Abstract
Cyclic-AMP response element binding protein (CREB) is often dysregulated in cancer cells and is an attractive cancer drug target. Previously, we described naphthol AS-E (1) as a small molecule inhibitor of CREB-mediated gene transcription. To understand its bioactive conformation, a series of conformationally constrained analogs of 1 were designed and synthesized. Biological evaluation of these analogs suggests that the global energy minimum of 1 is the likely bioactive conformation.
INTRODUCTION
Cyclic-AMP response element (CRE)-binding protein (CREB) is a stimulus-activated nuclear transcription factor enabling cells to respond to various extracellular stimuli.1 Its DNA-binding domain, located at the C-terminus, binds the consensus CRE sequence 5′-TGACGTCA-3′.2, 3 Even though CREB is able to bind CRE in the absence of any posttranslational modifications, its transcription activity is not activated until its activation domain called kinase-inducible domain (KID) is phosphorylated at Ser133.4 The thus phosphorylated CREB (p-CREB) allows for its subsequent binding with transcription co-activator CBP (CREB-binding protein).5 The CREB-CBP interaction is mediated by KID in CREB and KIX (KID-interacting) domain in CBP.5 A variety of protein serine/threonine kinases including protein kinase A (PKA),2 protein kinase B (PKB/Akt),6 protein p90 ribosomal S6 kinase (pp90RSK)7 and mitogen-activated protein kinase (MAPK)8, 9 could phosphorylate and activate CREB. Since these kinase signaling pathways are often deregulated in cancer cells, CREB was consistently found to be over-activated in cancer tissues from patients with non-small cell lung cancer (NSCLC),10 prostate cancer,11 breast cancer,12 acute myeloid leukemia and acute lymphoblastic leukemia.13, 14 Thus, CREB has been proposed as an intriguing target for the development novel cancer therapeutics15 and various strategies have been pursued to identify potential small molecule modulators of CREB-mediated gene transcription.15–17
We recently described naphthol AS-E (1, Figure 1) as a small molecule inhibitor of KIX-KID interaction and CREB-mediated gene transcription with potencies in the low μM range.18 However, its bioactive conformation, which would be very useful in facilitating the design of more potent analogs, is currently unknown. In this report, a series of conformationally constrained analogs of 1 were designed and synthesized to interrogate the bioactive conformation of 1 as inhibitors of KIX-KID interaction and CREB-mediated gene transcription.
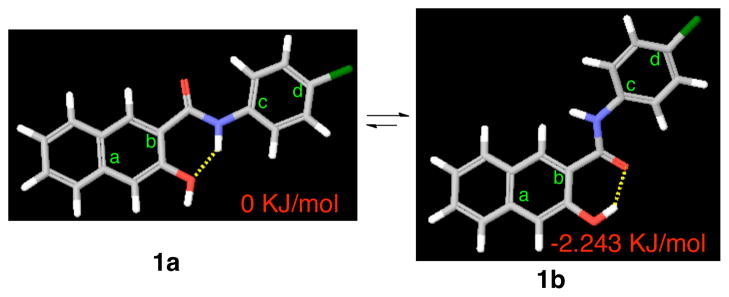
Figure 1.
Potential conformations of compound 1. The yellow dashed lines indicate hydrogen bonds. The dihedral angle of a-b-c-d in 1a is 0.1° while that in 1b is 10.6°.
RESULTS AND DISCUSSION
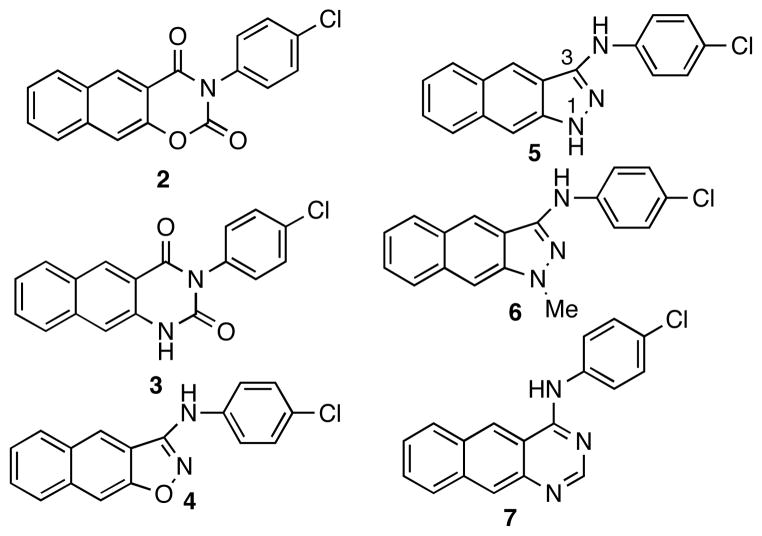
To investigate the conformational preference of compound 1, it was subjected to a systematic conformational search by rotating all the rotatable bonds in MacroModel. This search resulted in two alternatively hydrogen-bonded conformations 1a and 1b (Figure 1) as the energy minima. Conformer 1a, with the dihedral angle of a-b-c-d of 0.1°, is fully conjugated and adopts a co-planar conformation. On the other hand, the dihedral angle of a-b-c-d in conformer 1b is 10.6°. As a consequence, the naphthyl ring and the chlorophenyl ring are not co-planar. Counter-intuitively, conformer 1b is the global minimum and is energetically slightly more favorable than conformer 1a by 2.243 kJ/mol (Figure 1). In order to examine which conformation might represent the biologically active one, conformationally constrained analogs 2-7 (Chart 1) were designed. Compounds 2 and 3 were designed to mimic conformer 1a while compounds 4-7 were designed to mimic conformer 1b.
