Abstract
Association mapping of important traits of crop plants relies on first understanding the extent and patterns of linkage disequilibrium (LD) in the particular germplasm being investigated. We characterize here the genetic diversity, population structure and genome wide LD patterns in a set of asparagus bean (Vigna. unguiculata ssp. sesquipedialis) germplasm from China. A diverse collection of 99 asparagus bean and normal cowpea accessions were genotyped with 1127 expressed sequence tag-derived single nucleotide polymorphism markers (SNPs). The proportion of polymorphic SNPs across the collection was relatively low (39%), with an average number of SNPs per locus of 1.33. Bayesian population structure analysis indicated two subdivisions within the collection sampled that generally represented the ‘standard vegetable' type (subgroup SV) and the ‘non-standard vegetable' type (subgroup NSV), respectively. Level of LD (r2) was higher and extent of LD persisted longer in subgroup SV than in subgroup NSV, whereas LD decayed rapidly (0–2 cM) in both subgroups. LD decay distance varied among chromosomes, with the longest (≈5 cM) five times longer than the shortest (≈1 cM). Partitioning of LD variance into within- and between-subgroup components coupled with comparative LD decay analysis suggested that linkage group 5, 7 and 10 may have undergone the most intensive epistatic selection toward traits favorable for vegetable use. This work provides a first population genetic insight into domestication history of asparagus bean and demonstrates the feasibility of mapping complex traits by genome wide association study in asparagus bean using a currently available cowpea SNPs marker platform.
Keywords: asparagus bean, association mapping, cowpea, domestication history, linkage disequilibrium, population structure
Introduction
Linkage disequilibrium (LD) refers to association of particular allelic configurations at distinct loci in the genome of a sampled population (Weir, 1979). LD mapping or association mapping can be applied to natural populations, sets of germplasm accessions or cultivars developed recently (Rostoks et al., 2006; Robbins et al., 2011) and assumes that only markers in strong LD with a functionally important gene will be significantly associated to the variation in quantitative traits (Ardlie et al., 2002; Garris et al., 2003). LD mapping thus takes advantage of the much larger number of historical recombination events that have occurred over time compared with the relatively restricted levels of recombination inherent in biparental mapping populations where quantitative trait loci (QTL) mapping studies have frequently been used to unravel the genetic basis of traits (Cardon and Bell, 2001). In order to appropriately apply LD mapping in crop plants, it is a prerequisite to understand the extent and patterns of LD in the particular populations being investigated (Lander and Schork, 1994). It is also crucial to be able to distinguish between physical LD and the other different forces that can create LD in natural populations, to avoid the detection of spurious associations (Flint-Garcia et al., 2003).
LD can be caused by unknown population structure and several forces, including mutation, drift, genetic bottlenecks, founder effects, selection, and specifically for plants, level of inbreeding caused by their mating systems (Hartl and Clark, 1997). In contrast, physical LD tends to be continuously decreasing because of accumulated recombinations, which means that loci located far apart along chromosomes will generally remain in LD for shorter periods (Hartl and Clark, 1997; Mather et al., 2007). Outcrossing species generally exhibit low levels of LD because of many opportunities for effective recombination events in large highly heterozygous populations (Yan et al., 2009). In contrast, many crop species including asparagus bean, are inbreeders and would thus be expected to exhibit relatively higher levels of LD (Flint-Garcia et al., 2003; Morrell et al., 2005).
The extent and patterns of LD can be investigated on a haplotype, chromosome or whole genome level. Tenaillon et al. (2001) sequenced 21 loci located on the maize chromosome 1 and estimated that in landraces the LD decay distance was <1000 bp. In contrast, a study of 18 maize genes in 36 commercial inbred lines revealed extensive LD blocks as long as 100 Kb (Ching et al., 2002). Thus, as pointed out by Yan et al. (2009), LD decay estimation based on a single chromosome or a limited number of loci may be biased. Uncovering LD patterns of plant species of interest at a genome level is therefore essential for the optimal design of genotyping efforts before conducting LD mapping and for assessing the statistical power and resolution in whole-genome association studies (McNally et al., 2009).
Cowpea (Vigna unguiculata L. Walp., 2n=2x=22) is an important self-pollinating grain legume, fodder and vegetable crop in many tropical/subtropical regions of the world. Two main divisions of cultivated cowpea are the dominant subspecies unguiculata used primarily for dry grain and fodder and sesquipedialis, which is harvested at the immature green pod stage and used as a vegetable (Timko et al., 2007). The latter is also known as asparagus bean or ‘yard-long' bean, for it is characterized by its very long (0.5–1 m) pods with less fiber, narrow kidney-shaped seeds and climbing growth habit. Genetic similarity between the two subspecies is high as evidenced by fully fertile hybrids and synteny for most of the shared molecular markers between genetic maps (Muchero et al., 2009; Xu et al., 2011). Cowpea is thought to have undergone a severe bottleneck during domestication (Panella and Gepts, 1992). Although genetic diversity has not been comprehensively assessed in the asparagus bean germplasm, it is likely low, keeping the assumption that the crop may have derived from a limited sample of the wider cowpea gene pool that moved from Africa to east Asia, followed by a further genetic narrowing as strong selection for long pods gave rise to the present day ssp. sesquipedialis (Fang et al., 2007; Xu et al., 2010). Assessing both genetic variation levels and genome-wide LD patterns in asparagus bean thus would not only be important for LD mapping but also shed some light on the past effects of intensive human selection for vegetable use, compared with materials that would be closer to the ancestral species.
