Abstract
Extensive phenotypic variation is a common feature among village chickens found throughout much of the developing world, and in traditional chicken breeds that have been artificially selected for traits such as plumage variety. We present here an assessment of traditional and village chicken populations, for fine mapping of Mendelian traits using genome-wide single-nucleotide polymorphism (SNP) genotyping while providing information on their genetic structure and diversity. Bayesian clustering analysis reveals two main genetic backgrounds in traditional breeds, Kenyan, Ethiopian and Chilean village chickens. Analysis of linkage disequilibrium (LD) reveals useful LD (r2⩾0.3) in both traditional and village chickens at pairwise marker distances of ∼10 Kb; while haplotype block analysis indicates a median block size of 11–12 Kb. Association mapping yielded refined mapping intervals for duplex comb (Gga 2:38.55–38.89 Mb) and rose comb (Gga 7:18.41–22.09 Mb) phenotypes in traditional breeds. Combined mapping information from traditional breeds and Chilean village chicken allows the oocyan phenotype to be fine mapped to two small regions (Gga 1:67.25–67.28 Mb, Gga 1:67.28–67.32 Mb) totalling ∼75 Kb. Mapping the unmapped earlobe pigmentation phenotype supports previous findings that the trait is sex-linked and polygenic. A critical assessment of the number of SNPs required to map simple traits indicate that between 90 and 110K SNPs are required for full genome-wide analysis of haplotype block structure/ancestry, and for association mapping in both traditional and village chickens. Our results demonstrate the importance and uniqueness of phenotypic diversity and genetic structure of traditional chicken breeds for fine-scale mapping of Mendelian traits in the species, with village chicken populations providing further opportunities to enhance mapping resolutions.
Keywords: haplotype blocks, linkage disequilibrium, association mapping, selective sweep, SNP genotyping
Introduction
Traditional chicken breeds, Gallus gallus, exhibit a wide range of phenotypes and more than 200 ‘fancy' breeds are recognised (Scrivener, 2006, 2009). While some of these breeds are of recent origins (for example, Appenzeller Spitzhauben Bantams developed in the 1980s), many were developed in the 19th century (for example, Rhode Island Red date from 1890); and others like the Silkie and Dorking extend back for hundreds of years, or possibly even thousands of years. Birds resembling modern day Asil/Aseel breed, used for cock fighting, were described over 3000 years ago (Scrivener, 2006, 2009). Breeds recognised by the poultry community are characterised by breed-specific morphological and phenotypic traits (Roberts, 1997), which are often the result of artificial selection over several generations. However, occasional incorporation of alleles from other breeds occurs through crossbreeding with the intention of improving a particular phenotype, or to create new strains/varieties. The effective population sizes of most traditional breeds are considered to be small because enthusiastic breeders often maintain relatively few individuals of their favourite breed and exchange birds among a small group of enthusiasts leading to inbreeding (Wilkinson et al., 2011). Many traditional breeds display exceptional diversity in both qualitative and quantitative traits both within and between breeds (Crawford, 1990; Sheppy, 2011). Variation can be observed in traits such as plumage colour and pattern, feather structure and pigmentation, patterning, comb morphology, skin colour, number of toes and spurs, eggshell pigmentation, and production traits such as body mass and egg-laying capacity. This diversity has captured the imagination of both fancy and commercial breeders and biologists for centuries, resulting in a wealth of information on the genetic mechanisms behind the inheritance of several of these traits. As at February 2012, the Online Mendelian Inheritance in Animals (OMIA) database had listed 189 phenotypes in chicken, 34 of which have been characterised at the molecular level (http://omia.angis.org.au/).
Village chickens have dispersed across the world through trading networks, human migrations and expansion of agriculture, and so they carry the genetic legacy of past historic events (Mwacharo et al., 2011). Kenyan, Ethiopian and Chilean village chickens are found outside the putative centres of origin of domestic chicken in Asia. Both mitochondrial and microsatellite analyses of East African village chickens have revealed several distinct arrivals from Asia of founder stocks and subsequent admixture between them (Mwacharo et al., 2011). The earliest archaeological evidence of domestic chicken in Africa dates back to between 1300 BC and 1400 AD (Gifford-Gonzales and Hanotte, 2011). The origin of South American village chicken remains unclear with their eventual presence on the continent before the arrival of Europeans still the subject of debate (Storey et al., 2007; Gongora et al., 2008). Village chickens are typically considered as free-range panmictic birds (Dana et al., 2010a). The wide-ranging phenotypic variation observed in traditional breeds also occurs in indigenous village chicken found in the developing world (Dana et al., 2010b). While artificial selection may have been imposed on these birds for selected traits such as colour phenotypes or comb traits, village chicken phenotypes are expected to be largely shaped by natural selection. Village chicken populations are generally older than most traditional breeds, and their genome will have witnessed more recombination events, which theoretically makes them a valuable resource for association mapping. However, owing to panmixis and low human selection pressures, Mendelian traits will be expected to segregate within populations, rather than to become bred to fixation as is the case in traditional breeds.
Studies on genetic inheritance and mapping of quantitative, including Mendelian, traits have traditionally involved the establishment of pedigree resource populations. Typically, F1 populations are created by crossing breeds that are genetically diverse for the trait of interest. These are then either intercrossed (F2) or backcrossed (BC) to one of the parental lines. However, recent genome-wide studies involving dogs and cattle have illustrated the possibility of mapping Mendelian traits within and across breeds without creating experimental crosses. This approach, in the absence of pedigree information, takes full advantage of the phenotypic diversity found within and across different livestock breeds (Georges, 2007). By exploiting the greater number of recombination events that have occurred since breed divergence, as compared with a mapping resource pedigree, it is possible to map a phenotype to a smaller region than might be possible using a mapping pedigree. This approach has been particularly successful in dogs (Karlsson et al., 2007; Chase et al., 2009; Akey et al., 2010). The recent development and characterisation of a 60K single-nucleotide polymorphism (SNP) chip for domestic chicken (Groenen et al., 2011) provides an opportunity to demonstrate this approach using traditional chicken breeds. Moreover, the presence of a diversity of phenotypes in village chicken also provides the opportunity to apply the same approach in village chicken, with the expectation that these older chicken populations may provide enhanced mapping resolutions compared with the traditional chicken breeds of more recent origins.
Here, we present an assessment of genome-wide analysis of SNPs for genetic diversity and association mapping studies using non-pedigree traditional and village chicken populations. We report the extent of linkage disequilibrium (LD) and haplotype block partitioning in traditional and village chickens, and perform genome-wide association mapping of Mendelian traits in the former. We successfully remap the yellow skin phenotype to its correct position and fine-map chromosomal regions for blue eggshell (oocyan), rose and duplex comb phenotypes. Chromosomal regions associated with the earlobe pigmentation are identified for the first time. A critical assessment of the number of SNPs required to fine-map simple Mendelian traits on a genome-wide scale in traditional and outbred village chickens is provided.
Materials and methods
Chicken varieties and breeds
A total of 79 individuals representing 34 traditional breeds, together with 17, 24 and 10 village chickens from Kenya, Ethiopia and Chile, respectively, were studied. Sample details (breed origin, age, phenotype, and so on) for traditional breeds are indicated in Table 1a, while sampling locations for village chickens are indicated in Table 1b. Traditional chicken breeds included individuals from a single flock, individuals of a single variety but from different flocks, individuals of different varieties of the same breed but from different flocks and birds from different traditional breeds (Table 1a). Phenotypic data for traditional breeds were derived from Roberts (1997) and Scrivener, (2006, 2009), while phenotypic data for village chicken were recorded by questionnaires and/or photographs. In this study, only the yellow skin and oocyan phenotypes were analysed in the village chicken. Phenotypes that were ambiguous or otherwise difficult to accurately verify in the village chicken were recorded as unknown, as were phenotypes that are not fixed within traditional breeds according to the breed standards.
Table 1a. Traditional breeds and their phenotypes.