Chart 1.
Structures of designed conformationally constrained compounds 2-7.
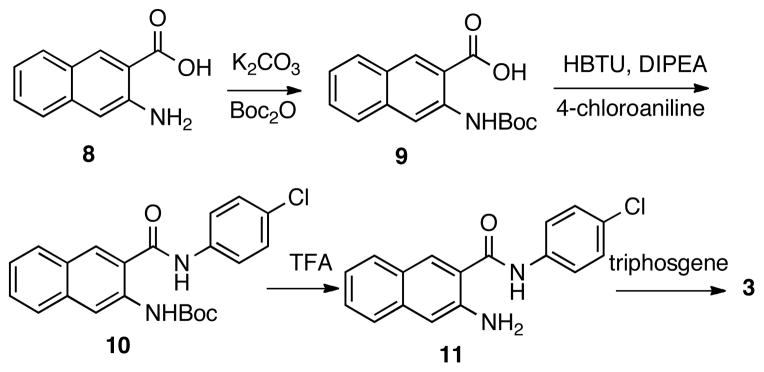
Compound 2 was synthesized in one step by treating 1 with oxaly chloride.19 The synthesis of benzo[g]quinazolinedione 3 would require precursor compound 11 (Scheme 1). Therefore, the amino group in 3-amino-2-naphthoic acid (8) was first protected by a Boc group to give acid 9, which was then coupled with 4-chloroaniline to provide amide 10. Removal of Boc under acidic condition (TFA) yielded amine 11, which was then treated with triphosgene to afford analog 3 uneventfully.
Scheme 1.
Synthesis of compound 3.
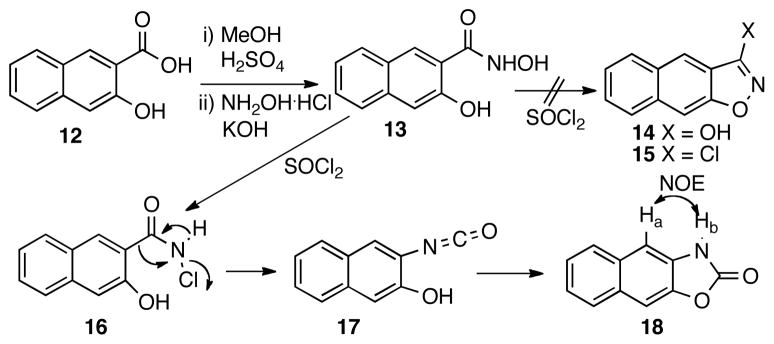
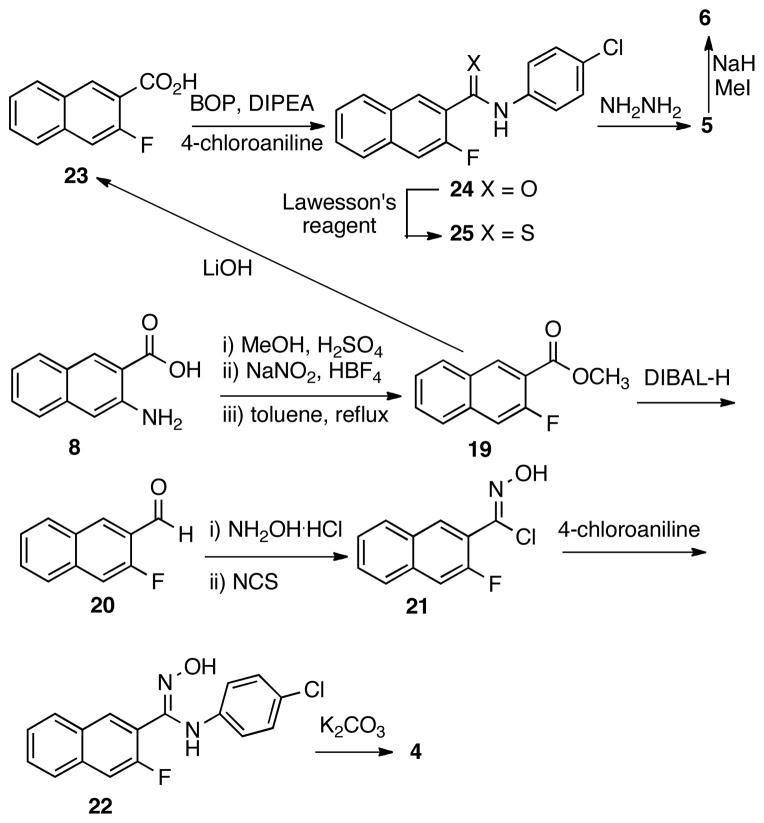
The synthesis of analog 4 was less straightforward. Initially, it was envisioned that an SNAr reaction between 4-chloroaniline and chloride 15 would deliver compound 4.20 Thus, a synthetic route depicted in Scheme 2 was designed. Hydroxamide 13 was prepared by condensing hydroxylamine with the methyl ester of acid 12. However, when 13 was treated with thionyl chloride,21 isoxazole 14 was not obtained as expected. Instead, oxazolone 18 was isolated in 37% yield, whose structure is consistent with the observed positive NOE between Ha and Hb. The formation of 18 is perhaps through a process analogous to the Lossen rearrangement22 of chloride 16 generated from chlorination of 13 to provide isocyanate 17, cyclization of which produced 18. The electron-rich nature of the naphthyl ring may facilitate the Lossen-type rearrangement pathway instead of direct ring closure to form 14. Other reagents including carbonyldiimidazole23 and Mitsunobu reagent (Ph3P/DEAD)24 were also tried, but they both failed to give compound 14.
Scheme 2.
Attempted synthesis of compound 14.
