Abstract
Expression variation is widespread between species. The ability to distinguish regulatory change driven by natural selection from the consequences of neutral drift remains a major challenge in comparative genomics. In this work, we used observations of mRNA expression and promoter sequence to analyze signatures of selection on groups of functionally related genes in Saccharomycete yeasts. In a survey of gene regulons with expression divergence between Saccharomyces cerevisiae and S. paradoxus, we found that most were subject to variation in trans-regulatory factors that provided no evidence against a neutral model. However, we identified one regulon of membrane protein genes controlled by unlinked cis- and trans-acting determinants with coherent effects on gene expression, consistent with a history of directional, nonneutral evolution. For this membrane protein group, S. paradoxus alleles at regulatory loci were associated with elevated expression and altered stress responsiveness relative to other yeasts. In a phylogenetic comparison of promoter sequences of the membrane protein genes between species, the S. paradoxus lineage was distinguished by a short branch length, indicative of strong selective constraint. Likewise, sequence variants within the S. paradoxus population, but not across strains of other yeasts, were skewed toward low frequencies in promoters of genes in the membrane protein regulon, again reflecting strong purifying selection. Our results support a model in which a distinct expression program for the membrane protein genes in S. paradoxus has been preferentially maintained by negative selection as the result of an increased importance to organismal fitness. These findings illustrate the power of integrating expression- and sequence-based tests of natural selection in the study of evolutionary forces that underlie regulatory change.
Keywords: regulatory variation, natural selection, Saccharomyces
Introduction
An outstanding question in comparative genomics is the evolutionary importance of gene expression differences between genetically distinct individuals. Comparative expression studies in many taxa have made clear that a large fraction of the transcriptome varies in expression level between species (Hodgins-Davis and Townsend 2009; Wilson and Odom 2009; Dowell 2010), but the vast majority of this divergence is expected to be the product of neutral genetic drift. Due in part to the complexities of detecting selection from observations of regulatory sequence and gene expression (Kohn et al. 2004; Blekhman et al. 2008; Torgerson et al. 2009; Bullard et al. 2010; Fraser et al. 2010; He et al. 2011), the prevalence and the mechanisms of natural selection on regulatory change remain incompletely understood.
In the search for an evolutionary logic underlying species changes in gene expression, many analyses have focused on trends across groups of functionally related genes. One powerful approach has been to trace gains or losses of a given cis-regulatory motif in the promoter sequences of genes in a pathway (Gasch et al. 2004; Ihmels et al. 2005; Ludwig et al. 2005; Tanay et al. 2005; Tsong et al. 2006; Borneman et al. 2007; Hinman et al. 2007; Hogues et al. 2008; Tuch et al. 2008; Perez and Groisman 2009; Piasecki et al. 2010), where each cis-regulatory change has arisen via an independent genetic event. Such analyses have highlighted a complex, polygenic mechanism for the evolution of a given regulatory program, involving a suite of subtly acting variants in unlinked genes that function together. As a complementary approach for the study of polygenic regulatory evolution, we recently developed an expression-based strategy (Bullard et al. 2010) to identify cases in which a species has accumulated cis-regulatory variants that predominantly up-regulate, or predominantly down-regulate, genes of common function. Given the neutral expectation of equivalent numbers of variants acting in one direction versus another (Orr 1998), an imbalance in the signs of cis-regulatory effects represents a key line of evidence for a change in selective pressure between species on the pathway: positive selection or a relaxation of purifying selection in a given lineage could give rise to concerted regulatory change across genes of related function (Bullard et al. 2010).
To date, a primary challenge of this and related expression-based tests for polygenic regulatory evolution (Bullard et al. 2010; Fraser et al. 2010, 2011) has been the ability to interpret the results in the context of classical sequence-based signatures of natural selection. In this work, we set out to develop an analysis pipeline to study cases of pathway-level regulatory evolution, harnessing both expression and DNA sequence data. To provide a tractable and data-rich model system for our analysis, we chose the well-characterized Saccharomycete yeasts.
Materials and Methods
Selection of Regulons
We downloaded regulons from Gasch et al. (2004) and, from each, eliminated genes annotated as dubious open reading frames (ORFs) (www.yeastgenome.org). We then filtered groups as follows. Any groups with fewer than 10 genes were eliminated from further analysis. Of the remainder, we compared the composition of each pair of gene groups and, if any two groups overlapped by more than 30%, we eliminated the smaller group from consideration. The final data set comprised 104 regulons.
