Abstract
RcnR (resistance to cobalt and nickel regulator) is a 40 kDa homotetrameric protein and metalloregulator that controls the transcription of the Co(II) and Ni(II) exporter, RcnAB, by binding to DNA as an apo-protein and releasing DNA in response to specifically binding Co(II) and Ni(II) ions. Using x-ray absorption spectroscopy (XAS) to examine the structure of metals bound, and lacZ reporter assays of the transcription of RcnAB in response to metal binding, in WT- and mutant proteins, the roles of coordination number, ligand selection and residues in the N-terminus of the protein were examined as determinants in metal ion recognition. The studies show that the cognate metal ions, Co(II) and Ni(II), which bind in (N/O)5S six-coordinate sites, are distinguished from non-cognate metal ions (Cu(I) and Zn(II)), which bind only three protein ligands and one anion from the buffer, by coordination number and ligand selection. Using mutations of residues near the N-terminus, the N-terminal amine is shown to be a ligand of the cognate metal ions that is missing in the complexes with non-cognate metal ions. The side chain of His3 is also shown to play an important role in distinguishing metal ions. The imidazole group is shown to be a ligand in the Co(II) RcnR complex, but not in the Zn(II) complex. Further, His3 does not appear to bind to Ni(II), providing a structural basis for the differential regulation of RcnAB by the two cognate ions. The Zn(II) complexes change coordination number in response to the residue in position three. In H3C-RcnR, the Zn(II) complex is five-coordinate, and in H3E-RcnR the Zn(II) ion is bound to six protein ligands. The metric parameters of this unusual Zn(II) structure resemble those of the WT-Ni(II) complex, and the mutant protein is able to regulate expression of RcnAB in response to binding the non-cognate ion. The results are discussed within a protein allosteric model for gene regulation by metalloregulators.
Introduction
E. coli requires nickel for three NiFe-hydrogenases that catalyze the reversible oxidation of hydrogen and are associated with different pathways in the anaerobic metabolism of this facultative anaerobe.1–3 Despite this specific requirement, nickel levels in the cell must be tightly regulated, as high concentrations of nickel in the cell are toxic.4 To control the level of nickel in the cell, E. coli utilizes two transcriptional regulators, NikR5 and RcnR.6 The first regulator, NikR, controls the expression of the Ni(II) importer, NikABCDE,5 while RcnR regulates the expression of the Ni(II) and Co(II) exporter, RcnA, and the periplasmic protein, RcnB.6,7 RcnAB expression does not occur until levels of Ni(II) ions are reached where NikABCDE expression is fully repressed.6 Thus, the roles of NikR and RcnR are complementary, and their functions are similar to those seen for the E. coli Zn-responsive regulatory proteins, Zur and ZntR.8 Collectively, the importer, exporter and the associated metalloregulators are responsible for maintaining the proper Ni(II) ion flux through the cell. There is no known role for Co(II) ions in E. coli, and RcnRAB are the only known proteins characterized so far that are known to be involved cobalt homeostasis.
E. coli RcnR is a 40 kDa tetrameric transcriptional repressor that responds to the binding of Ni(II) or Co(II) ions by releasing DNA, resulting in the expression of RcnAB.6,9,7 RcnR and CsoR, a Cu(I)-responsive transcriptional regulator,10,11 constitute a new structural family of transcriptional regulators that is characterized by an all α-helical structure comprised of a four-helix bundle.9,10 Both proteins bind to their respective operator/promoter regions, repressing the transcription of their corresponding exporter. Apo-RcnR recognizes a TACT-G6-N-AGTA sequence, two of which are located in the rcnA-rcnR intergenic region.12 It has also been determined that the protein interacts with flanking DNA regions (~50 base pairs), leading to DNA wrapping.12
RcnR binds a variety of metals in vitro but only Ni(II) and Co(II) de-repress the transcription of RcnA.9 Studies aimed at understanding the structural parameters involved in metal recognition in CzrA and NmtR show that coordination number is an important determinant in distinguishing cognate from non-cognate metals.13,14 In addition to differences in coordination number, ligand selection (from a set of seven invariant potential metal ligands) was shown to distinguish Ni(II) from other metals in NikR.15 NikR binds the cognate metal Ni(II) in a planar four-coordinate His3Cys complex15–19 and to non-cognate metals Co(II), Cu(I), Cu(II) and Zn(II) by adopting a variety of alternate geometries and ligands.15
Data obtained from XAS show that in contrast to NikR, RcnR forms six-coordinate complexes with its cognate metal ions (Ni(II) and Co(II)), which adopt a (N/O)5S ligand environment that involves coordination of Cys35, the only Cys residue in the protein.9 Mutagenesis studies coupled with lacZ reporter assays and UV-visible spectroscopy suggested that the Ni(II) and Co(II) sites might also be ligated by the N-terminal amine, as well as the side chains of His3, His64 and His60 for Co(II).9 In addition, the fact that response to Co(II) binding could be lost while retaining a response to Ni(II) in mutant RcnR proteins,9 plus the observation that the M-S distances observed in the Co(II) (2.3 Å) vs. the Ni(II) (2.6 Å) complexes are distinct,9 suggested that Ni(II) and Co(II) might be differentially recognized by RcnR.
Here we show that the binding of non-cognate metal ions to WT-RcnR results in complexes that incorporate only three protein ligands and typically one anion from the buffer. Additionally, the role of the N-terminus in discriminating cognate and non-cognate metals, as well as in distinguishing between Ni(II) and Co(II), was explored by examining the metal site structures of complexes formed with N-terminal mutant RcnR proteins. These studies show that the N-terminus plays a critical role in the recognition of cognate metals by involving the N-terminal amine as a ligand for cognate metals, and reveals an apparent differential role for the side chain of His3 in distinguishing Ni(II) from Co(II). In contrast, non-cognate metal binding is not dependent on ligands from the N-terminus.
Experimental Procedures
RcnR Mutagenesis
The N-terminal mutants A2*, an alanine insertion between Met1 and Ser2, and H3L were previously described.9 The H3C and H3E genes were made using the previously described9 procedure using the primers: gagatatacatatgtcttgtacaatccgtgataaacagaaactg; and gagatatacatatgtctgaaacaatccgtgataaacagaaactg, respectively, with the changes underlined.
RcnR Overexpression and Purification
Single colonies of E. coli DL41 (DE3) pLysS cells with the plasmid encoding wild-type protein were grown in 150 mL Luria-Bertani broth (LB) cultures, supplemented with 30 μg/mL chloramphenicol (cam) and 100 μg/mL ampicillin (amp) overnight at 37 °C with shaking. An aliquot (20 mL) of the overnight culture was added to 2 L of fresh LB with cam and amp. The cultures were grown to an OD600 of ~0.8 and then induced by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.8 mM. The cells were harvested after 3 hours by centrifugation, resuspended in residual media, and frozen at -80 °C. The cells were lysed upon thawing in a water bath at 37 °C and treated with 10 μL of a DNAse I solution (10 mg/ml DNAse I, 40% glycerol), 1.5 mM (final concentration) of phenylmethyl-sulfonylfluoride (PMSF) (MP Biomedicals) and 5 mM (final concentration) tris(2-carboxyethyl)-phosphine hydrochloride (TCEP) (Thermo Scientific). The mixture was then incubated at 37 °C for half an hour.
All chromatographic purifications employed an AKTA-FPLC system (Amersham Biosciences). The lysed cells were centrifuged and the lysate applied to a SP sepharose column (18 mL) equilibrated with 20 mM Hepes (pH 7.0), 1 mM TCEP, 5 mM EDTA, 10% glycerol, 50 mM NaCl (buffer A). The column was washed with 50 mL of buffer A followed by a linear gradient from 0–100% of buffer B (20 mM Hepes (pH 7.0), 1 mM TCEP, 5 mM EDTA, 10% glycerol, 1 M NaCl), total volume 117 mL at a flow rate of 2 mL/minute while collecting 5 mL fractions. A sodium dodecyl sulfate polyacrylamide electrophoresis (SDS-PAGE) gel was run to determine the fractions containing RcnR; RcnR eluted at ~35–55% buffer B. These fractions were combined, concentrated to 4 mL and loaded on a HiLoad 16/60 Superdex 200 (GE Life Sciences) column equilibrated with buffer C (20 mM Hepes (pH 7.0), 1 mM TCEP, 5 mM EDTA, 10% glycerol, 300 mM NaCl). One column volume (120 mL) of buffer C was run over the column at a flow rate of 0.7 mL/min, fractions were collected based on absorbance at 280 nM. Protein fractions with an absorbance > 10 mAU were collected in 2 mL fractions. Following this column, a SDS-PAGE gel was run to determine the fractions that contained RcnR; RcnR eluted as a single peak at a volume consistent with that of a tetramer. These RcnR fractions were concentrated and buffer exchanged into buffer A. A MonoS 5/50 GL column (GE Life Sciences) was equilibrated with buffer A before loading 20 mL of the protein in buffer A. The column was washed with 20 mL of buffer A, followed by a linear gradient of 0–100% buffer B over 40 mL at a flow rate of 1.5 mL/minute while collecting 2 mL fractions. The purity of the fractions was checked using SDS-PAGE gel; RcnR eluted as a single peak at ~15–30% buffer B. Pure fractions of RcnR were pooled together and stored at 4 °C. Molecular weights of the expressed proteins were determined by Electrospray ionization mass spectrometry (ESI-MS) using a Bruker Esquire instrument equipped with an HP-HPLC for the removal of salt present in the protein solutions. The molecular weights of WT-, A2*-, H3L-, H3C-, H3E-RcnR proteins are 10003 Da (calculated = 10134 Da), 10075 Da (calculated = 10205 Da), 9980 Da (calculated = 10110 Da), 9967 Da (calculated = 10100 Da), 9996 Da (calculated = 10126 Da), respectively. The molecular weights obtained are ~131 Da less than expected from the amino acid sequence, consistent the loss of the N-terminal methionine residue. N-terminal processing of the protein was also noted by N-terminal amino acid sequencing of WT-RcnR.9
Metal Complexes of RcnR Proteins
Wild-type Co(II)-RcnR samples in buffers containing 20 mM Hepes, 300 mM NaCl or 300 mM NaBr and 10 % glycerol, as well as Ni(II)-RcnR in the same buffer with 300 mM NaBr, were prepared as outlined previously.9 The rest of the samples were prepared as outlined below. Buffer containing NaBr or NaOAc was used to detect coordination positions on metal centers where Cl− ligands could not be discriminated from S-donor ligands because of the similar X-ray backscattering cross sections of elements differing by less than Z = 2. The protein concentrations were determined by using the experimentally determined extinction coefficient of €276 = 2530 M−1 cm−1 on protein denatured with 8 M guanidine hydrochloride.9 The protein was concentrated to ~150 μM and desalted twice using a buffer containing 20 mM Hepes (pH 7.0), 10% glycerol and 300 mM NaCl, NaBr or NaOAcsalts (buffer M) using Zebra spin desalting columns, 7K MWCO, 10 mL (Pierce) to remove EDTA and TCEP. In the case of the Cu(I) samples, the final buffer also contained 2 mM TCEP. Two to three equivalents of aqueous 10 mM stock solutions of CoCl2•6H2O, NiCl2•6H2O, Zn(CH3COO)2•2H2O (Fisher Scientific) or [(CH3CN)4Cu]PF6 (Aldrich) salts were then added to the RcnR solutions to prepare the respective complexes. These samples were allowed to incubate overnight before mixing with Chelex™ beads for ~30 minutes to remove any nonspecifically bound metal.