Recently, a high-throughput Illumina GoldenGate assay platform with 1536 expressed sequence tag-derived single nucleotide polymorphism markers (SNPs) became available for cowpea. Using this system, high-density consensus genetic linkage maps for the ssp. unguiculata were constructed (Muchero et al., 2009; Lucas et al., 2011). A comparable genetic linkage map of asparagus bean also was constructed in the authors' lab using the same system and additional SSR markers (Xu et al., 2011). The main objective of the present study was to analyze the genetic variation and LD patterns in asparagus bean based on a panel of 95 Chinese accessions genotyped using the above SNP markers.
Materials and methods
Plant materials
A sample set consisting of 95 asparagus bean accessions from a wide geographic origin across China and four ssp. unguiculata accessions (as a control) were used in the current study. Details of the plant materials including names, origins, subspecies assignment and morphological characteristics can be found in Supplementary Table 1.
DNA extraction
Genomic DNA was extracted from leaves of 2-week-old plants using a DNeasy Plant DNA miniprep kit (Qiagen, Hilden, Germany) according to the procedures described by the manufacturer.
SNP genotyping
Each of the 99 accessions was genotyped for SNPs using the KASPar (KBiosciences, Hoddeston, UK) 1127-SNP genotyping platform converted from the Illumina cowpea GoldenGate 1536-SNP assay system (Muchero et al., 2009). Names of the 1127 SNPs are listed in Supplementary Table 2 and their details including map positions information can be accessed at HarvEST (http://www.harvest-web.org/hweb/bin/wc.dll?hwebProcess~hmain~&versid=68). After fluorescence scanning of the reactions, the results were interpreted by the software KlusterCaller 1.1 (KBiosciences), which can be accessed at the Dryad repository (doi:10.5061/dryad.6tv35cc2).
Inference of structure and kinship
Population structure was calculated using a Bayesian model-based clustering method implemented in the software STUCTURE 2.3.3 (Pritchard et al., 2000) using data from 422 informative SNPs (see results below). We ran STRUCTURE under the ‘admixture model' with a burn-in period of 100 000 followed by 100 000 replications of Markov Chain Monte Carlo. Three independent runs each were performed with the number of clusters (K) varying from 1 to 10. A statistic ΔK based on the relative rate of change in the likelihood of the data between successive K values were used to determine the optimal number of clusters (Evanno et al., 2005). Lines with probability of membership >70% were assigned to a subgroup. No a priori population information was used. Pairwise genetic distances were calculated using the software Powermarker 3.25 under the Nei 1983 model (Liu and Muse, 2005). Relative kinship matrix was constructed using the software SPAGeDi and negative values between two individuals were changed to 0 (Hardy and Vekemans, 2002).
Analysis of LD
LD was measured by calculating the square value of correlation coefficient (r2) between each SNP pair with the software package TASSEL 2.1 (Bradbury et al., 2007). Only SNP loci with minor allele frequency values above 0.1 and having at least 80% successful calls among the sample set were included further for LD analyses. P-values for each r2 estimate were obtained with a two-sided Fisher's exact test as implemented in TASSEL. The critical r2 for LD decay was determined by taking the parametric 95th percentile of the distribution of r2 square root for all unlinked loci (Breseghello and Sorrells, 2006). LD plots against map distance for each linkage group (LG) were generated in Microsoft Excel (Redmond, WA, USA), where only r2 values with P<0.01 were included. Map position of each marker was obtained from the improved cowpea consensus map, allowing including the maximum number of markers (Lucas et al., 2011). The LD decay trend lines were generated using a window size based method (Yan et al., 2009).
Analysis of LD variance components
The computer program LinkDos (Garnier-gere and Dillmann, 1992) was employed to partition the variance of LD into within- (DIS2 and D'ST2) and between- (DST2 and D'IS2) subpopulation components, following Ohta's D-statistics estimation method (1982). The greater between-subpopulation than within-populations variance components would suggest that genetic drift has an important role in shaping observed patterns of LD. For each chromosome, all the marker pairs with an R2⩾0.2 and P<0.01 were included in Ohta's analysis and the results were averaged to give a comprehensive insight. Significance of the differences between within- and between-subpopulation variance components was tested by ANOVA.
