Abstract
Objective
The first genome-wide association study (GWAS) of systemic sclerosis (SSc) demonstrated three non-major histocompatibility complex (MHC) susceptibility loci. The goal of this study was to investigate the impact of these gene variants on survival and severity of interstitial lung disease (ILD) in SSc.
Methods
The authors examined 1443 Caucasian SSc patients enrolled in the Genetics versus Environment In Scleroderma Outcome Study (GENISOS) and Scleroderma Family Registry (n = 914 – discovery cohort) and The Johns Hopkins Scleroderma Cohort (n = 529 – replication cohort). Forced vital capacity (FVC)% predicted was used as a surrogate for ILD severity. Five single nucleotide polymorphisms, IRF5 (rs10488631, rs12537284, rs4728142), STAT4 (rs3821236), CD247 (rs2056626) reached genome-wide significance in the SSc-GWAS and were examined in the current study.
Results
Overall, 15.5% of the patients had died over the follow-up period of 5.5 years. The IRF5 rs4728142 minor allele was predictive of longer survival in the discovery cohort (p = 0.021) and in the independent replication cohort (p = 0.047) and combined group (HR: 0.75, 95% CI 0.62 to 0.90, p = 0.002). The association of this SNP with survival was independent of age at disease onset, disease type and autoantibody profile (anticentromere and antitopoisomerase antibodies). The minor allele frequency of IRF5 rs4728142 was 49.4%.
Moreover, IRF5 rs4728142 minor allele correlated with higher FVC% predicted at enrolment (p = 0.019). Finally, the IRF5 rs4728142 minor allele was associated with lower IRF5 transcript expression in patients and controls (p = 0.016 and p = 0.034, respectively), suggesting that the IRF5, rs4728142 SNP, may be functionally relevant.
Conclusion
An SNP in the IRF5 promoter region (rs4728142), associated with lower IRF5 transcript levels, was predictive of longer survival and milder ILD in patients with SSc.
INTRODUCTION
Scleroderma or systemic sclerosis (SSc) is a chronic, connective tissue disease characterised by wide-spread fibrosis of skin and internal organs, small-vessel vasculopathy and immune dysregulation with production of autoantibodies. SSc patients have markedly reduced survival rates compared with the age- and sex-matched general population. The overall pooled standardised mortality ratio of patients with SSc was 3.53 in a recently published meta-analysis.1 This is considerably higher than the standardised mortality ratio in other rheumatic diseases such as rheumatoid arthritis and Sjögren syndrome.2 Pulmonary involvement, including both interstitial lung disease (ILD) and pulmonary arterial hypertension, has become the primary cause of SSc-related death.3,4 Studies have shown that low forced vital capacity (FVC), a surrogate for severity of ILD, is highly predictive of mortality in SSc.5
Current therapies for SSc focus on treatment of specific complications while true disease-modifying agents targeting the underlying pathogenic mechanisms are lacking. Furthermore, the course of SSc is highly variable underscoring the need for identification of reliable prognostic markers in SSc.
The first SSc-genome-wide association study (GWAS) identified several highly robust susceptibility loci for SSc.6 Three non-major histocompatibility complex (MHC) loci in IRF5, STAT4 and CD247 regions were significantly associated with SSc. IRF57 and STAT48 had already been identified as SSc risk loci in case-control studies, whereas the association of a CD247 polymorphism with SSc was a novel finding which has recently been confirmed in an independent cohort.9 IRF5 belongs to a family of transcription factors in the type I interferon (IFN) pathway. STAT4 is also an important transcription factor for T-cell signalling and differentiation.10 Furthermore, STAT4 transduces type I IFN signals in activated monocytes.11 CD247 encodes the T cell receptor zeta subunit, and the low expression of this receptor can result in impaired immune responses.12 These recent breakthroughs in understanding the genetic basis of SSc can potentially lead to identification of novel therapeutic targets and development of prognostic biomarkers.
In a previous study conducted in the Genetics versus ENvironment In Scleroderma Outcome Study (GENISOS) cohort, we demonstrated that human leucocyte antigen (HLA) alleles DRB1*0802 and DQA1*0501 are independent predictors of mortality in SSc.5 However, the influence of the described non-MHC susceptibility loci on mortality in SSc patients has not been investigated.
Given the pivotal importance of the IRF5, STAT4 and CD247 in SSc susceptibility, we evaluated their associations with clinical outcomes of the disease in the current study. First, we demonstrated that the minor allele of IRF5 rs4728142 was associated with longer survival in two independent cohorts of SSc patients. Furthermore, this IRF5 variant was associated with milder ILD and correlated with a lower IRF5 gene expression.
METHODS
Study population
This study included 1443 SSc patients from three sources in the USA, which makes up 97% of US patients investigated in the first SSc-GWAS.6 The remainder of originally investigated patients (n = 43) were enrolled at the Fred Hutchinson Cancer Research Center and could not be included in the current study because of missing vital status information.
