Abstract
Background
Heterozygous SCN5A mutations have been associated with varied arrhythmia phenotypes; phenotype severity may range from asymptomatic ECG changes (mild phenotype) to symptomatic arrhythmias resulting in syncope, cardiac arrest and sudden cardiac death (severe phenotype) even among family members carrying the same mutation. Risk-stratification schemes for SCN5A mutation carriers remain uncertain.
Objective
We used a family based approach to determine the role of SCN5A promoter variants and DNA methylation in predicting phenotype severity in a kindred with loss-of-function SCN5A mutation.
Methods
In a large kindred with a heterozygous SCN5A loss-of-function mutation (1936delC, Q646RfsX5; 22 mutation carriers), we sought SCN5A promoter variants. In addition, we assessed SCN5A and genome-wide DNA methylation profiles on genomic DNA derived from blood (Illumina Human Methylation27 BeadChip).
Results
During systematic survey of 2.8kb SCN5A promoter region, we identified two SNPs in complete linkage disequilibrium (rs41310749 and rs41310239). These promoter variants were significantly associated with disease severity (mild vs. severe phenotype) (p=0.0007), as all three patients with severe phenotype carried the two-SNP variant on both mutant and wild-type alleles. Analysis did not support a role for methylation of SCN5A related genes.
Conclusion
These family-based genetic findings suggest that the presence of specific promoter variants increase the risk of a severe phenotype in heterozygous carriers of an SCN5A loss-of-function mutation.
Keywords: Arrhythmia, Promoter, Genetic polymorphisms, Sudden cardiac death
Introduction
Mutations in SCN5A, the cardiac sodium channel gene, are associated with varied arrhythmia phenotypes, e.g. long QT syndrome (LQTS), Brugada syndrome (BrS), sick sinus syndrome (SSS), progressive atrioventricular (AV) conduction system disease, ventricular fibrillation and dilated cardiomyopathy.1 Although these arrhythmia phenotypes were originally considered distinct entities, overlap in the clinical presentation has been frequently noted.2–4 The clinical heterogeneity associated with SCN5A mutations is partly explained by corresponding differences in the degree and characteristics of sodium channel dysfunction. For example, in LQTS the biophysical defects in mutant SCN5A protein lead to a molecular gain-of-function phenotype while loss-of-function defects result in BrS and AV conduction system disease.5
However, studies of channel dysfunction do not explain phenotype variation in family members who carry the same SCN5A mutation but have wide-ranging electrocardiographic (ECG) findings and different clinical manifestations. Varied phenotype severity, from asymptomatic to life-threatening, is a challenging clinical problem. For example, in loss-of-function mutation carriers, there is a consensus for device use for secondary prevention of symptomatic arrhythmias,6, 7 but despite extensive study of this problem,8–14 risk stratification of asymptomatic mutation carriers remains uncertain.
Phenotypic variability has been partially explained by the modifying effects of compound heterozygous mutations15 or other SCN5A variants on the mutant16 or wild-type allele.17 Among other genomic regions that modify regulatory functions, promoter elements play a central role in transcriptional regulation.18, 19 Previously, multiple genetic variants, some having altered in vitro activity, were identified in the SCN5A core promoter, supporting the concept of inter-individual variability in transcription of the cardiac sodium channel gene.20 In Asian subjects, Bezzina et al. identified six SCN5A promoter variants in near-complete linkage disequilibrium, that had a significant impact on channel expression in vitro and accounted for a large proportion of variance in ECG measures of conduction in both normal subjects and a cohort of BrS without an SCN5A mutation.21 Together, these studies support the potential biologic importance of SCN5A promoter variants.
Another factor, which may help explain phenotypic variability, is DNA methylation. Combined with protein modification, DNA methylation regulates transcription by establishing a silent chromatin state in a cell and development specific manner, and serves as a critical player in both normal and disease states.22 Such variation may help explain phenotypic variability as genetically identical monozygotic twins exhibit methylation differences.23 In arrhythmia disorders, methylation differences may help explain phenotypic variability although their ability to predict phenotype severity have not been reported until now.
To identify factors that influence phenotype severity associated with a heterozygous SCN5A loss-of-function mutation, we sought genetic variants in SCN5A promoter and assessed SCN5A and genome-wide DNA methylation profiles in a large kindred that exhibited a mixed phenotype (BrS and AV conduction disease) and marked variation in phenotype severity.