| Breed | Likely breed origina | Approximate time breed established | Sourceb/flock/ number | Variety | Yellow skin | Blue eggs | White earlobes | Rose comb | Duplex comb |
|---|---|---|---|---|---|---|---|---|---|
| Appenzellor | CHE | 1900s | RI/1/2 | - | - | Y | - | Y | |
| Araucana | CHL | 1920s | RI/1/2 | Cuckoo | ? | Y | - | - | - |
| RI/2/2 | Lavender | ? | Y | - | - | - | |||
| PB/3/1, RI/4/2 | Standard | ? | Y | - | - | - | |||
| Brahma | CHN | 1800s | RI/1/1 | Y | - | - | - | - | |
| Buff Orpington | GBR | 1800s | RI/1/1 | ? | - | - | - | - | |
| Cochin | CHN | 1800s | RI/1/1 | Y | - | - | - | - | |
| Cream Legbar | GBR | 1930s | PB/1/2 | Y | Y | ? | - | - | |
| Crevecoeur | FRA | 1800s | PB/1/3 | - | - | - | - | Y | |
| Croad Langshan | CHN | 1800s | RI/1/1 | ? | - | - | - | - | |
| Derbyshire Redcap | GBR | 1800s | RI/1/1 | - | - | - | Y | - | |
| Dorking | GBR | 1600s | RI/1/1, PB/2/2 | - | - | - | ? | - | |
| Hamburgh | NLD/DEU | 1700s | RI/1/2 | - | - | Y | Y | - | |
| Indian Game | GBR | 1500s | RI/1/1 | Y | - | - | - | - | |
| Ixworth | GBR | 1930s | RI/1/1 | - | - | - | - | - | |
| Leghorn | ITA | 1800s | RI/1/1 | Brown | Y | - | Y | ? | - |
| Lincolnshire Buff | GBR | 1800s | RI/1/1 | - | - | - | - | - | |
| Malay | ASIA | 1500s | RI/1/1 | Y | - | - | Yc | - | |
| Maran | FRA | 1920s | AD/1/2, RI/2/2 | Standard | - | - | - | - | - |
| RI/3/2 | Dark cuckoo | - | - | - | - | - | |||
| PB/4/2 | Cuckoo | - | - | - | - | - | |||
| PB/5/1 | Copper blue | - | - | - | - | - | |||
| PB/6/1 | Copper black | - | - | - | - | - | |||
| Marsh Daisy | GBR | 1880s | RI/1/1 | ? | -c | Y | Y | - | |
| Modern Langshan | CHN | 1890s | RI/1/1 | - | - | - | - | - | |
| Norfolk Grey | GBR | 1920s | RI/1/1 | - | -d | - | - | - | |
| Old English Pheasant Fowl | GBR | 1910s | RI/1/1 | - | -d | Y | Y | - | |
| Polish | POL | 1600s | RI/1/2 | - | - | Y | - | Y | |
| Rhode Island Red | USA | 1890s | RI/1/1 | Y | - | - | ? | - | |
| Scots Dumpy | GBR | 1800s | RI/1/1 | ? | - | - | - | - | |
| Scots Grey | GBR | 1800s | RI/1/1 | ? | - | - | - | - | |
| Silkie | CHN | 350BC | RI/1/2 | Blue | - | - | ? | Ye | - |
| RI/2/2 | Standard | - | - | ? | Ye | - | |||
| Spanish | ESP | 1500s | RI/1/1 | - | - | Y | - | - | |
| Sultan | TUR | 1800s | PB/1/1 | - | - | - | - | - | |
| Sussex | GBR | 1800s | RI/1/2 | Light | - | - | - | - | - |
| RI/2/2 | White | - | - | - | - | - | |||
| Totenko | JPN | 1600s | PB/1/7 | ? | -c | Y | - | - | |
| Villafranquina | ESP | 1970s | AD/1/2 | - | - | - | - | - | |
| Welsummer | NLD | 1920s | PB/1/4, PB/2/1 | Y | - | - | - | - | |
| White Star | ? | ? | PB/1/2 | ? | - | Y | - | - | |
| Yurlov | RUS | 1800s | AD/1/2 | Russia | ? | - | - | ? | - |
| AD/2/2 | Ukraine | ? | - | - | ? | - |
Likely breed origin and age ascertained from literature (Roberts, 1997; Scrivener, 2006, 2009), three-letter country code (ISO 3166-1 alpha-3).
AvianDiv Project (AD), Private Breeders (PB), Roslin Institute (RI), Olivier Hanotte (OH), Negussie Dana (ND) and José Alcalde (JA).
Breed documented as having blue-/black-/slate-coloured legs, and so the yellow skin allele is believed absent.
Breed documented as having willow-/olive-coloured legs, and so the yellow skin allele is believed present.
Breed documented as having a walnut comb, which is the result of having the alleles for both pea and rose comb.
Phenotype unknown (?), phenotype present (Y), phenotype absent (-).
Table 1b. Geographic origin of village chicken.
| Village | Region | Country | Sourcea/number | Yellow skin | Blue eggs |
|---|---|---|---|---|---|
| Simur East | Busia | Kenya | OH/1 | - | - |
| Bumala | Busia | Kenya | OH/1 | Y | - |
| Myanga | Busia | Kenya | OH/1 | Y | - |
| Ogallo | Busia | Kenya | OH/1 | ? | - |
| Luanda | Busia | Kenya | OH/1 | Y | - |
| Kidera | Busia | Kenya | OH/1 | Y | - |
| OH/1 | - | - | |||
| Busia | Busia | Kenya | OH/4 | - | - |
| OH/6 | Y | - | |||
| Gondar | Amhara | Ethiopia | ND/5 | Y | - |
| Guduru | Oromia | Ethiopia | ND/4 | Y | - |
| Konso | SNNP | Ethiopia | ND/1 | - | - |
| ND/4 | Y | - | |||
| Sheka | SNNP | Ethiopia | ND/1 | - | - |
| ND/5 | Y | - | |||
| Gumuze | Metekel | Ethiopia | ND/4 | Y | - |
| Cachagua | Valparaiso | Chile | JA/5 | Y | Yb |
| Pirque | Santiago | Chile | JA/2 | Y | - |
| JA/3 | Y | Yb |
Likely breed origin and age ascertained from literature (Roberts, 1997; Scrivener, 2006, 2009), three-letter country code (ISO 3166-1 alpha-3).
Includes both blue and green eggs as they are both the result of the oocyan locus.
Phenotype unknown (?), phenotype present (Y), phenotype absent (-).
A total of 43 DNA samples extracted from chicken embryos representing 26 traditional breeds were provided by the Roslin Institute, Edinburgh. The AvianDiv Project (http://aviandiv.tzv.fal.de/) provided eight DNA samples representing three traditional breeds. Hatching eggs was sourced from various private breeders and DNA was extracted from seven-day-old chicken embryos for 23 birds (Araucana=1, Cream Legbar=2, Crevecoeur=3, Dorking=2, Maran=10, Sultan=1, Totenko=7, Welsummer=5 and White Star=2). Blood samples from 17 village chickens from one region in Kenya (Busia), 24 village chickens from four different geographic regions in Ethiopia (Amhara (5); Oromia (4); Southern Nations, Nationalities and People's Region (10); and Benishangul-Gumuz (5)), and 10 village chickens from two regions in Chile (Valparaiso (5) and Santiago (5)) were collected on FTA cards. DNA extraction from all samples excluding those from the Roslin Institute and AvianDiv Project were performed at the University of Nottingham (UK) using in-house protocols.
Genotyping
Genotyping of the DNA samples was outsourced to a private sequencing company (DNA Landmarks Inc, Quebec, Canada) and was performed using the 60K SNP Illumina iSelect chicken array (http://www.illumina.com/). The chicken karyotype comprises 5 macrochromosomes (Gga 1–5), 5 intermediate chromosomes (Gga 6–10), 27 microchromosomes (Gga 11–38) and 1 pair of sex chromosomes (ZW female and ZZ male). The array has SNPs spanning 29 autosomes (Gga 1–28, and Gga 32), the sex chromosomes and across two linkage groups (that is, 148 SNPs on LGE22C19W28_E50C23 and 7 on LGE64) which are currently not assigned to any chromosome. Also included are several SNPs on the mitochondrial genome (n=7) and several others whose mapping remains unknown (n=1144).
Genotyping data were pruned with the GenABEL package (Aulchenko et al., 2007) for R (R Development Core Team, 2009), using the check.marker function (attributes: maf=0.005, call=0.9, perid.call=0.9, p.lev=0, ibs.mrk=‘ALL' and ibs.threshold=0.79). When calculating r2 values for LD, pruning was performed independently for traditional and village chickens, respectively. The ibs.threshold was increased to 0.99 to accommodate higher SNP identity-by-state (IBS) values found within flocks and between siblings. Pruning was performed on the overall dataset before STRUCTURE analysis, haplotype block inference and fine-scale mapping (Supplementary Tables 1 and 2).
Genetic relationships and structure
The extent of genetic relationships within and between breeds were assessed by calculating pairwise genetic distances from IBS scores using the ibs function of GenABEL based on 40 000 randomly selected autosomal SNPs (Gga 1 to 28). STRUCTURE analysis (Pritchard et al., 2000) was performed on these SNPs using one randomly selected bird per breed or per country (in the case of the village chicken), with the admixture unlinked loci model, a burn-in period of 50 000 followed by 100 000 MCMC repetitions, assuming one through to four clusters (K). The STRUCTURE output was analysed using Structure Harvester (Earl and Vonholdt, 2011), which identifies the optimal K based on the posterior probability of the data for a given K (Supplementary Figure 1a), and the ΔK (Supplementary Figure 1b) recommended by Evanno et al., 2005 (Supplementary Figure 1c). In addition, a neighbour-joining tree was constructed from the distance matrix using MEGA 5.05 (Kumar et al., 2008).
LD and haplotype blocks
For all SNPs on each chromosome, pairwise LD values of r2 (Hill and Roberston, 1968) were calculated using the r2fast function (Hao et al., 2007) of GenABEL. Based on the ΔK resulting from the STRUCTURE analysis (K=2), LD was calculated for 10 subsets of data as follows: (1) closely related birds from a single breed and flock (Totenko, n=7); (2) birds from a single breed from three different flocks (Araucana, n=7); (3) birds from a single breed from five different flocks (Marans, n=7); (4) eight birds representing different traditional breeds with ⩾80% inferred ancestry A; (5) eight birds representing different traditional breeds with ⩾80% inferred ancestry B; (6) eight birds representing different traditional breeds with ⩾30% ancestry inferred from either A or B (referred to as ‘A∼B'); (7) eight birds representing different traditional breeds from which four have at least 80% inferred A ancestry and four have at least 80% inferred B ancestry (referred to as ‘A+B'); (8) eight randomly selected Kenyan birds; (9) eight randomly selected Ethiopian birds; and (10) eight Chilean birds. The individual birds assigned to each group are detailed in Supplementary Tables 4 and 5. LD values were averaged across the genome (Gga 1–28) for eight arbitrarily defined physical distance bins (<10 Kb, 10–25 Kb, 25–50 Kb, 50–100 Kb, 100–250 Kb, 250–500 Kb, 500–750 Kb and 750 Kb-1 Mb) between pairs of markers. Chromosome 16 was excluded from the analysis owing to low SNP coverage and call rate.