The apparent difficulty in synthesizing 14 prompted us to investigate an alternative route to make compound 4 as presented in Scheme 3. Thus, 3-amino-2-naphthoic acid (8) was converted into fluoride 19 through a three-step sequence involving methyl esterification, diazonium salt formation and thermal decomposition of the diazonium tetra-fluoroborate. The ester group in compound 19 was then reduced with DIBAL-H to provide aldehyde 20, which was then transformed into the corresponding oxime. It was found that the oxime could be oxidized by NCS to give N-hydroxy-imidoyl chloride 21,25 which was then coupled with 4-chloroaniline to yield N-hydroxy-imidamide 22. Intramolecular SNAr reaction of 22 under basic condition (K2CO3/DMF) delivered analog 4.
Scheme 3.
Synthesis of compounds 4-6.
The intermediate ester 19 was also utilized to synthesize compounds 5 and 6 (Scheme 3). Saponification of ester in 19 gave acid 23, which was then coupled with 4-chloroaniline with BOP reagent to afford amide 24. The thioamide 25 was formed by treating 24 with Lawesson’s reagent. Ring closure between 25 and hydrazine26 delivered analog 5. Methylation of 5 with NaH/MeI afforded compound 6. The regiochemistry of the methyl group in 6 was confirmed by the positive NOE between the methyl protons and one of the naphthyl protons, while no NOE was observed between the protons in the chlorophenyl ring and the methyl group.
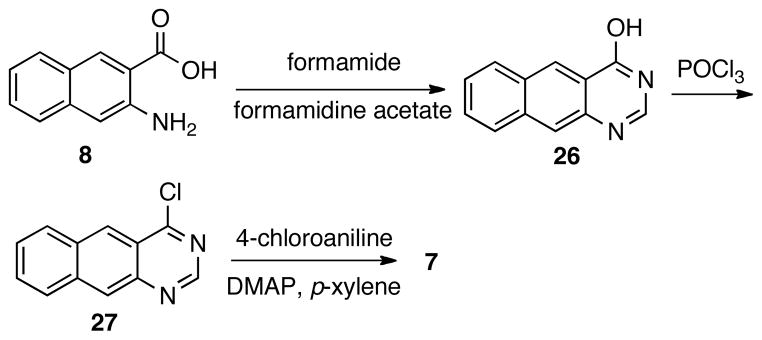
Finally, compound 7 was prepared by a sequence of reactions presented in Scheme 4. Intermediate 26 was prepared by condensing acid 8 with formamide under solvent-free condition.27 Compound 7 was made by an SNAr reaction between 4-chloroaniline and chloride 27, which was derived from 26 after being treated with POCl3.
