Abstract
Recently duplicated genes are believed to often overlap in function and expression. A priori, they are thus less likely to be essential. Although this was indeed observed in yeast, mouse singletons and duplicates were reported to be equally often essential. This contradiction can only partly be explained by experimental biases. We herein show that older genes (i.e., genes with earlier phyletic origin) are more likely to be essential, regardless of their duplication status. At a given phyletic gene age, duplicates are always less likely to be essential compared with singletons. The “paradoxical” high essentiality among mouse gene duplicates is then caused by different age profiles of singletons and duplicates, with the latter tending to be derived from older genes.
Keywords: gene essentiality, yeast, mouse, phyletic age, linking genotype to phenotype
In model organisms such as mouse and yeast, phenotypic changes caused by single-gene mutations were assayed on a genome-wide scale (Kelly et al. 2001; Blake et al. 2011). Of particular interest are essential genes, whose removal results in death or infertility. Many expressed genes performing important molecular functions are nonessential. In these cases, it is likely that the gene deletion can be partially compensated by another gene with overlapping function and expression.
Gene duplication is believed to be an important source of such functional redundancy (Ohno 1970). Accordingly, the proportion of essential genes (PE) among duplicates is much lower than among singletons in yeast (Gu et al. 2003). However, this expected trend was not confirmed in mouse, where the proportion of essentials among duplicates is comparable (Liao and Zhang 2007) or even lower (table 1) than among singletons.
Table 1.
Proportions of Essential Genes in Different Gene Categories in the Two Phenotypical Data Sets for Mouse.
| Proportion of Essential Genes (%) |
||
|---|---|---|
| Categories | Current Data Seta | Data from Makino et al. (2009) |
| All genes | 43.3 | 42.07 |
| Duplicates | 43.9 (41.6b) | 41.92 |
| Singletons | 41.1 | 42.61 |
| All developmental genes | 62.53 | 59.51 |
| Developmental duplicates | 64.75 | 60.9 |
| Developmental singletons | 53.1 | 53.36 |
| Old duplications (Ksc ≥ 2) | 47.31 | 44.94 |
a MGI 4.4 (October 2010).
b If only genes with valid phyletic ages are used.
c If a gene has multiple duplicates, all pairwise Ks (the number of synonymous substitutions per synonymous site) between this gene and its duplicates will be calculated, and the lowest Ks value is used. Synonymous substitutions in most genes with Ks ≥ 2 will have reached saturation, and hence, the corresponding genes will tend to be older than genes with Ks < 2.
The contradicting results in mouse were initially interpreted as evidence against widespread functional redundancy of duplicates (Liao and Zhang 2007); this interpretation was hotly disputed (Su and Gu 2008; Liang and Li 2009; Makino et al. 2009). At that time (Liao and Zhang 2007; Su and Gu 2008; Liang and Li 2009; Makino et al. 2009), only ∼5,000 mouse genes had been tested in knockout experiments. Biases were expected in this subset of mouse genes, as genes with known severe mutational phenotypes had been selected with higher priority. Two follow-up studies (Su and Gu 2008; Makino et al. 2009) discovered that the knockout data were further enriched in genes derived from old duplications and in developmental genes; after correcting these biases, the overall PE in duplicates became statistically significantly lower than that in singletons (Su and Gu 2008; Makino et al. 2009).
However, the authors did not explore two immediate conclusions from their studies (Su and Gu 2008; Makino et al. 2009): 1) genes derived from old duplications are more likely to be essential than singletons and 2) developmental duplicates are more likely to be essential than developmental singletons (or indeed singletons as a whole). Both conclusions hold true in the older as well as the latest versions of the mouse phenotype data sets (table 1). This appears to again contradict the duplication-functional redundancy concept, and we thus consider the issue unresolved.
What factors other than duplication status affect gene essentiality? Developmental genes are more likely to be essential than nondevelopmental genes (Makino et al. 2009), but this should apply to duplicates and singletons alike. It was also suggested that hubs in protein–protein interaction networks are more likely to be essential (Jeong et al. 2001); however, this observation probably reflects biases toward proteins in large essential protein complexes (Zotenko et al. 2008).
Previous studies indicated that the phyletic origin (age) of genes, defined by the evolutionarily most distant species group where homologs can be found (Wolf et al. 2009), is correlated with several gene features (Hao et al. 2010). Genes that originated early tend to be conserved across species, highly and broadly expressed, and broadly useful (Hao et al. 2010). Thus, we hypothesized that knocking out phyletically old genes is more likely to have severe phenotypic effects: old genes should be more often essential.
