Abstract
Objectives
To evaluate microRNA (miRNA) expression in pancreatic resection specimens and fine needle aspiration (FNA) biopsies and determine which, if any, miRNAs aid the distinction between benign and malignant pancreatic tumors in limited cytology material.
Methods
Resection specimens containing adenocarcinoma (n=17), intraductal papillary mucinous neoplasms (IPMN, n=11), and non-neoplastic tissues (n=15) were evaluated for miR-21, -221, -100, -155, and -181b expression by qRT-PCR, and a subset of carcinomas and IPMNs were analyzed with miRNA microarrays. Cellblocks containing carcinoma (n=26) or benign pancreatic lesions (n=11) from FNA biopsies were subjected to qRT-PCR for miR-21, miR-221, miR-181b, miR-196a and miR-217.
Results
Carcinomas showed higher expression of miR-21, -221, -155, -100, and -181b than benign lesions by qRT-PCR and overexpression of miR-21, -221, and-181b was confirmed by microarray analysis. Cellblocks containing carcinoma showed higher expression of miR-21, -221, and -196a than those from benign lesions (p<0.001, p=0.009, p<0.001, respectively).
Conclusions
Pancreatic ductal adenocarcinomas show differential expression of miRNAs compared to benign pancreatic lesions. A select panel of miRNAs aids the distinction between pancreatic lesions in cytology specimens.
Keywords: Intraductal papillary mucinous neoplasm, Pancreas, Cytology, Fine needle aspiration, Molecular
Introduction
Pancreatic cancer is the fourth leading cause of cancer-related death in the United States.1 Although surgical resection represents the best hope for cure, most patients present with tumors of advanced stage that are inoperable and associated with a poor prognosis. The preoperative evaluation of pancreatic tumors includes radiographic imaging to determine potential resectability and diagnostic biopsy, which is essential to guiding appropriate surgical and chemotherapeutic management. Biopsies of pancreatic tumors are usually performed via fine needle aspiration (FNA) biopsy at the time of endoscopic ultrasonographic evaluation. However, biopsy interpretation may be hindered by limited sampling and inflammatory changes that mask, or mimic, neoplasms. For this reason, many investigators have tried to identify biomarkers that reliably distinguish pancreatic adenocarcinoma from benign neoplasms and non-neoplastic pancreatic tissue. MicroRNAs (miRNAs) have received considerable attention as potential biomarkers of human malignancies. These molecules consist of 18-25 nucleotides and regulate gene expression at the post-transcriptional level. They promote oncogenesis by either inhibiting expression of tumor suppressor genes, or up-regulating expression of oncogenes.2-4 Emerging data suggest that miRNAs have characteristic expression profiles in some carcinomas and several species are dysregulated in pancreatic ductal adenocarcinoma.3, 5-9 For example, miR-21, miR-221, miR-181b, miR-155, miR-100 and miR-196a show significantly higher expression in pancreatic carcinoma when compared to the non-neoplastic pancreas; whereas miR-217 is normally found in the pancreas, but shows consistently decreased or absent expression in adenocarcinomas.10-13 The potential application of these markers to the preoperative evaluation of pancreatic lesions has not been extensively studied.
The purpose of this study was to assess the utility of miRNAs in distinguishing between benign and malignant pancreatic lesions. We first examined expression of 5 miRNAs (miR-21, miR-221, miR-181b, miR-100 and miR-155) in pancreatic resection specimens containing ductal adenocarcinoma (n=17), intraductal papillary mucinous neoplasms (IPMNs, n=11) and adjacent non-neoplastic pancreatic tissue (n=15) using qRT-PCR. A subset of these cases (8 pancreatic ductal adenocarcinomas and 8 IPMNs) was simultaneously evaluated for expression of 328 miRNA species using a miRNA microarray assay. Based on these results and those in the literature, we compiled a panel of 5 miRNA species (miR-21, miR-221, miR-181b, miR-196a and miR-217) for further evaluation in cytology aspirate specimens. We extracted miRNA from 49 cellblocks derived from FNA biopsy samples, including 38 pancreatic ductal adenocarcinomas and 11 benign lesions and performed qRT-PCR to amplify these 5 miRNAs, in order to determine whether miRNA expression profiles could be used to detect pancreatic carcinoma in limited biopsy samples.
Materials and Methods
Case Selection
Twenty-eight pancreatic resection specimens were identified from a search of the Department of Pathology and Laboratory Medicine surgical pathology files at Weill Cornell Medical College. The cases included 17 pancreatic adenocarcinomas and 11 IPMNs. Hematoxylin and eosin stained slides were prepared from routinely processed tissue sections (10% buffered formalin) and reviewed to confirm the diagnoses. Tumors were classified according to the WHO classification system and staged using the AJCC TNM Cancer Staging Manual.14-16 Four 5 μm sections were cut from a representative block of each tumor. Non-neoplastic tissues were manually removed from each slide and the remaining tumor tissue was subjected to RNA extraction. We also obtained tissue blocks of peritumoral non-neoplastic pancreatic tissue from 15 cases.