Expression-Based Tests for Directional Regulatory Evolution between Saccharomyces paradoxus and S. cerevisiae
We downloaded estimates of the cis- and trans-acting contributions to gene expression divergence between S. cerevisiae and S. paradoxus from Tirosh et al. (2009). These data derived from two measurements for each gene: one of the log2 of the ratio of expression of the S. cerevisiae allele to the expression of the S. paradoxus allele in an interspecific hybrid, reflecting the cis-acting contribution to expression variation between the species (RSc/Sp;cis), and the other of the total difference in expression between the parent species when grown independently in culture, RSc/Sp;total = log2(expressionScer/expressionSpar). As detailed in Tirosh et al. (2009), the trans-acting contribution to variation for a given gene, RSc/Sp;trans, is estimated as RSc/Sp;total − RSc/Sp;cis; thus, from the RSc/Sp;cis and RSc/Sp;trans values publicly available from Tirosh et al. (2009), we calculated RSc/Sp;total for each gene. To first survey regulons for directional expression change between species without distinguishing between cis- and trans-acting mechanisms, we used the sum of expression effects across a gene group as a statistic to assess the coherence of regulatory divergence. Specifically, for each regulon, we summed RSc/Sp;total across all genes in the regulon; to evaluate significance, we conducted a two-sided test against a null distribution of 5,000 groups comprised of randomly chosen genes from the genome, in which each such null group was of the same size as the regulon of interest. In analysis of table 1, the experiment-wise false positive count for this test was calculated as the product of the number of groups tested (104) and the P value threshold used (0.0204). For each regulon, we then conducted analogous tests for imbalance in cis- and trans-acting contributions to expression between the species using RSc/Sp;cis and RSc/Sp;trans, respectively, except that a one-sided P value was calculated in each case (upper tailed, if ΣRSc/Sp;total > 0 and lower tailed, if otherwise). The experiment-wise significance for the membrane protein group for the latter tests was calculated as the product of the number of groups tested (11) and the P value for the membrane protein regulon (0.0056 and 0.001, respectively). In supplementary table 1 (Supplementary Material online), we also converted each RSc/Sp;cis value to a sign statistic (+1, if S. cerevisiae was associated with higher expression and −1, otherwise) and repeated resampling tests as above, using a one-sided test in each case.
Table 1.
Tests for Directional Regulatory Change between Saccharomyces cerevisiae and S. paradoxus Expression in Coregulated Gene Groups.
| Namea | Number of Genes | ΣRSc/Sp;totalb | P Totalc | P cisd | P transe | Annotation |
|---|---|---|---|---|---|---|
| Cluster_adata-CalciumSpecific* | 47 | −25.8 | <0.0002 | 0.0056 | 0.001 | Membrane proteins |
| Node 7 | 183 | 85.8 | <0.0002 | 0.0506 | <0.0002 | Heat-shock proteins, stress |
| Cluster_PUF3 | 193 | −46.3 | <0.0002 | 0.2372 | <0.0002 | Mitochondrion |
| Node 73 | 392 | −140.1 | <0.0002 | 0.4644 | <0.0002 | Ribosome, translation |
| Cluster_ManualHAP1targets | 36 | −28.2 | <0.0002 | 0.5044 | <0.0002 | Respiration |
| Cluster_HAP5 | 24 | −12.6 | 0.0072 | 0.0114 | 0.108 | Respiration |
| Cluster_Vacuole | 56 | 17.1 | 0.0114 | 0.019 | 0.0434 | Vacuole |
| Cluster_PHD1 | 27 | −12.2 | 0.0114 | 0.0618 | 0.0232 | Stress, signaling |
| Cluster_SWI4 | 48 | −16.1 | 0.0136 | 0.071 | 0.0252 | Cell cycle |
| Node 6 | 10 | 7.0 | 0.0178 | 0.1296 | 0.003 | Protein modification and breakdown |
| Cluster_ARG80 | 18 | −9.1 | 0.0204 | 0.1622 | 0.01 | Amino acid biosynthesis |
a Gene groups defined by Gasch et al. (2004). Results from analysis of the membrane protein regulon are denoted with an asterisk.
b Sum, across genes in the indicated regulon, of the log2 ratio of expression in S. cerevisiae to expression in S. paradoxus, when each species was grown in independent culture (RSc/Sp;total).
c Significance of a two-sided resampling test relative to the genomic null for an extreme value of the sum, across genes in the indicated regulon, of the log2 ratio of expression in S. cerevisiae to expression in S. paradoxus, when each species was grown in independent culture (RSc/Sp;total). Shown are all groups meeting a significance level where 2.1 groups would be expected under the null.
d Significance of a one-sided resampling test relative to the genomic null for an extreme value of the sum, across genes in the indicated regulon, of the log2 ratio of the expression of the S. cerevisiae allele to that of the S. paradoxus allele in an interspecific hybrid.
e Significance of a one-sided resampling test relative to the genomic null for an extreme value of the sum, across genes in the indicated regulon, of the log2 ratio of the effects of trans-acting regulatory variation between S. cerevisiae and S. paradoxus, where the latter is derived from expression measurements of the interspecific hybrid and of parent strains grown independently (Tirosh et al. 2009).