Methods
Clinical studies
Relatives of a sudden cardiac death (SCD) victim were invited to participate in the study. Informed consent was obtained from all participants in accordance with the Cincinnati Children’s Hospital Medical Center Institutional Review Board. Participants were evaluated by history, review of medical records, and 12-lead ECG; no subject underwent pharmacologic testing or electrophysiologic study. To analyze phenotypic variability, arrhythmia phenotypes were classified as mild or severe. Individuals without cardiac symptoms were considered to have mild phenotype. Individuals with cardiac events, e.g. syncope, cardiac arrest or SCD, were considered to have severe phenotype. Clinical studies were performed without knowledge of genotype.
SCN5A mutation analysis
Genomic DNA was isolated from blood of study participants, and the polymerase chain reaction (PCR) was used to amplify the coding region and flanking intronic sequence of SCN5A, as described previously.24 Sequencing reactions were performed in the presence of fluorescence-labeled dideoxynucleotides and additional primer for exon-specific sequencing in both sense and antisense direction on isolated PCR product. The nucleotide and amino acid designations were based on the SCN5A transcript NM_198056.2.
Identification of SCN5A promoter variants
The SCN5A promoter has been identified as an ~2.8kb segment of DNA that extends into intron 1 of SCN5A and includes 2.1 kb of 5′ upstream sequence of exon 1, exon 1 (which is 142 bp and non-coding), and the proximal intron 1 regions that are relatively GC-rich (60.6% GC content) and highly conserved compared to mouse and rat.25 Several studies have identified single nucleotide polymorphisms (SNPs) that act as cis-acting elements in the promoter sequence.20, 21, 25 We considered three strategies to identify SCN5A promoter variants.
We performed a restriction enzyme map of SCN5A promoter of known SNPs but identified only two SNPs that altered restriction fragment length; we concluded that this approach would not be suitable for screening all known variants (Supplemental Table 1).20
To rapidly genotype promoter variants at low cost, we performed a high-resolution DNA melting (HRM) analysis followed by confirmation of variant sequences directly by Sanger sequencing. We designed primers to cover the promoter region by dividing it into overlapping amplicons with lengths <250 bp using Primer3 (v.0.4.0) (Supplemental Table 2). HRM analysis was performed using the 7500 Fast Real-Time PCR system (Applied Biosystems). All variants calls were confirmed with direct sequencing. Additionally, to reduce false negative calls, in each of 4 family members with and without the SCN5A mutation, we sequenced PCR products without variant calls. For direct sequencing, we used 9 overlapping primer sets with about twice the length of designed primers in HRM and one more primer set to give full coverage (Supplemental Table 3). As the screened SCN5A promoter gene segment is relatively GC-rich, 10 M betaine 5 μL (final 2 M) and 2 μL 1,2-propanediol (final 1.088 M) were used for enhanced amplification of GC-rich promoter region. Sequencing reactions were performed in the presence of fluorescence-labeled dideoxynucleotides and additional primer in both sense and antisense direction on isolated PCR product.
Haplotype construction
To determine the phase of the SCN5A alleles, haplotypes were manually constructed according to Mendelian inheritance of genetic variants in SCN5A promoter and coding sequence and compared with those of family members.
Analyzing SCN5A promoter variants against transcription factor-binding-site sequence database
We downloaded the upstream 4 kb region of human SCN5A using the UCSC Genome Browser.26 We used the wild type sequence and sequence containing the SNPs of interest to create an altered sequence (replacing the SNP affected base pairs). The wild type and altered sequences were then scanned for putative transcription factor binding sites (TFBSs) using MatInspector (http://www.genomatix.de).27 We then compared the list of putative TFBSs in both the sequences (with and without SNP) to identify potential loss or gain of TFBSs because of the SNPs.
DNA methylation profiling
DNA methylation was assayed in 31 family members using Infinium Human Methylation27 BeadChip (Illumina, Inc., San Diego, USA) to identify candidate genes that are differentially methylated. DNA was treated by sodium bisulfite administration to convert unmethylated cytosines to uracil using EZ DNA Methylation-Direct kit (Zymo research, Irvine, CA). Beta values were exported from the methylation module, and the quality of the data was checked using R packages Meth27QC, HumMeth27QCReport, and arrayQualityMetrics. Color adjustment, background correction, and normalization were then performed using R package lumi.