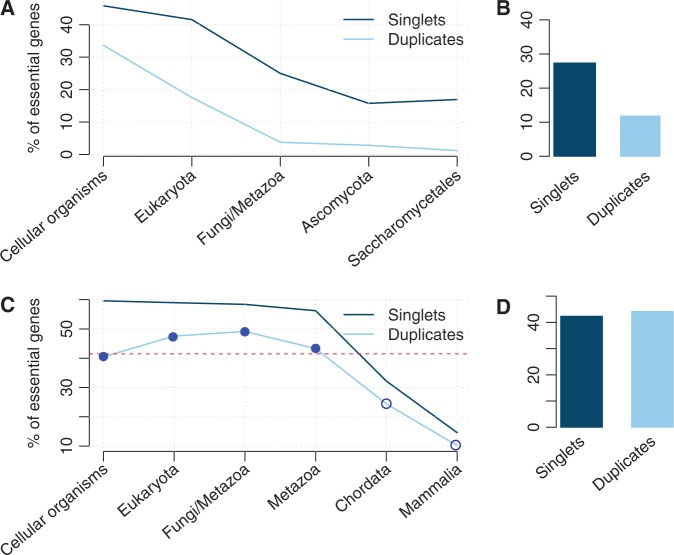
To test this idea, we classified mouse and yeast genes into different age groups according to their earliest phyletic origin (see Materials and Methods). We classified genes as specific to one of five taxonomic groups for yeast (fig. 1A) and six broad taxonomic groups for mouse (fig. 1C). Because of the large differences between yeast and mouse, we did not attempt any direct cross-species comparisons and did not attempt to map their histories onto a common timescale.
Fig. 1.
In both yeast (A) and mouse (C), genes with more recent phyletic origins are less likely to be essential, as are duplicated genes compared with singletons of the same phyletic age. However, ignoring age, the overall proportion of essential genes in singletons is higher in yeast (B) but lower in mouse (D) compared with duplications. Filled circles in (C) indicate that the proportion of essential genes in the corresponding duplication groups is higher than or closer to the overall PE in singletons (41.1%; the dashed horizontal line); whereas hollow circles indicate that PE is lower.
We found that within each age group, the PE among singletons is always higher than among duplicated genes; this is true both in mouse and in yeast (fig. 1). Thus, duplicated genes indeed tend to be less likely essential. Furthermore, for both singletons and duplicated genes, the fraction of essential genes increases with increasing age; thus, older genes are indeed more likely to be essential (fig. 1). The trends observed in figure 1 are reproduced when restricting the analysis either to developmental genes or to nondevelopmental genes (Supplementary figs. S1 and S2, Supplementary Material online; for the raw data, see Supplementary table, Supplementary Material online).
Gene duplicates have two ages: the age of the gene family (phyletic age; fig. 1) and the age of the most recent duplication event (duplication age). The effect of phyletic age is likely similar between duplicates and singletons. In addition, functional redundancy is expected to be strongly affected by the age of the duplication event, as duplicates derived from ancient duplications are more likely to be essential than genes derived from recent duplications (Su and Gu 2008). In mouse gene duplicates, essentiality reaches a plateau in the Fungi/Metazoa group and does not increase further in the two older age groups. According to the reasoning above, this plateau might be caused by a comparably young duplication age. We indeed find that the two oldest groups contain higher fractions of younger duplicates than the “Fungi/Metazoa” group (Supplementary fig. S3, Supplementary Material online).
In each phyletic age group, duplicates are less likely to be essential than singletons (fig. 1). Why then is the same not true when disregarding age, as done in previous studies (Liao and Zhang 2007; Su and Gu 2008; Liang and Li 2009; Makino et al. 2009)? This is in fact an instance of Simpson's paradox (Simpson 1951), which can arise when the dependence of two categorical variables (essentiality and duplication status) on a third variable (phyletic age) is disregarded. To illustrate the mathematics behind this paradox, we divided the six age groups of duplicated genes into two parts reflecting a very coarse definition of age: one including four age groups (the “old part,” filled circles in fig. 1C) that mostly have higher PEs than the overall singletons and other including the remaining two groups with lower PEs (the “young part,” open circles in fig. 1C). This partitioning results in a higher overall proportion of essential genes in the old duplicate part () are higher compared with the overall singletons (), whereas the corresponding proportion in the young duplicate part () is lower. The overall PE of duplicates regardless of age can be calculated from this as a weighted average:
where fold and fyoung are the fraction of duplicates contained in the “old” and “young” parts, respectively (with fold + fyoung = 1) (for more details, see Supplementary text and Supplementary table, Supplementary Material online). In theory, the overall PE could be as high as 44.89% or as low as 22.97%, depending on the values of fold and fyoung. In our study, we found that the vast majority of duplicates was derived from old gene families (fold = 84.66%), resulting in an overall PE of 41.6% for duplicates (see Supplementary table, Supplementary Material online). Thus, the surprising result of a higher essentiality among mouse duplicates compared with singletons is caused by a different age profile of singletons and duplicated gene families.