Subjects were categorised as discovery and replication cohorts. The discovery cohort consisted of 914 patients enrolled from two large North American SSc cohorts: (A) the National Institute of Health (NIH) Scleroderma Family Registry and DNA Repository, a nationwide registry13 and (B) the GENISOS.14 The impact of susceptibility genes on survival was further tested in the replication cohort, consisting of 529 patients with SSc enrolled at The Johns Hopkins University Scleroderma Center. Patients were enrolled if they met the following criteria: (1) age ≥18 years; and (2) diagnosis according to the American College of Rheumatology (ACR; formerly, the American Rheumatism Association) classification criteria for SSc,15 or had at least three of the five CREST (calcinosis, Raynaud’s phenomenon, oesophageal dysmotility, sclerodactyly and telangiectasias) features.16 All investigated patients were of self-reported white European descent. The institutional review boards of all participating sites approved the study, and written informed consent was obtained from all subjects, according to the declaration of Helsinki.
Demographic, clinical and serological data
Age, gender, disease type and duration, and the autoantibody profile were recorded at enrolment. Disease type was categorised as limited or diffuse based on the extent of skin involvement.16 Disease duration was calculated from the onset of the first non-Raynaud’s phenomenon symptom attributable to SSc. As previously described,5 the autoantibody determinations were conducted in the Division of Rheumatology at the University of Texas Health Science Center, Houston, Texas, USA. Pulmonary function tests were only available for the patients enrolled from the GENISOS cohort and Scleroderma Family Registry (discovery cohort). Although a variety of pulmonary function tests and imaging measures have been used to study ILD in SSc, only FVC has been validated as an outcome measure in randomised controlled trials.17 Therefore, FVC% predicted was used as a surrogate measure for severity of ILD. High-resolution chest CT results were not available in a large group of patients; thus, we cannot report on the prevalence of ILD based on this imaging modality.
Gene/polymorphism selection and genotyping
In the current study, we investigated all the non-MHC susceptibility loci that were identified/confirmed in the first large-scale GWAS in SSc.6 In this GWAS, all patients were genotyped by the Illumina Human610-Quad BeadChip, capturing 89% of the HapMap CEU variation at r2 >0.8. Five single nucleotide polymorphisms (SNPs) belonging to three genes (IRF5, STAT4 and CD247) were identified as genetic susceptibility loci for SSc. Three SNPs (rs10488631, rs12537284 and rs4728142) were located in the IRF5 region, while the SNP, rs3821236, was in an intronic region of STAT4, and the SNP, rs2056626, was in the intronic region of CD247. The genotype information for the current study was obtained from the GWAS described above.6 The linkage disequilibrium structure of the investigated IRF5 polymorphisms is shown in the online supplementary figure S1.
Vital status
The primary outcome was survival of the enrolled patients. The vital status was determined by the National Death Index, at Centers for Disease Control and Prevention and the Social Security Death Index. These two databases have very high sensitivity (up to 98%) for capturing mortality cases occurring within the USA.18 The censoring date was August 2010.
Determination of IRF5 transcript levels in monocytes
Purified monocyte cells were obtained from a subgroup of SSc patients enrolled in the GENISOS cohort and unaffected controls. The IRF5 gene expression levels in monocytes were examined on Illumina HumanHT-12 arrays. The transcript levels of the two IRF5 probes, representing two transcript variants of this gene were examined separately in SSc patients and controls, conditional on the three IRF5 polymorphisms (for further details on determination of transcript levels and investigated variants, see online supplementary text).
Statistical analysis
For survival analysis, the date of disease onset was used as the starting point for our time-to-event analysis. Using Cox proportional hazards regression models, the impact of three non-MHC SSc-susceptibility loci, namely IRF5, STAT4 and CD247 on survival was examined in the discovery cohort, and in the replication cohort. Finally, the association of investigated SNPs with survival was combined in the overall cohort by the inverse variance method. Furthermore, linear regression analysis was used to examine the effect of SSc-susceptibility loci on the severity of ILD (FVC% predicted value as a quantitative trait). Lastly, the correlation between the investigated IRF5 polymorphisms and IRF5 gene expression was examined by linear regression. The best fitting model was additive genetic inheritance mode which was used for all reported comparisons. All the statistical analyses were performed with STATA 11 (StataCorp, College Station, Texas, USA). The hypothesis testing was two-sided with a p ≤0.05 significance level.
RESULTS
Characteristics of the two SSc cohorts
The GWAS was conducted in 1443 white patients with SSc from the USA.6 The mean age of onset and disease duration at baseline was 45.6 and 9.7 years, respectively. Overall, 15.5% of the patients had died over the follow-up period of 5.5 years, 15.9% in the discovery, and 14.7% in the replication cohorts. The majority of patients (83.9%) fulfilled the ACR classification criteria for SSc.15 Further details about the demographic and clinical characteristics of discovery, replication and combined cohorts are provided in Table 1.