In addition to the Illumina chip analysis, the 12 CpG dinucleotides in the SCN5A promoter were examined by direct Sanger sequencing of PCR amplicons of bisulfite treated DNA. MethPrimer (http://www.urogene.org/methprimer/index1.html) program was used to design primers for PCR (Supplemental Table 4). PCR of bisulfite-converted DNA (100 ng/PCR reaction) was carried out using Taq RED DNA Polymerase Master Mix (Apex BioResearch products, Research Triangle Park, NC).
Statistical analyses
We examined the impact of both linkage and association on the cardiac phenotype in a large kindred. For linkage, we examined whether SCN5A variants of interest were linked with clinical phenotype severity. Using SUPERLINK v1.5 (http://cbl-fog.cs.technion.ac.il/superlink-online/), we performed two-point linkage analyses between the SCN5A coding variant and disease. Briefly, this program utilizes the framework of Bayesian networks to combine the Elston-Stewart and Lander-Green approaches for computing the likelihood of a pedigree exactly, even in large pedigrees.28, 29 The linkage model assumed a disease mutant gene frequency of 1% and a domina nt mode of inheritance with complete penetrance. We then used association analysis to examine not only the presence of disease but also severity. To account for the familial structure of the data, all association analyses were performed in SOLAR (Southwest Foundation for Biomedical Research, San Antonio, TX, USA).30 Briefly, this package uses mixed models where the fixed effects are the covariates (in this case the variants) and the random effects are defined by the kinship coefficient. To test for associations with severity, we restricted the analysis to individuals carrying an SCN5A mutation.
The primary analysis of data from the Human Methylation27 BeadChip focused on an assessment of a subset of 41 CpG loci located in SCN5A and SCN5A related genes. As a secondary analysis, we screened all probes. Differential methylation was assessed for each probe using R package Limma after probes on sex chromosomes and probes with detection p value>0.01 were excluded. After adjusting for age and sex, comparisons were performed between carriers and non-carriers of 1936delC and promoter SNP genotype, and between severe and mild phenotypes in 1936delC carriers. Significance was considered at alpha=0.0012 after Bonferroni correction for the subset of 41 probes. Multiple testing correction for the genome-wide secondary analysis resulted in an alpha of 2×10−6.
