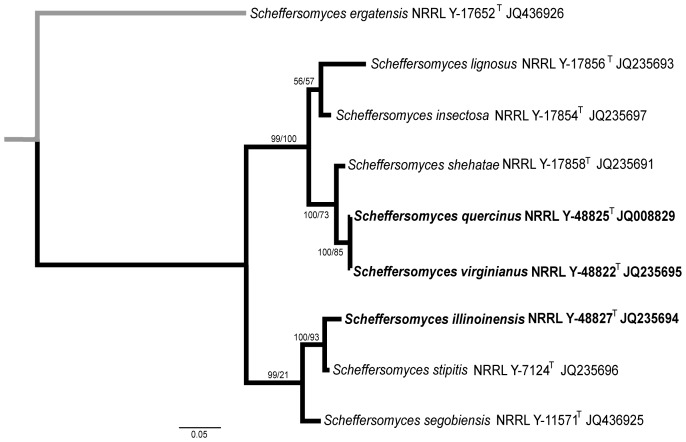
We amplified a ∼600 bp PCR product of XYL1 from X-F and non X-F yeasts tested (Fig. 5). The translated protein sequences at the N-terminal region have the conserved amino acids 49-D, 51-A, and 54-Y, described as part of the catalytic GX3DXAX2Y domain; the LX8DX4H and the GX3GXG domains, and the amino acids 83-K, 132-P, and 167-K that form the xylose-binding pocket previously reported [78], [79] (Fig. 6). Conserved amino acid substitutions at positions 64, 170, 194 (I = V); 65 (N = K); 69 (K = D); 70 (D = E); 80, 178 (I = L); 107 (L = V); 127 (N = E); 149 (L = I), 150 (A = L), 174 (T = P), 179 (Y = L), 184 (S = G), 186 (E = T), 188 (P = K), and 204 (S = P) and the percentage of conserved substitutions usually were higher than 50% at all sites compared with the non X-F fermenting yeast S. ergatensis (Fig. 6).
Figure 5. ML consensus tree based on XYL1 and the putative XR from of X-F members in S. stipitis subclade.
Scheffersomyces ergatensis was used as an outgroup taxon (in grey). Numbers above each branch refer to bootstrap values out of 1000 repetitions. ML scores are -1829.90 (DNA data) and -1284.79 (amino acid data).
Figure 6. Multiple sequence alignment of the N-terminal region of XR.
Identical residues are indicated by dots. Arrows indicate amino acids that constitute the active sites, stars indicate amino acids that form the xylose-binding pocket, and brackets indicate conserved domains.
Discussion
Yeasts Sampled from Rotted Wood
Most of the isolates from live oak and water oak were members of the Sugiyamaella clade: Candida boreocaroliniensis, Candida lignohabitans, and S. smithiae. The cosmopolitan genus Sugiyamaella is comprised of yeasts reported primarily from wood and frass of lignicolous beetles. It is of interest that, unlike other yeasts from these habitats, they are unable to ferment D-xylose [80], [81]. We also isolated other non X-F species Candida athensensis, Trichomonascus petasosporus, and a close relative of Candida anneliseae, ascomycete yeasts that previously were found associated with fungus-feeding beetles collected in Panama and the USA [56].
The yeast strains isolated from pecan wood, on the contrary, were dominated by the new X-F yeast species, S. illinoinensis, a close relative of S. stipitis. Only this wood was inhabited by Odontotaenius disjunctus (Passalidae, Coleoptera) in our study. Zhang et al. [82] recognized the relationship between S. stipitis and the lignicolous beetle commonly found inhabiting decayed hardwoods in the southeastern US, so the phylogenetic placement of these closely related species is consistent with the previous findings.
Other members of the Scheffersomyces clade (S. ergatensis, S. shehatae, and S. stipitis) have been reported frequently from associations with the gut of lignicolous beetles, including, not only the passalid beetle O. disjunctus but also beetles in the families Cerambycidae, Lucanidae, Buprestidiae, and Tenebrionidae [22], [83]. It is likely that the common gut yeasts are efficient at digesting components of the host diet, resisting toxic secondary metabolites, and adapting to the gut physiological environment (low oxygen and high carbon dioxide concentrations and extreme pH variation), characteristics that give them a greater chance to be horizontally transmitted to progeny. Consequently, in each host generation the symbiotic yeasts may be exposed to bottlenecks and positive selection driven by the host beetles, and these selective pressures increase when changes in the host diet occur. These evolutionary processes could have favored rapid speciation with morphological and other traditional characters lagging behind molecular changes in the Scheffersomyces yeast members. Evidence that supports our hypothesis is: 1) S. shehatae, S. lignicola, and S. insectosa, often found in association with insects, are indistinguishable by morphology and some molecular markers (e.g. SSU and D1/D2 LSU); 2) branch lengths are constrained in the phylogenetic tree (Fig. 2); and 3) gut morphology is modified to enhance the horizontal transmission of gut yeasts across generations, e.g. the posterior hindgut region of O. disjunctus is colonized mainly by filamentous yeasts attached by a holdfast [84]; in addition mycetomes occur in lignicolous cerambycid beetles colonized exclusively by closely related species of S. shehatae [22].