Cellblock specimens prepared from 49 FNA biopsy samples of pancreatic lesions were obtained. Thirty-eight patients had surgically, or clinically, confirmed pancreatic ductal adenocarcinoma, including 16 patients who underwent surgical resection and 22 who had unresectable disease at the time of diagnosis. All of the latter 22 patients ultimately developed disseminated pancreatic adenocarcinoma and died of disease. Eleven cellblocks contained benign epithelium and inflammatory tissue, and none of these patients developed any evidence of pancreatic carcinoma during follow-up (mean: 24 months, range: 1-46). Ten 10 μm whole sections from each case were subjected to RNA extraction as described below. Information regarding patient age, sex, and clinical outcome was obtained from the medical records and pathology reports. This study was performed following approval by the Institutional Review Board.
RNA Extraction
RNA was extracted using the Ambion RecoverAll Total Nucleic Acid Isolation kit (Applied Biosystems, Foster City, California). Briefly, tissue samples were deparaffinized in xylene and subjected to protease digestion. RNA was captured on a glass-fiber filter and eluted using 60 μL of nuclease-free water after ethanol wash. Total RNA concentrations were measured with the NanoDrop 8000 spectrophotometer (NanoDrop, Wilmington, DE). The surgical resection specimens yielded 0.42-42 μg of total RNA (mean: 8 μg, median: 3 μg). Cellblock specimens yielded considerably less total RNA and ranged from undetectable values in 5 cases to 2.7 μg (median: 0.2 μg).
Quantitative RT-PCR
Quantitative RT-PCR was performed in triplicate using the ABI 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, California). Reverse transcription was performed using species-specific primers (Applied Biosystems, Foster City, California) for each miRNA. RNA from resection specimens was amplified using 10 ng of RNA per 15 μl reverse transcription reaction, whereas 5 ng of RNA per 7.5 μl reverse transcription reaction was utilized for cellblock analysis. The maximum volume (4.6 μl) of extracted RNA was used for the 5 cellblock specimens with low RNA concentrations. Each PCR was performed using 1 μl of cDNA solution and the Taqman MicroRNA Assay kit (Applied Biosystems), and consisted of 45 amplification cycles (15 seconds at 95 °C and 1 minute at 60 °C). Control amplification for small endogenous RNA U6 was performed for all samples. The same post-amplification threshold was set for all samples after each experiment, in order to compare Ct values for each miRNA. Mean Ct values were calculated for each specimen and normalized against the corresponding RNA U6 Ct values (Ct miRNA − Ct U6B). All data are presented as normalized Ct values.
Flexmir™ MicroRNA Assay
A subset of 8 pancreatic ductal adenocarcinomas and 8 IPMNS was analyzed using the Flexmir™ miRNA assay (Luminex Corporation, Austin, TX). For each sample, 1 μg of biotin-labeled RNA was incubated with hybridization buffer and locked nucleic acid capture probes coupled to fluorescently dyed microspheres. Samples were incubated for 3 minutes at 95° C and then for 60 minutes at 60° C. Hybridized reactants were then transferred to a 96-well plate following the manufacturer’s instructions. Unbound sample was removed by washing with ethylenediaminetetraacetic acid. The reporter molecule, Streptavidin-phycoerythrin, was added to the beads (75 μl/sample of 1:300 dilution) and incubated for 30 minutes at room temperature. Fifty microliters of each sample were processed on the Luminex analyzer (Luminex Corporation, Austin TX) and median fluorescence intensity values were measured.
Statistical Analysis
The results from the initial experiments on pancreatic resection specimens were analyzed as follows. All samples were subjected to qRT-PCR to amplify each of 5 miRNA species. Average miRNA expression levels were calculated for cases of pancreatic adenocarcinoma and compared to those of non-neoplastic pancreatic tissues and IPMNs using Tukey’s test. Data generated from the Flexmir™ miRNA microarray assay were also evaluated. Mean and median expression levels of 328 miRNA species evaluated in pancreatic ductal adenocarcinomas and IPMNs were compared using the student t-test and Wilcoxin rank-sum test. The student t-test was used to evaluate mean miRNA expression in cellblock specimens that contained pancreatic ductal adenocarcinoma and those with benign tissue. An Akaike Information Criteria-based backward model selection procedure was subsequently used to develop a parsimonious logistic regression model to best identify malignancy in cytologic specimens.