Results
Genotype-phenotype: a family based analysis
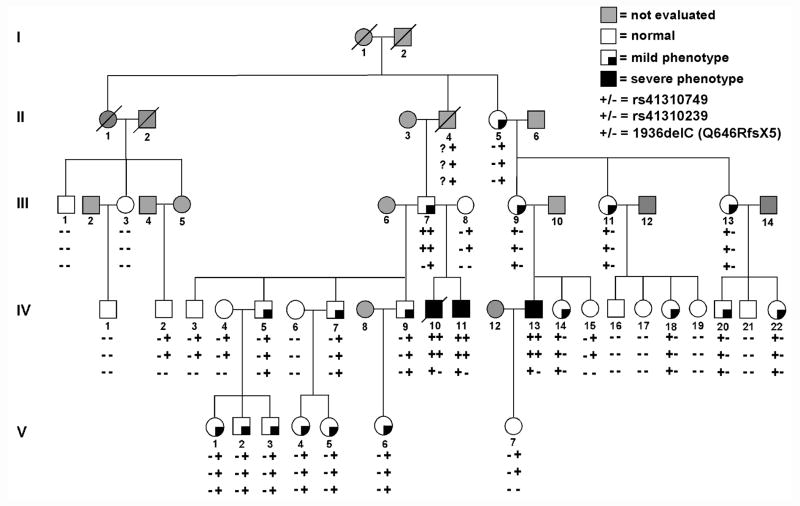
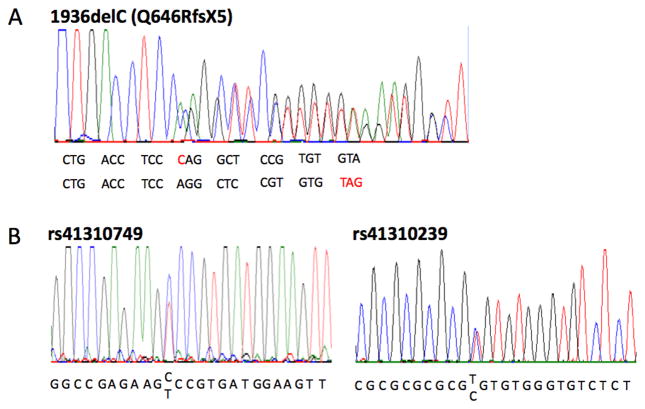
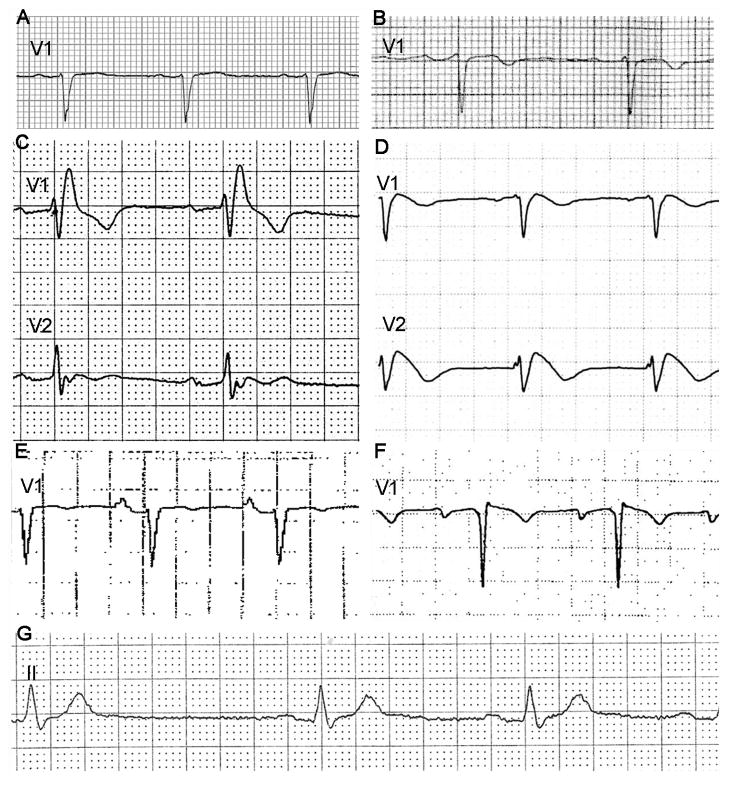
A large kindred was ascertained due to SCD of the proband (IV-10, age 21 years) (Figure 1). There were no abnormal findings on autopsy, but postmortem genetic analysis identified an SCN5A heterozygous mutation (1936delC, Q646RfsX5) (Figure 2A). The truncation mutation has been previously reported as a loss-of-function mutation causing BrS.31 SCN5A-Q646RfsX5 was identified in 21 other family members including two others with a severe phenotype: syncope in a brother (IV-11, age 21 years) (Figure 3G), and cardiac arrest in a cousin (IV-13, age 30 years) found to have BrS (Figure 3D). The proband’s grandfather (II-4) died suddenly at age 57 years; he was an obligate mutation carrier but could not be evaluated. Other mutation carriers had a mild phenotype characterized by absent symptoms and ECG findings of 1st degree AV block alone (n=11), intraventricular conduction delay (IVCD) alone (n=4) or in addition to 1st degree AV block (n=2) and right bundle branch block (RBBB) plus 1st degree AV block (n=1) (Figure 3). No individual had evidence of progressive AV block. All mutation carriers had an abnormal ECG, typically prolonged PR interval and/or QRS duration demonstrating that Q646RfsX5 exerted a functional effect.
Figure 1.
A large kindred segregating an SCN5A mutation (del1936C, Q646RfsX5) and two promoter variants (rs41310749 and rs41310239). Phenotypes (designated as mild or severe) and genotypes are indicated. Square symbols indicate males and round symbols indicate females. Slashed symbols indicate decreased individuals; Roman numerals indicate generations in the family. Arrhythmia phenotypes were classified as mild or severe. Individuals without cardiac symptoms were considered to have mild phenotype. Individuals with cardiac events, e.g. syncope, cardiac arrest or sudden cardiac death (SCD), were considered to have severe phenotype: SCD in a proband (IV-10, age 21 years), syncope in a brother (IV-11, age 21 years), and cardiac arrest in a cousin (IV-13, age 30 years).
Figure 2.
Sequencing of SCN5A mutation and SCN5A promoter variants. Automated DNA-sequencing electrophoretograph reveal heterozygous nucleotide change (A) and the two promoter variants (B).
Figure 3.