Results
Data quality and genome wide SNP diversity
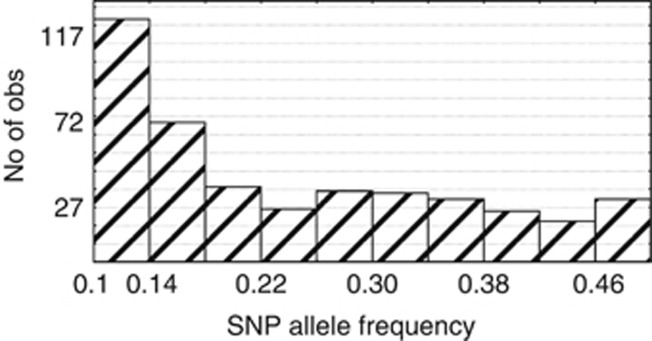
The KASPar SNP assay harboring 1127 SNPs gave a technical success rate of 97.7%, which means that 1102 SNPs were successfully called in the sample set. Of these, eight SNPs showed >20% missing data, 247 SNP loci were found to be monomorphic in all 99 lines and 420 SNPs had a minor allele frequency value below 0.1, which were removed from further analyses. Only five SNP loci were heterozygous in >10% of the accessions, consistent with the self-pollinating nature of asparagus bean and confirming the high level of homozygosity in the sample set. A final total of 422 SNPs were used for further data analysis. Of these, 415 SNP markers having known map positions and representing 370 unique loci were included for creating the LD plots (Supplementary Table 3). All the 11 LGs were covered by these SNPs, with the highest (76 SNPs) and lowest (23 SNPs) numbers observed in LG 3 and LG 10, respectively. The average marker distance was 1.58 cM, ranging from 1.01 to 2.69 cM across the 11 LGs. The allele frequency distribution of the SNPs, which are bi-allelic, showed a continuous pattern, with a peak position falling into the region of 0.1–0.18 (Figure 1).
Figure 1.
Allele frequency of the SNPs. Only SNPs with a minor allele frequency (MAF) >0.1 are shown. SNP frequencies >0.5 were shown as 0.5.
Identification of clusters and relative kinship in the sample set
Clustering inference performed with K from 1 to 10 showed that the most significant change of likelihood occurred when K increased from 1 to 2, and the highest ΔK value was observed at K=2 followed by a drastic decline of ΔK from K=3 (Table 1, Supplementary Figure 1). This suggested that the 99 genotypes could be assigned into two subgroups. Using a probability of membership threshold of 70%, 43 and 33 lines were assigned into the two subgroups, respectively. The remaining 23 lines were considered as intermediates (Supplementary Table 1, Supplementary Figure 2).
Table 1. Average Ln P(D), s.d. and ΔK values at K from 1 to 10 showing the population structure of the sample set.
| K | Ln P(D) | s.d. | ΔK |
|---|---|---|---|
| 1 | −39 598 | 1.0 | – |
| 2 | −33 986 | 2.4 | 1527.1 |
| 3 | −32 009 | 20.8 | 2.9 |
| 4 | −30 116 | 203.9 | 1.9 |
| 5 | −28 620 | 162.1 | 3.9 |
| 6 | −27 756 | 116.1 | 1.8 |
| 7 | −26 680 | 456.8 | 0.3 |
| 8 | −25 746 | 429.4 | 0.3 |
| 9 | −24 682 | 56.0 | 3.5 |
| 10 | −23 815 | 197.3 | – |
In general there was no association between subgroups inferred from structure and the geographic origin of the materials, reflecting the probable extensive exchange of parental lines by breeders nation-wide; however, we found strong associations of subgrouping with plant morphology and usage. For example, subgroup I was consisted entirely of those ‘standard vegetable' type lines, which means they all have very long tender pods (mean 50.5 cm, median 50.2 cm) typical of commercial varieties for vegetable use and strong climbing growth habit. In contrast, subgroup II (n=29 excluding the four ssp. unguiculata lines) mainly comprised the ‘non-standard vegetable' type lines, which in general have shorter pods (mean 30.7 cm, median 30.6 cm) with higher fiber content, oval-shaped seeds suitable for grain use and dwarf or bush-type plant architecture (Supplementary Table 1 and 4). The two subgroups were hereafter renamed as subgroup SV (standard vegetable) and NSV (non-standard vegetable), respectively. All the four ssp. unguiculata lines were grouped into the NSV subgroup, suggesting a closer genetic relationship between this subgroup and the subspecies unguiculata.
The pairwise genetic distance among the 99 genotypes ranged from 0.01 to 0.58, with an average of 0.32. The greatest genetic distance was observed between ‘Charleston Greenpack', an improved cowpea (ssp. unguiculata) cultivar from the southern United States (Fery, 1998) and ‘Prince Charming', an improved Chinese asparagus bean (ssp. sesquipedialis) cultivar. The average genetic distance within subgroup SV (0.22) was lower than that in subgroup NSV (0.37). Analysis of relative kinship showed that 3124 (69%) of all the pairwise kinship values were between 0 and 0.05, whereas 18.9 % of the values were above 0.25, indicating that most individual pairs are not or only weakly related whereas members of subsets have a considerable level of relatedness.