Results
Clinical and Pathologic Features of Resection Specimens
The clinical and pathologic features of 28 patients with resected pancreatic tumors are summarized in Table 1. Eight of 17 pancreatic ductal adenocarcinomas were low-grade and 9 were high-grade. Ten carcinomas were associated with lymph node metastases. At the time this study was performed, 9 patients had died from pancreatic adenocarcinoma, 5 were alive with disease, 2 had no evidence of disease, and 1 died from complications of another illness (mean follow up: 22.5 months, median: 11 months). Seven of 11 IPMNs, showed mild (n=3) or moderate (n= 4) dysplasia and 4 displayed severe dysplasia (carcinoma in situ).
Table 1.
Clinical and Pathologic Features of Resected Pancreatic Tumors
| Pancreatic Ductal Adenocarcinoma (n=17) |
Intraductal Papillary Mucinous Neoplasms (n=11) |
|
|---|---|---|
| Mean Age (years) | 66 | 62 |
| Male:Female Ratio | 9:8 | 5:6 |
| Tumor Location in Pancreas | ||
| Head | 14 | 7 |
| Body | 3 | 3 |
| Tail | 0 | 1 |
| Pathologic Tumor Stage | ||
| I | 13 | NA |
| II | 2 | NA |
| III | 2 | NA |
| IV | 0 | NA |
MicroRNA Expression in Resection Specimens
All 5 miRNA species were expressed at higher levels in pancreatic ductal adenocarcinoma than non-neoplastic pancreatic tissue (Figure 1). MicroRNA-21, miR-221, miR-155 and miR-100 were most significantly increased in pancreatic ductal adenocarcinoma versus non-neoplastic tissues (p=0.003, p=0.009, p<0.001, and p=0.04 respectively). MicroRNA-181b also showed a trend toward increased expression in carcinoma compared to non-neoplastic pancreas (p=0.07). Significantly higher expression of miR-21, miR-221, miR-155 and miR-181b was also observed in pancreatic ductal adenocarcinoma relative to IPMN (p<0.001, p<0.001, p<0.001 and p=0.019, respectively) and increased expression of miR-100 in the former was nearly significant (p=0.06) as depicted in Figure 1. Flexmir™ miRNA microarray analysis largely confirmed these results and demonstrated higher expression of miR-21, miR-221, and miR-181b in pancreatic ductal adenocarcinomas relative to IPMNs. The relationships between miRNA expression and tumor grade (low versus high), regional lymph node metastases, presence or absence of extrapancreatic extension, and development of metastatic disease during the follow up period were also examined. The mean expression levels of all 5 species were similar for all comparisons (fold differences ≤ 2).
Figure 1.
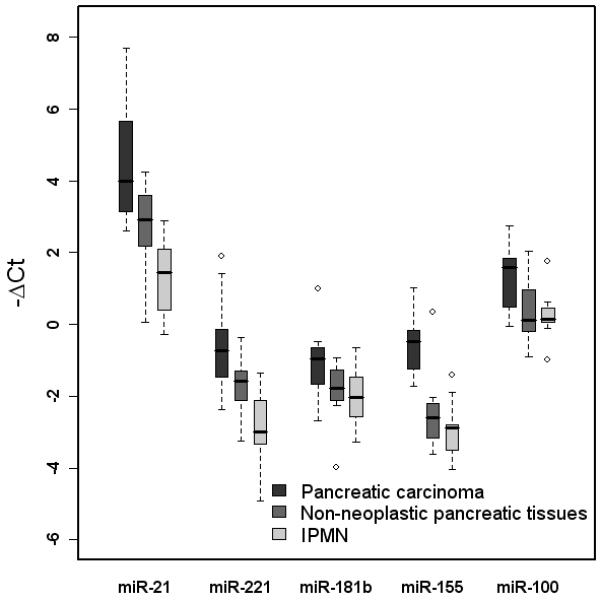
Box-and-whisker plot of miR-21, miR-221, miR-181b, miR-155, and miR-100 expression in resection specimens of pancreatic carcinoma, non-neoplastic pancreatic tissues, and IPMNs. Expression of miR-21, -221, -155, and -100 by qRT-PCR was significantly higher in carcinomas than non-neoplastic tissues (p=0.003, p=0.009, p<0.001, and p=0.04, respectively). There was also a trend toward increased miR-181b expression in cancers (p=0.07). Expression of miR-21, -221, -155, and -181b was also significantly higher in pancreatic carcinomas compared to IPMNs (p<0.001, p<0.001, p<0.001, and p=0.019, respectively) and a similar trend toward increased miR-100 expression in carcinomas was noted (p=0.06).