Electrocardiograms. (A) Lead V1 tracing showing intraventricular conduction delay (IVCD) in a 53 year old female (III-11). (B) Lead V1 tracing showing 1st degree atrioventricular (AV) block in a 50 year old female (III-9). (C) Lead V1 and V2 tracing showing right bundle branch block in a 69 year old male (III-7). (D) Lead V1 and V2 tracing showing typical features of Brugada syndrome in a 30 year old male (IV-13). (E) Lead V1 tracing showing 1st degree AV block and IVCD in a 56 year old female (III-13). (F) Lead V1 tracing showing 1st degree AV block in a 20 year old male (V-2). (G) Lead II tracing showing marked sinus bradycardia (cycle length >1500ms) and 1st degree AV block in a 21 year old male (IV-11). Electrocardiographic traces were obtained at standard recording conditions of 25 mm/s and 10 mm/mV.
Given the size of the family, we performed linkage analysis and found that 1936delC, exhibited significant evidence of both linkage (LOD=6.1) and association with disease (p<0.0001), but the 1936delC variant did not distinguish mild from severe phenotype (p=1.0).
SCN5A promoter variants
During systematic survey of 2.8kb promoter region of SCN5A, two SNPs (c.-194-1854 C>T, rs41310749; c.-194-865 T>C, rs41310239) (Figure 2B) were identified. As previously reported, these two SNPs were in strong linkage disequilibrium.20 The minor alleles of rs41310749 and rs41310239 segregated together in all of the 27 individuals who carried the variants (Figure 1).
The promoter variants exhibit lower evidence of linkage (LOD=3.0) compared to individuals carrying 1936delC but significant evidence of association with disease (p=0.00001). None of the six family members with the two SNP haplotype on one allele but without the 1936delC mutant variant showed a phenotype. Nearly all 18 family members with mild phenotype (except individual III-7) carried the two SNP haplotype only on the mutant allele with 1936delC (Figure 1). Restricting the analysis to all mutation carriers, the promoter variants are significantly associated with disease severity (mild vs. severe phenotype) (p=0.0007). All three members with the 1936delC mutant and severe phenotype carried the two SNP haplotype on both mutant and wild-type alleles. Although all three individuals with severe phenotype were young (< 40 years), it is possible that young individuals with a mild phenotype would develop the severe phenotype in time. Thus, using an age restrictive (>40 years) definition of mild phenotype, the two SNP promoter haplotype was still associated with phenotype severity (p = 0.003).
Effects of SCN5A promoter variants rs41310749 and rs41310239
Comparison of putative TFBSs in sequences with and without the SNPs with MatInspector, we found that two-SNP variant results in generation of novel binding sites for retinoid X receptor (RXR) heterodimer and vertebrate homolog of enhancer of split (Hes1) complex. Yang et al. compared activity of wild-type promoters to promoters modified by introducing minor allele SNP variants in both mouse neonatal cardiomyocytes and Chinese hamster ovary (CHO) cells.20 They identified a trend toward reduced activity for the two SNP haplotype (rs41310749 and rs41310239).20
Genome-wide DNA methylation profiles and SCN5A DNA methylation
Among 31 samples analyzed, chip data from one sample failed quality control and was excluded. For the 41 CpG loci in SCN5A, SCN5A related genes (FSHR, KCNE1, KCNQ1, NOS1, SCN1B, TCAP and SCN5A) or other BrS loci, e.g. GPD-1L, CACNA1C, CACNB2, SCN1B, KCNE3 and SCN3B, no significant differential methylation was detected between 20 carriers and 10 non-carriers of 1936delC. Further among 1936delC carriers, no differences were noted between 17 individuals with mild and 3 individuals with severe phenotypes (data not shown). When considering methylation patterns across the genome, we identified two CpG loci cg02288165 and cg08632701 in the genes SIGLEC-1 (sialic acid binding Ig-like lectin 1, member of the immunoglobulin superfamily) and SETD4 (Su(var)3–9, Enhancer-of-zeste domain-containing protein 4) which were differentially methylated between individuals with severe and mild phenotypes after adjusting for age and sex. Finally, analyses of methylation status of the 12 CpG dinucleotides in the SCN5A promoter by direct sequencing identified no differences between carriers and non-carriers of 1936delC, or between mild and severe phenotypes in 1936delC carriers (data not shown).
Clinical follow-up
In March 2007, 11 mutation carriers were identified among first degree relatives of the proband (Figure 1, center-left side of pedigree). Among mutation carriers, only individual IV-11 was under the care of a cardiologist for symptomatic sinus bradycardia and treated with a pacemaker (PM). Members of the family lived broadly in the United States. Given the results of the genetic testing and the family history of sudden death, most sought counsel of a cardiologist. Over the ensuing six months, 7 of the 11 living mutation carriers received an implantable cardiac defibrillator (ICD) based on advice of their cardiologist. In October 2009, individual IV-13 experienced a cardiac arrest and was treated with an ICD. Subsequently, 15 members of his family (descendants of II-5) underwent genetic testing and 9 SCN5A mutation carriers were identified (Figure 1, right side of pedigree). These family members also lived broadly in the United States. Given the experience of their relatives and more conservative use of ICD in recommendations for SCN5A mutation carriers no other family members underwent ICD implantation. In December 2011, Individual IV-13, a cardiac arrest survivor, experienced an appropriate ICD discharge.