Haplotype block sizes were calculated in Plink (Purcell et al., 2007) using the blocks function (SNPs within 1 Mb) for Gga 1–28 (excluding Gga 16) and Gga Z for 40 randomly selected traditional breeds and 40 village chickens (10 Chilean, 15 randomly selected Kenyan and 15 randomly selected Ethiopian). The number of segregating haplotypes per chromosome was estimated by dividing chromosome lengths in base pairs (WASHUC2 assembly 2.1, May 2006) by the median haplotype block size.
Association tests and selective sweeps
The qtscore function of GenABEL was used to perform tests of allelic association and the P-values corrected for false discovery rate (Benjamini and Hochberg, 1995) via the qvaluebh95 function. The corrected P-values were considered significant at P<0.05. Selective sweeps were detected by comparing the allele frequencies of samples exhibiting the trait of interest (TP) to those not exhibiting the trait (TA). Overlapping sliding windows of 20 Kb were used to detect sweeps. The starting point of overlapping windows was defined as zx+1=Z((yx+1)/2) where x is the window index, y is the number of SNPs in window x and z is the position of SNP y. The mean allele frequency (f) for the minor allele for SNPs within a window was calculated using the summary.snp.data function of GenABEL for the two groups of chicken. Allele frequencies were standardised using a similar method to that applied by Rubin et al. (2010), such that zTPf=(TPf–μf)/σf and zTAf=(TAf−μf)/σf in which μ and σ are the mean and s.d. of mean allele frequencies across the dataset. A final comparative Z score, indicating the number of deviations from the mean, was then derived for each of the windows using the equation ∣zTPf−zTAf∣. A significance threshold of Z⩾4 was used in this study; this number of deviations lies at the extremities of a normal distribution (0.006% outside the confidence interval). The significant results from all tests are available in the Supplementary Data.
Identical-by-descent (IBD) analysis
Where significant associations were identified, the genotypes at these SNPs were manually inspected in all birds in the raw dataset to check that the genotypic state was identical across all birds sharing the same phenotype. Surrounding SNPs were also inspected to see if there was a block pattern of conserved genotypes across birds sharing a given phenotype, which might be expected if there was a historical bottleneck, a consequence of inbreeding or a result of genetic hitchhiking.
Phenotypic traits
Yellow skin, oocyan, rose comb, duplex comb and earlobe pigmentation are traits inherited in a Mendelian fashion. As they are expressed differentially across breeds, they were chosen for mapping purposes. Eriksson et al. (2008) identified BCDO2 (Gga 24:6.26–6.28 Mb) to be the gene underlying the yellow skin phenotype, a recessive trait. The chromosomal region underlying the trait is thought to have introgressed into domestic chicken from the grey junglefowl Gallus sonneratii (Eriksson et al., 2008). Oocyan (O) is an autosomal dominant trait (Punnett, 1933) that occurs in some traditional chicken breeds. It gives rise to a blue or green eggshell colour depending on whether the base colour is white or brown. It has recently been mapped to a broad region on Gga 1 (67.3–69.1 Mb) in the Dongxiang chicken breed (Wang et al., 2010). The rose comb (R) is an autosomal dominant trait, which when combined with pea comb results in walnut comb. Dorshorst et al. (2010) recently mapped the rose comb at position 14.5–24.2 (Mb) of Gga 7. The duplex comb (D) is an autosomal incompletely dominant trait (Punnett, 1923). Studies have shown the trait to be multi-allelic with the most dominant allele (Dv) causing doubling of the comb and suppression of tissue mass. The less dominant allele (Dc) causes only doubling of the comb (Somes, 1991). These phenotypic variants have been described as duplex, horn or ‘V' shape (Dv), and the cup or buttercup (Dc). Dorshorst et al. (2010) mapped duplex comb to 33.6–39.8 Mb of Gga 2. Earlobe pigmentation is commonly red among traditional breeds, as is the case with comb and wattles. However, for some traditional breeds, especially those of Mediterranean origin, their earlobes are white (Warren, 1928). Warren, (1928) conducted crossing experiments involving several breeds to assess the inheritance of this phenotype. The results revealed that the trait was polygenic and it appeared to be sex-linked in some breeds. No clear conclusions could be drawn on the mode of inheritance of the trait, and any suggestion of dominance was always incomplete, a possible consequence that the trait may not have been fixed in the birds used for the crossings (Warren, 1928).
Candidate gene identification
For the identification of candidate genes, we used the ‘Ensembl Genes 60 (Sanger UK)' database according to BioMart (http://www.biomart.org/), which includes 17 934 genes annotated in G. gallus (WASHUC2, assembly 2.1, May 2006).
Results
Pruning the SNP data
Comprehensive statistics pre- and post-pruning of genotype data are available in Supplementary Tables 1 and 2 for each individual and chromosome. In summary, genotyping call rates averaged at 97.6% pre-pruning, but rose to 99.7% post pruning. Pruning of data was performed separately for the entire dataset and for subsets of data that were used for LD analysis. Across the entire dataset, out of 57 636 SNPs on the microarray 4143 were excluded from analysis (Supplementary Table 2). These included 934 SNPs that failed to call in all samples, 751 SNPs with a call rate below 90% across samples, 2209 SNPs that were monomorphic across the entire dataset and 249 SNPs that had a minor allele frequency <0.05. The single SNP assigned to Gga 32 failed to call in all individuals genotyped. The seven SNPs on Gga W were excluded as the sex of the birds were unknown; however, for those that did call in some of the samples (n=37), only one was monomorphic.
Overall, 36.8% of failed SNPs were on macrochromosomes, 13.3% on intermediate chromosomes, 29.6% on microchromosomes and the remaining 20.3% were on sex chromosomes, mitochondria or are yet to be mapped. One Ethiopian village chicken sample was excluded for having a call rate of <90% (Supplementary Table 1). Following the quality checks, 92.8% of SNPs (n=53 493) were suitable for analysis.
For pruning subsets of the data used for LD analysis, SNPs having call rates of <90% and a minor allele frequency of <5% were excluded (Supplementary Table 3). Given the expected first-degree relationship of Totenko samples, 70% of the SNPs for this breed had a minor allele frequency of <5% and were therefore excluded from analysis. With the exception of Totenko, an average of 45 000 SNPs were informative for analysis.
Genetic diversity and structure
The mean IBS score (0.65±0.03) and observed heterozygosity (Ho=0.21±0.07) for traditional breeds were lower than those of village chickens (IBS=0.72±0.03; Ho=0.31±0.02; Table 2). For traditional breeds, birds from the same flock showed higher IBS scores than those from different breeds/flocks (Table 2). However, large variations were observed with IBS scores ranging from 0.69 (between two Copper Blue Marans) to 0.91±0.02 (between Totenko). When comparing birds from different strains, IBS scores showed little variation with values ranging from 0.67 (between two strains of Sussex) to 0.74 (between two strains of Welsummer). For village chickens, higher heterozygosity values were observed in birds from Kenya (Ho=0.33±0.02) and Chile (Ho=0.32±0.02) than those from across Ethiopia (Ho=0.27±0.03), the difference, however, was not significant (P<0.05) (Table 2). Based on these results, to reduce the likelihood of spurious mapping associations, only birds showing an IBS score of <0.79 (this corresponds to the mean IBS of 0.68 found across the dataset plus four times the s.d. of 0.027) were included for association mapping and selective sweep analysis.
Table 2. Summary of identity-by-state (IBS) and observed heterozygosity (Ho) within breeds.