Scheme 4.
Synthesis of compound 7.
Compounds 2-7 were then evaluated for inhibition of KIX-KID interaction in vitro by a renilla luciferase complementation assay that we recently developed (Table 1).18 In this assay, compound 1 inhibits KIX-KID interaction with an IC50 of 2.90 μM.18 Compounds 2 and 3 were designed to mimic conformation 1a. Compound 3 also retains a hydrogen-bond donor as seen in 1. However, neither of these compounds showed any activity in inhibiting KIX-KID interaction in vitro, suggesting that conformation 1a may not be biologically relevant. This lack of activity also translates into low or no inhibitory activity against CREB-mediated gene transcription in HEK 293T cells using a CREB reporter assay.18 Compounds 4-7 were designed to interrogate if conformation 1b is a potential bioactive one. Although compound 4 was inactive in inhibiting KIX-KID interaction, compound 5 with a nitrogen linkage showed only slightly reduced activity (IC50 = 8.74 μM) compared to 1. The loss of activity with compound 4 does not seem to be due to elimination of a hydrogen-bond donor present in compound 1 because the methylated compound 6 (IC50 = 5.19 μM) and benzo[g]quinazoline compound 7 (IC50 = 9.46 μM) were in the same range of activity as compound 1. In the cell-based CREB reporter assay, the oxygen-linked compound 4 had very weak activity as expected from its lack of activity in the KIX-KID interaction assay. Compound 5, on the other hand, retained activity in inhibiting CREB-mediated gene transcription with an IC50 of 4.75 μM. The intermediate potency of inhibiting KIX-KID interaction by compound 7 was also reflected in its medium potency in CREB transcription reporter assay (IC50 = 18.96 μM). However, the methylated compound 6 was inactive in cell-based CREB reporter assay even though it was a reasonable inhibitor of KIX-KID interaction. This discrepancy could result from unfavorable cellular permeation and/or subcellular distribution of compound 6, which has a higher cLogP value (cLogP = 5.298) than that of compound 5 (cLogP = 4.433). Taken together, these results suggest that the likely bioactive conformation of compound 1 resembles conformer 1b.
Table 1.
The biological activities of compounds 1-7.a
| Compound | KIX-KID inhibition (μM)b | CREB inhibition (μM)c |
|---|---|---|
| 1 | 2.90 ± 0.81 | 2.29 ± 0.31 |
| 2 | >50 | 30.84 ± 8.40 |
| 3 | >50 | >50 |
| 4 | >50 | 24.45 ± 16.80 |
| 5 | 8.74 ± 1.68 | 4.75 ± 3.88 |
| 6 | 5.19 ± 2.20 | >50 |
| 7 | 9.46 ± 4.15 | 18.96 ± 6.09 |
All the biological activities are presented as IC50 with mean ± SD of at least two independent experiments or > 50 if IC50 was not reached at the highest tested concentration, which is 50 μM.
KIX-KID inhibition was evaluated by an in vitro renilla luciferase complementation assay.
CREB inhibition refers to inhibition of CREB-mediated gene transcription in HEK 293T cells using a CREB reporter assay.
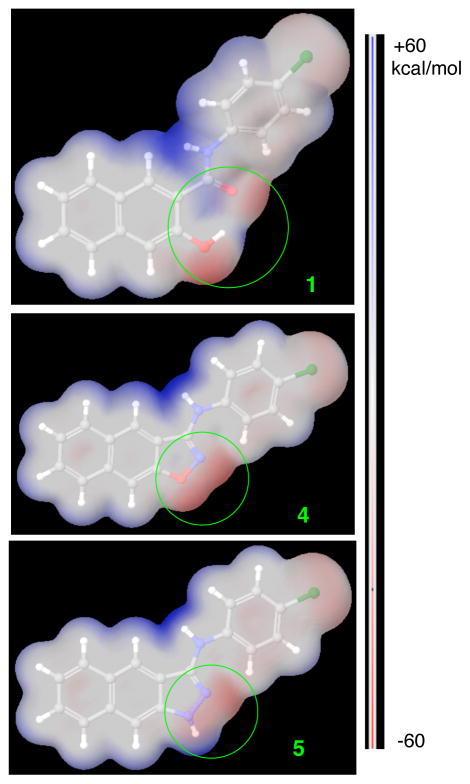
To further understand the potential reasons for the observed activity among 1, 4 and 5, these structures were geometrically optimized at the HF/6-31G** level of theory in Jaguar. We found that N1 atom in compound 5 (Chart 1) prefers a pyramidal configuration instead of a planar one even though it is planar when optimized at lower level of theory (e.g. HF/3-21G). Similar nitrogen pyramidization was also observed in other systems both experimentally28 and in theoretical calculations.29 With these optimized structures, we then generated molecular electrostatic potential (MEP) surface for each molecule from their respective electron densities (Figure 2). The major difference observed among compounds 1, 4 and 5 is located in the linkage section as highlighted in green circles in Figure 2. In both of the active compounds 1 and 5, this area is represented by both positively and negatively charges surfaces. However, only electrostatically negatively charged (−40–−50 kcal/mol) surfaces are present in the corresponding area in the inactive compound 4 (Figure 2). It could be that the binding site in KIX for the circled area is electrostatically negatively charged. As a consequence, compound 4 does not fit well in that binding site, contributing to its loss of inhibition of KIX-KID interaction. On the other hand, the mixed surfaces from 1 and 5 are well accommodated.