We evaluated the significance of the pattern of reinforcement between cis- and trans-acting variants impinging on the membrane protein regulon as follows. We assembled, by resampling from among all genes in the genome, 5,000 null sets of genes that mirrored the true regulon with respect to size and to the proportion of genes subject to trans-acting variants associated with elevated expression in S. paradoxus versus elevated expression in S. cerevisiae. In each such null group, we tabulated the number of genes subject to cis-regulatory variation at which the S. paradoxus allele was associated with higher expression. We then compared the count of such genes from the real regulon against this null distribution to obtain a one-sided P value.
Expression-Based Test for Directional Regulatory Evolution between S. paradoxus and S. bayanus
We downloaded measurements of RSb/Sc, the cis-acting contribution to regulatory variation between S. bayanus and S. cerevisiae, for each gene, from Bullard et al. (2010). From this data set, we eliminated from consideration all genes that were not represented in the data set comparing S. paradoxus and S. cerevisiae (Tirosh et al. 2009), as well as those annotated as dubious ORFs. We then calculated RSb/Sp, the sum of RSb/Sc and RSc/Sp, for each gene, and used this quantity in a one-sided resampling test for directional cis-regulatory evolution as above.
Expression-Based Test for Directional Regulatory Evolution between S. paradoxus and S. kudriavzevii
We mated S. cerevisiae BY4716 (Open Biosystems) and S. kudriavzevii strain JRY9187 (Scannell et al. 2011) to generate a diploid hybrid. To generate measurements of RSk/Sc, the log of the fold change in allele-specific expression levels between S. cerevisiae and S. kudriavzevii, for each gene, we conducted yeast culture, RNA isolation, and mapping as previously described (Bullard et al. 2010), except that one sequencing lane was used from one biological replicate, with the total RNA-seq data set comprising 7.04 million mapped reads. We eliminated from consideration all genes that were not represented in the data set comparing S. paradoxus and S. cerevisiae (Tirosh et al. 2009), as well as those annotated as dubious ORFs. We then formulated RSk/Sp, the sum of RSk/Sc and RSc/Sp, for each gene and used this quantity in a one-sided resampling test for directional cis-regulatory evolution as above.
Interspecific Sequence Analyses
Reference sequences for S. cerevisiae, S. paradoxus, S. bayanus, S. mikatae, and S. kudriavzevii were downloaded from www.saccharomycessensustricto.org (Scannell et al. 2011). For each species, we extracted the region 1,000 bp upstream of each gene for all genes not annotated as dubious ORFs, and we eliminated any gene that was not annotated in each of the five species in Scannell et al. (2011). For each such set of promoter sequences from a given gene, a five species alignment was generated using Fast Statistical Alignment (Bradley et al. 2009). Alignments in which >500 sites were gaps or ambiguous base calls were eliminated from analysis, yielding a final data set of promoters from 4,295 genes. For branch length inference, the aligned promoters from the genes of the membrane protein regulon were concatenated and used as input to the baseml module of PAML (Yang 2007) along with a fixed unrooted Saccharomycete tree topology (Scannell et al. 2011). A general time reversible substitution model was used without a molecular clock, and sites in the alignment that contained gaps or ambiguous data were not used in the analysis. To evaluate statistical significance of branch lengths for the membrane protein regulon in supplementary table 2 (Supplementary Material online), the concatenation of promoter alignments and branch length inference was repeated on each of 10,000 randomly sampled gene groups of the same size as the membrane protein group, and a one-sided empirical P value for each branch was then calculated as the proportion of such null groups with branch length greater than or equal to the true value inferred from the real membrane protein genes. A complementary analysis applying branch length inference to aligned sequences for each promoter separately yielded identical results (data not shown). For supplementary fig. S2 (Supplementary Material online), sequences of 5,167 open reading frames were downloaded from Scannell et al. (2011), aligned, and concatenated, and branch lengths were inferred as above; the tree image was generated using MEGA (Tamura et al. 2011).
Promoter Data from Saccharomyces Population Genomic Sequences and Estimation of Selection Coefficients
Chromosome alignments for strains of the European clade of S. paradoxus and the wine/European clade of S. cerevisiae populations were downloaded from Liti et al. (2009) and accessed using the alicat.pl script. Sites with an error probability of >0.0001 were eliminated from analyses, as were genes whose promoter alignments contained >5 segregating sites. Contig alignments for strains from the Portuguese clade of S. kudriavzevii were downloaded from Hittinger et al. (2010). For each species, we extracted the region 1,000 bp upstream of each gene, for all genes not annotated as dubious ORFs or in the case of S. kudriavzevii, the longest possible upstream region up to 1,000 bp for which sequence data were available. The final data sets comprised promoters for 5,284, 5,268, and 2,748 genes in S. paradoxus, S. cerevisiae, and S. kudriavzevii, respectively, harboring 2,670, 1,908, and 7,356 single-nucleotide polymorphisms, respectively.