Cu(I)-RcnR was prepared under anaerobic conditions. Air was removed from the protein solution on a Schlenk line by alternating between argon and vacuum five times. The protein was then placed into a Coy (Coy Laboratory Products Inc., Grass Lakes, MI) anaerobic chamber (90% N2 and 10% H2) and incubated with three equivalents of a 10 mM (CH3CN)4CuPF6 solution (nitrogen saturated anaerobic solution of 10 % acetonitrile in water) overnight. Chelex™ beads were added to the solution to remove any nonspecifically bound metal ions. The oxidation states of the Cu(I) samples were verified using X-band EPR (Bruker ELEXSYS E-500 X-band spectrometer) at 77 K on the samples frozen in XAS holders and inserted into a finger dewar. The percentage of Cu(II) present in the sample was determined by comparing the second integral of the Cu(I) protein spectra with that of a 1.03 mM Cu(II)-EDTA sample used as a standard. The Cu(I) sample in buffer M with NaBr contained ~7.4% Cu(II) while the sample in buffer M with NaCl had ~2.6% Cu(II) present.
Based on the previously determined protein concentrations, the volume of protein required to make 1 mL protein of solution containing 1 ppm of the respective metals was determined. Aliquots of the metal substituted proteins were diluted to 1 mL with deionized water for metal analysis. The protein metal content was determined using a Perkin-Elmer Optima DV4300 ICP-OES instrument. If the metal content was > 1.3 metals:protein, the sample was re-treated with chelex and the metal analysis was repeated. The metal:protein ratios for samples prepared in buffer M with 300 mM NaCl are as follows: Ni(II) 0.98:1, Cu(I) 1.24:1 and Zn(II) 0.84:1. The metal:protein ratios for samples prepared in buffer M with 300 mM NaBr are as follows: WT-RcnR: Cu(I) 1.14:1, Zn(II) 0.72:1; A2*-RcnR: Co(II) 1.28:1, Ni(II) 1.15:1, Zn(II) 1.27:1; H3L-RcnR: Co(II) 1.30:1, Ni(II) 0.99:1, Zn(II) 1.05:1; H3C-RcnR: Co(II) 0.59:1, Ni(II) 1.03:1, Zn(II) 0.97:1; H3E-RcnR: Co(II) 1.04:1, Ni(II) 0.44:1, Zn(II) 1.23:1. Ni(II)-substituted A2*- and H3L-RcnR in buffer M with NaCl had metal:protein ratios 0.67:1 and 0.84:1 respectively. A sample of Co(II) substituted H3E-RcnR in buffer M with NaCl and had metal:protein ratio 1.03:1. Samples prepared in NaOAc had the following metal:protein ratios: Ni(II) A2*-RcnR 0.88:1; Ni(II) H3L RcnR 0.66:1; Co(II) H3C-RcnR 1.23:1; Ni(II) H3E-RcnR 0.50:1.
β-Galactosidase Reporter Experiments
β-Galactosidase reporter experiments were conducted using two plasmids as follows:9 The chloramphenicol-resistant PrcnA-lacZ plasmid, pJI115,6 was transformed into the E.coli strain PC888 (ΔlacZ ΔrcnR) after which ampicillin-resistant plasmids expressing the WT, H3C, or H3E RcnR proteins were transformed into the strain. This strain background provides for low-level expression of the RcnR proteins. To assay reporter activity, the transformed cells were grown anaerobically in medium containing ampicillin and chloramphenicol with metals added up to the maximal concentration for Ni(II), Co(II), and Zn(II) (500 μM NiCl2, 150 μM CoCl2, and 150 μM ZnCl2) that resulted in < 10 % inhibition of growth (measured by final OD600) under these conditions.
X-ray Absorption Spectroscopy (XAS)
Samples of the metallated proteins were concentrated to 1–3.2 mM (50 μL) in 20 mM Hepes (pH 7.0), 300 mM NaCl or NaBr with 10% glycerol using a microspin concentrator (Vivascience). The samples were syringed into polycarbonate XAS holders that were wrapped in kapton tape, and rapidly frozen in liquid nitrogen. The final concentrating and freezing of Cu(I)-RcnR was conducted under an anaerobic atmosphere in a Coy chamber.
XAS data for the RcnR protein samples were collected as previously described.15 Data was collected under dedicated ring conditions on beamline 9-3 at Stanford Synchrotron Radiation Laboratory (SSRL) or at beam line X3b at the National Synchrotron Light Source (NSLS), Brookhaven National Laboratories. WT-RcnR + Co(II) in buffers containing NaBr or NaCl, + Ni(II) in NaBr buffer, + Zn(II) in buffers containing NaBr or NaCl, + Cu(I) in NaBr buffer, and H3L-RcnR + Ni(II) in NaOAc buffer were collected at SSRL. These data were collected at 10 K using a liquid helium cryostat (Oxford Instruments). The ring conditions were 3 GeV and 80–100 mA. Beam line optics consisted of a Si(220) double-crystal monochromator and two rhodium coated mirrors, a flat mirror before the monochromator for harmonic rejection and vertical columnation, a second toroidal mirror after the monochromator for focusing. X-ray fluorescence was collected using a 30-element Ge detector (Canberra) with the exception of Ni(II)-H3L in buffer with 300 mM NaOAc for which a 100-element detector was used. Soller slits with a Z-1 element filter were placed in between the sample chamber and the detector to minimize scattering.
The remaining samples were collected at NSLS on beam line X3b. Each sample was syringed into polycarbonate sample holders that were wrapped in kapton tape, and frozen in liquid nitrogen. The samples were loaded into an aluminum sample holder, which was cooled to ~50 K by using a He displex cryostat. Data were collected under ring conditions of 2.8 GeV and 120–300 mA using a sagitally focusing Si(111) double-crystal monochromator. Harmonic rejection was accomplished with a Ni-coated focusing mirror. X-ray fluorescence was collected using a 13-element Ge detector (Canberra). Scattering was minimized by placing a Z-1 filter between the sample chamber and the detector.
XANES was collected from ± 200 eV relative to the metal edge. The X-ray energy for each metal Kα-edge was internally calibrated to the first inflection point of the corresponding metal foil: Co, 7709.5 eV; Ni, 8331.6 eV; Cu, 8980.3 eV; and Zn, 9660.7 eV. EXAFS was collected to 13.5 – 16 k above the edge energy (Eo), depending on the signal:noise at high values of k.
Data Reduction and Analysis
The XAS data shown (vide infra) are the average of 4–10 scans. Each XANES spectrum used in the average was analyzed for edge energy shifts that might indicate redox chemistry in the beam. None of the samples showed any significant changes. XANES and EXAFS data were analyzed using EXAFS12320 and SixPack21 respectively. The SixPack fitting software builds on the ifeffit engine.22,23 XANES analysis was carried out as described previously15 by fitting a cubic function to the baseline in the pre-edge region of the spectra and using a 75% Gaussian and 25% Lorenzian function to fit the rise in fluorescence occurring at the edge. Transitions occurring at lower energy were fit using Gaussian functions and the areas of the Gaussians were taken to be the peak areas.
For the EXAFS analysis, each data set was background-corrected and normalized. The data were converted to k-space using the relationship k = [2me (E − Eo)/ℏ2]½ where me is the mass of the electron, ℏ is Plank’s constant divided by 2π, and Eo is the threshold energy of the absorption edge. The threshold energies chosen for the metals studied were 7723 eV for Co, 8340 eV for Ni, 8990 eV for Cu and 9670 eV for Zn.15 A Fourier-transform of the data was produced using the data range k = 2 – 12.5 Å−1, where the upper limit was determined by signal:noise. EXAFS was analyzed in r-space over the range r = 1 – 4 Å with four exceptions: wild-type RcnR + Ni(II) in buffer M with 300mM NaBr, wild-type RcnR + Cu(I) in buffer with 300 mM NaBr as well as wild-type RcnR + Zn(II) in buffer M with 300 mM NaBr, for which a Fourier-transform of the data was produced using data over the range k = 2 – 14 Å−1. The best fits for these data sets were also obtained using a Fourier-transform of the data over the range k = 2 – 12.5 Å−1 for direct comparison with the other data sets. Scattering parameters for EXAFS fitting were generated using FEFF 8.22 The first coordination sphere was determined by setting the number of scattering atoms in each shell to integer values and systematically varying the combination of N/O- and S-donors (Supporting Information, Tables S2 – S28).
Multiple-scattering parameters for various metals with histidine imidazole ligands were generated from previously published crystal structures10,24–27 using the FEFF8 software package.22 Paths of similar overall lengths were combined to make four imidazole paths.28 To compare different models of the same data set, ifeffit utilizes three goodness of fit parameters: χ2, reduced χ2, and the R-factor. χ2 is given by equation 1, where Nidp is the number of independent data points, Nε2 is the number of uncertainties to minimize, Re(fi) is the real part of the EXAFS function, and Im(fi) is the imaginary part of the EXAFS fitting function.
| (1) |
Reduced where Nvarys is the number of refining parameters and represents the degrees of freedom in the fit. Additionally Ifeffit calculates the R-factor for the fit, which is given by equation 2, and is scaled to the magnitude of the data making it proportional to χ2.
| (2) |
In comparing different models, the R-factor and reduced χ2 parameter were used to determine which model was the best fit for the data. The R-factor will always improve with an increasing number of adjustable parameters, while reduced χ2 will go through a minimum and then increase, indicating that the model is over-fitting the data.29
Circular Dichroism (CD)
CD experiments were carried out on a Jasco J-715 spectropolarimeter using 1 mm wavelength path length quartz cuvettes. Wavelength scans (20 °C, speed 20 nm/min, 2 nm step size) were performed on 15.5 μM apo WT-RcnR protein and 10 μM metal substituted WT-RcnR protein (in buffer with 10 mM Hepes, 150 mM NaCl, 5% glycerol at pH 7.0) that was allowed to equilibrate with two equivalents of metal for four hours. Each spectrum was the accumulation of four scans and the background buffer scans were subtracted from each spectrum and is shown in the Supporting Information Figure S2.
Results
RcnR Metal Specificity in vivo
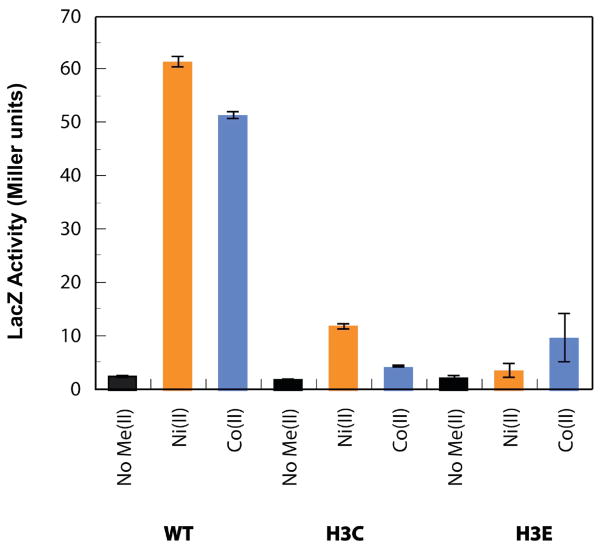
Using a LacZ reporter assay, in vivo metal-dependent transcription of PrcnA has been previously examined in WT-, A2*- and H3L-RcnR proteins.6,9 The studies of WT-RcnR showed that only in the presence of Ni(II) and Co(II) is PrcnA expression induced. Additionally, Mn(II), Fe(II), Cu(II), Zn(II), Cd(II) had no detectable effect on the expression of PrcnA. Thus, Ni(II) and Co(II) are the cognate metals and the rest are non-cognate metals. The A2*- and H3L-RcnR mutants were found to be unresponsive to all metals, suggesting an important role for the N-terminus and His3 in generating a response to the binding of cognate metals that would be consistent with roles as metal ligands.9 Results from the LacZ reporter assays for H3C- and H3E-RcnR with Ni(II) and Co(II) are shown in Figure 1 and show residual responses for Ni(II) and Co(II). The fact that the H3L mutant RcnR protein has no response,9 while substitution of His3 by potential metal ligands Cys or Glu leads to a residual response is further indication that His3 may be involved as a ligand in the metal binding site.
Figure 1.
The effects of H3C- and H3E-RcnR mutant proteins on Co(II) and Ni(II) responsiveness.