Clinical and Pathologic Features of Cellblock Specimens
Forty-nine cellblock specimens, including 38 adenocarcinomas and 11 benign lesions, prepared from pancreatic FNA biopsies were evaluated, all of which were obtained from a separate group of patients. Unfortunately, only 35/38 patients with pancreatic ductal adenocarcinoma had adequate material in their cellblocks for miRNA studies; the three patients without sufficient materials were excluded from further studies. All 35 patients with pancreatic cancer were adults (mean age: 67 years, male/female ratio: 12/23). A cytologic diagnosis of carcinoma was rendered in 26 (74%) cases and 9 were considered to be suspicious for malignancy. Eleven patients had benign pancreatic lesions, including chronic pancreatitis (n=2), serous cystadenomas (n=2) and pancreatic pseudocysts (n=7). The age and sex distributions of patients with benign lesions were similar to those with carcinoma (mean age: 67 years, male/female ratio: 3/8).
MicroRNA Expression in Cellblock Specimens
As previously noted, expression of three miRNAs (miR-21, miR-221 and miR-181b) was increased in pancreatic ductal adenocarcinoma compared to benign pancreatic lesions by both qRT-PCR and microarray. Results of a recent study also indicated that a panel of 2 miRNAs (miR-196a, miR-217) reliably detected carcinoma in FNA biopsy material.17 For this reason, we chose to evaluate cellblock material using a panel of 5 miRNAs (miR-21, miR-221, miR-181b, miR-196a, miR-217).
Forty-six cell blocks (35 carcinomas and 11 benign lesions) yielded sufficient RNA for analysis. Twenty-six cancers were definitively diagnosed by cytologic assessment and expression of miR-21, miR-221, and miR-196a in these cases was significantly higher than that of the 11 benign lesions (p<0.001, p=0.009, and p<0.001, respectively). No significant differences were observed with respect to expression of miR-181b or miR-217 between these two groups.
MicroRNA-21, miR-221 and miR-196a were overexpressed in pancreatic carcinomas compared to benign aspirates, but there was considerable overlap in the range of values between groups (Figure 2a) that precluded use of any single species as a reliable diagnostic tool. Expression levels of these 3 miRNAs were also assessed as additive values using all possible pairings, including a combination of all 3 markers. Although this approach resulted in greater separation between benign and malignant tumors (p<0.001 for all composite markers), the ranges of values still overlapped for all paired comparisons (Figure 2b).
Figure 2.
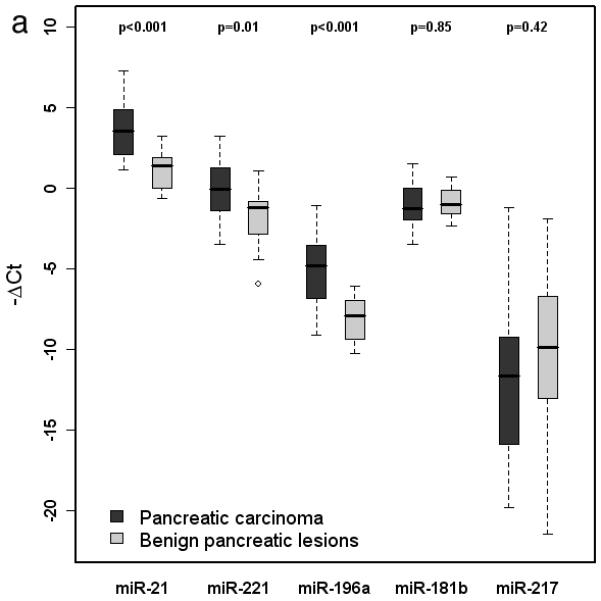
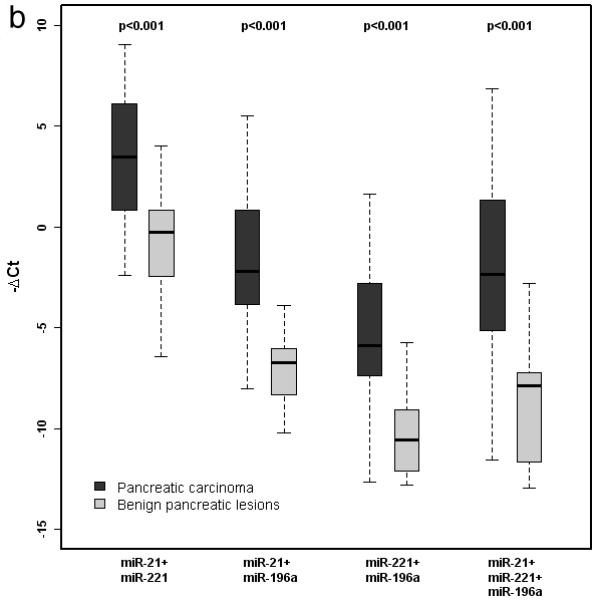
a. Box-and-whisker plot of miR-21, miR-221, miR-196a, miR-181b, and miR-217 expression in cellblock specimens derived from pancreatic carcinoma and benign pancreatic lesions. Expression of miR-21, -221, and -196a was significantly higher in carcinoma specimens. b. Box-and-whisker plot of additive miRNA values. Expression of all composite markers was significantly higher in pancreatic carcinoma than benign pancreatic lesions.