Kurtzman [58] recognized Candida shehatae var. shehatae, var. insectosa, and var. lignicola based on biochemical assays, a single nucleotide difference in the D1/D2 LSU region, and identical SSU rRNA among the varieties. He suggested that in order to understand the phylogenetic relationship between varieties of C. shehatae, analyses including ITS rRNA should be included. More recently, these varieties were raised to species level based on their distinctive electrokariotype profiles and the reinterpretation of the D1/D2 LSU locus [55], [57]. Yeast species often have been underestimated on the basis of only LSU and SSU rRNA data [85]-[87]. Therefore, the addition of more molecular markers in phylogenetic analyses has been used to increase the power of species recognition (see next section).
Phylogenetic Study of the Scheffersomyces Clade
Lachance et al. [88] proposed a method for species delimitation in yeasts based on parsimony networks. In our experience, the results obtained by using this method are difficult to interpret for several reasons: 1) the species are plotted as isolated entities with little information on phylogenetic relationship among members; 2) the method does not allow inclusion of a model of nucleotide selection for the analysis; and 3) node support values are lacking.
Because of these disadvantages, we implemented, instead, a multilocus phylogenetic analysis based on ML, commonly applied to fungi. We incorporated additional loci (ITS and RPB1) in order to increase the robustness of the phylogenetic analysis in the study of the Scheffersomyces members. We followed the recommendations of the Assembling the Fungal Tree of Life (AFTOL http://aftol.org/) research group in searching for orthologous genes in fungal genomes [89]. In addition results obtained by Schoch et al. [90] showed that RPB1, RPB2, and TEF1 (elongation factor 1 alpha) are more phylogenetically informative compared to rRNA genes in Ascomycota. More recently, Schoch et al. [59] proposed the use of ITS as a barcode gene for fungi, although RPB1 had higher species discriminatory power than ITS in Saccharomycotina. The authors [59] also pointed out that datasets containing combinations of at least three molecular markers (SSU, LSU, ITS or RPB1) showed the highest probability of correct identification for all Fungi.
As mentioned in the results section, the Scheffersomyces clade, including all known taxa reported in the literature, is comprised of three subclades: 1) an early diverged subclade of S. spartinae, the only member that lacks the ability to ferment both xylose and cellobiose, and S. gosingicus, a cellobiose-fermenting (C-F) yeast isolated from soil in southeastern Asia [91]; 2) a C-F subclade that includes Scheffersomyces sp. UFMG-HMD-26.3 (not yet validly published [20]), S. coipomoensis, S. ergatensis, S. lignicola, and S. queiroziae; and 3) the X-F subclade containing S. lignosus, S. illinoinensis, S. insectosa, S. quercinus, S. segobiensis, S. stipitis, S. shehatae, and S. virginianus (Fig. 2).
The phylogenetic outcome also suggests that the common ancestor of the Scheffersomyces clade may have shown the ability to ferment D-xylose and cellobiose. This hypothesis is supported by the phylogenetic analyses based on a multilocus dataset that places S. gosingicus, a C-F species, in the earliest derived subclade, and the results of the phylogenetic study showing XR presence in all Scheffersomyces clade members studied (Fig. 5). Moreover, the same tree topology of the multilocus analysis was recovered by using either XYL1 or XR, suggesting that X-F ability might have played a fundamental role in the speciation process of the X-F subclade (Figs. 2, 5). We did not include the single copy gene XYL1 in the multilocus phylogenetic analysis because the orthology of this locus has not been confirmed across several yeast taxa.
The ability to ferment both wood components (D-xylose and cellobiose) is exhibited by only a few yeasts: Ogataea polymorpha [92], Brettanomyces naardenensis [1], and Spathaspora passalidarum [18], and in these species fermentative abilities are weak or delayed. On the contrary, Scheffersomyces clade members exhibit only one or the other fermentative ability. The loss of fermentation capability could be a consequence of becoming more efficient in carrying out the fermentation of fewer sugars. In particular, the fermentation of cellobiose has an antagonistic effect against the fermentation of D-xylose, because during the extracellular fermentation of cellobiose, units of glucose are released by β-glucosidase, a sugar that has higher affinity for the pentose membrane transporter rather than D-xylose. Consequently, glucose is first incorporated into the cells to be fermented [93].