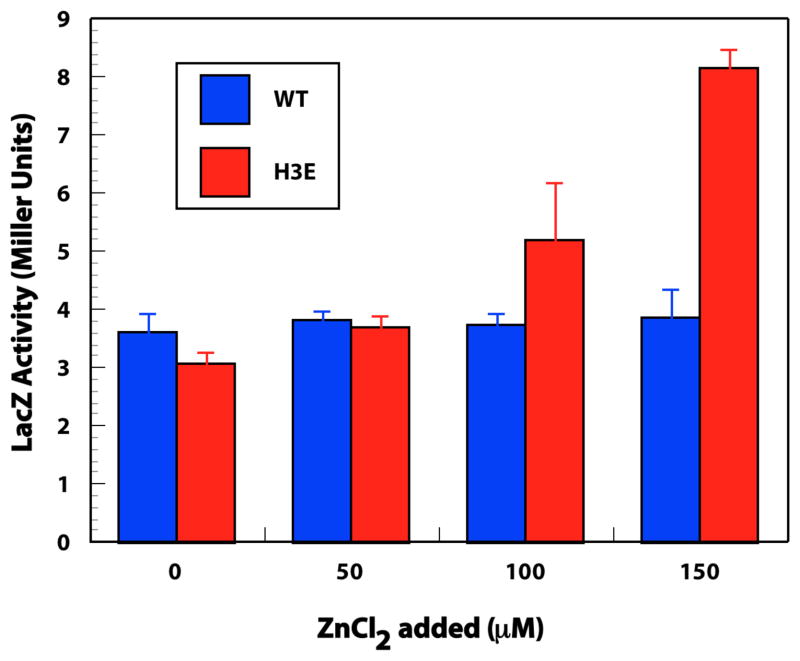
An unexpected result was obtained for H3E-RcnR in the presence of the non-cognate metal ion Zn(II). While WT-RcnR is unresponsive to Zn(II), the LacZ reporter assay data shown in Figure 2 demonstrate that H3E-RcnR has a response (albeit a weak one) to the binding of Zn(II). Thus, by changing one key residue, cognate status has been conferred on a non-cognate metal in the WT protein. The XAS data shows that the Zn(II) site is six/seven-coordinate in the H3E mutation (vide infra).
Figure 2.
LacZ reporter assay showing the expression of PrcnA with increased Zn(II) ion concentrations in the H3E mutant protein compared to the wild-type protein.
XAS Analysis
XAS was employed to determine the metal site structures for cognate and non-cognate metals in order to test the roles of coordination number and ligand selection in cognate metal recognition, to directly probe the role of the N-terminal amine and His3 side chain imidazole as ligands, and to explore the structural reasons for the response observed to the binding of Zn(II) in H3E-RcnR. In the case of the N-terminus, an insertion of Ala between the Met1 and Ser2 residue (A2*) was used to move the N-terminal amine one residue further from the metal site. In the case of His3, two types of mutations were involved: 1) where His3 was replaced by a non-liganding residue, such as Leu, and 2) where His3 was replaced by a potential S- or O-donor ligand (i.e.,Cys or Glu). The results of the XAS structural studies are summarized in and Tables 1–3 and Figures 3–9.
Table 1.
XANES Analysis of the Metal complexes of WT-, A2*-, H3L-, H3C- and H3E-RcnR in buffer with 20 mM Hepes, 300 mM NaBr or NaCl and 10% glycerol at pH7.0.
| Sample | 1s→3d Peak area (×10−2 eV) | 1s→4pz Observed | Coordination Number | Edge Energy (eV) |
|---|---|---|---|---|
| In buffer with NaCl | ||||
| WT | ||||
| Co(II)1 | 10.5(6) | No | 6 | 7719.8 |
| Ni(II) | 3(2) | No | 6 | 8343.3 |
| Cu(I) | NA | Yes | 3 | 8987.3 |
| Zn(II) | NA | NA | 4 | 9663.8 |
| In buffer with NaBr | ||||
| WT | ||||
| Co(II)1 | 9.9(6) | No | 6 | 7720.2 |
| Ni(II)1 | 3.4(7) | No | 6 | 8342.5 |
| Cu(I) | NA | Yes | 3 | 8987.0 |
| Zn(II) | NA | NA | 4 | 9663.6 |
| A2* | ||||
| Co(II) | 6.5(6) | No | 6 | 7720.2 |
| Ni(II) | 8.6(9) | Yes | 4 | 8343.9 |
| Zn(II) | NA | NA | 4 | 9663.0 |
| H3L | ||||
| Co(II) | 12.8(8) | No | 6 | 7721.4 |
| Ni(II) | 4.6(9) | No | 6 | 8344.5 |
| Ni(II)2 | 4.4(9) | No | 6 | 8344.1 |
| Zn(II) | NA | NA | 4 | 9662.7 |
| H3C | ||||
| Co(II) | 10(1) | No | 6 | 7720.6 |
| Ni(II) | 3(3) | No | 6 | 8343.5 |
| Zn(II) | NA | NA | 4 | 9663.9 |
| H3E | ||||
| Co(II) | 12(2) | No | 6 | 7720.8 |
| Ni(II) | 12(3) | No | 7 | 8343.7 |
| Zn(II) | NA | NA | 4 | 9665.6 |
From ref 9.
In buffer M with NaOAc.
Table 3.
Selected EXAFS Fits for the Metal Complexes of A2*-, H3L-, H3C-, H3E-RcnR Proteins in Buffer with 20 mM Hepes, 300 mM NaBr and 10% Glycerol at pH 7.0.
| Sample | Shell | r (Å) | σ2 (× 10−3 Å−2) | ΔEo (eV) | %R | Reduced χ2 |
|---|---|---|---|---|---|---|
| A2* | ||||||
| Co(II) | 4N/O (2Im) | 1.94(2) | 11(1) | −9(3) | 2.20 | 2.42 |
| 1S | 2.33(2) | 4(1) | ||||
| 1Br | 2.67(1) | 4(1) | ||||
| Ni(II) | 2N/O (2Im) | 1.85(2) | 6(2) | −7(2) | 1.85 | 4.01 |
| 1S | 2.09(3) | 11(4) | ||||
| 1Br | 2.41(2) | 6(1) | ||||
| Zn(II) | 2N/O (2Im) | 2.016(9) | 1.9(6) | −1(1) | 0.54 | 6.44 |
| 1S | 2.35(1) | 4(3) | ||||
| 1Br | 2.40(1) | 3.4(8) | ||||
| H3L | ||||||
| Co(II) | 4N/O (2Im) | 2.04(2) | 5(1) | −8(2) | 2.50 | 9.83 |
| 1S | 2.32(2) | 1(2) | ||||
| 1Br | 2.61(5) | 6(5) | ||||
| 4N/O (2Im) | 2.05(1) | 3(1) | −8(2) | 3.26 | 9.04 | |
| 1S | 2.33(1) | 3(1) | ||||
| Ni(II) | 2N/O | 1.98(2) | 0.3(17) | 2(2) | 3.15 | 14.77 |
| 3N/O (2Im) | 2.14(1) | 0(1) | ||||
| 1S | 2.62(4) | 7(5) | ||||
| Ni(II)2 | 2N/O | 2.01(2) | 0(1) | 0(2) | 2.19 | 109.34 |
| 3N/O (2Im) | 2.13(2) | 1(2) | ||||
| 1S | 2.61(4) | 8(5) | ||||
| Zn(II) | 2N/O (2Im) | 2.01(2) | 3(1) | −3(3) | 2.29 | 16.29 |
| 1S | 2.32(4) | 4(4) | ||||
| 1Br | 2.42(3) | 9(4) | ||||
| H3C | ||||||
| Co(II) | 3N/O | 2.10(2) | 1(1) | −1(3) | 4.19 | 10.47 |
| 2N/O (2Im) | 2.26(3) | 1(3) | ||||
| 1S | 2.68(2) | 3(3) | ||||
| Ni(II) | 5N/O (1Im) | 2.08(1) | 4.0(8) | 9(2) | 4.53 | 13.75 |
| 1S | 2.65(2) | 3(2) | ||||
| Zn(II) | 2N/O (1Im) | 2.00(2) | 4(1) | −8(2) | 1.05 | 5.07 |
| 2S | 2.35(2) | 4(2) | ||||
| 1Br | 2.36(2) | 3(1) | ||||
| H3E | ||||||
| Co(II) | 4N/O | 2.12(3) | 6(3) | 0(2) | 5.98 | 12.45 |
| 2N/O (1Im) | 2.25(5) | 5(6) | ||||
| 1S | 2.67(3) | 4(4) | ||||
| Ni(II) | 6N/O (1Im) | 2.080(9) | 5.3(8) | 9(1) | 3.79 | 11.52 |
| 1S | 2.66(2) | 0(2) | ||||
| Zn(II) | 3N/O | 2.06(1) | 2(1) | −2(1) | 1.60 | 9.25 |
| 2N/O (3Im) | 2.20(2) | 0(2) | ||||
| 1S | 2.57(4) | 9(4) | ||||
In buffer M with NaOAc.
Figure 3.
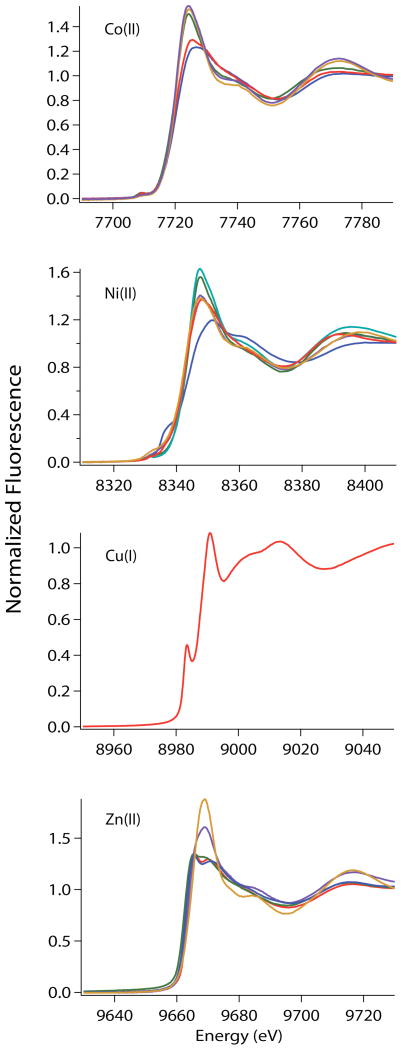
XANES overlay of metal complexes of RcnR proteins in buffer with 20 mM Hepes, 300 mM NaBr and 10% glycerol (except where noted): WT-RcnR (red), A2* (blue), H3L (green), H3L-OAc (cyan), H3C (purple) and H3E (orange).
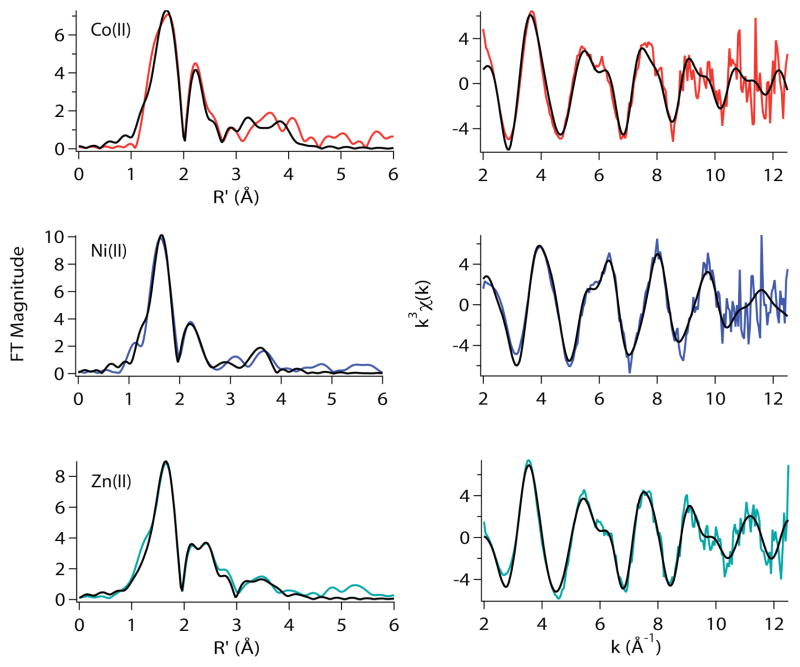
Figure 9.