Table 1.
Study population characteristics
| Discovery cohort (n = 914) |
Replication cohort (n = 529) |
Combined cohort (n = 1443) |
|
|---|---|---|---|
| Age, mean (±SD), years | 45.3 (13.6) | 45.9 (13.5) | 45.6 (13.6) |
| Gender, female (%) | 88.8 | 85.4 | 87.6 |
| Diffuse skin involvement (%) | 37.1 | 32.9 | 35.6 |
| Disease duration*, mean (±SD), years | 9.7 (8.7) | 9.6 (8.5) | 9.7 (8.6) |
| Deceased (%) | 15.9 | 14.7 | 15.5 |
| Antitopoisomerase antibody (%) | 17.5 | 15.9 | 17.0 |
| Anticentromere antibody (%) | 30.2 | 33.8 | 31.4 |
Disease duration at study enrolment.
IRF5 rs4728142 susceptibility locus is associated with better survival
The impact of three SSc-susceptibility loci, namely IRF5, STAT4 and CD247 on survival was examined. The number of minor alleles of IRF5 rs4728142 was predictive of longer survival in both discovery (p = 0.021) and replication (p = 0.047) cohorts. This SNP also was associated with better survival in the combined cohort (HR: 0.75, 95% CI 0.62 to 0.90, p = 0.002). The minor allele frequency of IRF5 rs4728142 in the combined sample was 49.4%.
The other two IRF5 SNPs, rs10488631 and rs12537284, were not consistently associated with survival in the discovery and replication cohorts. The STAT4 rs3821236 and CD247 rs2056626 did not correlate with survival neither in the discovery nor replication cohorts (Table 2).
Table 2.
Association of the SNPs with survival
| Discovery cohort (n = 914) |
Replication cohort (n = 529) |
Combined cohort (n = 1443) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | MA | MAF | HR (95% CI) | p | HR (95% CI) | p Value | HR (95% CI) | p Value |
| IRF5 | rs10488631 | C | 0.152 | 0.95 (0.68 to 1.33) | 0.760 | 0.59 (0.35 to 0.98) | 0.042 | 0.82 (0.62 to 1.09) | 0.171 |
| rs12537284 | A | 0.171 | 0.94 (0.69 to 1.29) | 0.726 | 0.84 (0.55 to 1.29) | 0.434 | 0.91 (0.71 to 1.17) | 0.455 | |
| rs4728142 | A | 0.494 | 0.76 (0.60 to 0.96) | 0.021 | 0.73 (0.54 to 0.99) | 0.047 | 0.75 (0.62 to 0.90) | 0.002 | |
| STAT4 | rs3821236 | A | 0.233 | 1.01 (0.77 to 1.32) | 0.943 | 1.45 (1.02 to 2.05) | 0.036 | 1.15 (0.94 to 1.43) | 0.179 |
| CD247 | rs2056626 | G | 0.379 | 1.23 (0.97 to 1.57) | 0.091 | 1.01 (0.74 to 1.39) | 0.926 | 1.14 (0.95 to 1.39) | 0.164 |
MA, minor allele; MAF, minor allele frequency; SNP, single nucleotide polymorphism.
To examine the independent predictive effect of IRF5 rs4728142 on survival, we conducted multivariable Cox regression analyses. The impact of IRF5 rs4728142 on survival was independent from the age at disease onset and gender (HR: 0.78, 95% CI 0.65 to 0.94, p = 0.008). Furthermore, we extended the analysis to include the disease type (limited versus diffuse) and autoantibody status (antitopoisomerase I autoantibody(ATA) and anticentromere autoantibody (ACA)) in the final model. In this multivariable model, IRF5 rs4728142 remained an independent predictor of better survival (HR: 0.8, 95% CI 0.66 to 0.98, p = 0.032) in SSc patients.
Furthermore, IRF5 rs4728142 (HR: 0.65, 95% CI 0.48 to 0.88, p = 0.006) was significantly associated with survival in a bivariable model that included the HLA DQA1*0501.5 The other HLA allele, DRB1*0802, previously reported to be associated with mortality, was not present in our sample because this allele is of Amerindian origin.
IRF5 rs4728142 also remained a significant predictor of survival after exclusion of patients who did not fulfil the ACR classification criteria (HR: 0.76, 95% CI 0.63 to 0.92, p = 0.005). Finally, subgroup analyses of the GWAS data6 did not indicate that rs4728142A was preferentially associated with limited and ACA subtypes of SSc (see online supplementary text).