LD in the asparagus bean genome
As the 95 asparagus bean accessions could be divided into two distinct subgroups, pairwise LD estimates were performed within each subgene pool. Globally, 3937 (4.6%) and 2909 (3.4%) of the total possible SNP locus pairs were in significant LD (P<0.01) in subgroup SV and NSV, respectively (Table 2). Of these, 756 and 628 of the significant associations were intrachromosomal, accounting for 8.6% and 7.1 % of the total possible intrachromosomal correlations. The proportion of significant LD among unlinked loci to the total possible interchromosomal correlations was lower, being 4.3% and 3% in the two subgroups, respectively. In both subgroups, an uneven distribution of LD among the 11 chromosomes was observed.
Table 2. Locus pairs in significant (P<0.01) LD, r2 values and extent of LD in the two subgene pools.
| LG | Gene pool | No. Loci | No. (%) of locus pair in LD | Range (mean, median) of r2 | Extent (mean, median) of LD (cM) |
|---|---|---|---|---|---|
| 1 | SV | 38 | 77 (12.2) | 0.05–1 (0.40, 0.37) | 30 (8, 6.6) |
| NSV | 40 (6.3) | 0.03–1 (0.33, 0.24) | 57 (13.7, 2.5) | ||
| 2 | SV | 56 | 57 (3.7) | 0.02–1 (0.33, 0.21) | 61 (24, 27) |
| NSV | 151 (9.8) | 0.04–1 (0.33, 0.24) | 72 (13.5, 6.5) | ||
| 3 | SV | 76 | 204 (7.2) | 0.01–1 (0.28, 0.2) | 78 (23, 19) |
| NSV | 136 (4.8) | 0.02–0.9 (0.25–0.2) | 73 (14.4, 6.4) | ||
| 4 | SV | 30 | 28 (6.4) | 0.05–0.9 (0.29, 0.21) | 35 (9.9, 8) |
| NSV | 27 (6.2) | 0.06–0.87 (0.29, 0.24) | 27 (6.2, 3.3) | ||
| 5 | SV | 28 | 62 (16.4) | 0.12–1 (0.57, 0.54) | 50 (11, 9.5) |
| NSV | 21 (5.6) | 0.09–1 (0.4, 0.39) | 38 (6.4, 1.2) | ||
| 6 | SV | 41 | 38 (4.6) | 0.12–1 (0.48, 0.44) | 42 (9.8, 7.3) |
| NSV | 67 (8.2) | 0.04–0.9 (0.3, 0.25) | 64 (17.3, 11.8) | ||
| 7 | SV | 32 | 71 (14.3) | 0.06–0.9 (0.4, 0.31) | 26 (10, 7.5) |
| NSV | 23 (4.6) | 0.04–0.6 (0.3, 0.26) | 32 (7.3, 0.6) | ||
| 8 | SV | 27 | 35 (10) | 0.21–1 (0.53, 0.48) | 37 (10.6, 6.9) |
| NSV | 32 (9.1) | 0.06–0.9 (0.39, 0.34) | 42 (5.1, 1.1) | ||
| 9 | SV | 36 | 39 (6.2) | 0.15–1 (0.46, 0.38) | 40 (12.7, 10) |
| NSV | 36 (5.7) | 0.02–1 (0.26, 0.17) | 29 (9.2, 6) | ||
| 10 | SV | 23 | 17 (6.7) | 0.05–0.95 (0.4, 0.36) | 42 (17.8, 8.8) |
| NSV | 13 (5.1) | 0.06–0.87 (0.4, 0.37) | 24 (4.4, 1.1) | ||
| 11 | SV | 28 | 71 (18.8) | 0.06–0.9 (0.46, 0.33) | 31 (7, 10) |
| NSV | 48 (12.7) | 0.18–0.87 (0.7, 0.86) | 13 (1.2, 0.5) | ||
| Genome-wide | SV | 415 | 3937 (4.6)a | 0.01–1 (0.39, 0.3) | 78 (15, 9.5) |
| NSV | 2909 (3.4)a | 0.02–1 (0.34, 0.27) | 66 (11.2, 3.5) |
Abbreviations: NSV, non-standard vegetable; SV, standard vegetable.
The numbers and proportions of locus pairs in linkage disequilibrium (LD) on the whole genome scale includes both intrachromosomal and interchromosomal associations.