To establish a data set for a given species in which the number of strain genomes without missing data was the same for each polymorphic site (Elyashiv et al. 2010), we set a cutoff strain count c equal to 15, 6, and 10 for S. paradoxus, S. cerevisiae, and S. kudriavzevii, respectively. If a given variant site had fewer than c strains with nonmissing allele calls, we eliminated it from consideration and if a site had more than c strains with nonmissing allele calls, we randomly subsampled alleles from c of these strains for inclusion in the analysis. Given the resulting allele set for each promoter for a given species, we tabulated allele frequencies at all sites of single-nucleotide polymorphism in the complete set of promoters for a given gene group of interest. We implemented the Poisson Random Field (PRF) method (Bustamante et al. 2001) in R (www.r-project.org), using the nlm function to find maximum-likelihood estimates of selection coefficients γ. For a given population, γ inferred from the whole-genome set of promoters was evaluated against the null hypothesis of a value of zero; γ from the membrane protein gene promoters was evaluated against the whole-genome estimate. In each case, likelihood ratio test P values were calculated assuming a chi-square distribution of the test statistic with one degree of freedom. As a complementary approach, we evaluated the γ inferred from the promoters of the membrane protein regulon via resampling. For this purpose, we generated 10,000 null gene sets by randomly selecting genes from the genome, and using the promoters of each such set, we estimated a maximum-likelihood selection coefficient as above; a one-sided empirical P value was then calculated as the proportion of null groups with γ as negative or more so than the true value inferred from the real membrane protein genes. We also tested for a distinction between raw allele frequencies in promoters of the membrane protein regulon and those of the rest of the genome, by computing the mean allele frequency across all segregating sites in the former and comparing against analogous means of 10,000 null gene sets as above. Conclusions from all analyses were unaffected by changes to the cutoff used in filtering promoters based on the number of segregating sites (data not shown).
Expression-Based Tests for Divergence in Stress Responsiveness between S. cerevisiae and S. paradoxus
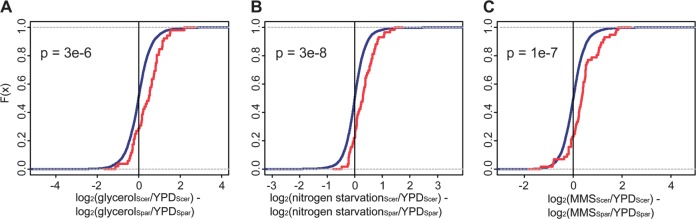
We downloaded microarray measurements of expression divergence between Saccharomyces species when grown in independent culture in stress conditions from Tirosh et al. (2006) and excluded dubious ORFs from consideration. We analyzed the transcriptional response to each of five conditions: growth on glycerol as a carbon source, nitrogen starvation, heat shock, and treatment with methyl methanesulfonate (MMS) and H2O2. For each gene and each stress, at each of six time points, we calculated the expression response as the log2 fold-change relative to the rich-medium yeast peptone dextrose, as an average across all probes on the microarray affiliated with the gene. For each species, in supplementary fig. S1 (Supplementary Material online), we compared the distributions of response values between the membrane protein regulon and the rest of the genome using a two-sided Wilcoxon test. In figure 3, we calculated the difference between species in the stress response for each gene as log2(expression in stressScer/expression in YPDScer) − log2(expression in stressSpar/expression in YPDSpar), and we compared these interspecies differences in responsiveness between the membrane protein regulon and the rest of the genome using a one-sided Wilcoxon test.
Fig. 3.
Interspecies differences between Saccharomycetes in stress responsiveness of the membrane protein regulon. Each trace represents a cumulative distribution function (F[x]) of the difference between Saccharomyces cerevisiae and S. paradoxus in induction of gene expression after stress exposure, relative to expression in the rich-medium YPD (Tirosh et al. 2006). In each panel, the x axis reports log2(expression in stressScer/expression in YPDScer) − log2(expression in stressSpar/expression in YPDSpar). The blue trace reports the distribution across all genes and the red trace reports that across the membrane protein regulon (fig. 1 and supplementary table 1, Supplementary Material online). (A) Switch to glycerol as the carbon source, 20-min time point. (B) Nitrogen starvation, 45-min time point. (C) Treatment with methyl methanesulfonate, 1.5-h time point. P, one-sided P value from a Wilcoxon test comparing expression changes in the membrane regulon to changes across the genome.