XR in the Scheffersomyces Clade Members
The metabolic constraint in the regeneration of the cofactor NAD+ to NADH+ has been described as a major constraint in the fermentation of D-xylose, therefore most studies have focused on the molecular characterization of the conserved domains involved in the uptake of this cofactor that is present in the C-terminal region of the XR, but few studies have been done on the characterization of the N-terminal region.
In the study in the N-terminal region of the XR of the X-F members of the S. stipitis subclade we were able to identify all of the conserved domains and amino acids in both X-F and non X-F yeasts. These findings indicated that the ability to ferment xylose does not rely solely on the presence of these conserved regions. We also found several biased nucleotide mutations that maintain the same polarity as the codified amino acid, and only the mutations on the residues 174, 179, 184, 188, and 204 showed a change in amino acid polarity. These mutations were found mainly surrounding conserved domains in comparison with the amino acid sequence of the non X-F yeast S. ergatensis (Fig. 6). The mutations could generate structural modifications that allow the fermentation of xylose in the Scheffersomyces clade members, results that could be supported by performing direct mutagenesis studies on the amino acids to characterize their individual roles in the performance of XR.
Although the X-F yeasts were dispersed throughout the Saccharomycotina, as we mentioned in the introduction, the Debaryomycetaceae includes the largest number of X-F yeasts, such as species of Scheffersomyces and Spathaspora found in association with lignicolous insects [21], [24]. This phylogenetic placement of the X-F yeasts supports two alternatives: the X-F ability was the result of convergent evolution in ascomycete yeasts, or the X-F ability was present in the earliest common ancestor in the Saccharomycotina and has been retained mainly in the yeasts associated with lignicolous habitats. Several independent lines of evidence favor the second premise: 1) classical biochemical studies have determined that the ability to assimilate D-xylose and xylitol is common throughout the yeasts; 2) several relatively early diverging yeasts, B. naardenensis, O. polymorpha, and P. tannophilus, ferment xylose [1], [92]; 3) many studies characterizing yeast diversity indicate that most X-F yeasts are associated with wood and the gut of lignicolous insects [21]; and 4) more recently, a report of the genome sequences of a diverse group of X-F and non X-F yeasts confirmed the presence of xylose genes in all of them [45].
Yeasts as a group, are known for their biochemical versatility in utilizing a wide variety of carbon sources. Individual strains, however, may be characterized by specific physiological profiles depending on their life style and environment. Wood substrates and the gut of lignicolous insects previously were unexplored environments for the isolation of X-F yeasts. The findings of this study further support the hypothesis that X-F yeasts and yeasts in the Sugiyamaella clade are common inhabitants of the wood substrates. The Scheffersomyces clade is comprised mainly of cellobiose- and D-xylose-fermenting yeasts isolated from distant geographical regions and associated with wood and insects that feed on plant tissues. The amino acid modifications present in the putative XR of X-F yeasts in the S. stipitis subclade, could be responsible for the enhanced rate of fermentation shown by the members of this clade. The addition of ITS and RPB1 loci in the phylogenetic studies on the Scheffersomyces clade dramatically increased the support of the phylogenetic relationships of the members. We have used the primers for XYL1 designed for this study across several yeast species (data not show), and they could be used to help understand how these genes have evolved in the members of Saccharomycotina. The phylogenetic reconstruction using only XYL1 or RPB1 was similar to the multilocus analysis, and these loci have potential for rapid identification of cryptic species in this clade.
Acknowledgments
We thank Robert Frank and Stephanie Gross, Department of Biological Sciences, Louisiana State University (LSU), for their skilled assistance in all phases of this study. Claire Reuter made the original isolates of the new strains as part of an LSU Honors College undergraduate thesis, and we acknowledge her participation in this project. Scott Redhead kindly helped with nomenclatorial questions. We thank Louisiana State University, the LSU AgCenter Burden Center Research Station, and Lawrence J. Rouse for permission to collect on their properties.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Science Foundation grants (NSF DEB-0417180 and 0732671), Department of Energy contract to Lawrence Livermore National Laboratory (DE-AC52-07NA27344), and the LSU Boyd Professor Research Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kurtzman CP, Fell JW, Boekhout T editors. The yeasts, a taxonomy study. 2011. 5 ed. Amsterdam: Elseveir xxii +1–289, xv +290–1335, xvi +1336–2080 p.