IRF5 rs4728142 minor allele correlates with higher FVC% predicted values
The correlation of the three non-HLA loci (IRF5, STAT4 and CD247) with severity of ILD was investigated. Among studied SNPs, the number of the IRF5 rs4728142 minor allele (A) associated with higher FVC% predicted value at enrolment (mean difference: 2.57, CI 0.38 to 4.76, p = 0.022). After including disease duration as a potential confounder in the model, the correlation between FVC% predicted value and IRF5 rs4728142 ‘A’ allele became more significant (mean difference: 2.64, CI 0.43 to 4.84, p = 0.019). The other two IRF5 SNPs, and the investigated STAT4 and CD247 SNPs did not correlate with FVC% predicted. Table 3 shows the details of correlation between SSc-susceptibility genes and FVC% predicted value.
Table 3.
Correlation of IRF5, STAT4 and CD247 number of minor allele with severity of interstitial lung disease in patients with systemic sclerosis
| Chromosome | Gene | SNP | Minor allele | Regression coefficient | p Value | p Value* |
|---|---|---|---|---|---|---|
| 7 | IRF5 | rs10488631 | C | 0.73 (−2.51 to 3.97) | 0.657 | 0.658 |
| rs12537284 | A | 1.27 (−1.64 to 4.18) | 0.391 | 0.390 | ||
| rs4728142 | A | 2.57 (0.38 to 4.76) | 0.022 | 0.019 | ||
| 2 | STAT4 | rs3821236 | A | −1.08 (−3.83 to 1.67) | 0.441 | 0.420 |
| 1 | CD247 | rs2056626 | G | 0.16 (−2.17 to 2.49) | 0.889 | 0.845 |
p Values were adjusted for disease duration.
SNP, single nucleotide polymorphism..
The IRF5 rs4728142 minor allele correlates with lower IRF5 expression in patients and controls
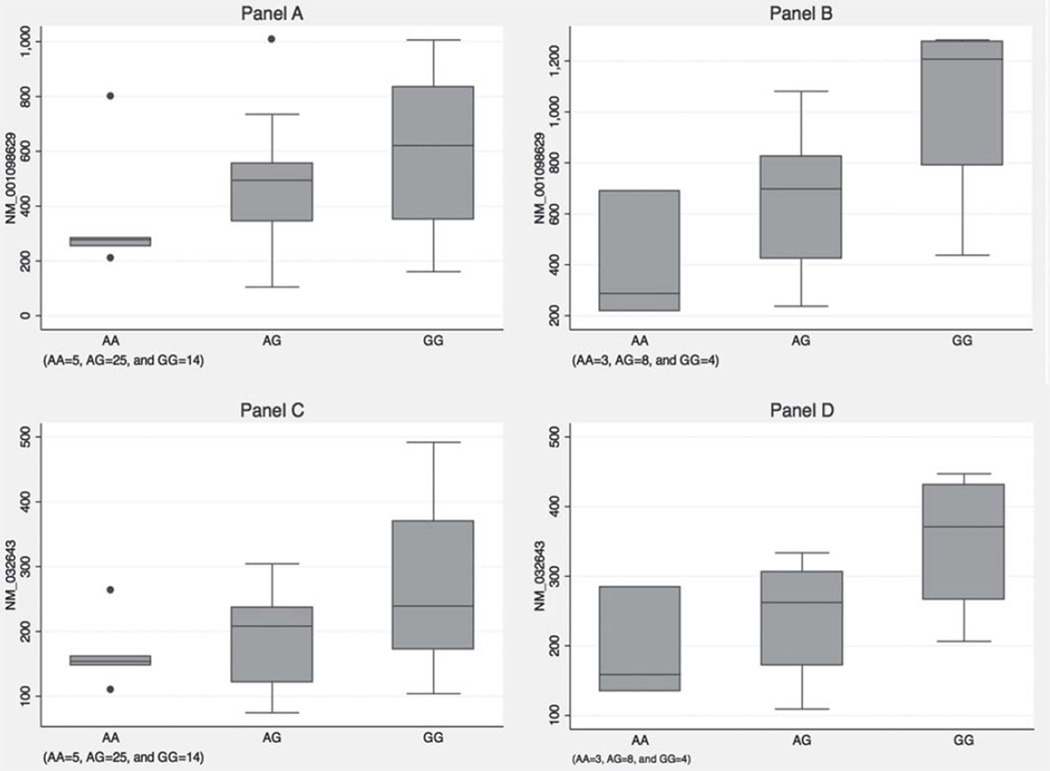
Gene expression microarray experiments were conducted in purified monocyte samples of 44 SSc patients and 15 unaffected controls. In SSc patients, the number of IRF5 rs4728142 minor alleles was associated with lower expression levels of both IRF5 transcript variants (p = 0.034 and p = 0.016). The other investigated IRF5 polymorphisms were not associated with differential expression of IRF5 transcripts.