Metal complexes of H3E-RcnR in buffer with 20mM Hepes, 300mM NaBr, and 10% glycerol at pH 7.0. Left: Fourier filtered XAS data (colored lines) and best fits (black lines) from Table 3. Right: Unfiltered k3-weighted EXAFS spectra and fits.
XANES Analysis
XANES analysis provides information on the coordination number and geometry of a metal center. The XANES analysis for various metal complexes of RcnR proteins are summarized in Table 1 and the spectra are shown in Figure 3. Metal ions such as Co(II) and Ni(II) have vacancies in the 3d manifold that give rise to features associated with bound transitions below the edge in energy in the XANES region. The intensity of these 1s→3d transitions (with shake-down contributions) is a measure of the centrosymmetricity of the metal site. For tetragonal geometries lacking one or more axial ligands, 1s→4pz electronic transitions may also be observed, and can be used to distinguish between different ;entrosymmetric geometries, e.g., square planar and octahedral.
In Co(II) complexes of WT-, A2*-, H3L-, H3C- and H3E-RcnR proteins, there is a single pre-edge feature at ~7710 eV (Tables 1, Figure 3) that is associated with the 1s→3d transition. The relatively small peak areas indicate that the Co(II) centers in all of these samples are centrosymmetric.20 In addition to the relatively small peak area, the absence of any other transition is consistent with an octahedral geometry.30 The 1s→3d peak areas for the Co(II) WT-, H3L-, H3C-, H3E-RcnR complexes is slightly larger than is typical for octahedral Co(II) complexes (0.069(8) eV)15 but smaller than that observed for five-coordinate complexes (~0.220(3) eV).20 The increase in the 1s→3d peak area seen for the Co(II) WT-RcnR complex is consistent with a distorted octahedral geometry that might result from a mixed-ligand donor atom set (e.g., (N/O)5S for WT-RcnR).31
The Ni(II) complexes of WT-, H3L-, and H3C-RcnR all have a small peaks associated with the 1s→3d transitions located at ~8330 eV (Table 1, Figure 3). The small areas of the 1s→3d transition in all samples except A2*-RcnR and H3E-RcnR suggest that the ligand arrangements are centrosymmetric, and the absence of a 1s→4pz transition in all the samples except A2*-RcnR is consistent with an octahedral Ni(II) center.32 The 1s→3d peak areas determined for A2*- and H3E-RcnR are larger, consistent with a non-centrosymmetric metal center and are within the range of peak areas expected for a tetrahedral complex (0.08–0.11).32 Only Ni(II) A2*-RcnR has a pre-edge shoulder at ~8338 eV that is associated with a 1s→4pz transition.32 This feature is not very well resolved, nevertheless its presence rules out octahedral, tetrahedral, or trigonal bipyramidal geometries.32,33 The intensity of the 1s→3d peak coupled with the shoulder at ~8338 eV is most consistent with a site that is a C2v distortion of a square planar geometry, in agreement with the four ligands found in EXAFS analysis (vide infra).33,34 The large 1s→3d peak area observed for Ni(II) H3E-RcnR is consistent with tetrahedral coordination.32 However, the best fit of the EXAFS indicates the presence seven ligands (vide infra), and so tetrahedral geometry is not supported. A seven-coordinate complex would also be non-centrosymmetric, and thus the data indicate that the Ni(II) site is seven-coordinate in H3E-RcnR, a geometry that might be supported by a bidentate carboxylate ligand in this complex.
The analysis of XANES from the copper complexes of RcnR was also used to determine the metal site geometry. No 1s→3d transition is expected for Cu(I) complexes, which feature a resolved peak between 8983 and 8984 eV associated with a 1s→4p transition. The shape, energy and intensity of this peak can be used to determine the coordination geometry of the Cu(I) ion.35 The 1s→4p transition in 2-, 3- and 4-coordinate model complexes have normalized absorption magnitudes of ~1, ~0.55 and between 0.6 and 0.9, respectively.35 Wild-type Cu(I)-RcnR has normalized absorption amplitude of 0.46 at 8983.6 eV. The intensity and energy of this peak suggests that Cu(I) forms a three-coordinate complex with RcnR.
In analogy with Cu(I), Zn(II) is a d10 metal and no 1s→3d transitions are possible. However, the electronic structure and ligand type has an effect on the intensity and shape of the XANES spectra, allowing for qualitative determination of the coordination number of the Zn(II) center as well as the type of ligands that are bound to the metal.36,37 Information regarding the coordination number of the Zn(II) center can be determined by the intensity of the XANES as well as the relative position of the edge in energy.37 The normalized intensity of the white line feature increases with increasing coordination number. For four-coordinate complexes the normalized intensity of the white line is ~1.3, five- and six-coordinate complexes have an intensity between 1.3 and 2, with six-coordinate being the most intense.37 The edge energy for six-coordinate complexes is ~2 eV higher than that of 4- or 5-coordinate complexes.37 Qualitative information about the type of ligand donor atoms present can be obtained from the two features in the XANES at ~9664 and ~9670 eV.36 If the Zn(II) site is made of purely thiolate ligands, then the first peak is most intense. As the ratio of nitrogen to sulfur ligands increases, the peak at ~9670 eV increases in intensity.36 In the Zn(II) complexes of WT-, A2*- and H3L-RcnR, the intensity of the white line is ~1.35 and the edge energy is ~9663 eV which is consistent with a four coordinate complex.37 There is an increase in the intensity of the white line features for Zn(II) H3C- and H3E-RcnR complexes to ~1.6 and ~1.9 respectively. The edge energy for the H3E-RcnR complex is ~9665.6 eV, which is 2 eV higher than the WT-, A2-* and H3L-RcnR Zn(II) complexes. Thus, the XANES analyses are consistent with wild-type, A2*- and H3L-RcnR forming four-coordinate tetrahedral complexes with Zn(II), whereas H3C- and H3E-RcnR proteins form complexes with higher coordination numbers, five- and six-coordinate, respectively. These results are also supported by the EXAFS analyses of the complexes (vide infra).
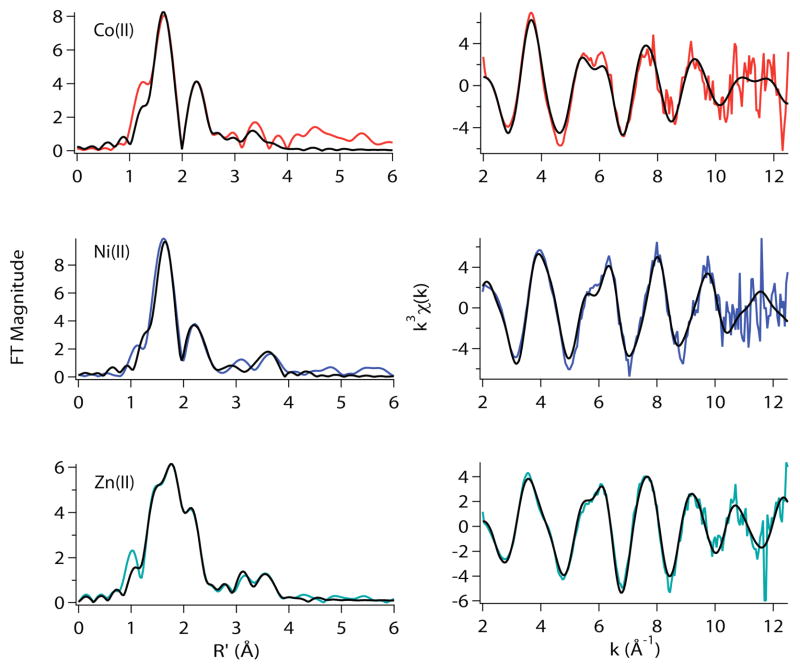
EXAFS analysis of WT-RcnR metal complexes
EXAFS analysis provides information regarding the atomic number of scattering atoms (Z ± 2), the distance between the absorbing and scattering atom (± 0.02 Å in the first coordination sphere), and an estimate of the number of similar and distinct ligands (± ~20% for the total number of ligands). Table 2, and Figures 4 and 5 compare the best fits, as judged by minimizing %R, minimizing reduced χ2, and acceptable σ2 values, for WT-RcnR metal complexes in buffer M containing 300 mM NaCl and NaBr. In cases where two fits are very close (and statistically indistinguishable), both are shown in Table 2. Additional fits are shown in the Supporting Information, Tables S2–S9.
Table 2.
EXAFS Analysis of the Cognate and Non-Cognate Metal Complexes of WT-RcnR in buffer with 20 mM Hepes, 300 mM NaCl or 300 mM NaBr and 10% Glycerol at pH 7.0.
| Sample | Shell | r (Å) | σ2(× 10−3 Å2) | ΔEo (eV) | %R | Reduced χ2 |
|---|---|---|---|---|---|---|
| NaCl | ||||||
| Co(II)1 | 5N/O (2Im) | 2.04(2) | 15(2) | −7(2) | 2.82 | 7.72 |
| 1S | 2.31(2) | 6(1) | ||||
| Ni(II) | 5N/O (1Im) | 2.076(7) | 5.1(6) | 8(1) | 2.15 | 7.78 |
| 1S | 2.62(3) | 10(3) | ||||
| Cu(I) | 2N/O (1Im) | 2.10(1) | 0.6(1) | 12(2) | 3.51 | 30.90 |
| 1S | 2.35(1) | 1(1) | ||||
| Zn(II) | 2N/O (2Im) | 1.98(1) | 3.0(8) | −8(2) | 1.64 | 19.63 |
| 2S | 2.27(1) | 6.2(7) | ||||
| NaBr | ||||||
| Co(II)1 | 5N/O (3Im) | 2.02(2) | 15(2) | −7(2) | 3.31 | 10.75 |
| 1S | 2.31(2) | 8(2) | ||||
| Ni(II)1 | 5N/O (1Im) | 2.063(8) | 6.6(6) | 5(1) | 2.40 | 19.83 |
| 1S | 2.62(3) | 10(3) | ||||
| Cu(I) | 2N/O | 2.13(2) | 2(2) | 3(1) | 1.69 | 25.30 |
| 1S | 2.291(7) | 0(0.8) | ||||
| 1Br | 2.62(1) | 7(1) | ||||
| Zn(II) | 2N/O (2Im) | 1.986(9) | 1.8(5) | −3(2) | 0.68 | 19.46 |
| 1S | 2.33(2) | 4(4) | ||||
| 1Br | 2.38(1) | 5(1) | ||||
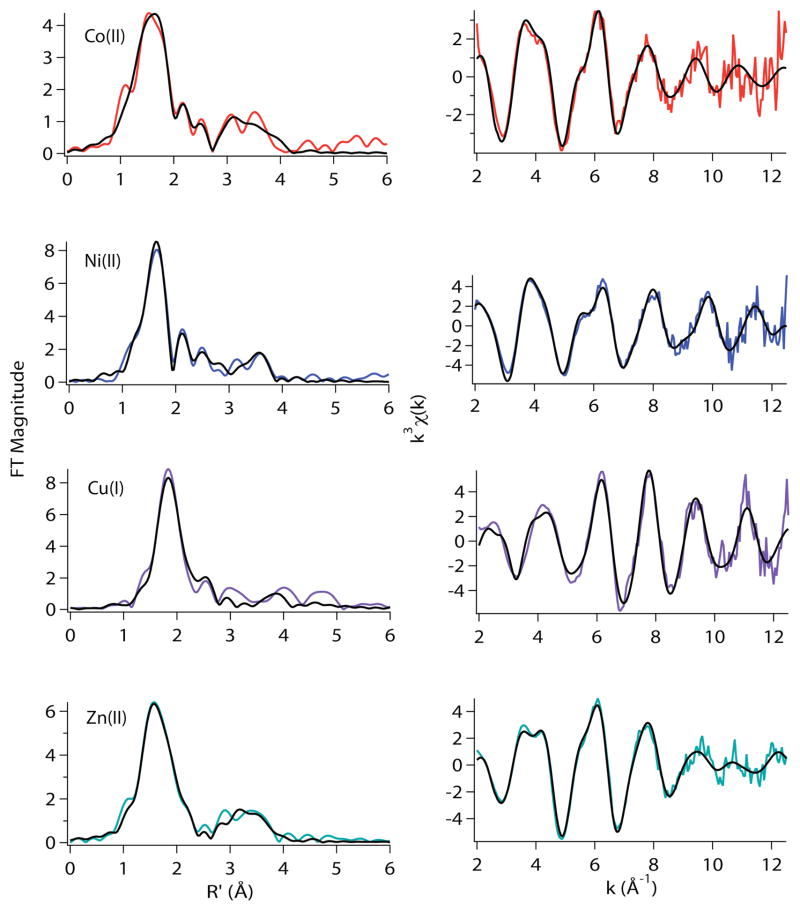
From ref 9.