- 2.Jeffries TW, Kurtzman CP. Strain selection, taxonomy, and genetics of close-fermenting yeasts. Enzyme Microb Tech. 1994;16:922–932. [Google Scholar]
- 3.Hahn-Hagerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol. 2007;108:147–177. doi: 10.1007/10_2007_062. [DOI] [PubMed] [Google Scholar]
- 4.Hahn-Hagerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- 5.Jeffries TW, Shi NQ. Genetic engineering for improved xylose fermentation by yeasts. Adv Biochem Eng Biotechnol. 1999;65:117–161. doi: 10.1007/3-540-49194-5_6. [DOI] [PubMed] [Google Scholar]
- 6.Shi NQ, Davis B, Sherman F, Cruz J, Jeffries TW. Disruption of the cytochrome c gene in xylose-utilizing yeast Pichia stipitis leads to higher ethanol production. Yeast. 1999;15:1021–1030. doi: 10.1002/(SICI)1097-0061(199908)15:11<1021::AID-YEA429>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 7.Hamacher T, Becker J, Gardonyi M, Hahn-Hagerdal B, Boles E. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology. 2002;148:2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
- 8.Jin YS, Ni H, Laplaza JM, Jeffries TW. Optimal growth and ethanol production from xylose by recombinant Saccharomyces cerevisiae require moderate D-xylulokinase activity. Appl Environ Microbiol. 2003;69:495–503. doi: 10.1128/AEM.69.1.495-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeffries TW, Grigoriev IV, Grimwood J, Laplaza JM, Aerts A, et al. Genome sequence of the lignocellulose-bioconverting and xylose-fermenting yeast Pichia stipitis. Nat Biotechnol. 2007;25:319–326. doi: 10.1038/nbt1290. [DOI] [PubMed] [Google Scholar]
- 10.Karhumaa K, Sanchez RG, Hahn-Hagerdal B, Gorwa-Grauslund MF. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb Cell Fact. 2007;6:5. doi: 10.1186/1475-2859-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karhumaa K, Fromanger R, Hahn-Hagerdal B, Gorwa-Grauslund MF. High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2007;73:1039–1046. doi: 10.1007/s00253-006-0575-3. [DOI] [PubMed] [Google Scholar]
- 12.Hughes SR, Sterner DE, Bischoff KM, Hector RE, Dowd PF, et al. Engineered Saccharomyces cerevisiae strain for improved xylose utilization with a three-plasmid SUMO yeast expression system. Plasmid. 2009;61:22–38. doi: 10.1016/j.plasmid.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Hector RE, Qureshi N, Hughes SR, Cotta MA. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl Microbiol Biotechnol. 2008;80:675–684. doi: 10.1007/s00253-008-1583-2. [DOI] [PubMed] [Google Scholar]
- 14.Kumar S, Gummadi SN. Metabolism of glucose and xylose as single and mixed feed in Debaryomyces nepalensis NCYC 3413: production of industrially important metabolites. Appl Microbiol Biotechnol. 2011;89:1405–1415. doi: 10.1007/s00253-010-2997-1. [DOI] [PubMed] [Google Scholar]
- 15.Shi NQ, Cruz J, Sherman F, Jeffries TW. SHAM-sensitive alternative respiration in the xylose-metabolizing yeast Pichia stipitis. Yeast. 2002;19:1203–1220. doi: 10.1002/yea.915. [DOI] [PubMed] [Google Scholar]
- 16.Jeffries TW. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol. 2006;17:320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Van Vleet JH, Jeffries TW, Olsson L. Deleting the para-nitrophenyl phosphatase (pNPPase), PHO13, in recombinant Saccharomyces cerevisiae improves growth and ethanol production on D-xylose. Metab Eng. 2008;10:360–369. doi: 10.1016/j.ymben.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen NH, Suh SO, Marshall CJ, Blackwell M. Morphological and ecological similarities: wood-boring beetles associated with novel xylose-fermenting yeasts, Spathaspora passalidarum gen. sp. nov. and Candida jeffriesii sp. nov. Mycol Res. 2006;110:1232–1241. doi: 10.1016/j.mycres.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Cadete RM, Santos RO, Melo MA, Mouro A, Goncalves DL, et al. Spathaspora arborariae sp. nov., a D-xylose-fermenting yeast species isolated from rotting wood in Brazil. FEMS Yeast Res. 2009;9:1338–1342. doi: 10.1111/j.1567-1364.2009.00582.x. [DOI] [PubMed] [Google Scholar]