| n | Mean IBS | Mean Ho | |
|---|---|---|---|
| Traditional (one per breed) | 33 | 0.65±0.03 | 0.21±0.07 |
| Appenzellor | 2 | 0.9 | 0.12±0.03 |
| Araucana | 7 | 0.7±0.05 | 0.25±0.04 |
| One per flock | 4 | 0.69±0.02 | 0.27±0.05 |
| Cuckoo (RI/1) | 2 | 0.9 | 0.21±0.02 |
| Lavender (RI/2) | 2 | 0.77 | 0.28±0.01 |
| Standard (RI/4) | 2 | 0.69 | 0.28±0.06 |
| Cream Legbar | 2 | 0.86 | 0.17±0.01 |
| Crevecoeur | 3 | 0.85±0.01 | 0.21±0.02 |
| Maran | 11 | 0.7±0.03 | 0.28±0.03 |
| One per flock | 7 | 0.7±0.02 | 0.29±0.03 |
| Standard (AD/1) | 2 | 0.77 | 0.27±0.01 |
| Standard (RI/2) | 2 | 0.84 | 0.33±0.01 |
| Dark Cuckoo (RI/3) | 2 | 0.83 | 0.26±0.01 |
| Copper Blue (PB/6) | 2 | 0.69 | 0.29±0.01 |
| Polish | 2 | 0.89 | 0.17±0.01 |
| Silkie | 4 | 0.75±0.04 | 0.26±0.06 |
| One per flock | 2 | 0.73 | 0.26±0.04 |
| Blue (RI/1) | 2 | 0.83 | 0.24±0.07 |
| Standard (RI/2) | 2 | 0.74 | 0.29±0.08 |
| Sussex | 4 | 0.73±0.1 | 0.19±0.04 |
| One per flock | 2 | 0.67 | 0.21±0.06 |
| Light (RI/1) | 2 | 0.82 | 0.23±0.02 |
| White (RI/2) | 2 | 0.9 | 0.16±0.00 |
| Totenko | 7 | 0.91±0.02 | 0.12±0.01 |
| Villafranquina | 2 | 0.77 | 0.27±0.01 |
| Welsummer | 5 | 0.78±0.04 | 0.18±0.07 |
| One per flock | 2 | 0.74 | 0.25±0.05 |
| Standard (PB/1) | 4 | 0.79±0.04 | 0.24±0.03 |
| White Star | 2 | 0.8 | 0.32±0.02 |
| Yurlov | 4 | 0.7±0.02 | 0.32±0.03 |
| One per flock | 2 | 0.68 | 0.35±0.02 |
| Russia (AD/1) | 2 | 0.72 | 0.34±0.01 |
| Ukraine (AD/2) | 2 | 0.71 | 0.32±0.05 |
|
Mean one per flock |
6 |
0.7±0.03 |
0.27±0.05 |
| All village | 40 | 0.72±0.03 | 0.31±0.04 |
| Kenya | 17 | 0.71±0.01 | 0.33±0.02 |
| Ethiopia | 23 | 0.74±0.02 | 0.27±0.03 |
| Chile | 10 | 0.71±0.03 | 0.32±0.02 |
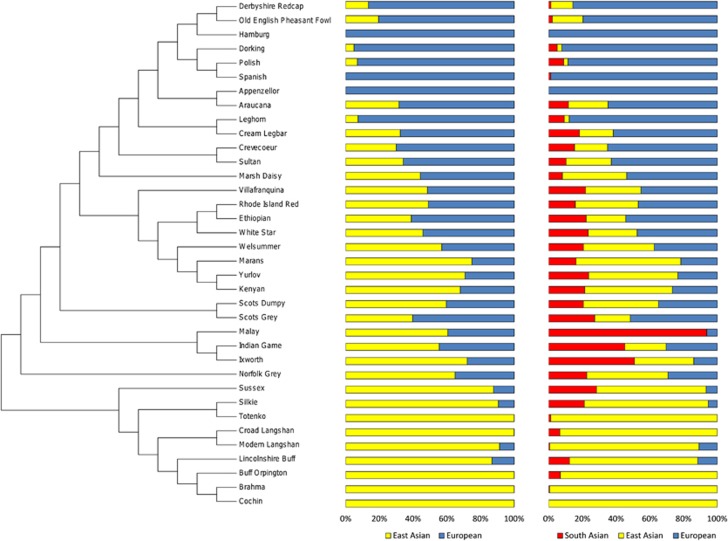
Following STRUCTURE analysis, the posterior probability (Ln P(D)) of the data indicated the most optimal K to be three while the Evanno et al. (2005) method indicated the optimal K to be two (Supplementary Figure 1), these genetic clusters are referred to as A and B (K=2); C, D and E (K=3). Birds with a large proportion of A (K=2) cluster together in the neighbour-joining tree. Similarly, birds with predominantly mixed ancestry (K=2 and K=3) are also clustering together. It includes the three representatives of village chicken (Ethiopian, Kenyan and Chilean) as well as some traditional breeds (Rhode Island Red, White Star, Marans and Yurlov) (Figure 1).
Figure 1.
Neighbour-joining tree and STRUCTURE analyses assuming K=2 and K=3 using one randomly selected bird for each traditional breed and each village chicken population.
Genome-wide SNP distribution and haplotype blocks
The number of SNPs when compared with the number of genes relative to the chromosome length was found to be proportionate across the genome (r2=0.526, P<0.01; Supplementary Figure 2). The mean spacing between pairs of adjacent SNPs within chromosomes was, however, uneven across chromosomes. It was largest for macrochromosomes (23.4±24.7 Kb) and the Z chromosome (24.7±30.3 Kb), followed by the intermediate chromosomes (16.8±25.3 Kb) and then the more gene-rich microchromosomes (9.7±10.3 Kb). However, variation was evident along individual chromosomes (Supplementary Figure 2).
SNPs in complete linkage allow for the calculation of haplotype block sizes, and the minimum number of SNPs required for whole-genome ancestry analysis by extrapolating the haplotype block sizes across the genome. Table 3 provides a summary of haplotype block distribution and sharing across traditional breeds and village chickens up to a pairwise marker distance of 1 Mb. A comparison of haplotype blocks along macro- and microchromosomes is also included in the table. Village chickens returned slightly fewer haplotype blocks than the traditional breeds, resulting in fewer SNPs per block and smaller median haplotype block sizes. However, the differences were not significant (P<0.05). The mode number of SNPs per block (n=2) and median block size (Gga 1–28: ∼11 Kb; Gga 1–5: ∼12 Kb; Gga 6–28: ∼9 Kb) were consistent in both groups of chicken. The median was preferred to the mean owing to the disproportionate number of haplotype blocks recorded below and above the mean. For example, for all chromosomes the number of haplotype blocks below the mean was 1831 while those above the mean were 322. The number of blocks above and below the median, however, remain relatively proportionate (1047 above and 1106 below), indicating it to be a more reliable estimate of block sizes. Approximately one-third of the haplotype blocks (316) are shared between traditional and village chickens (Table 3). The largest haplotype block (∼987 Kb) shared between traditional and village chickens is located on Gga 1 position 75860999–76848326 with 33 polymorphic SNPs and 7 monomorphic SNPs, which were excluded from analysis during pruning. The majority of blocks (91%) are <25 Kb in size.
Table 3. Haplotype block summary across traditional and village chickens for blocks up to 1 Mb in size.
| Chromosomes | Traditional breeds (n=40) | Village chickens (n=40) |
|---|---|---|
| All | ||
| Number of blocks | 894 | 792 |
| Number of SNPs in blocks | 2770 | 2568 |
| Mode SNPs per block | 2 | 2 |
| Max SNPs in block | 33 | 33 |
| Median block size (Kb) | 11.87 | 11.45 |
| Macro (1–5) | ||
| Number of blocks | 396 | 343 |
| Number of SNPs in blocks | 1157 | 1054 |
| Median block size (Kb) | 12.61 | 12.47 |
| Micro (11–28) | ||
| Number of blocks | 308 | 274 |
| Number of SNPs in blocks | 997 | 898 |
| Median block size (Kb) | 9.37 | 9.83 |
| Traditional breeds (n=40) | Village chickens (n=40) | Shared blocksa | |
|---|---|---|---|
| Number of haplotype blocks observed per block size bin | |||
| <10 Kb | 395 | 357 | 195 |
| 10–25 Kb | 342 | 274 | 93 |
| 25–50 Kb | 65 | 54 | 9 |
| 50–100 Kb | 58 | 56 | 7 |
| 100–250 Kb | 61 | 61 | 8 |
| 250–500 Kb | 11 | 21 | 3 |
| >500 Kb | 7 | 6 | 1 |
Abbreviation: SNP, single-nucleotide polymorphism.
Shared blocks are those containing the same SNPs in both traditional breed and village chicken datasets.
The number of SNPs required for whole-genome analysis of haplotype structure and mapping, assuming one SNP per haplotype block, was inferred from the median haplotype block size (11.87 Kb in traditional breeds and 11.45 Kb in village chicken) relative to chromosome length (Supplementary Table 6). The total number of SNPs required to provide full coverage of Gga 1–28 and Gga Z is ∼87K for traditional breeds and ∼90K for village chickens. From microchromosome analyses, a more conservative estimate assuming smaller haplotype block sizes (9.37 Kb in traditional breeds and 9.83 Kb in village chicken) indicated that at least 110k for traditional breeds and 105K for village chicken are required to provide full coverage across Gga 1–28 and Gga Z.