Discussion
Studies by Bezzina et al. suggested that SCN5A promoter variants, which slowed conduction in normal subjects and exacerbated conduction slowing in those with BrS, might be candidate modulators of variability in risk of SCD.21 Findings in our family-based genetic study implicate SCN5A promoter variants as modifiers of severe arrhythmia risk in carriers of SCN5A loss-of-function mutations. We found all mutation carriers had an abnormal ECG phenotype, but coinheritance of a two SNP promoter haplotype in the homozygous state was significantly associated with clinical phenoytpe severity. The presence of the promoter variants on either the mutant or wild-type allele alone did not predict disease severity, but all three family members with severe arrhythmia phenotype carried the two-SNP variant on both mutant and wild-type alleles suggesting the presence of these elements as risk factors for a severe phenotype in heterozygous carriers of an SCN5A loss-of-function mutation.
Compared to wild-type, a trend toward reduced promoter activity for the two SNP haplotype (rs41310749 and rs41310239) has been reported.20 In the case where promoter variants are present only on the wild-type allele, the reduction in wild-type protein abundance is predicted to be clinically insignificant. Similarly, the presence of the promoter variants only on the mutant allele would be predicted to be clinically insignificant as reduction in loss of function mutant protein is unlikely to alter the phenotype. However, when promoter variants are present on both wild-type and mutant alleles, we speculate that reduction in amount of wild-type channel is additive to the effect of the mutant loss of function allele thereby increasing the risk of a severe phenotype. This possibility is similar to findings by Leoni et al,32 who reported that variation in phenotype severity in heterozygous SCN5A knock-out mice, correlated with wild-type channel protein expression. Such a possibility was also suggested for a haplotype of 6 SCN5A promoter variants found in an Asian population,21 and 3′ untranslated region variants found in LQT1-associated KCNQ1 mutation carriers.33
The two SNP haplotype (rs41310749 and rs41310239) is common, occurring in 17–22% of white controls.20 Bioinformatic analysis identified that rs41310749 and rs41310239 result in generation of novel binding sites for RXR heterodimer and Hes1 complex, respectively. RXRs are expressed in virtually every tissue of the body, and they form obligate heterodimers with many other Nuclear Receptor superfamily members, e.g. peroxisome proliferator-activated receptor gamma (PPARγ).34 As such, RXRs play unique modulatory and integrative roles across multiple metabolic systems and are master regulators of gene networks that control cell growth, differentiation, survival and death. Because of the large number of heterodimer partners, creation of an RXR heterodimer binding site has the potential for wide-ranging biologic effects. Hes1 on the other hand belongs to the basic helix-loop-helix (bHLH) family of transcription factors and is part of Notch signaling pathway. As a transcriptional repressor of genes that require a bHLH protein for their transcription it too has the potential for wide-ranging biologic effects.
Our studies did not support a role for differential methylation of SCN5A or related genes in explaining variation in phenotype severity. However, we studied methylation profiles in blood derived DNA. We recognize that DNA methylation patterns may be tissue specific. Unfortunately, the optimal cell type, i.e. Purkinje cells, atrial myocytes or ventricular myocytes, were not available in the family members we studied. Additionally, our methylation studies were based on a small sample, thus we would only expect to detect differences of large effect. Indeed, we did identify statistically significant differential methylation of two genes, even with our small sample size. Nonetheless, given the large number of statistical comparisons and the lack of biologic plausibility of these genes, the significance of this finding is unknown.
Recently, Probst et al. studied large kindreds and found a lack of concordance between phenotype and SCN5A mutation carrier status;35 they interpreted their results as indicating that genetic background may play a powerful role in the pathophysiology of BrS. Results of our study and those of others20, 21 suggest that SCN5A promoter variants are a type of genetic background. However, genetic variation in the SCN5A promoter does not explain phenotype variation in all BrS families; for example, mutations in the SCN5A gene are identified in only ~20% of patients with BrS.30 These limitations aside, identification of predictors of phenotype variation among SCN5A mutation carriers would provide the opportunity to develop primary prevention strategies for individuals at high risk of cardiac events (SCD, cardiac arrest or syncope). Given that current treatment is limited to ICD implantation, with its inherent complications in young patients, risk stratification in asymptomatic mutation carriers would be of great clinical utility.