Parsimonious logistic regression modeling was performed utilizing the three most promising markers: miR-21, miR-221 and miR-196a. A composite equation using 2 markers (miR-221 + 2*miR-196a) was created and applied to the cytology samples. A resultant value of > −16.9 correctly classified 92% (24/26) of cytologically confirmed carcinomas as positive for malignancy and 73% (8/11) of the benign cases as negative for malignancy. This logistic regression model was applied to the 9 cellblocks obtained from patients with pancreatic ductal adenocarcinoma who had equivocal cytologic interpretations (suspicious for, but not diagnostic of, carcinoma). The model correctly predicted that 8 (89%) of these cases were malignant (Figure 3).
Figure 3.
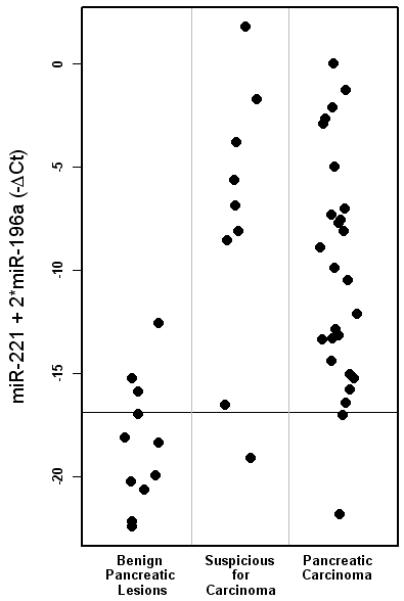
Scatter plot of cellblock specimens classified by logistic regression model. Using the equation, miR-221 + 2*miR-196a, a value of > −16.9 correctly classified 24/26 (92%) cytologically confirmed carcinomas as positive for malignancy and 8/11 (73%) benign lesions as negative for malignancy. When applied to 9 cases that were considered suspicious for, but not diagnostic of, pancreatic carcinoma, the model correctly predicted that 8 (89%) were malignant.
Discussion
In this study, we evaluated the utility of miRNA species as diagnostic markers of pancreatic ductal adenocarcinoma. We first used qRT-PCR to assess expression of miR-21, miR-221, miR-181b, miR-155 and miR-100 in pancreatic resection specimens that contained pancreatic ductal adenocarcinoma or IPMN, as well as non-neoplastic pancreatic tissues. We observed higher expression of all 5 miRNAs in adenocarcinomas compared to benign conditions. Sixteen of these cases, including 8 pancreatic ductal adenocarcinomas and 8 IPMNs were further evaluated using the Flexmir™ miRNA microarray assay, which confirmed higher expression of miR-21, miR-221 and miR-181b in cancers and these species were subsequently tested in cellblock specimens. Cellblock materials were also evaluated for miR-196a and miR-217, both of which are reportedly dysregulated in pancreatic adenocarcinomas.12, 17 We found increased expression of miR-21, miR-221 and miR-196a in cellblock specimens from patients with pancreatic ductal adenocarcinoma compared to those with benign lesions. We did not find miR-181b or miR-217 to reliably distinguish between these groups. Logistic regression modeling produced a formula (miR-221 + 2*miR-196a) that correctly predicted malignancy in 89% of FNA biopsy specimens that were equivocal based on cytologic evaluation alone.