Similar results were observed in unaffected controls. The IRF5 rs4728142 minor allele was associated with lower levels of IRF5 transcripts following the additive model (p = 0.016 and p = 0.034) while the other investigated IRF5 SNPs did not correlate with IRF5 expression in unaffected controls (tables 4 and 5 and Figure 1).
Table 4.
Correlation of IRF5 susceptibility loci with the IRF5 transcript variant, NM_001098629, in monocytes of patients and unaffected controls
| Patients (n = 44) | Controls (n = 15) | ||||
|---|---|---|---|---|---|
| IRF5 SNP | Minor allele |
Regression coefficient | p Value | Regression coefficient | p Value |
| rs10488631 | C | −66.89 (−225.26 to 91.48) | 0.399 | −118.69 (−487.33 to 249.95) | 0.499 |
| rs12537284 | A | −0.24 (−0.59 to 0.12) | 0.281 | 0.07 (−0.51 to 0.66) | 0.792 |
| rs4728142 | A | −122.67 (−235.52 to −9.82) | 0.034 | −321.78 (−573.02 to −70.54) | 0.016 |
SNP, single nucleotide polymorphism.
Table 5.
Correlation of IRF5 susceptibility loci with the IRF5 transcript variant, NM_032643, in monocytes of patients and unaffected controls
| Patients (n = 44) | Controls (n = 15) | ||||
|---|---|---|---|---|---|
| IRF5 SNP | Minor allele | Regression coefficient | p Value | Regression coefficient | p Value |
| rs10488631 | C | −42.88 (−103.36 to 17.59) | 0.160 | −27.24 (−129.54 to 75.06) | 0.575 |
| rs12537284 | A | −0.28 (−0.56 to 0.10) | 0.062 | 0.04 (−0.38 to 0.46) | 0.835 |
| rs4728142 | A | −53.72 (−96.78 to −10.66) | 0.016 | −80.27 (−153.23 to −7.30) | 0.034 |
SNP, single nucleotide polymorphism.
Figure 1.
(A) Box plot of IRF5 variant (NM_001098629) gene expression levels in patients with systemic sclerosis (SSc); (B) Box plot of IRF5 variant (NM_001098629) gene expression levels in unaffected subjects; (C) Box plot of IRF5 variant (NM_032643) gene expression levels in patients with SSc; (D) Box plot of IRF5 variant (NM_032643) gene expression levels in unaffected subjects. AA, AG, and GG represent genotypes of IRF5 rs4728142
Association of previously described IRF5 susceptibility loci with survival or severity of ILD
The IRF5 rs2004640 has been identified as a SSc-susceptibility locus and was associated with presence of fibrosing alveolitis on CT but not with reduced FVC in a previous study.7 In the current study, the IRF5, rs2004640, was neither significantly associated with survival (p = 0.115) nor with severity of ILD (p = 0.195). Similarly, a previously described SSc risk haplotype (rs3757385*C, rs2004640*T and rs10954213*A)19 that tags IRF5 CGGG insertion/deletion polymorphism20 was neither associated with survival (p = 0.122) nor severity of ILD (p = 0.271) in the current study (see online supplementary text).
DISCUSSION
We investigated the influence of recently described SSc-susceptibility loci on survival in two independent, well-defined SSc cohorts. We demonstrated that an IRF5 polymorphism was associated with survival and severity of ILD. This is the first study demonstrating a correlation of a non-MHC locus with survival and severity of ILD in SSc. The IRF5, rs4728142, had a high minor allele frequency of 49% in patients with SSc. This indicates that a high percentage of patients have at least one copy of the minor allele and this increases the likelihood that this SNP can be used as a prognostic marker.
In this study, the IRF5, rs4728142 variant which was associated with lower IRF5 transcript levels, was predictive of longer survival and milder ILD in SSc patients. Inversely, SSc patients with no copies of IRF5 rs4728142 minor allele had higher IRF5 expression levels and experienced more severe ILD and shorter survival. IRF5 as a transcription factor can affect the downstream type I IFN pathway. Presence of an IFN activation gene expression pattern in peripheral blood cells of SSc patients has been reported by several groups.21–23 In a follow-up study, we compared the peripheral blood gene expression profile of 72 patients with SSc to patients with systemic lupus erythematosus. This study demonstrated significant heterogeneity in transcriptomes of SSc patients. About 50% of SSc patients demonstrated a ‘systemic lupus erythematosus-like’ IFN activation pattern while the other half lacked this gene expression signature.24 It is conceivable that this observed heterogeneity is in part caused by genetic variants in the IFN pathway. However, the actual role of IRF5 in development of SSc has not yet been elucidated. Future mechanistic studies are needed to examine the contribution of IRF5 to the pathogenesis of SSc.