- 20.Cadete RM, Melo MA, Lopes MR, Pereira GM, et al. Candida amazonensis sp. nov., an ascomycetous yeast isolated from rotting wood in Amazonian Forest, Brazil. Int J Syst Evol Microbiol. 2011. (In press). [DOI] [PubMed]
- 21.Suh SO, Marshall CJ, McHugh JV, Blackwell M. Wood ingestion by passalid beetles in the presence of xylose-fermenting gut yeasts. Mol Ecol. 2003;12:3137–3145. doi: 10.1046/j.1365-294x.2003.01973.x. [DOI] [PubMed] [Google Scholar]
- 22.Grunwald S, Pilhofer M, Holl W. Microbial associations in gut systems of wood- and bark-inhabiting longhorned beetles [Coleoptera: Cerambycidae]. Syst Appl Microbiol. 2010;33:25–34. doi: 10.1016/j.syapm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Tanahashi M, Kubota K, Matsushita N, Togashi K. Discovery of mycangia and the associated xylose-fermenting yeasts in stag beetles (Coleoptera: Lucanidae). Naturwissenschaften. 2010;97:311–317. doi: 10.1007/s00114-009-0643-5. [DOI] [PubMed] [Google Scholar]
- 24.Suh SO, Blackwell M, Kurtzman CP, Lachance MA. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia. 2006;98:1006–1017. doi: 10.3852/mycologia.98.6.1006. [DOI] [PubMed] [Google Scholar]
- 25.Toivola A, Yarrow D, van den Bosch E, van Dijken JP, Scheffers WA. Alcoholic fermentation of D-xylose by yeasts. Appl Environ Microbiol. 1984;47:1221–1223. doi: 10.1128/aem.47.6.1221-1223.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delgenes JP, Moletta R, Navarro JM. Fermentation of D-xylose, D-glucose, L-arabinose mixture by Pichia stipitis: Effect of the oxygen transfer rate on fermentation performance. Biotechnol Bioeng. 1989;34:398–402. doi: 10.1002/bit.260340314. [DOI] [PubMed] [Google Scholar]
- 27.Delgenes JP, Moletta R, Navarro JM. The effect of aeration on D-xylose fermentation by Pachysolen tannophilus, Pichia stipitis, Kluyveromyces marxianus and Candida shehatae. Biotechnol Lett. 1986;8:897–900. [Google Scholar]
- 28.Du Preez JC, Bosch M, Prior BA. Xylose fermentation by Candida shehatae and Pichia stipitis, effects of pH, temperature and substrate concentration. Enzyme Microb Tech. 1986;8:360–364. [Google Scholar]
- 29.Parekh SR, Yu S, Wayman M. Adaptation of Candida shehatae and Pichia stipitis to wood hydrolysates for increased ethanol production. Appl Microbiol Biotechnol. 1986;25:300–304. [Google Scholar]
- 30.Parekh S, Wayman M. Fermentation of cellobiose and wood sugars to ethanol by Candida shehatae and Pichia stipitis. Biotechnol Lett. 1986;8:597–600. [Google Scholar]
- 31.Sreenath HK, Chapman TW, Jeffries TW. Ethanol production from D-xylose in batch fermentations with Candida shehatae: Process variables. Appl Microbiol Biotechnol. 1986;24:294–299. [Google Scholar]
- 32.Alexander MA, Chapman TW, Jeffries TW. Xylose metabolism by Candida shehatae in continuous culture. Appl Microbiol Biotechnol. 1988;28:478–486. [Google Scholar]
- 33.Alexander MA, Chapman TW, Jeffries TW. Continuous ethanol production from D-xylose by Candida shehatae. Biotechnol Bioeng. 1987;30:685–691. doi: 10.1002/bit.260300515. [DOI] [PubMed] [Google Scholar]
- 34.Prior BA, Alexander MA, Yang V, Jeffries TW. The role of alcohol-dehydrogenase in the fermentation of D-xylose by Candida shehatae ATCC-22984. Biotechnol Lett. 1988;10:37–42. [Google Scholar]
- 35.Ho NW, Lin FP, Huang S, Andrews PC, Tsao GT. Purification, characterization, and amino terminal sequence of xylose reductase from Candida shehatae. Enzyme Microb Tech. 1990;12:33–39. doi: 10.1016/0141-0229(90)90177-r. [DOI] [PubMed] [Google Scholar]
- 36.Palnitkar S, Lachke A. Effect of nitrogen sources on oxidoreductive enzymes and ethanol production during D-xylose fermentation by Candida shehatae. Can J Microbiol. 1992;38:258–260. doi: 10.1139/m92-043. [DOI] [PubMed] [Google Scholar]