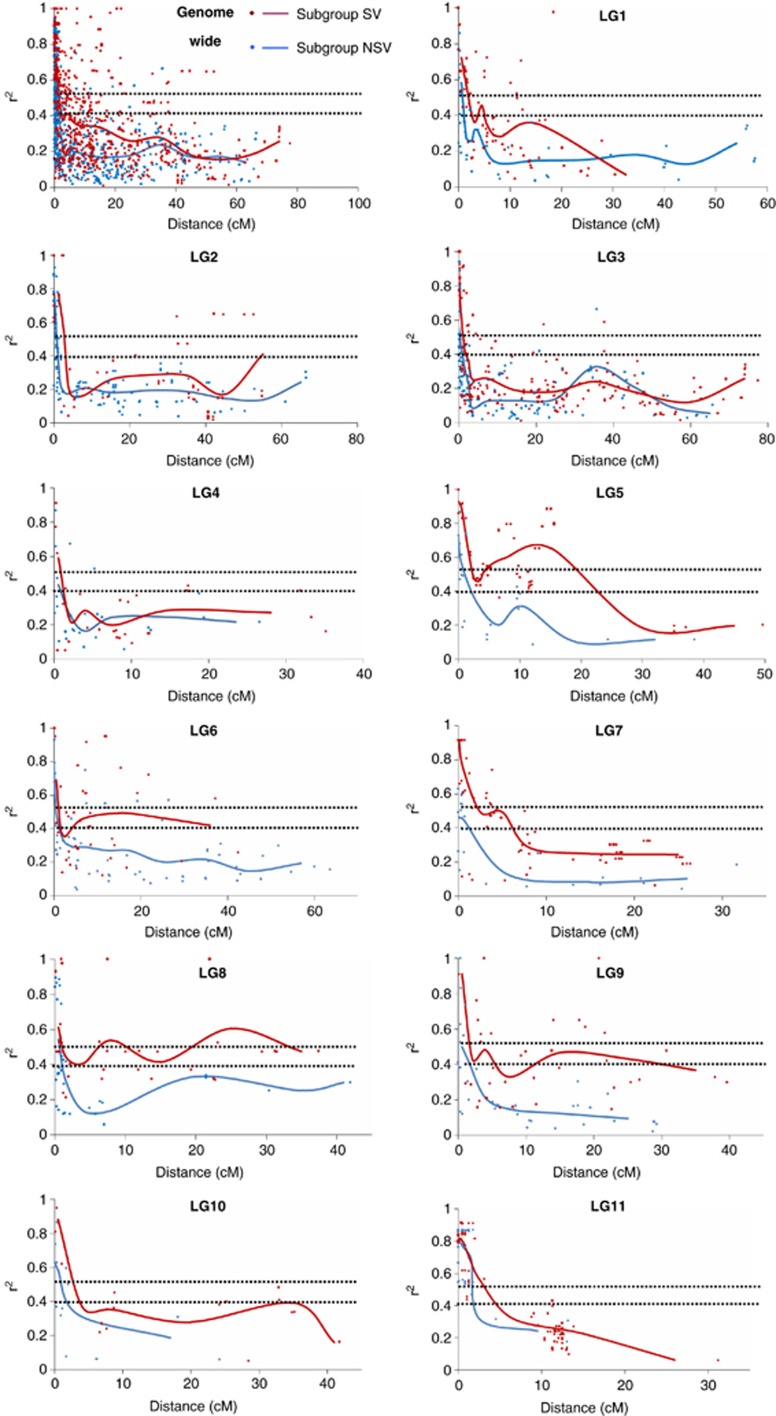
Intrachromosomal LD extended to a relatively long distance in asparagus bean, with the means of 15 (median 9.5) and 11.2 (median 3.5) cM in the two subgene pools, respectively. Significant LD was even observed at a distance over 60 cM, despite the very small proportions (2.4% and 1.7% of all the significant intrachromosoaml associations in subgroup SV and NSV, respectively). The strength of LD was high in asparagus bean, as reflected by the mean r2 of 0.39 and 0.34 in subgroup SV and NSV, respectively. Albeit variable among LGs, the level (r2) and extent of LD were generally higher in subgroup SV than in subgroup NSV (Table 2, Figure 2).
Figure 2.
Decay of LD (r2) as a function of genetic distance (cM) between pairs of loci on individual LGs and the whole genome in asparagus bean. Only r2 values with P<0.01 are shown.
Based on the parametric 95th percentile of the distribution of square-rooted r2 values for unlinked markers, r2 thresholds of 0.5 and 0.4 were adopted for estimating LD decay in subgroup SV and NSV, respectively. It was showed that, though high in r2, LD decayed rapidly with increasing genetic distance in both subgene pools. On the whole genome scale, LD decayed within 2 cM (Figure 2). At the chromosome level, the LD for LG 3, 4 and 6 all decayed at 0–2 cM in subgroup SV, and for LG 1, 2, 7, 9, 10 and 11 within 2.5–5 cM. LD appeared more complicated for LG 5 and LG 8, as indicated by the uneven decay along the chromosomes. In subgroup NSV, LD decayed more rapidly. Six of the eleven chromosomes (LG 1, 2, 4, 6, 7 and 8) had their LD decay distances near or shorter than 1 cM, whereas LD for the other five chromosomes all decayed within 2–3 cM.
LD partitioning
To allow for discrimination between genetic drift and epistatic selection, the different possible causes that create and maintain LD in natural populations, Ohta's (1982) analysis was performed by comparing the variance components within- (DIS2 and D'ST2) and between- (DST2 and D'IS2) subpopulations. As shown in Table 3, DST2 was significantly greater than DIS2 on the whole genome scale, whereas D'IS2 was significantly smaller than D'ST2, suggesting that globally the effect of epistatic selection and genetic drift cannot be simply compared. Similar results also were observed at the chromosome level; however, LD in at least two of the 11 LGs, that is, LG 5 and 7 may primarily be because of epistatic selection, because the within-subpopulations variances components were consistently greater than those between subpopulations (see also discussion below).