Figure 4.
Metal complexes of wild-type RcnR in buffer with 20 mM Hepes, 300 mM NaCl, and 10 % glycerol at pH 7.0. Left: Fourier-filtered XAS data (colored lines) and best fits (black lines) from Table 2. Right: Unfiltered k3-weighted EXAFS spectra and fits.
Figure 5.
Metal complexes of wild-type RcnR in buffer with 20 mM Hepes, 300 mM NaBr, and 10% glycerol at pH 7.0. Left: Fourier filtered XAS data (colored lines) and best fits (black lines) from Table 2. Right: Unfiltered k3-weighted EXAFS spectra and fits.
The best EXAFS fits obtained for the cognate metals, Co(II) and Ni(II) are six-coordinate, in agreement with the XANES analysis (vide supra) and feature a ligand environment comprised of 5 N/O- and 1 S-donor ligands, consistent with prior results.9 There are two features that distinguish the Co(II) and Ni(II) site in WT-RcnR, the M-S distance and the number of His ligands. The Ni(II) site features a long Ni-S bond (2.62 Å) compared to the Co(II) sulfur distance of 2.31 Å (Table 2). The number of His ligands found for the Co(II) site is consistently greater by ~1 than that found for Ni(II) WT-RcnR. The best fit for the Co(II) site in buffer containing 300 mM NaCl features two imidazoles. In buffer with 300 mM NaBr the best fit has three imidazole ligands (Table 2). A fit for the Co(II) WT-RcnR (in buffer M with NaBr) data with two imidazoles fit is possible but this fit is inferior as it results in an increase of 19% in %R and reduced χ2 (Supporting Information, Table S6). Thus the best estimate for the His ligation in the Co(II) complex of WT-RcnR is two to three. In contrast, the best fits for the Ni(II) site feature one imidazole in both buffers. The fit involving two imidazole ligands is results in a 36% increase in %R and reduced χ2 for the sample made in buffer with NaCl (Table 2). For the sample made in buffer with NaBr the two imidazole fit resulted in an increase in %R of 19% and 13% in reduced χ2 when compared with the fit featuring one imidazole (Table 2).
The WT-RcnR complexes of non-cognate metals (Cu(I) and Zn(II)) also clearly feature ligation to a S-donor ligand, indicating that the only cysteine residue present in the protein, Cys35 is bound in all these cases, and that these metals therefore bind in the same locus in the protein as the cognate metals. The non-cognate metals are distinguished from the cognate metals by lower coordination numbers (three protein ligands), and the fact that they can bind an exogenous buffer ligand.
These trends are most clearly seen for the Zn(II)-RcnR complex. The best EXAFS fits obtained using N/O and S parameters for the sample prepared in buffer containing 300 mM NaCl indicate the presence of two N/O-donors from His ligands with an average Zn-N/O distance of 1.98 Å, and two S-donors at an average distance of 2.27 Å (Table 2). Considering that RcnR has only one cysteine, the additional S-donor is likely due to the binding of a Cl− anion from the buffer, which is not easily distinguishable from an S-donor by Zn K-edge EXAFS analysis. Replacement of NaCl by NaBr in the buffer shows that one of the Zn-S(Cl) vectors is replaced by a Zn-Br vector, but the structure is otherwise not significantly altered. The best fit in buffer containing NaBr corresponds to two N-donors from imidazole ligands with an average Zn-N distance of 1.99 Å, one S-donor at 2.33 Å and a Br− ligand at 2.38 Å.
The EXAFS data obtained for Cu(I)-RcnR in buffer M containing NaCl (Table 2) indicates that the best fit consists of two N/O-donors at 2.10 Å (one can be fit using imidazole multiple-scattering parameters) and one S-donor at 2.35 Å. This three-coordinate fit is consistent with the XANES analysis in both NaCl- and NaBr-containing buffers. However, in buffer M with NaBr, EXAFS fits for Cu(I)-RcnR indicate a four-coordinate site with two N/O-donors at 2.13 Å, one S-donor at 2.29 Å and a Br− ligand at 2.62 Å (Table 2), which appears at face value to be inconsistent with the XANES analysis (vide supra). The addition of a weak 2.62 Å Cu-Br interaction is apparently not sufficient to perturb the electronic structure of Cu(I) enough to change the prediction of geometry based on XANES features from three to four coordinate. The Cu(I) complex also shows a UV-vis transition at 236 nm (11100 M−1cm−1, see supporting information), similar to that seen for the Cu(I) CsoR complex.10,11 The appearance of Br−, but not Cl− in the coordination sphere of Cu(I) is not surprising considering the fact that in contrast to Zn(II), which binds Cl−, Cu(I) often occupies three-coordinate sites and is a softer acid that would have a higher affinity for Br− than for Cl−. The Cu(I)-RcnR complex is the only WT-RcnR complex where changing the anion present in the buffer appears to influence the coordination number of the metal center.
EXAFS analysis of mutant RcnR metal complexes
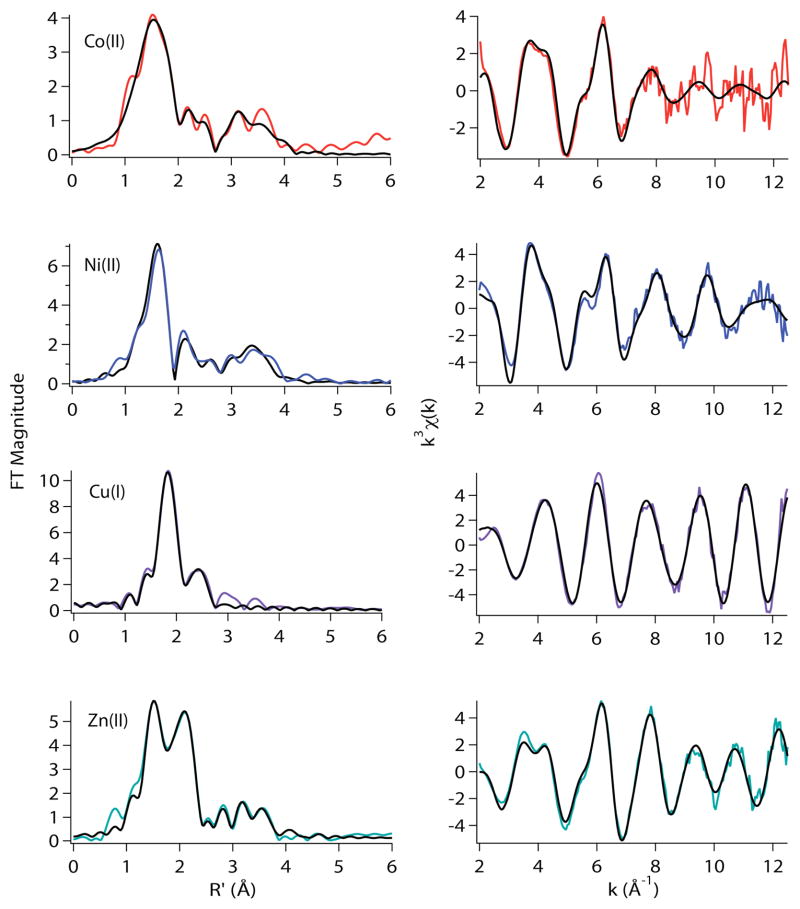
The results of XAS studies of the metal site structures in A2*-RcnR and mutations involving His3 are summarized in Table 3 and Figures 6 – 9. The A2* mutation involves the insertion of an alanine residue at the second position of the gene, which results in an alanine addition to the N-terminus of the mature protein. This mutation leads to a loss of metal binding response for both Ni(II) and Co(II).9 The Co(II) complex of A2*-RcnR is predicted by XANES analysis be six-coordinate, the same coordination number as the WT-RcnR complex. However, EXAFS analysis reveals that the ligand environment is altered and consists of four N/O-donor ligands at an average distance of 1.94 Å, of which approximately two can be modeled as His imidazole ligands, one S-donor at 2.33 Å (Cys35) and one is a Br− anion at 2.67 Å (Table 3). Thus, loss of the N-terminal amine results in an open coordination position on the Co(II) center that is occupied by an anion from the buffer, confirming that the N-terminal amine is a ligand for Co(II).
Figure 6.
Metal complexes of A2*-RcnR in buffer with 20 mM Hepes, 300 mM NaBr, and 10% glycerol at pH 7.0. Left: Fourier filtered XAS data (colored lines) and best fits (black lines) from Table 3. Right: Unfiltered k3-weighted EXAFS spectra and fits.
The Ni(II) A2*-RcnR complex shows a more dramatic change in structure. XANES analysis predicts a four-coordinate structure (vide supra) and the EXAFS analysis shows that this structure is best modeled by a combination of two N-donors from His ligands at an average Ni-N distance of 1.85 Å, one S-donor at 2.09 Å and a Br− at 2.41 Å (Table 3). The Ni-S bond distance determined by EXAFS is too short, even for a square-planar Ni(II) complex, where Ni-S distances fall in the range 2.13 – 2.22 Å.38–42 For this reason, XAS spectra were also run in buffers containing NaCl, where the anion and the S-donor ligand may not be distinguishable, and in buffers containing NaOAc, where an extra O/N-donor ligand would be expected and is found. These data gave more reasonable Ni-S bond distances of 2.19 Å and 2.14 Å respectively (Supporting Information, Tables S12 and S13). The EXAFS analysis agrees with the prediction of a four-coordinate structure that is a C2v distortion of a planar complex where the variation of the Ni-S distances in the different anion complexes suggest that the degree of distortion may be dependent on the nature of the anion. The structure of the Ni(II) site in A2*-RcnR that emerges from the analysis of the XAS data is reminiscent of that found for the Zn(II) site in WT-RcnR, His2CysBr (vide supra). Thus, loss of the N-terminal amine results in a nickel complex whose structure is more typical of a non-cognate metal, and is also consistent with ligation of the N-terminal amine Ni(II) WT-RcnR.
In contrast to the cognate metal complexes, the structure of the Zn(II) site in A2*-RcnR is unperturbed by the mutation (Table 3). The EXAFS fit obtained is indistinguishable from the fit obtained for the Zn(II) WT-RcnR sample. The N-terminus is therefore one of three protein ligands missing in complexes of non-cognate metals, and is thus a key structural element that distinguishes cognate from non-cognate metal binding.