Our data suggest that higher IRF5 levels are associated with a worse prognosis in SSc, providing further evidence for type I IFN pathways as a therapeutic target in SSc. Our findings are supported by previous reports of deleterious effects of type I IFN in SSc. A randomised, placebo-controlled trial of subcutaneous IFN α in patients with early SSc showed that treatment with IFNα was significantly associated with worsening in lung function and a trend toward skin deterioration.25 Furthermore, a type I IFN gene expression signature in peripheral blood of patients with SSc correlated with higher modified Rodnan skin scores.26
We examined the effect of IRF5 susceptibility SNPs on transcript levels of this gene in purified monocyte samples from patients and matched controls. The minor allele of IRF5 rs4728142 was associated with lower IRF5 expression in patients and controls, indicating that the influence of this SNP on IRF5 expression is not conditional on disease status. The SNP, rs4728142, is located in the promoter region of the IRF5 gene. Further studies are needed to investigate the interplay of IRF5 expression in monocytes with other cell types and its overall influence on the observed IFN signature in patients with SSc.
The minor allele of IRF5, rs4728142, was associated with susceptibility to SSc.6 Our data indicate that this susceptibility variant predisposes patients to the milder forms of disease. Further studies are needed to explore how various SSc-susceptibility loci interact with each other and affect the observed heterogeneity in the gene expression profile of patients with SSc. On the other hand, it is possible that the IRF5, rs4728142, is enriched among SSc patients in comparison with controls secondary to survival bias, because the investigated SSc samples with mean disease duration of 9.7 years were prevalent cohorts.
In the current study, IRF5 rs4728142 was not associated with age at disease onset. Furthermore, the relationship between the IRF5, rs4728142, and survival remained significant even after adjustment for age at disease onset and gender, indicting an independent association of this SNP with survival. The IRF5, rs4728142, was also associated with survival even after adjustment for disease type (limited versus diffuse) and serological characteristics (ATA and ACA). This indicates an association of IRF5 rs4728142 independent of disease type and autoantibody status.
The relationship between this SNP and investigated outcomes followed the additive rather than dominant or recessive inheritance mode. This means that the number of minor alleles was associated with longer survival, milder ILD and lower IRF5 transcript levels, implying a dose-response effect for this SNP.
In the current study, the IRF5, rs4728142, SNP was associated also with FVC% predicted which is the only validated surrogate for severity of ILD in SSc.17 Specifically, the number of IRF5 rs4728142 minor alleles was associated with higher FVC% predicted and milder ILD. This finding may explain in part the association of this SNP with survival because pulmonary involvement is presently the leading cause of SSc-related mortality.3–5 This finding can have important implications for development of predictive biomarkers, as routinely obtained demographic and clinical variables did not predict the course of ILD in a prospective study of the GENISOS cohort.27 Further longitudinal studies are needed to investigate whether this SNP or other genetic loci are reliably predictive of ILD course in SSc.
Two previously described susceptibility loci (IRF5 rs2004640 and the above mentioned risk haplotype) were not associated with survival or severity of ILD in the current study. A different IRF5 SNP, rs2280714, was associated with SSc in a study of Japanese SSc patients.28 This SNP was preferentially associated with diffuse and ATA-positive subtypes of SSc. Of note, this polymorphism is in linkage disequilibrium with IRF5 rs4728142.
Our study has some limitations. The lack of sufficient clinical data or reliable clinical instruments precluded us from investigating the association of other important organ manifestations, such as pulmonary arterial hypertension and gastrointestinal involvement with the susceptibility loci identified in the SSc-GWA study. Furthermore, longitudinal studies with comprehensive collection of clinical data are needed to determine whether IRF5 rs4728142 can be included in prognostic models of survival for clinical use. Another limitation of our study is that we did not investigate polymorphisms in the IRF5 gene in a comprehensive manner. IRF5 rs4728142 might be tagging another polymorphism that is the actual causal variant. Moreover, rare variants (<1%) are not investigated in GWA studies. Given that there may be multiple risk polymorphisms within one gene, other IRF5 genetic polymorphisms might also contribute to the severity of disease in SSc. Finally, we examined the overall mortality because reliable data on causes of death were not available in our sample. Future studies are needed to investigate the relationship of susceptibility loci to specific causes of mortality in SSc.
In summary, an SNP in the IRF5 promoter region (rs4728142), associated with lower IRF5 transcript levels, was predictive of longer survival and milder ILD in patients with SSc. This finding underscores the pivotal role of IRF5 and related type I IFN pathways in SSc that could lead to identification of novel therapeutic targets and development of prognostic biomarkers.