Table 3. Overall and linkage group (LG)-based Ohta's variance components of LD.
| LG | DIS2 | DST2 | DIS2/DST2 | D'ST2 | D'IS2 | D'ST2/D'IS2 |
|---|---|---|---|---|---|---|
| 1 | 0.060 | 0.067 | 0.89 | 0.241 | 0.052 | 4.63* |
| 2 | 0.031 | 0.089 | 0.34* | 0.316 | 0.033 | 9.57* |
| 3 | 0.032 | 0.085 | 0.37* | 0.319 | 0.036 | 8.86* |
| 4 | 0.035 | 0.072 | 0.48* | 0.263 | 0.037 | 7.1* |
| 5 | 0.083 | 0.047 | 1.76* | 0.214 | 0.058 | 3.68* |
| 6 | 0.024 | 0.131 | 0.18* | 0.486 | 0.040 | 12.15* |
| 7 | 0.092 | 0.039 | 2.35* | 0.160 | 0.065 | 2.46* |
| 8 | 0.035 | 0.143 | 0.24* | 0.493 | 0.074 | 6.66* |
| 9 | 0.018 | 0.068 | 0.26* | 0.261 | 0.019 | 13.73* |
| 10 | 0.018 | 0.131 | 0.12* | 0.456 | 0.040 | 11.4* |
| 11 | 0.177 | 0.036 | 4.91* | 0.157 | 0.160 | 0.98 |
| Overall | 0.041 | 0.087 | 0.47* | 0.322 | 0.044 | 7.31* |
*Significant at P=0.01.
Discussion
Genetic diversity and domestication history of Chinese asparagus bean germplasm
Unlike staple food crops such as rice and maize where abundant germplasm resources are available for LD analysis, the genetic resources of asparagus bean is limited. In the current study, 99 genotypes were selected, based on morphology, geographic origin and pedigree (if known), from the available Chinese asparagus bean germplasm to represent a putative core collection. Despite their diverse morphological characteristics and origins, the genetic diversity within the sample set is low, as reflected by the low level of SNP polymorphism and genetic distance. This result is consistent with previous phylogenetic studies based on AFLP or SSR markers (Fang et al., 2007; Xu et al., 2010). However, these results should be viewed with some caution because of potential ascertainment bias in the original selection of SNPs put on the platform used in the assays (Muchero et al., 2009). Although highly variable in morphology, several authors provide evidence that cowpea went through a severe genetic bottleneck during domestication and therefore has lower inherent genetic diversity (Panella and Gepts 1992; Pasquet 1999). Asparagus bean (ssp. unguiculata), as a subspecies of cowpea originating from only a small portion of the genetic variation of domesticated ssp. unguiculata would have gone through an additional bottleneck due to founder effects and intense selection for pod characteristics favorable for vegetable use (Fang et al. 2007). Therefore, the low level of genetic diversity observed in this study across the Chinese asparagus bean germplasm fits well with its likely domestication history.
The classification of the 99 accessions into two subgroups that differ mainly in pod length and growth habit indicates the impact of long-term human selection toward traits for vegetable use on the differentiation of ssp. unguiculata/asparagus bean, as lines with pods shorter than 36 cm or dwarf/semi-dwarf architecture were grouped together with the four ssp. unguiculata accessions. The analysis of LD variance components also suggested that epistatic selection might be the main force shaping the current pattern of LD in at least part of the genome. Our results therefore provide solid evidence proving the long-existing hypothesis of the domestication history of cultivated asparagus bean as mentioned above from a population genetic perspective. We also note that three landrace accessions (No.7, 18 and 80) morphologically more fitting the ‘standard vegetable' type were classified into the NSV subgroup. This observation, though common, indicates that genetic factors controlling morphological variations of pod length and growth habit are not the only forces dominating the stratification of Chinese asparagus bean germplasm. An interesting task in the near future is to more thoroughly address the mechanisms behind the domestication of cultivated asparagus bean.
Comparison of LD patterns between asparagus bean and other plant species
The LD level appears high in asparagus bean; however, decay of LD is still rapid (0–2 cM genome wide). Given the genome size of 630 Mb and the genetic linkage map of 680 cM (Lucas et al., 2011; Xu et al., 2011), this approximately equals 1.84 Mb of physical distance. Such level and extent of LD are comparable to those observed in the European barley collection (Rostoks et al., 2006), and the LD decay distance is even shorter than that in a small population of common bean (Phaseolus vulgaris L.) from America, a close relative of asparagus bean, where LD decays to 0.1–0.2 within 6–12 cM (McConnell et al., 2010). Collectively, these observations indicate that in inbreeding species including asparagus bean, the LD decay distances are fairly long. In contrast, LD declines very rapidly in outcrossing species where physical recombination is more effective. For instance, LD decays within only a few kilobases in maize (Remington et al., 2001; Yan et al., 2009) and only 200 bp in a wild sunflower population (Liu and Burke, 2006).