Results
We first sought to screen groups of functionally related genes for coherent patterns of regulatory change between S. cerevisiae and S. paradoxus. For this purpose, we used measurements of gene expression in each species grown in rich medium (Tirosh et al. 2009). In each of a set of gene groups defined on the basis of coregulation in S. cerevisiae (Gasch et al. 2004), we assessed the tendency for one species to express the genes of the pathway at predominantly higher, or predominantly lower, levels relative to the other species. The results revealed 11 gene groups with evidence for coherent regulatory variation between S. cerevisiae and S. paradoxus, at a level where ∼2 groups were expected under a null model of independent evolution across genes (table 1). Top-scoring gene groups in this analysis included heat-shock and stress-response genes, translation genes, cell cycle factors, and protein processing genes, indicating that a range of stress-response and housekeeping functions have been subject to directional expression change between the two yeasts.
Because coherent differential expression between species in a group of functionally related genes does not provide evidence for natural selection per se, we next analyzed the mechanisms of expression change between S. cerevisiae and S. paradoxus in regulons. For this purpose, we used expression measurements from a stable hybrid diploid formed by mating the two species (Tirosh et al. 2009). Combining observations of allele-specific expression in the hybrid with analysis of the species when grown independently allows regulatory divergence to be partitioned into cis- and trans-acting contributions for each gene. In each top-scoring gene group from our screen for coherent expression change, we tested a model in which one species' cis-regulatory alleles drove expression in the same direction relative to the allele from the other species (Bullard et al. 2010), and we also carried out an analogous test using trans-regulatory effects. Across the groups, statistical significance measures were more striking for the latter test method (table 1); thus, in the divergence between this pair of Saccharomycetes, detectable cases of coherent regulatory variation most often followed a model in which genetic change at trans-acting factors affects expression of a set of downstream targets in the same direction.
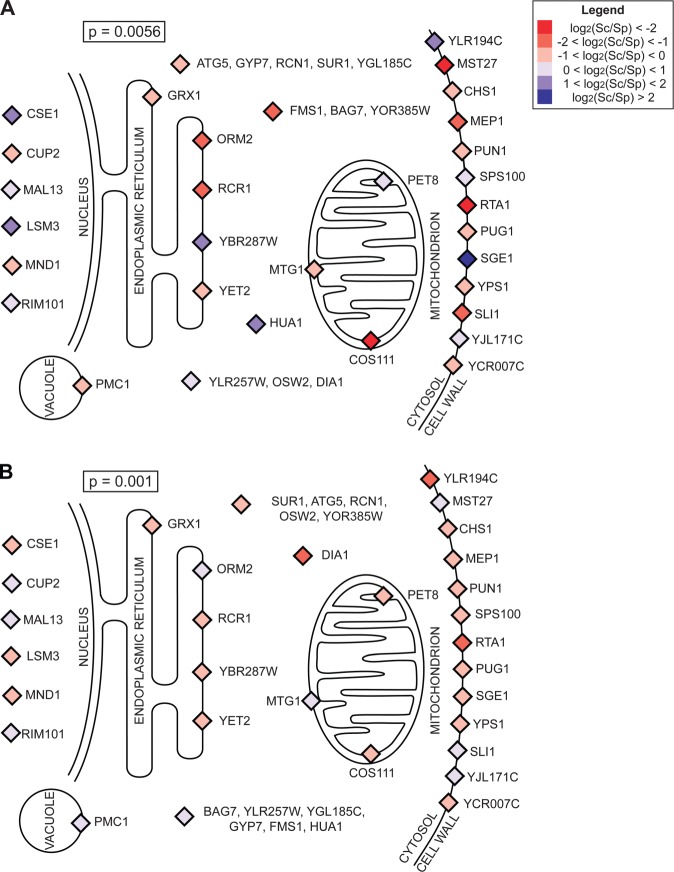
To investigate the role of nonneutral evolutionary forces underlying expression variation between S. cerevisiae and S. paradoxus, we focused on a set of genes mediating membrane protein trafficking and function and membrane lipid composition (fig. 1 and supplementary table 1, Supplementary Material online). This gene group showed strong evidence for coherent regulatory change of cis-acting factors and of those acting in trans (reaching a significance level in each case where <0.1 group would be expected under the null; table 1). For both cis- and trans-acting determinants of expression of this gene group, the S. paradoxus allele at the respective locus conferred high expression relative to that of S. cerevisiae (fig. 1), a pattern of reinforcement between the two mechanisms of regulatory change rarely observed in null data (resampling P = 0.04; see Materials and Methods). The presence of many unlinked cis-regulatory variants between species acting in the same direction is unlikely under a neutral model (Bullard et al. 2010), and indicative of a change in selective pressure on the regulation of the membrane protein set. Additionally, the reinforcement between cis- and trans-mediated changes impinging on this regulon provides evidence against a model in which compensatory variants in a given species have arisen, in regulators and their targets, to preserve a constant degree of DNA-binding activity or transcription.
Fig. 1.