Immunohistochemical markers may be used to distinguish benign from malignant tumors in pancreatic FNA biopsy specimens. Itoi et al. evaluated 62 FNA specimens and found p53 staining in 67% of carcinomas, but none of the chronic pancreatitis cases.18 Mucin histochemical stains are also helpful in this differential diagnosis: MUC1 and MUC4 are expressed in >90% of pancreatic carcinoma FNA specimens but <10% of benign pancreatic aspirates, whereas clusterin is more commonly expressed in benign ductal epithelium (90%) compared to carcinoma (7%).19,20 Most recently, insulin-like growth factor-II mRNA binding protein 3 (IMP3) expression was shown to be a highly sensitive marker of pancreatic ductal adenocarcinoma in cellblock material, being present in 88-92% of adenocarcinomas compared to 0% of chronic pancreatitis cases and benign tumors.21, 22, 23
Available molecular markers have limited utility in distinguishing between benign and malignant lesions in FNA biopsy samples from pancreatic tumors. K-ras mutations are detected in patients with benign and malignant pancreatobiliary diseases. Yan et al. found k-ras mutations in aspirates from 31/57 (54%) pancreatic ductal adenocarcinomas, 23/67 (34%) chronic pancreatitis cases, and 13/61 (21%) patients with biliary stones who did not have cancer.24 Shi et al. improved the sensitivity (94%) and specificity (89%) of k-ras testing by using DNA ligation and PCR amplification to compare ratios of mutated versus wild-type k-ras in surgically-collected pancreatic duct juice samples.25 Routine assessment for p53 mutations is challenging because the gene is large and mutations occur in multiple exons.26 Yan et al. used a functional assay for p53 wherein PCR-amplified p53 DNA from pancreatic aspirates was transformed into yeast colonies yielding white colonies for wild-type p53 and red colonies for mutant p53. This technically complicated assay correctly identified only 20/48 (42%) carcinomas and failed to detect p53 mutations in 4/12 (33%) tissue-matched cases that harbored mutations in the primary tumor.24 Epigenetic alterations, such as DNA methylation, may represent future targets, but methylation specific PCR assays are problematic.27
Fukushima et al. evaluated 45 patients with pancreatic adenocarcinoma and found that 67% had methylated ppENK in their pancreatic juice, but also found methylated ppENK in duodenal secretions from patients without cancer and in samples from histologically normal duodenum.28 Matsubayashi et al. evaluated 11 pancreatic ductal adenocarcinoma samples for hypermethylation using a panel of 5 genes (cyclin D2, TFP12, ppENK, NPTX2, and FOXE1). Using quantitative methylation-specific PCR, they found that 9 (82%) adenocarcinomas had at least 1% methylation of 2 or more genes, compared to none of 64 benign lesions.29 Future validation and improved detection methods may advance the clinical utility of these gene- and protein-based assays, but they are of limited value currently.
MicroRNAs have gained recent attention as diagnostic markers because they are dysregulated in several types of cancer. Up-regulation of miR-21 occurs in tumors of the pancreas, lung, breast, stomach, prostate, colon, brain, thyroid, head and neck, and esophagus.10, 11, 30-34 Increased miR-196a expression occurs in carcinomas of the endometrium, breast, and esophagus, and coincides with progression from PanIN2 to PanIN3 in the pancreas.12, 35, 36 MicroRNA-181b and miR-221 overexpression occurs in cancers of the brain, thyroid, colon, breast, prostate and in several types of leukemia.11, 37-42 The mechanisms by which miRNAs promote oncogenesis are not entirely clear, but the frequency with which they are dysregulated in carcinomas suggests their potentially important role in diagnosis, prognosis, and management of cancer patients.7, 30, 35, 43 Some miRNA species may distinguish pancreatic ductal adenocarcinomas from benign pancreatic lesions. MicroRNA-21 is increased early in the progression of pancreatic intraepithelial neoplasia (PanIN) to carcinoma and its presence by in-situ hybridization is associated with decreased survival among patients with node-negative pancreatic carcinoma.44, 45 Bloomston et al. examined 65 pancreatic adenocarcinomas and benign adjacent pancreatic tissue as well as 42 cases of chronic pancreatitis for miRNA expression. They found 21 miRNAs to be dysregulated in cancer and noted that a panel of 11 species (miR148a, miR-148b, miR-155, miR-181a, miR-181b, miR-181b-1, miR-181c, miR-181d, miR-21, miR-221, miR-375) distinguished adenocarcinoma from benign pancreatic tissue. They also found that increased miR-196a was associated with a 2-year survival of 17%, compared with 64% among tumors with low expression.10 Lee et al. profiled 28 pancreatic ductal adenocarcinomas and 21 non-neoplastic pancreatic tissues. They found 100 miRNAs to be aberrantly expressed in cancer, including miR-100 and 4 species (miR-21, miR-221, miR-181b and miR-155) that were identified by Bloomston et al.11 Similarly, we found all 5 of these miRNA species to be increased in pancreatic adenocarcinomas compared to non-neoplastic tissues.