Linkage disequilibrium
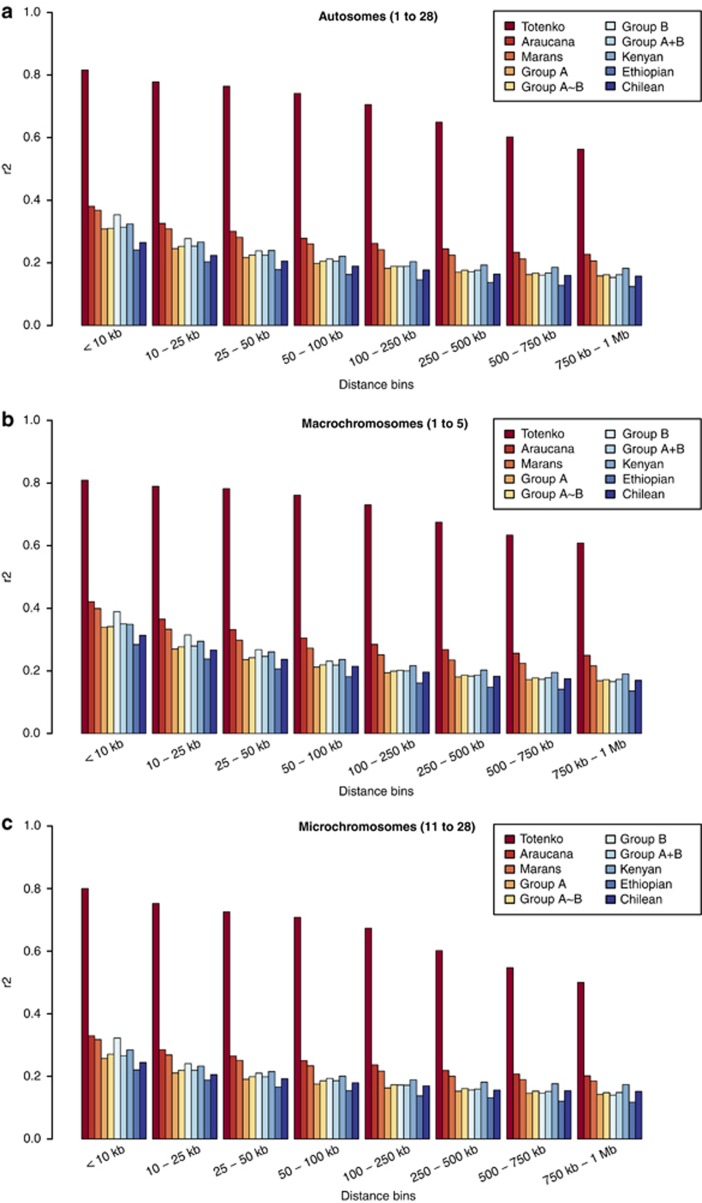
Previous studies on LD in chicken have reported observable differences between macro- and microchromosomes at similar physical distances (Megens et al., 2009; Qanbari et al., 2010). Figure 2 presents r2 values binned over short distances (<250 Kb) for all autosomes, macro- and microchromosomes for each subset of data. LD was on average 1.2 times lower across microchromosomes compared with macrochromosomes. For traditional breeds in all distance bins, Totenko has the highest r2 values, followed by those obtained for individual breeds (Araucana and Marans). Similar r2 values are obtained for each of the A, B, A∼B and A+B subsets. Across the admixed subsets (A, B, A∼B and A+B) and village chickens, group B generally showed the highest values for distances <100 Kb and the lowest values for distances exceeding 500 Kb. Beyond 50 Kb, the village chickens, sampled from Kenya, always show higher r2 values compared with the other admixed subsets. This might be expected when sampling birds from a panmictic population in a single region, where genetic exchange is likely to be more frequent than across birds from multiple regions. This also appears to be reflected in the relative r2 values observed in the Chilean and Ethiopian birds given that the Chilean birds were sampled from two regions and the Ethiopian birds from four.
Figure 2.
Mean binned recombination rates (r2). Values were calculated from SNP data for seven or eight randomly selected birds in each group using background genetic information from STRUCTURE (K=2) as relevant. Totenko (n=7, all first-degree relatives); Araucana (n=7, birds from three distinct flocks); Marans (n=7, birds from five distinct flocks); group A (n=8, birds with 80% background A); group B (n=8, birds with 80% background B); group A∼B (n=8, birds with at least 30% background A and B); group A+B (n=8, four birds with 80% background A and four birds with 80% background B); and village chicken (Kenya, Ethiopia or Chile (n=8 for each country)). (a) All autosomes (Gga 1–28), (b) macrochromosomes (Gga 1–5) and (c) microchromosomes (Gga 11–28).
Mapping of Mendelian traits
Yellow skin
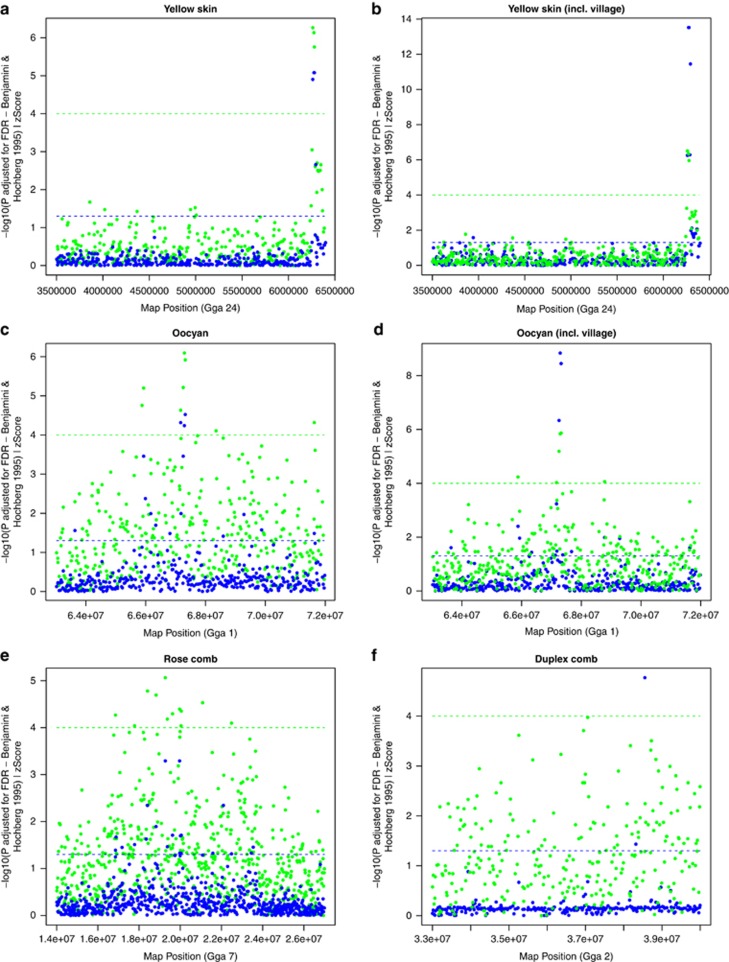
The yellow skin phenotype was analysed both in traditional breeds only and with the inclusion of village chickens. Post pruning, in the absence of village chickens, 13 birds exhibited the phenotype while 37 birds did not; whereas in the presence of village chickens, 51 birds exhibited the phenotype while 32 did not. The phenotypes of all birds are detailed in Tables 1a and b. BCDO2 has been identified as the gene controlling the yellow skin pigmentation (Eriksson et al., 2008). It therefore provides an excellent control trait for the fine mapping strategy presented here. There are three intronic SNPs within the BCDO2 gene on the SNP chip. One of these, GgaluGA193893 (Gga 24: 6.26 Mb), was monomorphic across the dataset and was filtered out during pruning. Figures 3a and b illustrate the results of the association tests and selective sweeps. From genome-wide association tests in the absence of village chicken, the two most significant SNPs (Gga_rs13725199 and Gga_rs13725203, P=8.32 × 10−6) were both located on Gga 24 at 6.27 Mb and 6.28 Mb, respectively. Following the inclusion of village chicken the significance of these SNPs rises to P=3.02 × 10−14. Both SNPs occur within the BCDO2 gene, and are also contained within the highest scoring sweep window Gga 24: 6.26–6.28 Mb (Z=6.27 traditional breeds only and Z=6.51 including village chicken). These results confirm the association mapping of the yellow skin phenotype to the BCDO2 gene.
Figure 3.
Significant plots for association tests and selective sweeps for phenotypes at previously known mapping intervals. Yellow skin was tested in traditional breeds (a) and then inclusively with the village chicken (b), as was oocyan (c and d respectively). Due to ambiguous comb phenotypes in village chicken, the rose comb (e) and duplex comb (f) were tested in the traditional breeds only. The −log10(FDR-adjusted P) from association tests are plotted in blue, while Z scores from selective sweeps are plotted in green along the same axes. The dashed lines indicate the significance threshold for each of the respective tests.
Blue egg—oocyan
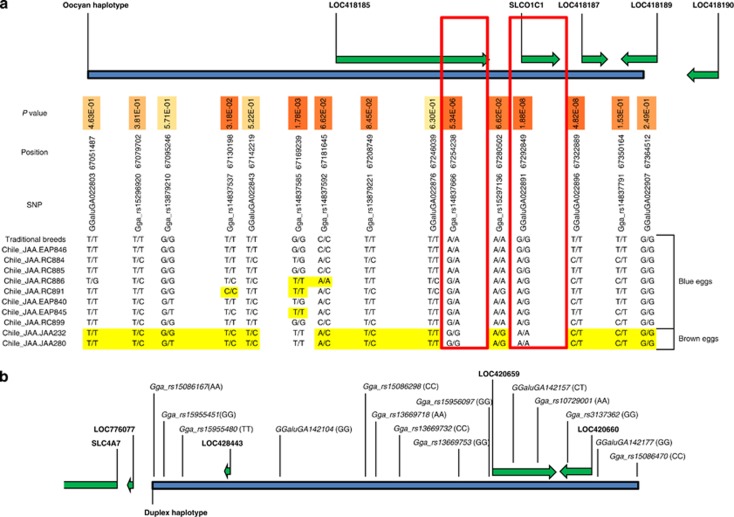
As with yellow skin, the oocyan phenotype was analysed both in the presence and absence of village chicken. Post pruning, in the absence of village chicken, 7 birds exhibit the phenotype while 55 birds do not; whereas in the presence of village chicken, 14 birds exhibit the phenotype and 93 birds do not. Previous information from an F2 resource population has mapped the genetic control for oocyan to Gga 1 within interval 67.3–69.1 Mb (Wang et al., 2010). There are 84 SNPs on the chip within this mapping interval. Figures 3c and d illustrate the results of the association tests and selective sweeps within the mapped region. In the absence of village chicken, the most significant SNP following the association test is located within the previously known mapping interval—GGaluGA022896 (67.32 Mb; P=3.0 × 10−5, increasing to P=3.45 × 10−9 with village chicken), although with the inclusion of village chicken this is superseded by GGaluGA022891 (67.29 Mb; P=1.45 × 10−9, P=5.76 × 10−5 before the inclusion of village chicken). Similar results were obtained from selective sweep analysis in which 4 of the 11 significant windows (Z⩾4) were located within Gga 1: 67.17–67.32 Mb, the most significant of which contained a single SNP (GgaluGA022896) at position 67.32 Mb (Z=5.86 traditional breeds only and Z=5.87 with village chicken). IBD analysis in this region reveals a 313Kb conserved haplotype with 15 SNPs across all the traditional breed birds (n=9 pre-pruning) expressing the oocyan phenotype (Figure 4a). Including the Chilean village chicken allows to fine-map this region by identifying in birds exhibiting the phenotype alternate homozygote genotypes, and subsequently by identifying in birds not exhibiting the phenotype, homozygote genotypes shared with the birds exhibiting the phenotype and heterozygote genotypes (Figure 4a). This leads to the exclusion of the region upstream and inclusive of GGaluGA022876, together with that downstream and inclusive of GGaluGA022896, resulting in two small candidate regions: 34.5 Kb (GGaluGA022876–Gga_rs15297136) and 42.4 Kb (Gga_rs15297136–GGaluGA022896). Two relevant candidate genes occur within the oocyan haplotype: SLCO1C1 (solute carrier organic anion transporter family, member 1C1) found within the 42.4 Kb region and located 1.5 Kb downstream of GgaluGA022891 (Z=6.09 traditional breeds only and Z=5.84 with village chicken) and SLCO1B3 (solute carrier organic anion transporter family, member 1B3; LOC418187) located 5.5 Kb from GgaluGA022896. Analysis of eggshell pigments by mass spectrometry in several wild avian and Dongxiang chicken eggshells confirm the principal eggshell pigments to be protoporphyrin and the bile pigment biliverdin, with the accumulation of the latter being responsible for the oocyan phenotypes (Gorchein et al., 2009; Wang et al., 2009). Organic anion transporters have been shown to be involved in bile acid transport (for review see Klaassen and Aleksunes, 2010), adding credibility to these genes as possible candidates for the trait.