Directional regulatory change between Saccharomycetes in the membrane protein regulon. Each colored element represents regulatory changes between Saccharomyces cerevisiae and S. paradoxus in rich-medium (Tirosh et al. 2009) at one membrane protein gene, overlaid on a cartoon localization of the encoded protein. P, proportion of randomly sampled gene groups showing regulatory changes whose directions were at least as coherent as those observed in the membrane regulon (table 1). (A) Signs of cis-regulatory effects. Orange, the allele from S. paradoxus was the most highly expressed in an interspecific hybrid; blue, the S. cerevisiae allele was most highly expressed. (B) Signs of trans-regulatory effects, which derive from expression measurements of interspecific hybrids and of parental strains grown independently (Tirosh et al. 2009). Orange, the regulatory allele from S. paradoxus was associated with higher expression; blue, the S. cerevisiae allele was associated with higher expression.
We next aimed to trace the evolutionary history of expression change in the membrane protein regulon across Saccharomycetes. We developed an analysis scheme harnessing allele-specific expression data sets from multiple interspecies yeast hybrids grown in rich medium. We first used RNA-seq to assess allele-specific expression for each gene in turn, in a diploid hybrid formed from the mating of S. kudriavzevii and S. cerevisiae, and we also tabulated the analogous measurements from a hybrid of the latter and S. bayanus (Bullard et al. 2010). We then integrated these data with allele-specific expression measurements from the hybrid between S. cerevisiae and S. paradoxus (Tirosh et al. 2009), and, for each species comparison involving S. paradoxus, we tested for an imbalance in the direction of cis-regulatory changes in the membrane protein regulon. The results mirrored our analysis of S. cerevisiae (supplementary table 1, Supplementary Material online): cis-regulatory variants at the membrane protein genes were associated with high expression in S. paradoxus relative to S. bayanus (P = 0.007) and to S. kudriavzevii (P = 0.01). Conclusions were unchanged when we analyzed the qualitative signs of cis-regulatory variation between species, rather than the quantitative effects (supplementary table 1, Supplementary Material online). We conclude that the cis-regulatory alleles harbored by S. paradoxus at the genes of the membrane protein regulon confer elevated expression relative to three other Saccharomycete species, providing strong evidence for a history of distinct selective pressure at these loci in S. paradoxus.
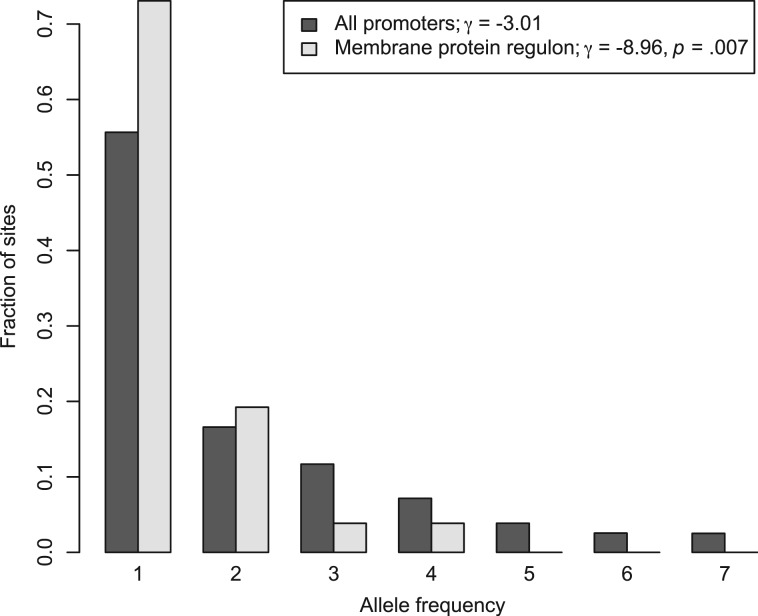
Given the regulatory divergence between S. paradoxus and other yeasts in the membrane protein group, we hypothesized that sequence-based signatures of selection would exhibit distinct characteristics across the genes of the regulon in the former species. To test this, we first sought to estimate the strength of selection on promoters of the membrane protein regulon, using genome sequences of isolates from the European population of S. paradoxus (Liti et al. 2009), the wine/European population of S. cerevisiae (Liti et al. 2009), and the Portuguese population of S. kudriavzevii (Hittinger et al. 2010). For each population, we tabulated allele frequencies at single-nucleotide polymorphisms in all gene promoters, and we used the resulting folded site frequency spectra as input into the PRF method for estimation of population-scaled selection coefficients γ (Bustamante et al. 2001). As expected (Arthur and Ruvinsky 2011; Vishnoi et al. 2011), for the whole-genome set of promoters in each species, likelihood ratio testing strongly rejected a model of neutrality in favor of an inference of purifying selection (S. paradoxus: γ = −2.64, P = 7.87 × 10−122; S. cerevisiae: γ = −1.1, P = 3.2 × 10−7; S. kudriavzevii: γ = −0.72, P = 4.1 × 10−17). In S. paradoxus, the membrane protein gene promoters harbored an excess of low-frequency alleles compared with the genomic promoter set, reflecting the stronger action of purifying selection in culling variants from the population (fig. 2). Likelihood ratio testing confirmed the more negative selection coefficient for the membrane protein gene promoters as a better fit to the data than the parameter value inferred from the whole-genome promoter set (fig. 2). As an independent test, we conducted a resampling-based analysis of selection coefficients (see Materials and Methods); the results confirmed the difference between the membrane protein group and the rest of the genome for S. paradoxus (P = 0.03). By contrast, in S. cerevisiae and S. kudriavzevii, selection coefficients inferred from promoters of the membrane protein regulon were indistinguishable from those of the rest of the genome (likelihood ratio test P = 0.4 and 0.94 and resampling P = 0.2 and 0.54, respectively). Analyses of raw allele frequencies rather than selection coefficients (see Materials and Methods) also confirmed the difference between the membrane protein regulon and the rest of the genome in S. paradoxus (resampling P = 0.03) but not S. cerevisiae or S. kudriavzevii (P = 0.66 and 0.43, respectively). Thus, in S. paradoxus alone, allele frequencies were indicative of tight constraint on the membrane protein group, reflecting a particular importance of regulation at these loci in the niche occupied by this species.