Figure 2.
Molecular electrostatic potential (MEP) surfaces of compounds 1, 4 and 5 mapped to their electron densities calculated at the HF/6-31G** level of theory in Jaguar. The blue indicates electrostatically positively charged while the red is negatively charged. All the surfaces are normalized from −60 to +60 kcal/mol.
CONCLUSION
In conclusion, a series of compounds (2-7) were designed and synthesized to mimic two alternative conformations of 1 to derive its potential bioactive conformation in inhibiting KIX-KID interaction. These compounds were then biologically evaluated for their capability to inhibit KIX-KID interaction in vitro and CREB-mediated gene transcription in HEK 293T cells. These results suggest that the energetically favored conformer 1b may represent the bioactive conformation. Molecular modeling studies suggest that the electrostatic potential surface of the newly created heterocycle needs to be at least partially positively charged. Further investigations are underway to utilize these concepts to design more potent derivatives in inhibiting KIX-KID interaction and CREB-mediated gene transcription.
EXPERIMENTAL SECTION
General
Please see Supporting Information for details. All final compounds were confirmed to be of > 95% purity based on HPLC analysis.
3-(4-Chlorophenyl)-2H-naphtho[2,3-e][1,3]oxazine-2,4-(3H)-dione (2)
Compound 1 (100 mg, 0.34 mmol) was dissolved in p-xylene (4.0 mL) and the solution was stirred for 5 min at room temperature. Oxalyl chloride (150 μL, 1.7 mmol) in p-xylene (1.0 mL) was then added and the reaction mixture was heated at 120°C for overnight. The precipitate was collected by filtration and washed with p-xylene to give a pink-white solid 17 (57 mg, 52%): m.p. 306–308 °C (dec). 1H NMR (400 MHz, CDCl3) δ 8.77 (s, 1 H), 8.06 (d, J = 8.4 Hz, 1 H), 7.96 (d, J = 8.4 Hz, 1 H), 7.78 (s, 1 H), 7.72 (t, J = 9.6 Hz, 1 H), 7.61 (t, J = 7.3 Hz, 1 H), 7.55 (d, J = 8.8 Hz, 2 H), 7.33 (d, J = 8.4 Hz, 2 H); 13C NMR (100 MHz, DMSO-d6) δ 161.3, 148.5, 148.1, 137.0, 134.8, 133.9, 131.1, 130.3, 130.1, 130.1, 129.6, 127.9, 126.9, 114.9, 112.6.
3-(4-Chlorophenyl)-benzo[g]quinazoline-2,4-(1H,3H)-dione (3)
Triphosgene (7.5 mg, 0.025 mmol) was added to a stirred solution of 11 (15 mg, 0.05 mmol) in THF (1.5 mL) at room temperature. The resulting mixture was heated under reflux for 2 h. The reaction mixture was then cooled to room temperature and diluted with THF (5 mL). The solid was collected by filtration to give a white solid (11 mg, 69%): m.p. > 400 °C. 1H NMR (400 MHz, DMSO-d6) δ 11.67 (s, 1 H), 8.70 (s, 1 H), 8.14 (d, J = 7.5 Hz, 1 H), 7.95 (d, J = 7.7 Hz, 1 H), 7.63 (dt, J = 7.5, 1.3 Hz, 1 H), 7.59 (s, 1 H), 7.57 (d, J = 9.0 Hz, 2 H), 7.47 (dt, J = 7.6, 1.3 Hz, 1 H), 7.43 (d, J = 8.8 Hz, 2 H); 13C NMR (100 MHz, DMSO-d6) δ 162.2, 150.1, 136.39, 135.41, 134.7, 132.7, 131.2, 129.6, 129.6, 129.4, 128.8, 128.5, 126.7, 125.0, 115.1, 110.1; ESI-MS m/z 320.7 (M−H)−; HRESI-MS for C18H11ClN2O2-H, calcd 321.0425, found 321.0416.