Figure 4.
Schematic diagrams illustrating the chromosomal locations of the conserved haplotypes identified in traditional breeds for (a) oocyan and (b) duplex phenotypes, and the surrounding genes. The P-value at each SNP within the oocyan haplotype is included as are the genotypes for the Chilean village chicken; genotypes incompatible with the dominant inheritance of the phenotype are shaded. The two SNPs concordant with the phenotype in the village chicken are outlined. The genotypes for the duplex haplotype are indicated in brackets after the SNP identifier.
Rose comb
Owing to the ambiguous nature of the comb phenotype in village chicken, analyses for this trait were performed using the traditional breeds only. From published literature (Roberts, 1997), rose comb was expected to be present in 9 birds (from 6 different breeds) but absent in 42 birds (from 22 breeds) post pruning. Figure 3e illustrates the results of association mapping and selective sweep. At the genome-wide level, two of the three most significant SNPs are located on Gga 7 spanning the region 19.27–19.97 Mb (Gga_rs15854456 at 19.27 Mb and Gga_rs15855521 at 19.97 Mb). These share the most significant P-value (P=5.10 × 10−4) with GGaluGA301302 on Gga 6 at 22.53 Mb which, lying outside the known mapping interval, is a false-positive for the phenotype. Similarly, the most significant selective sweep window was located on Gga 7 at 19.27 Mb (Z=5.06). The majority of the significant results found on Gga 7 are located between 18.4 Mb (Gga,_rs14611492, P=4.52 × 10−3, Z=4.77) and 22.1 Mb (GGaluGA315119, P=4.52 × 10−3, Z=2.18). Gga_rs15854456 is located 27.9 Kb upstream of DLX1 (homeodomain transcription factor distal-less homeobox 1). Variation in the expression of the DLX gene repertoire has been linked to craniofacial dysmorphology in several species, most recently in cichlid fish (Renz et al., 2011). Subfertility has been reported in birds homozygous for rose comb phenotype (McLean and Froman, 1996). Givens et al. (2005) found evidence to support the role of DLX1 in regulating the GnRH promoter during development in mice. Several recent studies have associated the misexpression of GnRH with infertility in mice (Larder et al., 2011). DLX1 might therefore be associated with the rose comb phenotype and/or the subfertility linked to this trait.
Duplex comb
As with the rose comb, the analyses performed on this trait did not include the village chickens. Post pruning, the duplex comb was expected to be found in 3 birds from 3 traditional breeds but absent in 59 birds from 31 breeds. Dorshorst et al. (2010) mapped duplex comb to Gga 2 interval 33.6–39.8 Mb. Figure 3f illustrates the results of association mapping and selective sweep. Genome-wide, the two most significant SNPs (P=1.7 × 10−5) identified in the association test were Gga_rs14293190, which lies outside of the previously mapped region being located on Gga 24 at 1.51 Mb, and Gga_rs15086167, which is located within the mapped region at Gga 2 at 38.55 Mb (P=1.7 × 10−5). The selective sweep analysis identified 48 significant (Z⩾4) windows genome-wide, the most significant of which was on Gga Z at 6.04 Mb (Z=5.14), however, none were identified on Gga 2 within the known mapping interval. IBD analysis reveals a conserved haplotype of 14 SNPs commencing from Gga_rs15086167 and spanning 331 Kb in all the birds (n=8) expressing the duplex comb phenotype (Figure 4b). This region contains the AZI2 (5-azacytidine induced 2; LOC420660), CMC1 (COX assembly mitochondrial protein homologue (Saccharomyces cerevisiae); LOC420659) and EOMES (eomesodermin homologue (Xenopus laevis); LOC428443) genes. Also of note is the RARB (retinoic acid receptor beta, Gga 2: 37.41–37.83 Mb) gene located 723 Kb upstream of Gga_rs15086167. Exogenously applied retinoic acid acts on the mesenchyme and has been found to affect craniofacial development in chicken embryos (Richman, 1992). In addition, sonic hedgehog (Shh, Gga 2: 7.7–8.1 Mb) has been shown to participate in craniofacial morphogenesis and can be downregulated through doses of retinoic acid (Helms et al., 1997).
Earlobe pigmentation
As with the comb phenotypes, earlobe pigmentation can be difficult to record accurately in village chicken and so they have not been included in these analyses. Earlobe pigmentation is often a breed standard, and post-pruning 10 birds (9 breeds) show white earlobes while 47 birds (23 breeds) had red earlobes. The Silkie has turquoise earlobes; however, it is not yet understood whether or not this is an interaction of earlobe pigmentation on black skin—and if this is the case, what the base earlobe colour might be. Studies by Warren (1928) indicate earlobe pigmentation to be sex-linked and polygenic possibly under incomplete dominance. Taking both the mapping and selective sweep analyses into consideration, concordant significances (P<0.05 and Z>4) were identified for seven SNPs (Table 4). These results are lower in statistical significance than those returned for the other traits analysed in this study; however, they do support the fact that the trait is polygenic and sex-linked. One in particular, Gga_rs14762712 located on Gga Z at 32.08 Mb (P=1.57 × 10−2, Z=4.73) lies ∼13 Kb downstream of the basonuclin 2 (BNC2) gene. A study by Lang et al. (2009) involving zebrafish bonaparte mutants found BNC2 to be an important mediator of pigment pattern formation. An interesting feature of the bonaparte mutants is a ventroanterior patch of iridophores, which are pigment cells containing purine-rich reflecting platelets (Lang et al., 2009). The mapping of an association of white earlobe pigmentation to a gene mediating the formation of purine-rich pigment cells is consistent with Louvier's (1934) observations that the pigment of white earlobes is a compound made up of purine bases.
Table 4. Results of earlobe pigmentation association test (P<0.05) with sweep Z scores (Z>4).
| SNP | Chromosome | Pos | P | Z |
|---|---|---|---|---|
| GGaluGA024632 | 1 | 71435269 | 1.13E-03 | 4.265564 |
| Gga_rs14762712 | Z | 32081988 | 1.57E-02 | 4.7293776 |
| Gga_rs14170217 | 2 | 41692774 | 3.70E-02 | 4.8662406 |
| Gga_rs14801862 | 1 | 27785677 | 3.74E-02 | 4.204736 |
| GGaluGA008940 | 1 | 26684319 | 3.93E-02 | 4.143908 |
| Gga_rs14170463 | 2 | 41892291 | 4.67E-02 | 4.1515115 |
| Gga_rs14487048 | 4 | 70367646 | 4.67E-02 | 4.5621006 |
Discussion
We characterised here at the genome-wide level using the 60K chicken SNP chip (Groenen et al., 2011) the diversity, haplotype blocks and LD structure of traditional and village chicken genomes, addressing the usefulness of non-pedigree chicken populations for mapping Mendelian traits. The use of a high-density SNP chip allows us first to investigate the genetic make-up of the breeds/populations studied. STRUCTURE analysis at K=2 and K=3 reveals genetic backgrounds of different proportion to be present in the Kenyan, Ethiopian and Chilean village chickens. These genetic backgrounds are also present across the traditional breeds although with large variation in their proportions, reflecting the admixed origins of some of the breeds and/or the effect of selective breeding. Also, a common feature across some breeds is the apparent relationship between their supposed geographic origin (Scrivener, 2006, 2009) and their genetic background. For example the Brahma, Buff Orpington, Cochin, Croad Langshan, Lincolnshire Buff, Modern Langshan, Silkie and Totenko breeds supposedly of Asian origin cluster together and reveal little admixture with the genetic background that dominates breeds with a more Mediterranean origin (Appenzellor, Hamburgh, Leghorn, Polish and Spanish).