Supplementary Material
Acknowledgements
The authors thank Marilyn Perry, Alison Z. Brown, Samuel Theodore, Adrianne Woods and Barbara A. Boyle for their assistance in data collection. This study was supported by the National Institute of Health (NIH)/NIAMS Scleroderma Family Registry and DNA Repository (N01-AR-2251), NIH/NIAMS-R01-AR055258 (Mayes); NIH/NIAMS Center of Research Translation P50-AR-054144 (Arnett and Mayes); NIH-KL2-RR-024149 and K23-AR-061436 (Assassi); NIH T32-AR-052283-03(Reveille); University Clinic Research Center Grants: M01-RR-00073 (UTMB) and M01-RR-01346 (UT-HSC-SA); NIH Clinical and Translational Sciences Award UL1-RR-024148 and TL1-RR-024147 from the National Center for Research Resources. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Funding The study was supported by the National Institute of Health. N01-AR-2251; R01-AR055258; P50-AR-054144; KL2-RR-024149; K23-AR-061436; T32-AR-052283; M01-RR-00073; M01-RR-01346; UL1-RR-024148; TL1-RR-024147.
Footnotes
Contributors All authors contributed and were involved in the conduct of this study, in study design and/or subject recruitment, analysis and manuscript preparation.
Competing interests LKH is a consultant for NexMed and Amira; receives grant support from Actelion, United Therapeutics, Novartis and Medimmune. FMW is a consultant for Amira, Novartis and Orion; advisory board member for Orion Phamaceuticals; received grant support from United Therapeutics and Actelion. DK has received grant support from Takeda, URL and Savient; consultant for Novartis, Ardea, Takeda and Savient; on the speakers bureau of Takeda and Savient; received support for travel expenses from Takeda, Novartis and Savient. DEF has received grants from, is a consultant and scientific review board member for Actelion, Gilead and Roche/Genetech. Actelion, Pfizer and Medimmune have provided grants to the institution where AAS works. RS, MDM, FKT, OYG, JM, LB-C, EBG, HTD, JY, SKA, JDR, FCA and SA declare no competing interest.
Ethics approval Institutional Review Board of the pariticipating sites.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Elhai M, Meune C, Avouac J, et al. Trends in mortality in patients with systemic sclerosis over 40 years: a systematic review and meta-analysis of cohort studies. Rheumatology (Oxford) doi: 10.1093/rheumatology/ker269. Published Online First: 7 September 2011. [DOI] [PubMed] [Google Scholar]
- 2.Thomas E, Symmons DP, Brewster DH, et al. National study of cause-specific mortality in rheumatoid arthritis, juvenile chronic arthritis, and other rheumatic conditions: a 20 year followup study. J Rheumatol. 2003;30:958–965. [PubMed] [Google Scholar]
- 3.Steen VD, Medsger TA. Changes in causes of death in systemic sclerosis, 1972–2002. Ann Rheum Dis. 2007;66:940–944. doi: 10.1136/ard.2006.066068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyndall AJ, Bannert B, Vonk M, et al. Causes and risk factors for death in systemic sclerosis: a study from the EULAR Scleroderma Trials and Research (EUSTAR) database. Ann Rheum Dis. 2010;69:1809–1815. doi: 10.1136/ard.2009.114264. [DOI] [PubMed] [Google Scholar]
- 5.Assassi S, Del Junco D, Sutter K, et al. Clinical and genetic factors predictive of mortality in early systemic sclerosis. Arthritis Rheum. 2009;61:1403–1411. doi: 10.1002/art.24734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radstake TR, Gorlova O, Rueda B, et al. Genome-wide association study of systemic sclerosis identifi es CD247 as a new susceptibility locus. Nat Genet. 2010;42:426–429. doi: 10.1038/ng.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieudé P, Guedj M, Wipff J, et al. Association between the IRF5 rs2004640 functional polymorphism and systemic sclerosis: a new perspective for pulmonary fibrosis. Arthritis Rheum. 2009;60:225–233. doi: 10.1002/art.24183. [DOI] [PubMed] [Google Scholar]
- 8.Rueda B, Broen J, Simeon C, et al. The STAT4 gene influences the genetic predisposition to systemic sclerosis phenotype. Hum Mol Genet. 2009;18:2071–2077. doi: 10.1093/hmg/ddp119. [DOI] [PubMed] [Google Scholar]
- 9.Dieudé P, Boileau C, Guedj M, et al. Independent replication establishes the CD247 gene as a genetic systemic sclerosis susceptibility factor. Ann Rheum Dis. 2011;70:1695–1696. doi: 10.1136/ard.2010.147009. [DOI] [PubMed] [Google Scholar]
- 10.Ross JA, Nagy ZS, Cheng H, et al. Regulation of T cell homeostasis by JAKs and STATs. Arch Immunol Ther Exp (Warsz) 2007;55:231–245. doi: 10.1007/s00005-007-0030-x. [DOI] [PubMed] [Google Scholar]