N-(4-Chlorophenyl)-naphtho[2,3-d]isoxazol-3-amine (4)
K2CO3 (7.9 mg, 0.057 mmol) was added to a stirred solution of compound 22 (6.0 mg, 0.019 mmol) in DMF (1 mL) at room temperature. The resulting mixture was then heated at 110 °C for 4 h. The reaction mixture was cooled to room temperature and DMF was removed in vacuo. The residue was dissolved in EtOAc (50 mL), which was then washed with H2O (2 × 10 mL) and brine (2 × 10 mL). The organic solution was dried over anhydrous Na2SO4, filtered and concentrated. The residue was then subjected to column chromatography eluting with hexanes : EtOAc (10:1-4:1) to give a white solid (2.7 mg, 48%): m.p. 218–220 °C. 1H NMR (400 MHz, CDCl3) δ 8.12 (s, 1 H), 7.98 (d, J = 8.8 Hz, 1 H), 7.95 (d, J = 8.4 Hz, 1 H), 7.86 (s, 1 H), 7.60 (d, J = 8.8 Hz, 2 H), 7.56 (td, J = 8.4, 1.2 Hz, 1 H), 7.47 (td, J = 8.0, 1.2 Hz, 1 H), 7.35 (d, J = 8.8 Hz, 2 H), 6.60 (brs, 1 H); ESI-MS m/z 295 (M+H)+; HRESI-MS for C17H11ClN2O-H, calcd 293.0476, found 293.0470.
N-(4-Chlorophenyl)-1H-benzo[f]indazol-3-amine (5)
A mixture of thioamide 24 (60 mg, 0.19 mmol) in anhydrous hydrazine/DMSO (0.5/0.5 mL) was stirred at 150 °C for 1 h. The reaction mixture was cooled to room temperature and poured into water (10 mL). The organic compound was extracted with dichloromethane (3 × 10 mL). The combined organic layers were washed with brine (2 × 10 mL), dried with Na2SO4, filtered and concentrated. The residue was purified by silica gel flash column chromatography, eluting with hexanes : ethyl acetate (4 : 1) to yield a yellow solid (10 mg, 18%): m.p. 220–221 °C. 1H NMR (400 MHz, DMSO-d6) δ 11.95 (s, 1 H), 9.37 (s, 1 H), 8.62 (s, 1 H), 7.98 (d, J = 8.0 Hz, 1 H), 7.96 (d, J = 8.0 Hz, 1 H), 7.82 (s, 1 H), 7.81 (d, J = 8.8 Hz, 2 H), 7.44 (t, J = 8.0 Hz, 1 H), 7.34 (d, J = 8.8 Hz, 2 H), 7.32 (t, J = 8.4 Hz, 1 H); 13C-NMR (100 MHz, DMSO-d6) δ 145.2, 142.1, 139.9, 133.3, 129.5, 129.0, 127.9, 127.5, 126.3, 123.0, 122.9, 119.1, 117.9, 104.3; ESI-MS m/z 292 (M−H)−; HRESI-MS for C17H12ClN3-H, calcd 292.0636, found 292.0629.
N-(4-Chlorophenyl)-1-methyl-1H-benzo[f]indazol-3-amine (6)
NaH (60% in mineral oil, 6 mg, 0.15 mmol) was added to a stirred solution of compound 5 (40 mg, 0.136 mmol) in DMF (0.5 mL) at 0 °C under Ar. The mixture was stirred at 0 °C for 30 min, when CH3I (8.6 μL, 0.136 mmol) was added. The mixture was stirred at room temperature for another 1 h and poured into cooled water (10 mL). The solid was precipitated and collected by filtration. The yellow solid was purified by silica gel flash column chromatography, eluting with hexanes : ethyl acetate = 4 : 1 to yield a yellow solid (19 mg, 45%): mp: 121–122 °C. 1H NMR (400 MHz, DMSO-d6) δ 9.47 (s, 1 H), 8.64 (s, 1 H), 8.00 (d, J = 8.8 Hz, 1 H), 7.96 (d, J = 8.4 Hz, 1 H), 7.91 (s, 1 H), 7.83 (d, J = 8.8 Hz, 2 H), 7.54 (t, J = 8.4 Hz, 1 H), 7.37 (d, J = 8.8 Hz, 2 H), 7.32 (t, J = 8.0 Hz, 1 H), 3.98 (s, 3 H); 13C-NMR (100 MHz, DMSO-d6) δ 143.8, 141.3, 139.3, 132.9, 129.1, 128.6, 127.3, 126.9, 126.1, 122.7, 122.6, 119.1, 117.7, 117.5, 103.1, 35.3; ESI-MS m/z 305.9 (M−H)−; HRESI-MS for C18H14ClN3-H, calcd 306.0793, found 306.0785.