Fig. 2.
Increased strength of purifying selection in the membrane protein regulon in Saccharomyces paradoxus. Each shade of gray represents analysis of single-nucleotide polymorphisms in one set of promoter sequences from European strains of S. paradoxus (Liti et al. 2009). Each set of bars represents a histogram of frequencies of minor alleles. All promoters, analysis of promoter regions from all genes in the genome. Membrane protein regulon, analysis of promoter regions from the membrane protein gene group (fig. 1 and supplementary table 1, Supplementary Material online). γ, population-scaled selection coefficient inferred from the allele frequency spectrum from the indicated set of promoters. P, significance of the distinction between the genomic selection coefficient and the coefficient inferred from the membrane protein group, according to likelihood ratio testing. Results are based on a data set of resampled alleles to ensure fully informative sequence data at each site (see Materials and Methods).
To substantiate this conclusion by an independent method, we next applied a phylogenetic strategy, using promoter sequences from five Saccharomycete-type strains (Scannell et al. 2011). We inferred evolutionary rates for each branch of the Saccharomyces phylogeny (supplementary fig. S2, Supplementary Material online) with PAML (Yang 2007), for promoters of genes in the membrane protein regulon, and we compared the rates along the terminal branches to those inferred from promoters of randomly sampled gene groups. In S. paradoxus, but none of the other Saccharomycetes, the genes of the membrane protein regulon exhibited shorter branch lengths than the genomic null, corresponding to a slower rate of evolution (supplementary table 2, Supplementary Material online). Additionally, branch-specific rates for the membrane protein regulon were elevated in the distantly related species S. bayanus, suggesting a separate trend for a change in selection pressure in the latter (supplementary table 2, Supplementary Material online). We conclude that the rate of evolution has been constrained at the promoters of the membrane protein genes in S. paradoxus, echoing results from population genetic analyses (fig. 2) and lending further support to the inference that regulation of these genes has been particularly important in the life history of S. paradoxus.
The membrane protein regulon includes a number of genes known to mediate response to cell wall and salt stress in S. cerevisiae (supplementary table 1, Supplementary Material online). We expected that expression of the regulon was likely to be responsive to stress, and given the change in expression between species in rich-medium conditions (fig. 1 and supplementary table 1, Supplementary Material online), we hypothesized that aspects of stress regulation of the membrane protein genes would have diverged between S. paradoxus and other yeasts. To test these hypotheses, we used gene expression measurements from S. cerevisiae and S. paradoxus grown in nutrient starvation conditions and during exposure to toxic agents (Tirosh et al. 2006). As predicted, the membrane protein regulon stood out relative to the rest of the genome for its expression regulation across stress conditions (fig. 3 and supplementary fig. S1, Supplementary Material online): both S. cerevisiae and S. paradoxus induced the regulon under nitrogen starvation and growth on glycerol, and repressed it in response to the DNA-damaging agent MMS. Critically, however, expression response to environmental treatments was markedly different between the species (fig. 3). Saccharomyces paradoxus repressed membrane gene expression to a greater extent in MMS than did S. cerevisiae and induced the membrane genes less in other conditions than did S. cerevisiae. Standard sequence search analyses had little power to detect sequence motif changes between species in promoters of the membrane protein regulon (data not shown), as expected if the complexity of regulatory inputs at these loci served to obscure the signal of gain or loss of any given motif. In light of our observation of elevated expression levels of the membrane protein genes in rich medium in S. paradoxus (fig. 1), we conclude that the two species ultimately achieve similar regulatory programs for this gene group in stress conditions, starting from distinct set points in the rich-medium basal state. Since the membrane protein genes are expressed in rich medium at a higher level in S. paradoxus than in S. cerevisiae, it follows that S. paradoxus requires more dramatic repression upon a switch to MMS treatment and less avid induction during nitrogen limitation and growth on glycerol. Such patterns of divergent stress response further highlight the evolutionary dynamics of regulation of the membrane protein gene group between yeast species.