Conclusion
Based on findings of this study, further examination of the relationship between genetic variants known or suspected to modulate sodium channel expression and clinical phenotypes resulting from SCN5A loss-of-function mutations seems warranted. Ultimately, these data might be useful in risk stratification in individual patients.
Supplementary Material
Acknowledgments
The authors thank the family members for their participation. The studies were facilitated by the assistance of Eric L. Vey, MD, Kerry A. Shooner, MS and Nessie Hicks-Johnson in collecting patient material. This study was supported in part by a grant from the National Institute of Health (NIH HL69712, DWB).
Abbreviations
- AV
atrioventricular
- bHLH
basic helix-loop-helix
- BrS
Brugada syndrome
- ECG
electrocardiogram
- Hes1
vertebrate homolog of enhancer of split
- HRM
high resolution melting
- ICD
implantable cardiac defibrillator
- IVCD
intraventricular conduction delay
- LQTS
long QT syndrome
- PCR
polymerase chain reaction
- PM
pacemaker
- RBBB
right bundle branch block
- RXR
retinoid X receptor
- SCD
sudden cardiac death
- SNP
single nucleotide polymorphism
- SSS
sick sinus syndrome
- TFBS
transcription factor binding site
Footnotes
No authors declare any conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lehnart SE, Ackerman MJ, Benson DW, Jr, et al. Inherited arrhythmias: a National Heart, Lung, and Blood Institute and Office of Rare Diseases workshop consensus report about the diagnosis, phenotyping, molecular mechanisms, and therapeutic approaches for primary cardiomyopathies of gene mutations affecting ion channel function. Circulation. 2007;116:2325–2345. doi: 10.1161/CIRCULATIONAHA.107.711689. [DOI] [PubMed] [Google Scholar]
- 2.Remme CA, Wilde AA, Bezzina CR. Cardiac sodium channel overlap syndromes: different faces of SCN5A mutations. Trends Cardiovasc Med. 2008;18:78–87. doi: 10.1016/j.tcm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Ruan Y, Liu N, Priori SG. Sodium channel mutations and arrhythmias. Nat Rev Cardiol. 2009;6:337–348. doi: 10.1038/nrcardio.2009.44. [DOI] [PubMed] [Google Scholar]
- 4.Meregalli PG, Tan HL, Probst V, et al. Type of SCN5A mutation determines clinical severity and degree of conduction slowing in loss-of-function sodium channelopathies. Heart Rhythm. 2009;6:341–348. doi: 10.1016/j.hrthm.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 5.Amin AS, Asghari-Roodsari A, Tan HL. Cardiac sodium channelopathies. Pflugers Arch. 2010;460:223–237. doi: 10.1007/s00424-009-0761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Priori SG, Aliot E, Blomstrom-Lundqvist C, et al. Task force on sudden cardiac death of the European Society of Cardiology. Eur Heart J. 2001;22:1374–1450. doi: 10.1053/euhj.2001.2824. [DOI] [PubMed] [Google Scholar]
- 7.Remme CA, Wever EF, Wilde AA, Derksen R, Hauer RN. Diagnosis and long-term follow-up of the Brugada syndrome in patients with idiopathic ventricular fibrillation. Eur Heart J. 2001;22:400–409. doi: 10.1053/euhj.2000.2366. [DOI] [PubMed] [Google Scholar]
- 8.Brugada J, Brugada R, Antzelevitch C, Towbin J, Nademanee K, Brugada P. Long-term follow-up of individuals with the electrocardiographic pattern of right bundle-branch block and ST-segment elevation in precordial leads V1 to V3. Circulation. 2002;105:73–78. doi: 10.1161/hc0102.101354. [DOI] [PubMed] [Google Scholar]
- 9.Brugada J, Brugada R, Brugada P. Determinants of sudden cardiac death in individuals with the electrocardiographic pattern of Brugada syndrome and no previous cardiac arrest. Circulation. 2003;108:3092–3096. doi: 10.1161/01.CIR.0000104568.13957.4F. [DOI] [PubMed] [Google Scholar]
- 10.Priori SG, Napolitano C, Gasparini M, et al. Natural history of Brugada syndrome: insights for risk stratification and management. Circulation. 2002;105:1342–1347. doi: 10.1161/hc1102.105288. [DOI] [PubMed] [Google Scholar]