N-(4-Chlorophenyl)benzo[g]quinazolin-4-amine (7)
A mixture of 4-chlorobenzo[g]quinazoline (27) (50 mg, 0.23 mmol), 4-chloroaniline (36 mg, 0.28 mmol) and DMAP (29 mg, 0.23 mmol) in p-xylene (1 mL) was stirred at 140 °C for 10 min. The reaction mixture was cooled to room temperature. Evaporation of p-xylene resulted in a residue which was subjected to silica gel flash column chromatography, eluting with hexanes : ethyl acetate (2 : 1) to yield a yellow solid (31 mg, 44%): mp: 251–252 °C. 1H NMR (400 MHz, CDCl3) δ 10.26 (s, 1 H), 9.32 (s, 1 H), 8.67 (s, 1 H), 8.47 (s, 1 H), 8.21 (d, J = 8.8 Hz, 1 H), 8.19 (d, J = 8.4 Hz, 1 H), 8.08 (d, J = 8.8 Hz, 2 H), 7.42 (t, J = 7.2 Hz, 1 H), 7.67 (t, J = 7.2 Hz, 1 H), 7.54 (d, J = 8.8 Hz, 2 H); 13C NMR (100 MHz, CDCl3) δ 158.6, 154.4, 145.5, 138.6, 135.9, 131.4, 129.3, 128.9, 128.5, 128.3, 128.0, 126.8, 125.5, 124.3, 123.7, 115.5; ESI-MS m/z 303.9 (M−H)−; HRESI-MS for C18H12ClN3+H, calcd 306.0793, found 306.0805.
Supplementary Material
Acknowledgments
This work was made possible by financial supports from NIH (R01GM087305) and Susan G. Komen for the Cure (KG100458). We thank Jenny Luo, Dr. Andrea DeBarber and Dr. Dennis Koop for running the mass spectroscopic analyses.
ABBREVIATIONS
- CRE
cyclic AMP-response element
- CREB
CRE-binding protein
- CBP
CREB-binding protein
- KID
kinase-inducible domain
- KIX
KID-interacting
- p-CREB
phosphorylated CREB
- PKA
protein kinase A
- PKB
protein kinase B
- MAPK
mitogen-activated protein kinase
- pp90RSK
protein p90 ribosomal S6 kinase
- NSCLC
non-small cell lung cancer
- NOE
nuclear Overhauser effect
- MEP
molecular electrostatic potential
- SNAr
nucleophilic aromatic substitution
Footnotes
Supporting Information Available. Experimental procedures for synthesis of and characterization data for compounds 9-11, 19-20, and 22-27. Details for the biological assays and molecular modeling. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Shaywitz AJ, Greenberg ME. CREB: A stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 2.Montminy MR, Bilezikjian LM. Binding of a nuclear-protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 3.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci U S A. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 5.Radhakrishnan I, PerezAlvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:Coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 6.Du KY, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 7.Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 8.Tan Y, Rouse J, Zhang AH, Cariati S, Cohen P, Comb MJ. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 9.Ginty DD, Bonni A, Greenberg ME. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 10.Seo HS, Liu DD, Bekele BN, Kim MK, Pisters K, Lippman SM, Wistuba, Koo JS. Cyclic AMP response element-binding protein overexpression: A feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer Res. 2008;68:6065–6073. doi: 10.1158/0008-5472.CAN-07-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W, Zhau HE, Huang WC, Iqbal S, Habib FK, Sartor O, Cvitanovic L, Marshall FF, Xu Z, Chung LWK. cAMP-responsive element-binding protein regulates vascular endothelial growth factor expression: implication in human prostate cancer bone metastasis. Oncogene. 2007;26:5070–5077. doi: 10.1038/sj.onc.1210316. [DOI] [PubMed] [Google Scholar]
- 12.Chhabra A, Fernando H, Watkins G, Mansel RE, Jiang WG. Expression of transcription factor CREB1 in human breast cancer and its correlation with prognosis. Oncol Rep. 2007;18:953–958. [PubMed] [Google Scholar]
- 13.Crans-Vargas HN, Landaw EM, Bhatia S, Sandusky G, Moore TB, Sakamoto KM. Expression of cyclic adenosine monophosphate response-element binding protein in acute leukemia. Blood. 2002;99:2617–2619. doi: 10.1182/blood.v99.7.2617. [DOI] [PubMed] [Google Scholar]