- 37.Palnitkar SS, Lachke AH. Efficient simultaneous saccharification and fermentation of agricultural residues by Saccharomyces cerevisiae and Candida shehatae. The D-xylose fermenting yeast. Appl Biochem Biotechnol. 1990;26:151–158. doi: 10.1007/BF02921531. [DOI] [PubMed] [Google Scholar]
- 38.Agbogbo FK, Wenger KS. Production of ethanol from corn stover hemicellulose hydrolyzate using Pichia stipitis. J Ind Microbiol Biotechnol. 2007;34:723–727. doi: 10.1007/s10295-007-0247-z. [DOI] [PubMed] [Google Scholar]
- 39.Agbogbo FK, Coward-Kelly G. Cellulosic ethanol production using the naturally occurring xylose-fermenting yeast, Pichia stipitis. Biotechnol Lett. 2008;30:1515–1524. doi: 10.1007/s10529-008-9728-z. [DOI] [PubMed] [Google Scholar]
- 40.Agbogbo FK, Haagensen FD, Milam D, Wenger KS. Fermentation of acid-pretreated corn stover to ethanol without detoxification using Pichia stipitis. Appl Biochem Biotechnol. 2008;145:53–58. doi: 10.1007/s12010-007-8056-4. [DOI] [PubMed] [Google Scholar]
- 41.Agbogbo FK, Wenger KS. Effect of pretreatment chemicals on xylose fermentation by Pichia stipitis. Biotechnol Lett. 2006;28:2065–2069. doi: 10.1007/s10529-006-9192-6. [DOI] [PubMed] [Google Scholar]
- 42.Jeffries TW. Regulation of the xylose fermentation in Candida shehatae and Pachysolen tannophilus. Abstr Pap Am Chem S. 1986;192:51–BTEC. [Google Scholar]
- 43.Jeffries TW, Van VleetJR. Pichia stipitis genomics, transcriptomics, and gene clusters. FEMS Yeast Res. 2009;9:793–807. doi: 10.1111/j.1567-1364.2009.00525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JW, Zhu JY, Scordia D, Jeffries TW. Evaluation of ethanol production from corncob using Scheffersomyces (Pichia) stipitis CBS 6054 by volumetric scale-up. Appl Biochem Biotechnol. 2011;165:814–822. doi: 10.1007/s12010-011-9299-7. [DOI] [PubMed] [Google Scholar]
- 45.Wohlbach DJ, Kuo A, Sato TK, Potts KM, Salamov AA, et al. Comparative genomics of xylose-fermenting fungi for enhanced biofuel production. Proc Natl Acad Sci U S A. 2011;108:13212–13217. doi: 10.1073/pnas.1103039108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeppsson M, Bengtsson O, Franke K, Lee H, Hahn-Hagerdal B, et al. The expression of a Pichia stipitis xylose reductase mutant with higher K(M) for NADPH increases ethanol production from xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng. 2006;93:665–673. doi: 10.1002/bit.20737. [DOI] [PubMed] [Google Scholar]
- 47.Fromanger R, Guillouet SE, Uribelarrea JL, Molina-Jouve C, Cameleyre X. Effect of controlled oxygen limitation on Candida shehatae physiology for ethanol production from xylose and glucose. J Ind Microbiol Biotechnol. 2010;37:437–445. doi: 10.1007/s10295-009-0688-7. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Yang M, Tian S, Zhang Y, Yang X. Co-expression of xylose reductase gene from Candida shehatae and endogenous xylitol dehydrogenase gene in Saccharomyces cerevisiae and the effect of metabolizing xylose to ethanol. Prikl Biokhim Mikrobiol. 2010;46:456–461. [PubMed] [Google Scholar]
- 49.Bajwa PK, Phaenark C, Grant N, Zhang X, Paice M, et al. Ethanol production from selected lignocellulosic hydrolysates by genome shuffled strains of Scheffersomyces stipitis. Bioresour Technol. 2011;102:9965–9969. doi: 10.1016/j.biortech.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 50.Khattab SM, Watanabe S, Saimura M, Kodaki T. A novel strictly NADPH-dependent Pichia stipitis xylose reductase constructed by site-directed mutagenesis. Biochem Biophys Res Commun. 2011;404:634–637. doi: 10.1016/j.bbrc.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 51.Hughes SR, Gibbons WR, Bang SS, Pinkelman R, Bischoff KM, et al. Random UV-C mutagenesis of Scheffersomyces (formerly Pichia) stipitis NRRL Y-7124 to improve anaerobic growth on lignocellulosic sugars. J Ind Microbiol Biotechnol. 2012;39:163–173. doi: 10.1007/s10295-011-1012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kreger-van Rij NJW. Lodder J, editor. Pichia Hansen. 1970. pp. 455–554. editor. The Yeasts, A taxonomic study. 2 ed. Amsterdam: North-Holland.