Variation in LD magnitudes across different germplasm and chromosomes
The extent of LD is usually affected by population structure. For example, Morrell et al (2005) reported a rapid decay of LD in wild barley compared with domesticated barley, even though both are highly self-pollinated. Similar results were reported between the indica and japonica groups of rice (Mather et al., 2007), the wild and domesticated subspecies of grapevine (Barnaud et al., 2010) and the landrace and modern varieties groups of wheat (Hao et al., 2011). Our results here showed that LD patterns in the two asparagus bean subdivisions are also different and that the rate of interchromosomal LD was reduced to, on average, 3.6% compared with 8% when using the combined gene pool (data not shown). The subgroup SV globally had a higher level and longer decay distance of LD, suggesting relatively limited historical recombination events and less genetic diversity in this more typical vegetable-type subdivision perhaps because of a more severe historical population bottleneck. The distances and patterns of LD decay are most different on LG 5, 7 and 11 between the two subgroups, suggesting that these chromosomes may carry more genes/QTLs related to agronomic traits favorable for vegetable use that have been strongly selected by cultivators and breeders of this crop. This is in good agreement with the Ohat's LD variance component analysis. An important future task is to fine-dissect the LD patterns between the two subgroups of asparagus bean germplasm using broader populations that may also include more ssp. unguiculata accessions to get better understanding of the genetic architecture of the cowpea natural resources globally.
Implications on genome wide association study in asparagus bean
For genome wide association mapping, long-range LD will reduce the number of markers needed to cover the genome, although the resolution for mapping is expected to be low (Yu and Buckler, 2006). The level of LD decay at a −cM level in the Chinese asparagus bean germplasm we observed suggests that a genome wide detection of QTLs is feasible using the currently available SNP maps and marker system. Assuming that one marker is needed per 1.5 cM along the 680 cM asparagus bean genome, ∼450 informative markers evenly distributed across the genome would be sufficient to uncover any significant associations using genome wide association studies. Given the expressed sequence tag-derived nature of the SNP markers employed here, the power for association mapping is expected to be higher than using other marker systems such as genomic SSRs, of which the majority are located in the non-coding regions of the genome (Yan et al., 2009). In addition, the presence of population stratification and kinship among a subset of the plant materials, as well as the 3.6% interchromosomal associations, all should be taken into account in association analyses (Yan et al., 2009; Hao et al., 2011).
To further complement the possible low mapping resolution caused by high-level LD in asparagus bean, a two-step procedure as proposed by Barnaud et al. (2010) would be considered for future work. Based on the discovery here that landrace asparagus bean lines (and especially the ssp. unguiculata), are genetically more diverse and thus might should exhibit shorter LD decay distance, QTL locations determined from ‘standard' asparagus bean populations could be refined by studying populations of more ‘rustic' asparagus bean or ssp. unguiculata accessions with much more limited LD. In addition, bi-parental QTL mapping using large RIL populations or NIL sets would also provide higher resolution for QTLs of interest. In our lab, a project that aims to uncover the genetic factors controlling the pod length and climbing ability of asparagus bean using the combined two-step association mapping and bi-parental QTL mapping approach is now underway.
Data archiving
Data have been deposited at Dryad: doi:10.5061/dryad.6tv35cc2.
Acknowledgments
This study was partially supported by the Science Foundation of Zhejiang Province (No. Y3090146), the National Science Foundation of China (No. 30900985) and the Sino-US joint project funded by Zhejiang Academy of Agricultural Sciences, China. We are grateful to Pauline Garnier-Géré, for advising us on Ohta's LD variance components analysis and for valuable comments on earlier versions of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- Barnaud A, Laucou V, This P, Lacombe T, Doligez A. Linkage disequilibrium in wild French grapevine, Vitis vinifera L. subsp. Silvestris. Heredity. 2010;104:431–437. doi: 10.1038/hdy.2009.143. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Breseghello F, Sorrells ME. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L. cultivars) Genetics. 2006;172:1165–1177. doi: 10.1534/genetics.105.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Ching A, Caldwell KS, Jung M, Dolan M, Smith OS, Tingey S, et al. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genetics. 2002;3:19. doi: 10.1186/1471-2156-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fang JG, Chao CCT, Roberts PA, Ehlers JD. Genetic diversity of cowpea [Vigna unguiculata. L. Walp.] in four West African and USA breeding programs as determined by AFLP analysis. Genet Resour Crop Ev. 2007;54:1197–1209. [Google Scholar]
- Fery RL. Charleston Greenpack' a pinkeye-type southernpea with a green cotyledon phenotype. HortSci. 1998;33:907–908. [Google Scholar]