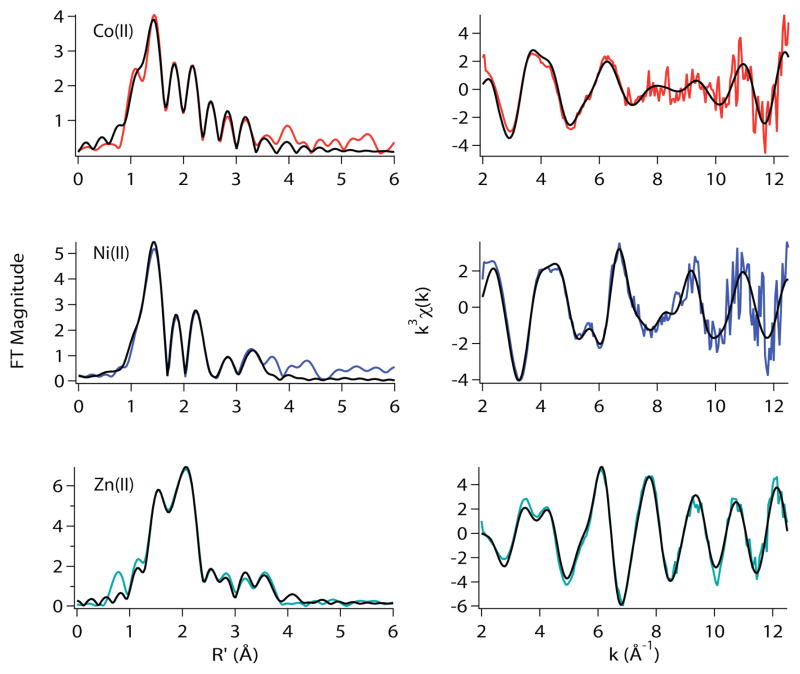
Mutating His3 to a Leu is known to lead to a loss of function for both Co(II) and Ni(II) ions.9 In terms of metal site structure, mutating His3 to a Leu affects only the structure of the Co(II) site, and neither the Ni(II) nor the Zn(II) site structures. (Table 3, Figure 7). The best EXAFS fit for the Co(II) site of H3L-RcnR indicates a six-coordinate site that is consistent with the XANES analysis (vide supra) and is composed of four N/O-donors at an average distance of 2.04 Å, of which approx. two are His imidazole ligands, one S-donor at 2.32 Å and one Br−-ligand at 2.61 Å. Compared with a five-coordinate fit sans the Br− anion (Table 3), this fit results in a better %R, but the value of reduced χ2 increases relative to the five-coordinate fit by ~ 9%, indicating that the data is being slightly over-fit by the model. In either case, the effect of the mutation is to open a coordination site on the Co(II) center that may be occupied by an anion from the buffer. It is possible that the ambiguity in the fits with respect to the presence of Br− indicates a mixture that results from an equilibrium between Br− on and off complexes.
Figure 7.
Metal complexes of H3L-RcnR in buffer with 20mM Hepes, 300mM NaBr /1NaOAc, and 10% glycerol at pH 7.0. Left: Fourier filtered XAS data (colored lines) and best fits (black lines) from Table 3. Right: Unfiltered k3 weighted EXAFS spectra and fits.
In contrast, the Ni(II) site structure is not significantly perturbed by the H3L mutation (Table 3), indicating that His3 is not a required ligand for Ni(II). While it is possible that another His residue substitutes for His3 in the H3L-RcnR complex, given that the average bond distances for the N/O- and S-donors vary by only 0.01 Å from the distances found in the WT-RcnR complex makes a protein ligand substitution very unlikely. This result confirms that Ni(II) and Co(II) are differentially recognized by RcnR, as might also be inferred by the difference in the M-S distances in the complexes formed with WT-RcnR and the tendency for Co(II) to coordinate one more His imidazole than Ni(II), assigned on the basis of this data as His3. In addition, the fact that both metals are six-coordinate and differ in coordination of His3 indicates that ligand selection is involved in discriminating between the two metals. The best fit of the Ni(II) EXAFS data for the H3L-RcnR complex is consistent with the six-coordinate site predicted by XANES analysis, with two shells comprised of a total of five N/O-donors at average distances of 1.98 Å and 2.14 Å, of which two are His ligands, and the SCys35 ligand at 2.62 Å (Table 3). [An acceptable fit was also obtained involving a Br-donor instead of a S-donor that had a similar coordination environment except for substitution of the long S-donor by a Br− ligand with a very long Ni-Br distance = 2.75(7) Å (Supporting Information, Table S16). Data obtained on the Ni(II) H3L-RcnR complex in NaOAc removes this ambiguity. The best fit for the Ni(II)-H3L in NaOAc buffer retains the S-donor at a Ni-S distance of 2.61 Å (Table 3).]
As was the case in A2*-RcnR, the Zn(II) complex of H3L-RcnR is unperturbed by the H3L mutation. The best fit for the data is two N/O-donors modeled as two histidine imidazole ligands, a S-donor and a Br−-ligand. This mutation identifies a second metal binding residue in the wild-type RcnR protein that is present for a cognate metal but does not coordinate the non-cognate Zn(II) ion.
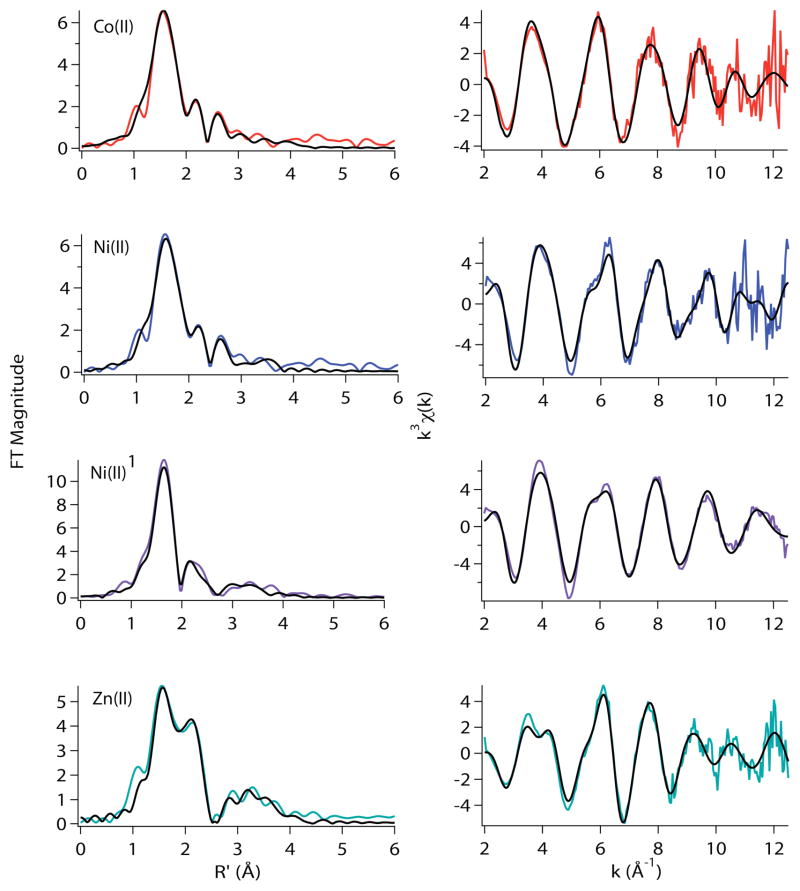
If His3 is a ligand for Co(II) and not for Ni(II) or Zn(II), then the structures of the metal sites would be expected to respond differently when mutations involving substitution of His3 by other potential metal ligands are employed, such as H3C and H3E (Table 3, Figures 8 and 9). XANES and EXAFS analyses of the H3C-RcnR Co(II) complex are consistent with a six-coordinate Co(II) site. However, in contrast to the Co(II) H3L-RcnR site, the XAS data obtained for Co(II) H3C-RcnR are not sensitive to the nature of the anion present, indicating that there is no open coordination site on the Co(II) centers. (Data obtained in buffers containing NaBr and NaOAc are essentially identical, Table 3 and Supporting Information Tables S20, and S21). The best EXAFS model has only one S-donor ligand (Table 3) and ligation of Cys3 vs. Cys35 cannot be unambiguously distinguished by XAS alone. However, the structure of the Co(II) site in H3C-RcnR resembles that of the Ni(II) site WT-RcnR in that it is an (N/O)5S site that has a long (2.68 Å) M-S distance and ~two His ligands. Thus, despite the apparent ambiguity, the data are consistent with Co(II) adopting the ligand set found for the Ni(II) complex of WT-RcnR, which does not include a ligand from the amino acid residue in position three (vide supra).
Figure 8.
Metal complexes of H3C-RcnR in buffer with 20mM Hepes, 300mM NaBr, and 10% glycerol at pH 7.0. Left: Fourier filtered XAS data (colored lines) and best fits (black lines) from Table 2. Right: Unfiltered k3-weighted EXAFS spectra and fits.
The Ni(II) site in H3C-RcnR is not significantly altered by the H3C mutation, and is therefore similar to sites determined for Ni(II) complexes of WT- and H3L-RcnR, and consistent with the notion that His3 is not a ligand for Ni(II) in WT-RcnR. The site is six-coordinate, and the best EXAFS fit is comprised of five N/O-donors at an average distance of 2.08 Å and one S-donor at 2.65 Å. The best fit is obtained for a model featuring one His imidazole ligand (Table 3), but an acceptable fit is also obtained with two His ligands (Supporting Information, Table S22).
The H3C-RcnR mutant is the first Zn(II) complex examined where the structure of the Zn(II) site is altered. Based on the EXAFS analysis (Table 3 and Supporting Information Table S23), the Zn(II) site is five-coordinate with two N/O-donors at an average distance of 2.00 Å, including ~ one His ligand, two S-donors at an average distance of 2.35 Å and a Br− ligand at 2.36 Å. The change in coordination number from four in Zn(II) WT-RcnR to five in Zn(II) H3C-RcnR is in agreement with the XANES analysis presented above.
The EXAFS analyses of the H3E-RcnR complexes reveal that the Co(II), Ni(II) and Zn(II) sites are best modeled as seven-coordinate structures (Table 3). Although acceptable six-coordinate fits can also be obtained (Supporting Information, Tables S24–S28), in each case seven-coordination leads to lower values of reduced χ2 and %R without leading to unreasonably large values of σ2. Further, no other RcnR samples exhibit a preference for seven-coordinate fits, and the non-centrosymmetric geometry is in agreement with the more intense feature associated with a 1s→3d transition in the XANES spectra from Ni(II) and Co(II) complexes of H3E-RcnR (vide supra). A seven-coordinate geometry is a bit misleading since bidentate coordination of the Glu3 carboxylate would lead to seven scattering atoms in the EXAFS but a minor perturbation in an octahedral arrangement of six protein ligands.
The best EXAFS models for the two cognate metal sites in H3E-RcnR are virtually identical and feature a (N/O)5–6S ligand donor atom set, involving one – two His ligands and featuring the long Ni-S (2.6 Å) distance that are characteristics of the WT-RcnR Ni(II) site (Table 2). The best six-coordinate fits arise from the loss of one N/O donor from the best seven-coordinate fit (Supporting Information, Tables S24–S28). As was the case for the H3C mutation, data obtained in the presence of NaBr indicated that there is no evidence of an open coordination position on the Co(II) or Ni(II) center in the H3E-RcnR complexes (Supporting Information, Tables S24–S28).
The structure of the Zn(II) site in the H3E-RcnR mutant is also perturbed from the structure found for Zn(II) WT-RcnR structure. The best model found from EXAFS analysis is consistent with either a six- or seven-coordinate (N/O)5–6S structure including two - three His imidazole ligands, and one SCys-donor at 2.57 Å. This structure, which is associated with the gain of cognate function, also most closely resembles the metal site structure in Ni(II) WT-RcnR, and does not coordinate a Br− ligand in buffers containing NaBr.