- 11.Gehi AK, Duong TD, Metz LD, Gomes JA, Mehta D. Risk stratification of individuals with the Brugada electrocardiogram: a meta-analysis. J Cardiovasc Electrophysiol. 2006;7:577–583. doi: 10.1111/j.1540-8167.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- 12.Probst V, Veltmann C, Eckardt L, et al. Long-term prognosis of patients diagnosed with Brugada syndrome: results from the FINGER Brugada Syndrome Registry. Circulation. 2010;121:635–643. doi: 10.1161/CIRCULATIONAHA.109.887026. [DOI] [PubMed] [Google Scholar]
- 13.Raju H, Papadakis M, Govindan M, et al. Low prevalence of risk markers in cases of sudden death due to Brugada syndrome relevance to risk stratification in Brugada syndrome. J Am Coll Cardiol. 2011;57:2340–2345. doi: 10.1016/j.jacc.2010.11.067. [DOI] [PubMed] [Google Scholar]
- 14.Priori SG, Gasparini M, Napolitano C, et al. Risk Stratification in Brugada Syndrome Results of the PRELUDE (PRogrammed ELectrical stimUlation preDictive valuE) Registry. J Am Coll Cardiol. 2012;59:37–45. doi: 10.1016/j.jacc.2011.08.064. [DOI] [PubMed] [Google Scholar]
- 15.Benson DW, Wang DW, Dyment M, et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viswanathan PC, Benson DW, Balser JR. A common SCN5A polymorphism modulates the biophysical effects of an SCN5A mutation. J Clin Invest. 2003;111:341–346. doi: 10.1172/JCI16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinlapawittayatorn K, Du XX, Liu H, Ficker E, Kaufman ES, Deschênes I. A common SCN5A polymorphism modulates the biophysical defects of SCN5A mutations. Heart Rhythm. 2011;8:455–462. doi: 10.1016/j.hrthm.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler JE, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 19.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 20.Yang P, Koopmann TT, Pfeufer A, et al. Polymorphisms in the cardiac sodium channel promoter displaying variant in vitro expression activity. Eur J Hum Genet. 2008;16:350–357. doi: 10.1038/sj.ejhg.5201952. [DOI] [PubMed] [Google Scholar]
- 21.Bezzina CR, Shimizu W, Yang P, et al. Co mmon sodium channel promoter haplotype in asian subjects underlies variability in cardiac conduction. Circulation. 2006;113:338–344. doi: 10.1161/CIRCULATIONAHA.105.580811. [DOI] [PubMed] [Google Scholar]
- 22.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Li Z, Shen J, Keating MT. Genomic organization of the human SCN5A gene encoding the cardiac sodium channel. Genomics. 1996;34:9–16. doi: 10.1006/geno.1996.0236. [DOI] [PubMed] [Google Scholar]
- 25.Yang P, Kupershmidt S, Roden DM. Cloning and initial characterization of the human cardiac sodium channel (SCN5A) promoter. Cardiovasc Res. 2004;61:56–65. doi: 10.1016/j.cardiores.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 26.Fujita PA, Rhead B, Zweig AS, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silberstein M, Tzemach A, Dovgolevsky N, Fishelson M, Schuster A, Geiger D. Online system for faster multipoint linkage analysis via parallel execution on thousands of personal computers. Am J Hum Genet. 2006;78:922–935. doi: 10.1086/504158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics. 2002;18:S189–S198. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 30.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapplinger JD, Tester DJ, Alders M, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leoni AL, Gavillet B, Rougier JS, et al. Variable Na(v)1.5 protein expression from the wild-type allele correlates with the penetrance of cardiac conduction disease in the Scn5a(+/−) mouse model. PLoS One. 2010;5:e9298. doi: 10.1371/journal.pone.0009298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amin AS, Giudicessi JR, Tijsen AJ, et al. Variants in the 3′ untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. Eur Heart J. doi: 10.1093/eurheartj/ehr473. Epub 2011 Dec 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamza MS, Pott S, Vega VB, et al. De-novo identification of PPARgamma/RXR binding sites and direct targets during adipogenesis. PLoS One. 2009;4:e4907. doi: 10.1371/journal.pone.0004907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Probst V, Wilde AA, Barc J, et al. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome. Circ Cardiovasc Genet. 2009;2:552–557. doi: 10.1161/CIRCGENETICS.109.853374. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.