- Flint-Garcia SA, Thornsberry JM, Buckler ES. Structure of linkage disequilibrium in plants. Ann Rev Plant Biol. 2003;54:357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- Garnier-Gere P, Dillmann C. A computer program for testing pairwise linkage disequilibria in subdivided populations. J Hered. 1992;83:239. doi: 10.1093/oxfordjournals.jhered.a111204. [DOI] [PubMed] [Google Scholar]
- Garris AJ, McCouch SR, Kresovich S. Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L) Genetics. 2003;165:759–769. doi: 10.1093/genetics/165.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao C, Wang L, Ge H, Dong Y, Zhang X. Genetic diversity and linkage disequilibrium in Chinese bread wheat (Triticum aestivum L.) revealed by SSR markers. PLoS One. 2011;6:e17279. doi: 10.1371/journal.pone.0017279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy OJ, Vekemans X. Spagedi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes. 2002;2:618–620. [Google Scholar]
- Hartl DL, Clark AG. Principles of population genetics. Sinauer Associates: Sunderland, MA; 1997. [Google Scholar]
- Lander ES, Schork NJ. The genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- Liu A, Burke JM. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics. 2006;173:321–330. doi: 10.1534/genetics.105.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KJ, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lucas MR, Diop N-N, Wanamaker S, Ehlers JD, Roberts PA, Close TJ. Cowpea-soybean synteny clarified through an improved genetic map. Plant Genome. 2011;4:218–225. [Google Scholar]
- Mather KA, Caicedo AL, Polato NR, Olsen KM, McCouch S, Purugganan MD. The extent of linkage disequilibrium in rice (Oryza sativa L. Genetics. 2007;177:2223–2232. doi: 10.1534/genetics.107.079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell M, Mamidi S, Lee R, Chikara S, Rossi M, Papa R, McClean P. Syntenic relationships among legumes revealed using a gene-based genetic linkage map of common bean (Phaseolus vulgaris L.) Theor Appl Genet. 2010;121:1103–1116. doi: 10.1007/s00122-010-1375-9. [DOI] [PubMed] [Google Scholar]
- McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. PNAS. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell PL, Toleno DM, Lundy KE, Clegg MT. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. PNAS. 2005;102:2442–2447. doi: 10.1073/pnas.0409804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchero W, Diop NN, Bhat PR, Fenton RD, Wanamaker S, Pottorff M, et al. A consensus genetic map of cowpea (Vigna unguiculata L. Walp) and synteny based on EST-derived SNPs. PNAS. 2009;106:18159–18164. doi: 10.1073/pnas.0905886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. Linkage disequilibrium due to random genetic drift in finite subdivided populations. PNAS. 1982;79:1940–1944. doi: 10.1073/pnas.79.6.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panella L, Gepts P. Genetic relationship within Vigna unguiculata. L. Walp. based on isozyme analyses. Genet Resour Crop Ev. 1992;39:71–88. [Google Scholar]
- Pasquet RS. Genetic relationships among subspecies of Vigna unguiculata. L. Walp. based on allozyme variation. Theor Appl Genet. 1999;98:1104–1119. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure from multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Thornsberry JM, Matsuoka Y, Wilson LM, Whitt SR, Doebley J, et al. Structure of linkage disequilibrium and phenotypic associations in the maize genome. PNAS. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins MD, Sim SC, Yang W, Deynze AV, Knaap E, Joobeur T, et al. Mapping and linkage disequilibrium analysis with a genome-wide collection of SNPs that detect polymorphism in cultivated tomato. J Exp Bot. 2011;62:1831–1845. doi: 10.1093/jxb/erq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostoks N, Ramsay L, MacKenzie K, Cardle L, Bhat PR, Roose ML, et al. Recent history of artificial outcrossing facilitates whole-genome association mapping in elite inbred crop varieties. PNAS. 2006;103:18656–18661. doi: 10.1073/pnas.0606133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon MI, Sawkins MC, Long AD, Gaut RL, Doebley JF, Gaut BS. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.) PNAS. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timko MP, Ehlers JD, Roberts PA.2007CowpeaIn: Kole C (ed)Pulses, sugar and tuber crops, Genome mapping and molecular breeding in plantsvol 3. Springer-Verlag: Berlin Heidelberg; 49–67. [Google Scholar]
- Weir B. Inferences about linkage disequilibrium. Biometrics. 1979;35:235–254. [PubMed] [Google Scholar]
- Xu P, Wu X, Wang B, Liu Y, Ehlers JD, Close TJ, et al. A SNP and SSR based genetic map of asparagus bean (Vigna. unguiculata ssp. sesquipedialis) and comparison with the broader species. PLoS One. 2011;6:e15952. doi: 10.1371/journal.pone.0015952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Wu XH, Wang BG, Liu YH, Qin DH, Ehlers JD, et al. Development and polymorphism of Vigna unguiculata ssp. unguiculata microsatellite markers used for phylogenetic analysis in asparagus bean (Vigna unguiculata ssp. sesquipedialis. L. Verdc.) Mol Breeding. 2010;25:675–684. [Google Scholar]
- Yan J, Shah T, Warburton ML, Buckler ES, McMullen MD, Crouch J. Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS One. 2009;4:e8451. doi: 10.1371/journal.pone.0008451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Buckler ES. Genetic association mapping and genome organization of maize. Curr Opin Biotech. 2006;17:155–160. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.