Discussion
In E. coli, Ni(II) is essential for the production of three NiFe-hydrogenases.43,44 No specific target for Co(II) has been identified in E. coli, which cannot synthesize vitamin-B12 de novo.45,46 The Co(II) ion is thought to enter the cell through non-specific metal importers,9,47,48 and is presumably treated as a toxic metal. Cellular levels of the two metal ions must be regulated to avoid becoming toxic to the bacterium. The transcriptional regulator RcnR is responsible for controlling the expression of the exporter RcnA in response to cellular levels of Ni(II) and Co(II) by binding to the rcnA-rcnR intergenic region in the absence of metals and releasing DNA when the cognate metals are bound, allowing transcription of PrcnA.6,12 Thus, one plausible role for cognate metals is to disrupt DNA binding by causing the protein to adopt a conformation that disfavors interaction with DNA, or altering the protein dynamics ;o disfavor the DNA-binding conformation, as is the case for CsoR, NikR, CzrA and NmtR.10,13,14,49,50 In any case, how the RcnR protein distinguishes among many transition metals, many of which have a +2 charge and similar ionic radii, is not well-defined.
XAS studies of WT-RcnR complexes were employed to examine the metal site structures of cognate and non-cognate metals. These studies reveal that coordination number plays an important role in recognition of cognate metal ions by RcnR. While cognate metals bind to RcnR with a six-coordinate geometry and (N/O)5S ligand donor set, non-cognate metals are coordinated by three protein ligands with (N/O)2S donor atoms, and have one open coordination position that can be occupied by an anion from the buffer. All WT-RcnR metal complexes have one S-donor. Since Cys35 is the only Cys residue in the protein, all the metals bind in the same locus in the protein, but with different numbers of protein ligands. The six-coordinate structures of the Ni(II) and Co(II) sites differ in the M-S distance (2.31 Å for Co(II) and 2.62 Å for Ni(II)) and in the number of His imidazole ligands (~ 3 for Co(II) and ~ 2 for Ni(II)), suggesting that the two cognate metals are differentially recognized. This feature likely gives rise to the different binding constants for the two metals (Kd of 5 nM for Co(II) and 25 nM for Ni9), the fact that mutants can be produced that lose response to one metal and not the other,9 as well as providing a mechanism for different sensitivity to the two metals.9
Despite being members of the same family of transcriptional regulators, RcnR and CsoR appear to recognize their cognate metals by distinct mechanisms. Crystal structures show that like RcnR, CsoR is a tetrameric helical protein.10,51 Crystallographic and XAS studies show that the cognate metal, Cu(I), is coordinated in a trigonal N2S complex by two cysteine residues, Cys36 from one subunit and Cys65 from another subunit, as well as histidine residue, His61, from the same subunit as Cys65.10,11 Although these residues correspond to Cys35, His60 and His64 in the RcnR sequence, no ligands in the N-terminus of CsoR are bound to the cognate metal. B. subtilis CsoR binds Co(II), Ni(II) and Zn(II) with a binding affinity ≤ 105 M−1, 109 M−1 and 108 M−1 respectively,11 and binds Cu(I) the tightest with a binding affinity ≥ 1021 M−1,11 suggesting that metal selectivity may be under more thermodynamic control in CsoR than in RcnR. UV-vis spectroscopy showed Co(II) forms a complex with tetrahedral geometry with CsoR, while Ni(II) binds with square planar geometry (not six-coordinate as in RcnR),11 although the identity of the additional ligands is not apparent.
A few other nickel-responsive transcriptional regulators, including the E. coli NikR,15 bind their cognate metals with one particular geometry and non-cognates with another. NikR can bind to a variety of other transition metals in vitro.15,52 The relative metal binding affinities follow the Irving-Williams series (Co(II) < Ni(II) < Cu(II) > Zn(II)), with Cu(II) binding 1000x tighter than Ni(II)).52 However, in vivo, the protein only responds to the presence of Ni(II) ions.15 Metal ion selectivity in NikR appears to be achieved by the coordination number/geometry of the metal-protein complex as well as ligand selection.15 XAS studies showed that the NikR metal sites feature a four-coordinate planar geometry for cognate Ni(II) and non-cognate Cu(II).15 These two sites are distinguished by the metal-S bond distances, which are 2.13 Å in the Ni(II) complex and 2.21Å in the Cu(II) complex.15 The Co(II) ion binds to NikR in a site with octahedral geometry that lacks the S-donor from Cys95, while Cu(I) forms a three-coordinate trigonal geometry and Zn(II) forms a tetrahedral structure and both include the Cys95 ligand.15
The role of coordination number in metal recognition is also apparent in CzrA and NmtR,13 and in CsoR.11,53 CzrA and NmtR are members of the ArsR/SmtB family of transcriptional regulators, and like RcnR release DNA in response to cognate metal binding. S. aureus CzrA regulates the expression of the czr operon that encodes a Co(II)/Zn(II)-facilitated pump, CzrB.54 M. tuberculosis NmtR controls the expression of the P-type ATPase metal efflux transporter, NmtA, by regulating the nmt operon in response to Ni(II) and Co(II).14,13 Despite the fact that CzrA and NmtR share 30 % sequence identity they respond to different metals that bind with distinct coordination numbers/geometries. UV-vis and XAS studies show that Zn(II) and Co(II) bind to CzrA forming four-coordinate tetrahedral structures while Ni(II) adopts an octahedral geometry. NmtR binds Co(II) and Ni(II) in a five/six-coordinate and a six-coordinate geometry respectively.13 Thus, the two metallosensors are designed to respond to cognate metals that bind in different geometries, and Ni(II) adopts a six-coordinate geometry in both proteins.
The situation in the two E. coli nickel metalloregulators, RcnR and NikR is somewhat different in that the same cognate metal adopts different coordination numbers/geometries in the two metalloregulators, a high coordination number in the former and a low-coordination number geometry (four-coordinate planar) in the latter, in order to produce metalloregulators that de-repress and repress transcription in response to Ni(II) binding, respectively. The metallosensor roles of the two proteins are complementary and similar to that seen for the E. coli Zn-responsive regulatory proteins, Zur and ZntR, which work to control Zn(II) concentrations in the cell.8 Zur and ZntR function by repressing the transcription of ZnuABC, an ABC-like Zn2+ uptake protein and activating the Zn2+ exporter, ZntA in the presence of excess zinc respectively.8,55–57
In addition to coordination number/geometry, ligand selection also appears to play a role in discriminating cognate from non-cognate metals in RcnR. Results from LacZ reporter assays that monitor the transcription of PrcnA point to the importance of the N-terminus in allosterically coupling cognate metal binding in RcnR to a decrease in the protein’s affinity for DNA. Mutations that affect the position of the N-terminal amine (A2*-RcnR) and availability of His3 (H3L-RcnR) both lead to a loss of response to both metals.9
XAS studies of A2*-RcnR complexes show that the Co(II) and Ni(II) site structures are both altered, but in different ways. For Co(II) A2*-RcnR, the alanine insertion, which moves the N-terminal amine one residue further away from the metal binding site, opens a coordination position on the metal ion that is occupied by Br− in buffer containing NaBr. For Ni(II) A2*-RcnR, the mutation causes the Ni to adopt a His2Cys35Br ligand set that resembles the coordination environment of the non-cognate Zn(II) complex of WT-RcnR. In contrast, the structure of the Zn(II) complex of A2*-RcnR is unaffected by this mutation. Thus, the coordination of the N-terminal amine is one key structural determinant in cognate metal recognition. Although somewhat unusual in metalloproteins, coordination of the N-terminal amine has been observed in the nickel-dependent superoxide dismutase enzyme (NiSOD) which carries out the disproportionation of superoxide to molecular oxygen and hydrogen peroxide,58,59 The binding of the N-terminus was also observed for the binding Ni(II) and Cu(II) to serum albumin.60 Although the N-terminal amine is not a ligand of the Cu(I) site in CsoR, the crystal of the Cu(I) complex of WT-CsoR supports the potential for the coordination of the N-terminus to metals, as the N-terminus is positioned directly above the Cu(I) binding site.10
XAS studies of H3L-RcnR also reveal the different structural roles played by the imidazole side chain of His3. For Co(II)RcnR, His3 is clearly a ligand, as the H3L mutation opens a coordination position on the Co(II) site that is occupied by Br− in buffer containing NaBr. However, the Ni(II) and Zn(II) sites are unaffected by this mutation. To further examine the differential use of His3 in the two cognate metal complexes, His3 was substituted by two potential metal-binding residues, Cys and Glu. These mutations restored some residual responsiveness to Co(II) and Ni(II) binding, suggesting that having a metal ligand or H-bonding residue in position three is important to cognate metal recognition. However, the structural effects of the mutations that lead to partially restored function was not anticipated. In both mutations, the Co(II) site has six protein ligands (no evidence of an open coordination site). However, this was not generally due to the coordination of the residue in position three. Rather, the structure obtained from EXAFS resembles that of Ni(II) WT-RcnR, featuring a long Co-S bond distance of 2.68 Å (compared to a Ni-S bond-distance of 2.61 Å in WT-RcnR), suggesting, at least in as far as the M-S distance indicates the nature of the metal bound, that Co(II) may now be recognized as Ni(II).
The general loss of response to Ni(II) binding associated with mutations of His3 indicates that His3 may be playing some other role in allosterically coupling Ni(II) binding to a decrease in the protein’s affinity for DNA. One such role might be in participating in structurally important H-bonding interactions, as seen for His61 in CsoR. His61 coordinates the cognate metal, Cu(I), in CsoR and also participates in a H-bonding network with Tyr35 and Glu81 that stabilizes the allosterically inhibited conformation.10 In elegant work incorporating unnatural amino acids that retain the ligand function but disrupt the H-bond, substitutions of His61 with Nε2-methyl-histidine (MeH) or 4-(thiazolyl)-L-alanine (Thz) showed that while Cu(I) binding to the protein was unaffected, DNA-binding was no longer strongly regulated by cognate metal binding.61 Similar results were also obtained for His97 mutations in CzrA and His117 mutations in SmtB, where disruption of H-bonds set up by coordination of the His residues to the cognate metals were shown to be essential for coupling cognate metal binding with the allosteric response.50
The fact that the structures of the Co(II) and Ni(II) complexes are distinct provides a mechanism for adjusting the binding constants so that cobalt binds tighter than nickel, opposite of what is expected from the Irving-Williams series, thus providing a mechanism for tuning the sensitivity of the metallosenor to the two metals. That His3 is a ligand for Co(II) and not for Ni(II), suggests that different protein conformations might exist that can be distinguished by the M-S distance in the complex. Further, whether the Co(II) site or the Ni(II) site is occupied depends on whether His3 is utilized as a ligand. These conformations might perhaps be stabilized by different H-bonding interactions involving His3.
The Zn(II) complexes showed unusual structural changes in response to the His3 mutations. For H3L-RcnR, the Zn(II) site is indistinguishable from that in WT-RcnR, Zn(His)2CysBr, and thus His3 is not a ligand in the WT-RcnR Zn(II) site. However, the H3C-RcnR mutation resulted in a five-coordinate Zn(N/O)2S2Br site that appears to arise from the coordination of the second the Cys residue. Most unusual is the characterization of a six- (or seven) coordinate Zn(N/O)5–6S complex for H3E-RcnR that gives rise to transcription of PrcnA in response to the binding of a non-cognate metal, Zn(II), in the lacZ reporter assay. A six-coordinate Zn(II) center is very unusual in biology, but was observed in the crystal structure of P. aeruginosa ferric uptake regulator (Fur), where the zinc site at the dimerization domain had a distorted octahedral geometry.62 The resemblance between the Zn(II) structure in H3E-RcnR, and the cognate metal sites in WT- and H3E-RcnR suggests that Zn(II) is being recognized by a similar mechanism. The H3E-RcnR Zn(II) site has a Zn-S distance of 2.61 Å (like the WT- and H3E-RcnR complexes of Ni(II)), plus binds the third amino acid residue, E3, a feature of the WT- and H3E-RcnR Co(II) site, suggesting that the Zn(II) site has adopted the coordination number, and geometry associated with the cognate metals, with the protein ligand selection similar to WT-RcnR Co(II) ,which also has a weak transcriptional response in this mutant. This is a strong confirmation of the hypothesis that metal recognition is governed by coordination number and ligand selection in RcnR.