- 11.Frucht DM, Aringer M, Galon J, et al. Stat4 is expressed in activated peripheral blood monocytes, dendritic cells, and macrophages at sites of Th1-mediated infl ammation. J Immunol. 2000;164:4659–4664. doi: 10.4049/jimmunol.164.9.4659. [DOI] [PubMed] [Google Scholar]
- 12.Krishnan S, Warke VG, Nambiar MP, et al. Generation and biochemical analysis of human effector CD4 T cells: alterations in tyrosine phosphorylation and loss of CD3zeta expression. Blood. 2001;97:3851–3859. doi: 10.1182/blood.v97.12.3851. [DOI] [PubMed] [Google Scholar]
- 13.Assassi S, Fritzler MJ, Arnett FC, et al. Primary biliary cirrhosis (PBC), PBC autoantibodies, and hepatic parameter abnormalities in a large population of systemic sclerosis patients. J Rheumatol. 2009;36:2250–2256. doi: 10.3899/jrheum.090340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reveille JD, Fischbach M, McNearney T, et al. Systemic sclerosis in 3 US ethnic groups: a comparison of clinical, sociodemographic, serologic, and immunogenetic determinants. Semin Arthritis Rheum. 2001;30:332–346. doi: 10.1053/sarh.2001.20268. [DOI] [PubMed] [Google Scholar]
- 15.Subcommittee for Scleroderma Criteria of the American Rheumatism Association Diagnostic and Therapeutic Criteria Committee. Preliminary criteria for the classifi cation of systemic sclerosis (scleroderma) Arthritis Rheum. 1980;23:581–590. doi: 10.1002/art.1780230510. [DOI] [PubMed] [Google Scholar]
- 16.LeRoy EC, Black C, Fleischmajer R, et al. Scleroderma (systemic sclerosis): classifi cation, subsets and pathogenesis. J Rheumatol. 1988;15:202–205. [PubMed] [Google Scholar]
- 17.Furst D, Khanna D, Matucci-Cerinic M, et al. Systemic sclerosis - continuing progress in developing clinical measures of response. J Rheumatol. 2007;34:1194–1200. [PubMed] [Google Scholar]
- 18.Cowper DC, Kubal JD, Maynard C, et al. A primer and comparative review of major US mortality databases. Ann Epidemiol. 2002;12:462–468. doi: 10.1016/s1047-2797(01)00285-x. [DOI] [PubMed] [Google Scholar]
- 19.Dieude P, Dawidowicz K, Guedj M, et al. Phenotype-haplotype correlation of IRF5 in systemic sclerosis: role of 2 haplotypes in disease severity. J Rheumatol. 2010;37:987–992. doi: 10.3899/jrheum.091163. [DOI] [PubMed] [Google Scholar]
- 20.Kristjansdottir G, Sandling JK, Bonetti A, et al. Interferon regulatory factor 5 (IRF5) gene variants are associated with multiple sclerosis in three distinct populations. J Med Genet. 2008;45:362–369. doi: 10.1136/jmg.2007.055012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duan H, Fleming J, Pritchard DK, et al. Combined analysis of monocyte and lymphocyte messenger RNA expression with serum protein profi les in patients with scleroderma. Arthritis Rheum. 2008;58:1465–1474. doi: 10.1002/art.23451. [DOI] [PubMed] [Google Scholar]
- 22.Tan FK, Zhou X, Mayes MD, et al. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology (Oxford) 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- 23.York MR, Nagai T, Mangini AJ, et al. A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis Rheum. 2007;56:1010–1020. doi: 10.1002/art.22382. [DOI] [PubMed] [Google Scholar]
- 24.Assassi S, Mayes MD, Arnett FC, et al. Systemic sclerosis and lupus: points in an interferon-mediated continuum. Arthritis Rheum. 2010;62:589–598. doi: 10.1002/art.27224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Black CM, Silman AJ, Herrick AI, et al. Interferon-alpha does not improve outcome at one year in patients with diffuse cutaneous scleroderma: results of a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 1999;42:299–305. doi: 10.1002/1529-0131(199902)42:2<299::AID-ANR12>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 26.Higgs BW, Liu Z, White B, et al. Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann Rheum Dis. 2011;70:2029–2036. doi: 10.1136/ard.2011.150326. [DOI] [PubMed] [Google Scholar]
- 27.Assassi S, Sharif R, Lasky RE, et al. GENISOS Study Group. Predictors of interstitial lung disease in early systemic sclerosis: a prospective longitudinal study of the GENISOS cohort. Arthritis Res Ther. 2010;12:R166. doi: 10.1186/ar3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito I, Kawaguchi Y, Kawasaki A, et al. Association of a functional polymorphism in the IRF5 region with systemic sclerosis in a Japanese population. Arthritis Rheum. 2009;60:1845–1850. doi: 10.1002/art.24600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.