One other study to date has applied miRNA expression analysis to pancreatic cytology specimens. Szafranska et al. evaluated 16 FNA biopsies obtained from 10 pancreatic ductal adenocarcinomas, 1 patient with suspected pancreatic ductal adenocarcinoma, 1 pancreatic neuroendocrine tumor, 1 aspirate that contained atypical epithelial cells, and 3 aspirates of non-pathologic tissue obtained after pancreatectomy. They found that a combination of miR-196a over-expression and decreased miR-217 expression correctly identified 9/10 pancreatic ductal adenocarcinomas and classified 1 suspicious case as malignant.17 We included these two species in our cellblock analysis. We also observed significantly higher miR-196a expression in cancers, but failed to confirm a relationship between decreased miR-217 expression in pancreatic ductal adenocarcinoma relative to samples of non-neoplastic tissues. We believe that this discrepancy can be explained as follows. MicroRNA-217 is necessary for pancreatic development in animal models and is highly expressed in normal pancreatic parenchyma.17, 46 Szafranska et al. used ex vivo aspirated material from non-lesional tissue in pancreatic resection specimens, which presumably contained abundant normal pancreatic tissue. In contrast, the benign cytology specimens used in our study were derived from cases of chronic pancreatitis, serous cystadenomas and pseudocysts, and all largely consisted of lesional, rather than acinar, elements. The lack of agreement between the two studies could reflect the relative amounts of ductal and acinar tissue present in the samples used in each study. We also found a high level of concordance between miR-21, miR-221 and miR-196a expression profiles in resection and cellblock specimens, but noted increased miR-181b only in cancer resection specimens. It is possible that miR-181b is expressed in desmoplastic stroma, which is more abundant in resection specimens than cytology aspirates.
One interesting aspect of our study is the use of logistic regression modeling to classify cytology samples. We established a miRNA expression cutoff threshold using two markers (miR-221 and miR-196a) that accurately predicted the presence of malignancy in 89% of cancer specimens that were not definitively classified based on cytologic evaluation alone. Although this model misclassified 3 benign cytology specimens as positive for malignancy and 1 of 9 cancers as negative for malignancy, its sensitivity and specificity may be improved by using fresh aspirates and extending the analysis to include additional miRNA species (e.g. miR-155).
In summary, our data show that most cytology cellblocks contain adequate material for quantitative analysis of miRNA expression. Differential miRNA expression in resection specimens is reliably reproduced in cellblock material and a select panel of miRNA species (miR-21, miR-221, miR-196a) distinguishes most pancreatic adenocarcinomas from benign lesions. Continued evaluation of other miRNAs may lead to a revised miRNA panel with improved sensitivity and specificity for diagnosing pancreatic ductal adenocarcinoma. Prospective studies that utilize other miRNA targets, as well as both cellblock and freshly collected aspirate material should further refine the utility of this tool in the assessment of patients with pancreatic tumors.
Acknowledgments
Contribution from XK Zhou is supported in part by National Institutes of Health Clinical and Translational Science Award UL1-RR024996.
Footnotes
Disclosure: The authors have no conflicts of interest to disclose.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tam W. The emergent role of microRNAs in molecular diagnostics of cancer. J Mol Diagn. 2008;10(5):415–423. doi: 10.2353/jmoldx.2008.080067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171:728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szafranska AE, Davidson T, Shingara J, et al. Accurate molecular characterization of formalin-fixed, paraffin-embedded tissues by microRNA expression profiling. J Mol Diagn. 2008;10:415–423. doi: 10.2353/jmoldx.2008.080018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeffrey SS. Cancer biomarker profiling with microRNAs. Nat Biotechnol. 2008;26:400–401. doi: 10.1038/nbt0408-400. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 9.Bloom G, Yang IV, Boulware D, et al. Multi-platform, multi-site, microarray-based tumor classification. Am J Pathol. 2004;164:9–16. doi: 10.1016/S0002-9440(10)63090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297(17):1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 11.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2006;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene. 2007;26:4442–4452. doi: 10.1038/sj.onc.1210228. [DOI] [PubMed] [Google Scholar]
- 13.Zhu Z, Gao W, Qian Z, Miao Y. Genetic variation of miRNA sequence in pancreatic cancer. Acta Biochim Biophys Sin. 2009;41(5):407–413. doi: 10.1093/abbs/gmp023. [DOI] [PubMed] [Google Scholar]
- 14.Longnecker DS, Adler G, Hruban RH, et al. World Health Organization Classification of Tumours. Pathology and genetics of tumours of the digestive system. IARC Press; Lyon: 2000. Intraductal papillary-mucinous neoplasms of the pancreas. [Google Scholar]
- 15.Wittenkind C, Greene FL, Hutter RVP, Klimpfinger M, Sobin LH, editors. 5th ed John Wiley & Sons, Inc.; Hoboken: 2005. TNM Atlas: Illustrated Guide to the TNM Classification of Malignant Tumors. [Google Scholar]