- 53.Vaughan-Martini A. Comparazione dei genomi del lievito Pichia stipitis e di alcune specie imperfette affini. Ann. Fac. Agrar. Univ. Perugia. 1984;38:331–335. [Google Scholar]
- 54.Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 55.Kurtzman CP, Suzuki M. Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience. 2010;51:2–14. [Google Scholar]
- 56.Suh SO, White MM, Nguyen NH, Blackwell M. The status and characterization of Enteroramus dimorphus: a xylose-fermenting yeast attached to the gut of beetles. Mycologia. 2004;96:756–760. doi: 10.1080/15572536.2005.11832923. [DOI] [PubMed] [Google Scholar]
- 57.Passoth V, Hansen M, Klinner U, Emeis CC. The electrophoretic banding pattern of the chromosomes of Pichia stipitis and Candida shehatae. Curr Genet. 1992;22:429–431. doi: 10.1007/BF00352445. [DOI] [PubMed] [Google Scholar]
- 58.Kurtzman CP. Candida shehatae genetic diversity and phylogenetic relationships with other xylose-fermenting yeasts. Antonie Van Leeuwenhoek. 1990;57:215–222. doi: 10.1007/BF00400153. [DOI] [PubMed] [Google Scholar]
- 59.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, the Barcoding Consortium. The internal transcribed spacer as a universal DNA barcode marker for Fungi. Fungal Barcoding Consortium. Proc Natl Acad Sci U S A. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suh SO, McHugh JV, Blackwell M. Expansion of the Candida tanzawaensis yeast clade: 16 novel Candida species from basidiocarp-feeding beetles. Int J Syst Evol Microbiol. 2004;54:2409–2429. doi: 10.1099/ijs.0.63246-0. [DOI] [PubMed] [Google Scholar]
- 61.White TJ, Bruns TD, Lee S, Taylor JW. Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. 1990. pp. 315–322. PCR protocols, a guide to methods and applications. San Diego: Academic Press.
- 62.Hibbett DS. Phylogenetic evidence for horizontal transmission of group I introns in the nuclear ribosomal DNA of mushroom-forming fungi. Mol Biol Evol. 1996;13:903–917. doi: 10.1093/oxfordjournals.molbev.a025658. [DOI] [PubMed] [Google Scholar]
- 63.Matheny PB, Liu YJJ, Ammirati JF, Hall BD. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am J Bot. 2002;89:688–698. doi: 10.3732/ajb.89.4.688. [DOI] [PubMed] [Google Scholar]
- 64.Tanabe Y, Saikawa M, Watanabe MM, Sugiyama J. Molecular phylogeny of Zygomycota based on EF-1 alpha and RPB1 sequences: limitations and utility of alternative markers to rDNA. Mol Phylogenet Evol. 2004;30:438–449. doi: 10.1016/s1055-7903(03)00185-4. [DOI] [PubMed] [Google Scholar]
- 65.Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141 (Pt. 1995;7):1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 66.Fadda ME, Mossa V, Pisano MB, Deplano M, Cosentino S. Occurrence and characterization of yeasts isolated from artisanal Fiore Sardo cheese. Int J Food Microbiol. 2004;95:51–59. doi: 10.1016/j.ijfoodmicro.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 67.Fadda ME, Viale S, Deplano M, Pisano MB, Cosentino S. Characterization of yeast population and molecular fingerprinting of Candida zeylanoides isolated from goat's milk collected in Sardinia. Int J Food Microbiol. 2010;136:376–380. doi: 10.1016/j.ijfoodmicro.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 68.Fuentefria AM, Suh SO, Landell MF, Faganello J, Schrank A, et al. Trichosporon insectorum sp. nov., a new anamorphic basidiomycetous killer yeast. Mycol Res. 2008;112:93–99. doi: 10.1016/j.mycres.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Yarrow D. Kurtzman CP, Fell JW, editors. Methods for the isolation, maintenance and identification of yeasts. 1998. pp. 77–100. The Yeasts, A taxonomic study. Amsterdam: Elsevier.
- 70.Barnett JA, Payne RW, Yarrow D. Yeasts: Characteristics and identification. Cambridge: Cambridge University Press. x+1139 p. 2000.