Conclusion
Discrimination of cognate vs. non-cognate metal ions by the coordination number and the ligands adopted by the metal is emerging as a common theme in metal recognition by many metallosensor proteins.63 The Ni(II) and Co(II) responsive transcriptional regulator in M. tuberculosis, NmtR, also possesses a His ligand in the position three.14,64 Mutagenesis and metal binding studies identified His3 as an important Ni(II) ligand. The H3Q-NmtR mutant protein showed more sensitivity to non-cognate Zn(II) regulation, relative to the cognate metals Co(II) and Ni(II). However, unlike RcnR, H3Q-NmtR could regulate protein-DNA interactions in response to Ni(II) as well as the WT-NmtR protein. These results also point to the importance of N-terminal residues in metal recognition that may be common features in Co(II)- and Ni(II)-responsive metalloregulators.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01-GM69696 to MJM and NSF MCB0520877 to PTC. XAS data collection at the National Synchrotron Light Source at Brookhaven National Laboratory was supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences. Beam line X3B at NSLS is supported by the NIH. This publication was made possible by the Center for Synchrotron Biosciences grant, P30-EB-009998, from the National Institute of Biomedical Imaging and Bioengineering (NIBIB). Portions of this research were carried out at the Stanford Synchrotron Radiation Light source, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
Footnotes
EPR spectra of Cu(I) WT-RcnR and Cu(II)-EDTA. CD spectra of Apo, Co(II), Ni(II) and Zn(II) WT-RcnR. Tables with additional XAS fits for wild-type and mutant RcnR proteins. This material is available free of charge via the Internet at http://pubs.avs.org.
References
- 1.Olson JW, Maier RJ. Science. 2002;298:1788. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- 2.Böck A, King PW, Blokesch M, Posewitz MC. Adv Microb Physiol. 2006;51:1. doi: 10.1016/s0065-2911(06)51001-x. [DOI] [PubMed] [Google Scholar]
- 3.Forzi L, Sawers RG. Biometals. 2007;20:565. doi: 10.1007/s10534-006-9048-5. [DOI] [PubMed] [Google Scholar]
- 4.Macomber L, Hausinger RP. Metallomics. 2011;3:1153. doi: 10.1039/c1mt00063b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF. J Bacteriol. 1999;181:670. doi: 10.1128/jb.181.2.670-674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwig JS, Rowe JL, Chivers PT. Mol Microbiol. 2006;62:252. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 7.Blériot C, Effantin G, Lagarde F, Mandrand-Berthelot MA, Rodrigue A. J Bacteriol. 2011;193:3785. doi: 10.1128/JB.05032-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Outten CE, O'Halloran TV. Science. 2001;292:2488. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 9.Iwig JS, Leitch S, Herbst RW, Maroney MJ, Chivers PT. J Am Chem Soc. 2008;130:7592. doi: 10.1021/ja710067d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. Nat Chem Biol. 2007;3:60. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 11.Ma Z, Cowart DM, Scott RA, Giedroc DP. Biochemistry. 2009;48:3325. doi: 10.1021/bi900115w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwig JS, Chivers PT. J Mol Biol. 2009;393:514. doi: 10.1016/j.jmb.2009.08.038. [DOI] [PubMed] [Google Scholar]
- 13.Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Proc Natl Acad Sci U S A. 2003;100:3713. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavet JS, Meng W, Pennella MA, Appelhoff RJ, Giedroc DP, Robinson NJ. J Biol Chem. 2002;277:38441. doi: 10.1074/jbc.M207677200. [DOI] [PubMed] [Google Scholar]
- 15.Leitch S, Bradley MJ, Rowe JL, Chivers PT, Maroney MJ. J Am Chem Soc. 2007;129:5085. doi: 10.1021/ja068505y. [DOI] [PubMed] [Google Scholar]
- 16.Schreiter ER, Wang SC, Zamble DB, Drennan CL. Proc Natl Acad Sci U S A. 2006;103:13676. doi: 10.1073/pnas.0606247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chivers PT, Sauer RT. Protein Sci. 1999;8:2494. doi: 10.1110/ps.8.11.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dian C, Schauer K, Kapp U, McSweeney SM, Labigne A, Terradot L. J Mol Biol. 2006;361:715. doi: 10.1016/j.jmb.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 19.Benini S, Cianci M, Ciurli S. Dalton Trans. 2011;40:7831. doi: 10.1039/c1dt11107h. [DOI] [PubMed] [Google Scholar]
- 20.Padden KM, Krebs JF, MacBeth CE, Scarrow RC, Borovik AS. J Am Chem Soc. 2001;123:1072. doi: 10.1021/ja003282b. [DOI] [PubMed] [Google Scholar]
- 21.Webb SM. Physica Scripta. 2005;T115:1011. [Google Scholar]
- 22.Ankudinov AL, Ravel B, Rehr JJ, Conradson SD. Physical Review B. 1998;58:7565. [Google Scholar]
- 23.Zabinsky SI, Rehr JJ, Ankudinov A, Albers RC, Eller MJ. Phys Rev B: Condens Matter. 1995;52:2995. doi: 10.1103/physrevb.52.2995. [DOI] [PubMed] [Google Scholar]
- 24.Guncar G, Wang CI, Forwood JK, Teh T, Catanzariti AM, Ellis JG, Dodds PN, Kobe B. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:209. doi: 10.1107/S1744309107004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chivers PT, Tahirov TH. J Mol Biol. 2005;348:597. doi: 10.1016/j.jmb.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 26.Crane BR, Di Bilio AJ, Winkler JR, Gray HB. J Am Chem Soc. 2001;123:11623. doi: 10.1021/ja0115870. [DOI] [PubMed] [Google Scholar]
- 27.Gasper R, Scrima A, Wittinghofer A. J Biol Chem. 2006;281:27492. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- 28.Costello A, Periyannan G, Yang KW, Crowder MW, Tierney DL. J Biol Inorg Chem. 2006;11:351. doi: 10.1007/s00775-006-0083-z. [DOI] [PubMed] [Google Scholar]
- 29.Herbst RW, Guce A, Bryngelson PA, Higgins KA, Ryan KC, Cabelli DE, Garman SC, Maroney MJ. Biochemistry. 2009;48:3354. doi: 10.1021/bi802029t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheuring EM, Clavin W, Wirt MD, Miller LM, Fischetti RF, Lu Y, Mahoney N, Xie AH, Wu JJ, Chance MR. J Phys Chem. 1996;100:3344. [Google Scholar]
- 31.Wirt MD, Sagi I, Chen E, Frisbie SM, Lee R, Chance MR. J Am Chem Soc. 1991;113:5299. [Google Scholar]
- 32.Colpas GJ, Maroney MJ, Bagyinka C, Kumar M, Willis WS, Suib SL, Baidya N, Mascharak PK. Inorg Chem. 1991;30:920. [Google Scholar]
- 33.Cramer SP, Eidsness MK, Pan WH, Morton TA, Ragsdale SW, Dervartanian DV, Ljungdahl LG, Scott RA. Inorg Chem. 1987;26:2477. [Google Scholar]
- 34.Tan GO, Ensign SA, Ciurli S, Scott MJ, Hedman B, Holm RH, Ludden PW, Korszun ZR, Stephens PJ, Hodgson KO. Proc Natl Acad Sci U S A. 1992;89:4427. doi: 10.1073/pnas.89.10.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kau LS, Spirasolomon DJ, Pennerhahn JE, Hodgson KO, Solomon EI. J Am Chem Soc. 1987;109:6433. [Google Scholar]
- 36.Clark-Baldwin K, Tierney DL, Govindaswamy N, Gruff ES, Kim C, Berg J, Koch SA, Penner-Hahn JE. J Am Chem Soc. 1998;120:8401. [Google Scholar]
- 37.Jacquamet L, Aberdam D, Adrait A, Hazemann JL, Latour JM, Michaud-Soret I. Biochemistry. 1998;37:2564. doi: 10.1021/bi9721344. [DOI] [PubMed] [Google Scholar]
- 38.Mirza SA, Day RO, Maroney MJ. Inorg Chem. 1996;35:1992. doi: 10.1021/ic950162+. [DOI] [PubMed] [Google Scholar]
- 39.Mirza SA, Pressler MA, Kumar M, Day RO, Maroney MJ. Inorg Chem. 1993;32:977. [Google Scholar]
- 40.Colpas GJ, Kumar M, Day RO, Maroney MJ. Inorg Chem. 1990;29:4779. [Google Scholar]
- 41.Kruger HJ, Holm RH. Inorg Chem. 1989;28:1148. [Google Scholar]
- 42.Kruger HJ, Peng G, Holm RH. Inorg Chem. 1991;30:734. [Google Scholar]
- 43.Sawers RG, Ballantine SP, Boxer DH. J Bacteriol. 1985;164:1324. doi: 10.1128/jb.164.3.1324-1331.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ballantine SP, Boxer DH. J Bacteriol. 1985;163:454. doi: 10.1128/jb.163.2.454-459.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roth JR, Lawrence JG, Bobik TA. Annu Rev Microbiol. 1996;50:137. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 46.Lawrence JG, Roth JR. Genetics. 1996;142:11. doi: 10.1093/genetics/142.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niegowski D, Eshaghi S. Cell Mol Life Sci. 2007;64:2564. doi: 10.1007/s00018-007-7174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. J Bacteriol. 2005;187:1604. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Musiani F, Bertosa B, Magistrato A, Zambelli B, Turano P, Losasso V, Micheletti C, Ciurli S, Carloni P. J Chem Theory Comput. 2010;6:3503. doi: 10.1021/ct900635z. [DOI] [PubMed] [Google Scholar]
- 50.Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. J Mol Biol. 2003;333:683. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Sakamoto K, Agari Y, Agari K, Kuramitsu S, Shinkai A. Microbiology. 2010;156:1993. doi: 10.1099/mic.0.037382-0. [DOI] [PubMed] [Google Scholar]
- 52.Wang SC, Dias AV, Bloom SL, Zamble DB. Biochemistry. 2004;43:10018. doi: 10.1021/bi049405c. [DOI] [PubMed] [Google Scholar]
- 53.Ma Z, Jacobsen FE, Giedroc DP. Chem Rev. 2009;109:4644. doi: 10.1021/cr900077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuroda M, Hayashi H, Ohta T. Microbiol Immunol. 1999;43:115. doi: 10.1111/j.1348-0421.1999.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 55.Patzer SI, Hantke K. Mol Microbiol. 1998;28:1199. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 56.Outten CE, Outten FW, O'Halloran TV. J Biol Chem. 1999;274:37517. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- 57.Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP. Mol Microbiol. 1999;31:893. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 58.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Biochemistry. 2004;43:8038. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- 59.Wuerges J, Lee JW, Yim YI, Yim HS, Kang SO, Carugo KD. Proc Natl Acad Sci U S A. 2004;101:8569. doi: 10.1073/pnas.0308514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laussac JP, Sarkar B. Biochemistry. 1984;23:2832. doi: 10.1021/bi00307a046. [DOI] [PubMed] [Google Scholar]
- 61.Ma Z, Cowart DM, Ward BP, Arnold RJ, DiMarchi RD, Zhang L, George GN, Scott RA, Giedroc DP. J Am Chem Soc. 2009;131:18044. doi: 10.1021/ja908372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pohl E, Haller JC, Mijovilovich A, Meyer-Klaucke W, Garman E, Vasil ML. Mol Microbiol. 2003;47:903. doi: 10.1046/j.1365-2958.2003.03337.x. [DOI] [PubMed] [Google Scholar]
- 63.Giedroc DP, Arunkumar AI. Dalton Trans. 2007:3107. doi: 10.1039/b706769k. [DOI] [PubMed] [Google Scholar]
- 64.Reyes-Caballero H, Lee CW, Giedroc DP. Biochemistry. 2011;50:7941. doi: 10.1021/bi200737a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.