We observed no significant differences in haplotype block structure between traditional and village chicken breeds/populations. Both shared a mode number (two) of SNPs per block and an overall median block size of 11–12 Kb. A reduction in block size along the microchromosomes compared with the macrochromosomes was observed in both cases. This was expected given the higher rate of recombination along the microchromosomes as indicated by our LD results. Similar observations have been reported previously (Megens et al., 2009). In calculating blocks of up to 1 Mb in size, we observed that 77% of the blocks were <25 Kb in size and ∼91% of these were shared by both traditional breeds and village chickens. The sharing of haplotype blocks between traditional and village chickens, and the presence of haplotype blocks of different size is a strong indication that recombination rate varies along chromosomes, and that some chromosomal regions witness little or no recombination.
Genome-wide calculations of haplotype block sizes have been reported in other domestic species. The ancestral haplotype block size across different breeds of dog is ∼10 Kb (Lindblad-Toh et al., 2005). Within breeds blocks can extend from 500 Kb to 1 Mb (Sutter et al., 2004; Lindblad-Toh et al., 2005). The median block size of 11–12 Kb across the populations/breeds studied here is consistent with the ancestral block size observed in dogs, despite both domestic species having very distinct genetic histories with the dog having been domesticated several thousands of years before the chicken (Lindblad-Toh et al., 2005; Gifford-Gonzales and Hanotte, 2011).
The size of the assembled reference sequence for the chicken genome is 1.05 Gb (ICGSC, 2004). Considering haplotype block sizes of 9 and 11 Kb, our data indicate that at least 90–110K SNPs would be required to construct a haplotype map of the chicken genome for traditional and village chickens, assuming each haplotype block is represented by a single SNP. Such higher-density chips will optimise association fine mapping of Mendelian and quantitative traits in non-pedigree chicken populations. While 60K SNPs may be suitable for use with commercial poultry (Qanbari et al., 2010), our findings are in agreement with those of Megens et al. (2009) who studied LD for four ∼1-cM genome regions in macro- and microchromosomes and suggested that at least 100K SNPs are required for effective genome-wide association studies. However, the design of a universal SNP chip will require the inclusion of many more SNPs for haplotype block analysis, with a certain proportion of SNPs expected to be uninformative in some populations, and as shown here with differences in haplotype block structure between sets of populations. Of importance in this respect in the development of higher-density SNP arrays would be to include chickens of different genetic backgrounds for screening and selection of informative SNPs.
All subsets of samples show a decrease in r2 over physical distance (Figure 2) and, with the exception of Totenko, mean r2 decays rapidly over pairwise distances. For all subsets of samples analysed, with the exception of the single-breed subsets (Totenko, Araucana and Marans), r2 at pairwise distances >10 Kb are below the threshold for ‘useful LD' of r2>1/3 suggested for association analysis (Ardlie et al., 2002). The extent and range of LD along the genome can be affected by many factors such as breeding history (effective population size, number of generations, degree of admixture, and so on) and recombination rate. Besides its importance for mapping traits, measurement of LD allows an estimation of effective population size (Ne) over past generations (Pritchard and Przeworski, 2001) and can be used to date the time of divergence between two populations (McEvoy et al., 2011). Both might be relevant to study the history of village chickens across the world. While to date, the former has commonly been applied in domestic species, the latter has only recently been performed in humans (McEvoy et al., 2011). Excluding Totenko, the highest r2 values were observed across Araucana and Marans. This likely reflects their recent (<100 years ago) standardisation as breeds compared with other traditional breeds studied. Similar LD values are observed in village chickens and traditional breeds in agreement with the results from the haplotype block structure analysis.
Traditional breeds are the result of hundreds of years of artificial selection for utility and ‘fanciful' phenotypes giving rise to an array of morphological diversity in colour, shape, size, form and function. Such diversity is also evident in panmictic populations of village chicken (Dana et al., 2010b). Our results indicate that many Mendelian traits currently uncharacterised at the molecular level could be fine mapped using association analysis in non-pedigree chicken populations. Assuming an average haplotype block size of ∼11–12 Kb, the genetic control of Mendelian phenotypes could theoretically be mapped to this resolution, opening the door towards identifying possible causative mutations.
The across-breed mapping approach takes explicit advantage of the cumulative number of recombination events that have occurred since breed divergence. Birds that are closely related will share a large proportion of genomic information, and a mapping resource pedigree would seek to explore the regions that differ within the pedigree. Conversely, birds that are distantly related share less genomic information, and so the across-breed mapping approach seeks to exploit regions of similarity across birds sharing a particular phenotype. We validate this approach in chicken with the successful remapping of the yellow skin to the BCDO2 gene, after which we applied the approach to three other phenotypes (oocyan, rose comb and duplex comb) that have previously been mapped to large chromosomal intervals, and to the unmapped earlobe pigmentation trait. For this purpose, we only included birds with pairwise IBS scores below 0.79. This included birds from different strains as well as birds from different traditional breeds, but excluded birds that are likely to be closely related so as to minimise exposure to false-positives due to the limited sample size.
Mapping results were obtained from single-marker associations and selective sweep analysis. In spite of limitations in the extent of genome coverage, informativeness of the SNP chip and sample availability, we have refined mapping for the oocyan locus using traditional breeds from a region of ∼1.8 Mb to one of ∼330 Kb—within which two small candidate regions (34.5 Kb and 42.4 Kb) were identified using village chickens. In addition, we have refined the mapping for rose comb from a region of ∼9.5 Mb to one of 3.7 Mb, and the duplex comb from a region of ∼6 Mb to one of ∼330 Kb. The results are supported by a large number of SNPs suggesting that the inclusion of more genetically distant case and control individuals may improve the mapping resolution. Having mapped associations to regions previously identified to specific phenotypes, we used the same approach to identify chromosomal regions associated with earlobe pigmentation. In particular, we identify significant associations on Gga 22 and Z supporting earlier conclusions by Warren (1928) that the trait is sex-linked and polygenic. However, the inheritance and biochemistry of the phenotype is poorly understood, lacking the level of investigation, which has been afforded to the other phenotypic traits present in chicken. More research would hence be required to substantiate a link from the phenotype to any of the associations identified in this study.
There are limitations to the mapping approach used in this study. More particularly, it requires access to a large number of unrelated case and control birds, the more distantly related the better. Of benefit to the scientific community would be the public availability of databases that report genotyping data from high-density SNP chips, together with detailed phenotypic descriptions of the birds genotyped. As with any genome-wide association study, increasing the number of samples increases the statistical significance of any associations mapped, and reduces the likelihood of false-positives due to chance associations. Mapping across traditional breeds takes advantage of the genetic diversity expected from many recombination events. An important benefit of accurate phenotype recording is that genotyped individuals may support multiple association studies targeting different phenotypic traits. This is illustrated here with the inclusion of the same bird, either as case or control, for the mapping of different traits. The segregation of several phenotypes in traditional chicken breeds and among village chickens offers the opportunity to independently map such traits in different sets of unrelated populations therefore increasing the statistical confidence of the results.
Accurate recording of phenotypes is important in all cases. While some traits are fixed within breeds, for example, hookless feathering in Silkie, others have been diluted as breeders reinvigorate their flocks by occasionally introducing new blood from other breeds to enhance particular traits. Unfortunately, this can often occur at the expense of another trait. Furthermore, breed standards can vary between countries, for example, French Marans have lightly feathered shanks whereas British Marans are clean-legged (Scrivener, 2009). Rather than depending on the adherence to such standards by breeders, recording directly the presence or absence of traits of interest is beneficial for accurate mapping.
Conclusions
The analysis presented here indicates that at least 90–110K SNPs are required for effective genome-wide association studies in traditional breeds and village chickens. This has been shown to be the case by extrapolating the median block sizes of ∼9 Kb (along microchromosomes) and ∼11 Kb (along Gga 1–28) across the 1.05 Gb chicken genome, and through r2 values <0.3 for all pairwise distances over and above 10 Kb.
The across-breed mapping strategy presented here has successfully remapped the yellow skin phenotype and generated refined mapping intervals for oocyan, rose and duplex combs. Initial mapping of the more complex earlobe pigmentation phenotype also identified several significant associations supporting previous studies suggesting the trait to be sex-linked and polygenic. The key value in such a strategy over traditional mapping using pedigree resource populations is that the recorded genotypic and phenotypic data can support several mapping and/or diversity studies.
A major caveat of current ‘genome-wide' studies in chicken using the 60K SNP chip (Groenen et al., 2011) is that they are not entirely genome-wide. Chickens have 38 pairs of autosomes, and of the ∼1.05 Gb reference genome only 95% has been anchored to some but not all chromosomes (Gga 1–28, Gga 32, Gga Z and Gga W). Additionally, there are nine microchromosomes that remain elusive to geneticists. Reducing the scope yet further, the 60K SNP chip has very few informative SNPs located on Gga 16, Gga 32 and Gga W, while a large number of SNPs still remain unmapped. Ultimately, genome-wide association studies incorporating the currently available 60K SNP chip may have limited success owing to the absence of SNPs on the chip in LD with phenotypes of interest.
Data archiving
Data have been deposited at Dryad: doi:10.5061/dryad.996cr8f0.