- 16.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. Springer; New York: 2010. [Google Scholar]
- 17.Szafranska AE, Doleshal M, Edmunds HS, et al. Analysis of microRNA in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clin Chem. 2008;54(10):1716–1724. doi: 10.1373/clinchem.2008.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoi T, Takei K, Sofuni A, et al. Immunohistochemical analysis of p53 and MIB-1 in tissue specimens obtained from endoscopic ultrasonography-guided fine needle aspiration biopsy for the diagnosis of solid pancreatic masses. Oncol Rep. 2005;13:229–234. [PubMed] [Google Scholar]
- 19.Chhieng DC, Benson E, Eltoum I, et al. MUC1 and MUC2 expression in pancreatic ductal carcinoma obtained by fine-needle aspiration. Cancer. 2003;99:365–371. doi: 10.1002/cncr.11857. [DOI] [PubMed] [Google Scholar]
- 20.Jhala N, Jhala D, Vickers SM, et al. Biomarkers in diagnosis of pancreatic carcinoma in fine-needle aspirates. Am J Clin Pathol. 2006;126:572–579. doi: 10.1309/cev30be088cbdqd9. [DOI] [PubMed] [Google Scholar]
- 21.Yantiss RK, Cosar E, Fischer AH. Use of IMP3 in identificaton of carcinoma in fine needle aspiration biopsies of pancreas. Acta Cytol. 2008;52(2):133–138. doi: 10.1159/000325470. [DOI] [PubMed] [Google Scholar]
- 22.Zhao H, Mandich D, Cartun RW, Ligato S. Expression of K homology domain containing protein overexpressed in cancer in pancreatic FNA for diagnosing adenocarcinoma of pancreas. Diagn Cytopathol. 2007;35(11):700–704. doi: 10.1002/dc.20739. [DOI] [PubMed] [Google Scholar]
- 23.Ligato S, Zhao H, Mandich D, Cartun RW. KOC (K homology domain containing protein overexpressed in cancer) and S100A4-protein immunoreactivity improves the diagnostic sensitivity of biliary brushing cytology for diagnosing pancreatobiliary malignancies. Diagn Cytopathol. 2008;36(8):561–567. doi: 10.1002/dc.20836. [DOI] [PubMed] [Google Scholar]
- 24.Yan L, McFaul C, Howes N. Molecular analysis to detect pancreatic ductal adenocarcinoma in high-risk groups. Gastroenterology. 2005;128:2124–2130. doi: 10.1053/j.gastro.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Shi C, Fukushima N, Abe T, et al. Sensitive quantitative detection of KRAS2 gene mutations in pancreatic duct juice differentiates patients with pancreatic cancer from chonic pancreatitis, potential for early detection. Cancer Biol Ther. 2008;7(3):353–360. doi: 10.4161/cbt.7.3.5362. [DOI] [PubMed] [Google Scholar]
- 26.Hollstein M, Sidransky D, Vogelstein B. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 27.Goggins M. Identifying molecular markers for the early detection of pancreatic neoplasia. Semin Oncol. 2007;34(4):303–310. doi: 10.1053/j.seminoncol.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukushima N, Walter KM, Ueki T. Diagnosing pancreatic cancer using methylation specific PCR analysis pancreatic juice. Cancer Biol Ther. 2003;2:78–83. doi: 10.4161/cbt.183. [DOI] [PubMed] [Google Scholar]
- 29.Matsubayashi H, Canto M, Sato N, et al. DNA methylation alterations in pancreatic juice of patients with suspected pancreatic disease. Cancer Res. 2006;66(2):1208–1217. doi: 10.1158/0008-5472.CAN-05-2664. [DOI] [PubMed] [Google Scholar]
- 30.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 31.Roldo C, Missiaglia E, Hagan JP, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24(29):4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 32.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mardin WA, Mees ST. MicroRNAs: novel diagnostic and therapeutic tool for pancreatic ductal adenocarcinoma? Ann Surg Oncol. 2009;16(11):3183–3189. doi: 10.1245/s10434-009-0623-1. [DOI] [PubMed] [Google Scholar]
- 34.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 35.Luthra R, Singh RR, Luthra MG, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 36.Maru DM, Singh RR, Hannah C, et al. MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174(5):1940–1948. doi: 10.2353/ajpath.2009.080718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pallante P, Visone R, Ferracin M, et al. MicroRNA deregulation in human thyroid papillary carcinoma. Endocr Relat Cancer. 2006;13(2):497–508. doi: 10.1677/erc.1.01209. [DOI] [PubMed] [Google Scholar]
- 38.Zanette DL, Rivadavia F, Molfetta GA, et al. MiRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40(11):1435–1440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 39.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan LX, Huang XF, Shao Q, et al. MicroRNA miR-21 expression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ciafre SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 42.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102(52):19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.leSage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26(15):3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.duRieu MC, Torrisani J, Selves J, et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56(4):1–10. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 45.Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176. doi: 10.1007/s11605-008-0584-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuckenholz C, Lu L, Thakur P, Kaminski N, Bahary N. FACS-Assisted microarray profiling implicates novel genes and pathways in zebrafish gastrointestinal tract development. Gastroenterology. 2009;137:1321–1332. doi: 10.1053/j.gastro.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]