- 14.Shankar DB, Cheng JC, Kinjo K, Federman N, Moore TB, Gill A, Rao NP, Landaw EM, Sakamoto KM. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell. 2005;7:351–362. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 15.Xiao X, Li BX, Mitton B, Ikeda A, Sakamoto KM. Targeting CREB for Cancer Therapy: Friend or Foe. Curr Cancer Drug Targets. 2010;10:384–391. doi: 10.2174/156800910791208535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aggarwal S, Kim SW, Ryu SH, Chung WC, Koo JS. Growth suppression of lung cancer cells by targeting cyclic AMP response element-binding protein. Cancer Res. 2008;68:981–988. doi: 10.1158/0008-5472.CAN-06-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc Natl Acad Sci U S A. 2004;101:17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li BX, Xiao X. Discovery of a Small-Molecule Inhibitor of the KIX-KID Interaction. ChemBioChem. 2009;10:2721–2724. doi: 10.1002/cbic.200900552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mu F, Lee DJ, Pryor DE, Hamel E, Cushman M. Synthesis and investigation of conformationally restricted analogues of lavendustin A as cytotoxic inhibitors of tubulin polymerization. J Med Chem. 2002;45:4774–4785. doi: 10.1021/jm0202270. [DOI] [PubMed] [Google Scholar]
- 20.Yevich JP, New JS, Smith DW, Lobeck WG, Catt JD, Minielli JL, Eison MS, Taylor DP, Riblet LA, Temple DL., Jr Synthesis and biological evaluation of 1-(1,2-benzisothiazol-3-yl)- and (1,2-benzisoxazol-3-yl)piperazine derivatives as potential antipsychotic agents. J Med Chem. 1986;29:359–369. doi: 10.1021/jm00153a010. [DOI] [PubMed] [Google Scholar]
- 21.Kalkote UR, Goswami DD. New synthesis of 1,2-benzisoxazole derivatives. Aust J Chem. 1977;30:1847–1850. [Google Scholar]
- 22.Yale HL. The Hydroxamic Acids. Chem Rev. 1943;33:209–256. [Google Scholar]
- 23.Deng BL, Hartman TL, Buckheit RW, Jr, Pannecouque C, De Clercq E, Fanwick PE, Cushman M. Synthesis, anti-HIV activity, and metabolic stability of new alkenyldiarylmethane HIV-1 non-nucleoside reverse transcriptase inhibitors. J Med Chem. 2005;48:6140–6155. doi: 10.1021/jm050452s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi G. The first general synthesis of N-substituted 1,2-benzisoxazolin-3-ones. Tetrahedron Lett. 2000;41:2295–2298. [Google Scholar]
- 25.Pettus LH, Xu S, Cao GQ, Chakrabarti PP, Rzasa RM, Sham K, Wurz RP, Zhang D, Middleton S, Henkle B, Plant MH, Saris CJM, Sherman L, Wong LM, Powers DA, Tudor Y, Yu V, Lee MR, Syed R, Hsieh F, Tasker AS. 3-Amino-7-phthalazinylbenzoisoxazoles as a Novel Class of Potent, Selective, and Orally Available Inhibitors of p38α Mitogen-Activated Protein Kinase. J Med Chem. 2008;51:6280–6292. doi: 10.1021/jm8005405. [DOI] [PubMed] [Google Scholar]
- 26.Burke MJ, Trantow BM. An efficient route to 3-aminoindazoles and 3-amino-7-azaindazoles. Tetrahedron Lett. 2008;49:4579–4581. [Google Scholar]
- 27.Oerfi L, Waczek F, Pato J, Varga I, Hegymegi-Barakonyi B, Houghten RA, Keri G. Improved, high yield synthesis of 3H-quinazolin-4-ones, the key intermediates of recently developed drugs. Curr Med Chem. 2004;11:2549–2553. doi: 10.2174/0929867043364423. [DOI] [PubMed] [Google Scholar]
- 28.Acheson RM, Benn MH, Jacyno J, Wallis JD. Pyramidal Bonding about Nitrogen in 1-Benzoyloxyindole Determined by X-Ray Diffraction. J Chem Soc Perkin Trans II. 1983:497–500. [Google Scholar]
- 29.Gould IR, Kollman PA. Theoretical Investigation of the Hydrogen-Bond Strengths in Guanine-Cytosine and Adenine-Thymine Base-Pairs. J Am Chem Soc. 1994;116:2493–2499. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.