Acknowledgments
We would like to extend our sincere gratitude to the Rare Breeds Survival Trust through the Roslin Institute, AVIANDIV project and Negussie Dana (Ethiopian Institute of Agricultural Research) for providing some of the samples used in this study. Graeme Robertson (Roslin) and Karen Troup (ArkGenomics) provided invaluable technical assistance by collating, curating and extracting DNA from the Roslin samples. We would also like to thank Phil McGowan of the World Pheasant Association, and Jaime Gongora of the University of Sydney for their assistance in locating breeders for some of the chicken populations used in this study. Finally, we wish to acknowledge the owners of the village chickens for their help and cooperation during sampling. Funding for this study was made possible through a BBSRC PhD fellowship to the first author, and a BBSRC research grant (BB/H009051/1) to the last author. The Roslin Institute is supported by a core strategic grant from the BBSRC.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Akey JM, Ruhe AL, Akey DT, Wong AK, Connelly CF, Madeoy J, et al. Tracking footprints of artificial selection in the dog genome. Proc Natl Acad Sci USA. 2010;107:1160–1165. doi: 10.1073/pnas.0909918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Society. 1995;57:289–300. [Google Scholar]
- Chase K, Jones P, Martin A, Ostrander EA, Lark KG. Genetic mapping of fixed phenotypes: disease frequency as a breed characteristic. J Hered. 2009;100 (Suppl 1:S37–S41. doi: 10.1093/jhered/esp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RD.1990Poultry Breeding and Genetics Elsevier: Amsterdam, Oxford; xvi, p1123. [Google Scholar]
- Dana N, Dessle T, van der Waaij LH, van Arendonk JA. Morphological features of indigenous chicken populations of Ethiopia. Anim Genet Resources. 2010b;46:11–23. [Google Scholar]
- Dana N, van der Waaij LH, Dessie T, van Arendonk JA. Production objectives and trait preferences of village poultry producers of Ethiopia: implications for designing breeding schemes utilizing indigenous chicken genetic resources. Trop Anim Health Prod. 2010a;42:1519–1529. doi: 10.1007/s11250-010-9602-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorshorst B, Okimoto R, Ashwell C. Genomic regions associated with dermal hyperpigmentation, polydactyly and other morphological traits in the silkie chicken. J Hered. 2010;101:339–350. doi: 10.1093/jhered/esp120. [DOI] [PubMed] [Google Scholar]
- Earl D, Vonholdt B.2011STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method Conservation Genetics Resources(doi: 10.1007/s12686-011-9548-7 [DOI]
- Eriksson J, Larson G, Gunnarsson U, Bed'hom B, Tixier-Boichard M, Stromstedt L, et al. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 2008;4:e1000010. doi: 10.1371/journal.pgen.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Georges M. Mapping, fine mapping, and molecular dissection of quantitative trait loci in domestic animals. Annu Rev Genomics Hum Genet. 2007;8:131–162. doi: 10.1146/annurev.genom.8.080706.092408. [DOI] [PubMed] [Google Scholar]
- Gifford-Gonzales D, Hanotte O. Domesticating animals in Africa: implications of genetic and archaeological findings. J World Prehistory. 2011;24:1–23. [Google Scholar]
- Givens ML, Rave-Harel N, Goonewardena VD, Kurotani R, Berdy SE, Swan CH, et al. Developmental regulation of gonadotropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J Biol Chem. 2005;280:19156–19165. doi: 10.1074/jbc.M502004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongora J, Rawlence NJ, Mobegi VA, Jianlin H, Alcalde JA, Matus JT, et al. Indo-European and Asian origins for Chilean and Pacific chickens revealed by mtDNA. Proc Natl Acad Sci USA. 2008;105:10308–10313. doi: 10.1073/pnas.0801991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchein A, Lim CK, Cassey P. Extraction and analysis of colourful eggshell pigments using HPLC and HPLC/electrospray ionization tandem mass spectrometry. Biomed Chromatogr. 2009;23:602–606. doi: 10.1002/bmc.1158. [DOI] [PubMed] [Google Scholar]
- Groenen MA, Megens HJ, Zare Y, Warren WC, Hillier LW, Crooijmans RP, et al. The development and characterization of a 60K SNP chip for chicken. BMC Genomics. 2011;12:274. doi: 10.1186/1471-2164-12-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao K, Di X, Cawley S. LdCompare: rapid computation of single- and multiple-marker r2 and genetic coverage. Bioinformatics. 2007;23:252–254. doi: 10.1093/bioinformatics/btl574. [DOI] [PubMed] [Google Scholar]
- Helms JA, Kim CH, Hu D, Minkoff R, Thaller C, Eichele G. Sonic hedgehog participates in craniofacial morphogenesis and is down-regulated by teratogenic doses of retinoic acid. Dev Biol. 1997;187:25–35. doi: 10.1006/dbio.1997.8589. [DOI] [PubMed] [Google Scholar]
- Hill WG, Roberston AR. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- ICGSC Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62:1–96. doi: 10.1124/pr.109.002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MR, Patterson LB, Gordon TN, Johnson SL, Parichy DM. Basonuclin-2 requirements for zebrafish adult pigment pattern development and female fertility. PLoS Genet. 2009;5:e1000744. doi: 10.1371/journal.pgen.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder R, Clark DD, Miller NL, Mellon PL. Hypothalamic dysregulation and infertility in mice lacking the homeodomain protein Six6. J Neurosci. 2011;31:426–438. doi: 10.1523/JNEUROSCI.1688-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Louvier R.1934Rechersches chimiques sur la pigmentation de l'oreillon du coq domestiqueIn: Crawford RD (ed).Poultry Breeding and Genetics1st edn.Elsevier: Oxford,p153 [Google Scholar]
- McEvoy BP, Powell JE, Goddard ME, Visscher PM. Human population dispersal ‘Out of Africa' estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 2011;21:821–829. doi: 10.1101/gr.119636.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean DJ, Froman DP. Identification of a sperm cell attribute responsible for subfertility of roosters homozygous for the rose comb allele. Biol Reprod. 1996;54:168–172. doi: 10.1095/biolreprod54.1.168. [DOI] [PubMed] [Google Scholar]
- Megens HJ, Crooijmans RP, Bastiaansen JW, Kerstens HH, Coster A, Jalving R, et al. Comparison of linkage disequilibrium and haplotype diversity on macro- and microchromosomes in chicken. BMC Genet. 2009;10:86. doi: 10.1186/1471-2156-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwacharo JM, Bjornstad G, Mobegi V, Nomura K, Hanada H, Amano T, et al. Mitochondrial DNA reveals multiple introductions of domestic chicken in East Africa. Mol Phylogenet Evol. 2011;58:374–382. doi: 10.1016/j.ympev.2010.11.027. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnett RC.1923Heredity in Poultryvol. 8. Macmillan: London [Google Scholar]
- Punnett RC. Genetic studies in poultry - IX. The blue egg. Genetics. 1933;27:465–470. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanbari S, Hansen M, Weigend S, Preisinger R, Simianer H. Linkage disequilibrium reveals different demographic history in egg laying chickens. BMC Genet. 2010;11:103. doi: 10.1186/1471-2156-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing: Vienna, Austria; 2009. [Google Scholar]
- Renz AJ, Gunter HM, Fischer JM, Qiu H, Meyer A, Kuraku S. Ancestral and derived attributes of the dlx gene repertoire, cluster structure and expression patterns in an African cichlid fish. Evodevo. 2011;2:1. doi: 10.1186/2041-9139-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman JM. The role of retinoids in normal and abnormal embryonic craniofacial morphogenesis. Crit Rev Oral Biol Med. 1992;4:93–109. doi: 10.1177/10454411920040010701. [DOI] [PubMed] [Google Scholar]
- Roberts V.(ed) (1997British Poultry Standards5th edn.Blackwell Science: Oxford, viii; p368. [Google Scholar]
- Rubin CJ, Zody MC, Eriksson J, Meadows JR, Sherwood E, Webster MT, et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Scrivener D. Rare Poultry Breeds. Crowood: Ramsbury, Wiltshire; 2006. [Google Scholar]
- Scrivener D. Popular Poultry Breeds. Crowood: Ramsbury; 2009. [Google Scholar]
- Sheppy A. The colour of domestication and the designer chicken. Opt Laser Technol. 2011;43:295–301. [Google Scholar]
- Somes RG., Jr Duplex comb in the chicken: a multi-allelic trait. J Hered. 1991;82:169–172. doi: 10.1093/oxfordjournals.jhered.a111054. [DOI] [PubMed] [Google Scholar]
- Storey AA, Ramirez JM, Quiroz D, Burley DV, Addison DJ, Walter R, et al. Radiocarbon and DNA evidence for a pre-Columbian introduction of Polynesian chickens to Chile. Proc Natl Acad Sci USA. 2007;104:10335–10339. doi: 10.1073/pnas.0703993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, Kruglyak L, et al. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XT, Bai JR, Zhao CJ, Zhang H, Bao HG, Xu GY, et al. Localisation of the genomic sequence interval for the blue eggshell gene using an F2 resource population of Dongxiang chickens. Br Poult Sci. 2010;51:507–509. doi: 10.1080/00071668.2010.502520. [DOI] [PubMed] [Google Scholar]
- Wang XT, Zhao CJ, Li JY, Xu GY, Lian LS, Wu CX, et al. Comparison of the total amount of eggshell pigments in Dongxiang brown-shelled eggs and Dongxiang blue-shelled eggs. Poult Sci. 2009;88:1735–1739. doi: 10.3382/ps.2008-00434. [DOI] [PubMed] [Google Scholar]
- Warren DC. Inheritance of earlobe color in poultry. Genetics. 1928;13:470–487. doi: 10.1093/genetics/13.6.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S, Wiener P, Teverson D, Haley CS, Hocking PM.2011Characterization of the genetic diversity, structure and admixture of British chicken breeds Anim Genet(e-pub ahead of print December 2011; doi: 10.1111/j.1365-2052.2011.02301.x [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.