Our results differ significantly from a recent publication on Drosophila melanogaster (Chen et al. 2010). Based on RNAi knockdowns of ∼440 genes, Chen et al. found that ∼30% of young genes (<35 myr) were essential compared with ∼35% of old genes (>40 myr). The authors concluded that “young genes are as essential as old genes in terms of viability” (Chen et al. 2010). We reanalyzed their data using our methods, which differ from those of Chen et al. in age classification and in the separate analysis of duplicates and singletons (for the raw data, see Supplementary table, Supplementary Material online). We found that the proportion of essential genes in both singletons and duplicates in general increases with increasing age in the five age groups, with some fluctuations (Supplementary fighttp://www.mbe.oxfordjournals.org/lookup/suppl/doi:10.1093/molbev/mss014/-/DC1. S4, Supplementary Material online). However, the PE in duplicates is not always lower than in singletons of similar age, and the differences are not statistically significant (Fisher's exact test, all comparisons P > 0.05; Supplementary table, Supplementary Material online), probably due to the small size of the data set. Since only a small number (∼3.2%) of D. melanogaster genes have been tested (Chen et al. 2010), our findings regarding D. melanogaster are not yet conclusive.
Materials and Methods
We determined the phyletic origins of genes from yeast, mouse, and fly using a method described in Wolf et al. (2009) with modifications (for more details, see Supplementary text, Supplementary Material online, and for the results, Supplementary table, Supplementary Material online). We separated genes into singletons and duplicates as previously described (Liao and Zhang 2007; Makino et al. 2009). We grouped duplicates into gene families using a clustering-based method (Markov cluster algorithm [MCL]; Enright et al. 2002) and then used the most ancient origin of all members as the age of the corresponding family.
We obtained the phenotypic data for the three species from online gene essentiality database (Chen et al. 2012), which were originally published by the Saccharomyces Genome Deletion Project (Cherry et al. 1997), the Mouse Genome Informatics (Blake et al. 2011), and the authors of Chen et al. (2010), respectively. We restricted further analyses to genes that were tested in these phenotypic data sets.
Supplementary Material
Supplementary text, Supplementary figures, and Supplementary table are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
SysteMtb grant #241587 to P.B. provided funding for the open access license.
References
- Blake JA, Bult CJ, Kadin JA, Richardson JE, Eppig JT Mouse Genome Database G. The Mouse Genome Database (MGD): premier model organism resource for mammalian genomics and genetics. Nucleic Acids Res. 2011;39:D842–D848. doi: 10.1093/nar/gkq1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M. New genes in Drosophila quickly become essential. Science. 2010;330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WH, Minguez P, Lercher MJ, Bork P. OGEE: an online gene essentiality database. Nucleic Acids Res. 2012;40:D901–D906. doi: 10.1093/nar/gkr986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Ball C, Weng S, et al. (11 co-authors) Genetic and physical maps of Saccharomyces cerevisiae. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Hao L, Ge X, Wan H, Hu S, Lercher M, Yu J, Chen W-H. Human functional genetic studies are biased against the medically most relevant primate-specific genes. BMC Evol Biol. 2010;10:316. doi: 10.1186/1471-2148-10-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Kelly DE, Lamb DC, Kelly SL. Genome-wide generation of yeast gene deletion strains. Comp Funct Genomics. 2001;2:236–242. doi: 10.1002/cfg.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Li WH. Functional compensation by duplicated genes in mouse. Trends Genet. 2009;25:441–442. doi: 10.1016/j.tig.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B-Y, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Makino T, Hokamp K, McLysaght A. The complex relationship of gene duplication and essentiality. Trends Genet. 2009;25:152–155. doi: 10.1016/j.tig.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Simpson EH. The interpretation of interaction in contingency tables. J R Stat Soc Ser B. 1951;13:238–241. [Google Scholar]
- Su Z, Gu X. Predicting the proportion of essential genes in mouse duplicates based on biased mouse knockout genes. J Mol Evol. 2008;67:705–709. doi: 10.1007/s00239-008-9170-9. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, Koonin EV, Lipman DJ. Inaugural article: the universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc Natl Acad Sci U S A. 2009;106:7273–7280. doi: 10.1073/pnas.0901808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotenko E, Mestre J, O'Leary DP, Przytycka TM. Why do hubs in the yeast protein interaction network tend to be essential: reexamining the connection between the network topology and essentiality. PLoS Comput Biol. 2008;4:e1000140. doi: 10.1371/journal.pcbi.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.