- 71.Maddison W, Maddison D. Mesquite: A modular system for evolutionary analysis. Evolution. 2005;62:1103–1118. [Google Scholar]
- 72.Liu K, Warnow TJ, Holder MT, Nelesen SM, Yu J, et al. SATe-II: Very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Syst Biol. 2012;61:90–106. doi: 10.1093/sysbio/syr095. [DOI] [PubMed] [Google Scholar]
- 73.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 74.Ohama T, Suzuki T, Mori M, Osawa S, Ueda T, et al. Non-universal decoding of the leucine codon CUG in several Candida species. Nucleic Acids Res. 1993;21:4039–4045. doi: 10.1093/nar/21.17.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Muller T, Vingron M. Modeling amino acid replacement. J Comput Biol. 2000;7:761–776. doi: 10.1089/10665270050514918. [DOI] [PubMed] [Google Scholar]
- 76.Milan EP, Kallas EG, Costa PR, da Matta DA, Lopes Colombo A. Oral colonization by Candida spp. among AIDS household contacts. Mycoses. 2001;44:273–277. [PubMed] [Google Scholar]
- 77.Knapp S, McNeill J, Turland NJ. Changes to publication requirements made at the XVIII International Botanical Congress in Melbourne: What does e-publication mean for you? Taxon. 2011;60:1498–1501. doi: 10.1186/1471-2148-11-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chu BC, Lee H, et al. Investigation of the role of a conserved glycine motif in the Saccharomyces cerevisiae xylose reductase. Curr Microbiol. 2006;53:118–123. doi: 10.1007/s00284-005-0325-2. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Q, Shirley NJ, Burton RA, Lahnstein J, Hrmova M. The genetics, transcriptional profiles, and catalytic properties of UDP-alpha-D-xylose 4-epimerases from barley. Plant Physiol. 2010;153:555–568. doi: 10.1104/pp.110.157644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurtzman CP, Robnett CJ. Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov. and 14 new species combinations. FEMS Yeast Res. 2007;7:141–151. doi: 10.1111/j.1567-1364.2006.00157.x. [DOI] [PubMed] [Google Scholar]
- 81.Houseknecht JL, Hart EL, Suh SO, Zhou JJ. Yeasts in the Sugiyamaella clade associated with wood-ingesting beetles and the proposal of Candida bullrunensis sp. nov. Int J Syst Evol Microbiol. 2011;61:1751–1756. doi: 10.1099/ijs.0.026427-0. [DOI] [PubMed] [Google Scholar]
- 82.Zhang N, Suh SO, Blackwell M. Microorganisms in the gut of beetles: evidence from molecular cloning. J Invertebr Pathol. 2003;84:226–233. doi: 10.1016/j.jip.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 83.Gaster PF. Rosa CA, Gabor P, editors. Yeast and invertebrate associations. 2006. pp. 303–370. Biodiversity and ecophysiology of yeasts. Berlin: Springer-Verlag.
- 84.Nardi JB, Bee CM, Miller LA, Nguyen NH, Suh SO, et al. Communities of microbes that inhabit the changing hindgut landscape of a subsocial beetle. Arthropod Struct Dev. 2006;35:57–68. doi: 10.1016/j.asd.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 85.Rokas A, Williams BL, King N, Carroll SB. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- 86.Cadez N, Raspor P, Smith MT. Phylogenetic placement of Hanseniaspora-Kloeckera species using multigene sequence analysis with taxonomic implications: descriptions of Hanseniaspora pseudoguilliermondii sp. nov. and Hanseniaspora occidentalis var. citrica var. nov. Int J Syst Evol Microbiol. 2006;56:1157–1165. doi: 10.1099/ijs.0.64052-0. [DOI] [PubMed] [Google Scholar]
- 87.Liti G, Barton DB, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lachance MA, Dobson J, Wijayanayaka DN, Smith AM. The use of parsimony network analysis for the formal delineation of phylogenetic species of yeasts: Candida apicola, Candida azyma, and Candida parazyma sp. nov., cosmopolitan yeasts associated with floricolous insects. Antonie Van Leeuwenhoek. 2010;97:155–170. doi: 10.1007/s10482-009-9399-3. [DOI] [PubMed] [Google Scholar]
- 89.Robbertse B, Reeves JB, Schoch CL, Spatafora JW. A phylogenomic analysis of the Ascomycota. Fungal Genet Biol. 2006;43:715–725. doi: 10.1016/j.fgb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 90.Schoch CL, Sung GH, Lopez-Giraldez F, Townsend JP, Miadlikowska J, et al. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- 91.Chang CF, Yao CH, Young SS, Limtong S, Kaewwichian R, et al. Candida gosingica sp. nov., an anamorphic ascomycetous yeast closely related to Scheffersomyces spartinae. Int J Syst Evol Microbiol. 2011;61:690–694. doi: 10.1099/ijs.0.020511-0. [DOI] [PubMed] [Google Scholar]
- 92.Ryabova OB, Chmil OM, Sibirny AA. Xylose and cellobiose fermentation to ethanol by the thermotolerant methylotrophic yeast Hansenula polymorpha. FEMS Yeast Res. 2003;4:157–164. doi: 10.1016/S1567-1356(03)00146-6. [DOI] [PubMed] [Google Scholar]
- 93.Han JH, Park JY, Yoo KS, Kang HW, Choi GW, et al. Effect of glucose on xylose utilization in Saccharomyces cerevisiae harboring the xylose reductase gene. Arch Microbiol. 2011;193:335–340. doi: 10.1007/s00203-011-0678-9. [DOI] [PubMed] [Google Scholar]