Discussion
Distinguishing between natural selection and neutral drift as forces underlying regulatory variation remains a major challenge in evolutionary biology. Analysis of directional expression change in gene groups of common function can be a powerful tool toward this end. To date, however, methods have been at a premium for the incorporation of expression-based tests for selection with those based on DNA sequence. In this work, we have used Saccharomycete yeasts as a test bed for these complementary paradigms in the dissection of pathway-level regulatory change.
We have shown that, in a comparison of S. cerevisiae and S. paradoxus, most instances of directional regulatory change in groups of functionally related genes can be explained by variation in trans-acting factors. For a given such pathway, an appealing mechanism invokes a variant in a single upstream factor, or a small number of such loci, driving directional changes in expression of downstream targets. Under this model, the relatively short waiting time required for a species to accumulate mutations in a small number of trans-acting factors would be consistent with the prevalence of trans-acting variation we observe across pathways. Importantly, any case of simple Mendelian trans-acting regulatory change provides no a priori means to reject a neutral model (Denver et al. 2005; Rockman and Kruglyak 2006). We hypothesize that many of the trans-regulatory changes in the pathways we study may be present in yeast genomes as a consequence of drift.
As a model system for the study of nonneutral regulatory evolution, we focused on a group of genes involved in membrane protein function and trafficking and membrane lipid composition. Cis- and trans-regulatory variants harbored by S. paradoxus at these loci were associated with upregulation in rich-medium conditions and altered stress responsiveness relative to other yeasts. Our observation of multiple independent cis-regulatory variants driving expression of the membrane protein genes in the same direction is unlikely under neutrality (Bullard et al. 2010) and provides evidence for a change in selective pressure in S. paradoxus. Additionally, the fact that alleles of cis- and trans-acting factors in S. paradoxus affect this gene set in the same direction argues against a model in which target genes have accumulated locally acting variants to compensate for changes in a soluble regulatory factor. Instances of such compensation have been common during the divergence of yeast species (Tirosh et al. 2009; Kuo et al. 2010; Lavoie et al. 2010; Baker et al. 2011), consistent with the hypothesis that directional selection rarely underlies these patterns (Takahasi et al. 2011). By contrast, an inference of a change in selective pressure in a given species becomes strongest when cis- and trans-acting variants impinging on a suite of genes drive expression in a consistent direction, as has been reported in a handful of previous studies (Tsong et al. 2006; Fraser et al. 2010) and in the present observations of the membrane protein group.
A priori, polygenic directional expression change across the membrane protein regulon could be the result of either adaptation or relaxed purifying selection in S. paradoxus. Our findings provide strong evidence against the latter hypothesis. Inference using membrane protein promoters from S. paradoxus revealed short branch lengths on the Saccharomyces phylogenetic tree, and a strongly negative selection coefficient within species, relative to genomic controls. These findings dovetail with previous reports of shifts in the strength of purifying selection between gene groups (Vishnoi et al. 2011) and between populations (Elyashiv et al. 2010) in yeast. How might we reconcile the distinct expression program of the membrane protein genes in S. paradoxus with the evidence for tight selective constraint? Our results are consistent with either of two possible interpretations. The S. paradoxus expression program could represent an ancestral state from which all other species have diverged; under this model, ancestral alleles have been subject to tight constraint in S. paradoxus as a consequence of particular importance to organismal fitness relative to that in the niches of other yeasts. Alternatively, the S. paradoxus expression program could represent a derived state that arose through a series of selective sweeps and since then has been preferentially maintained by negative selection. As we show in the Appendix, the latter model could manifest as a reduced number of substitutions along the S. paradoxus lineage if the period of adaptive evolution were sufficiently ancient, a plausible scenario for the membrane protein group given that S. paradoxus diverged from its closest relative, S. cerevisiae, ∼7 Ma (Scannell et al. 2011). Under either model, it is tempting to speculate that the distinct regulatory program we have uncovered in S. paradoxus affects acute tolerance to environmental insults in the wild, as Saccharomycetes respond to many stress treatments by upregulating the membrane protein gene group (supplementary fig. S1, Supplementary Material online). The elevated constitutive expression of this regulon in S. paradoxus is in keeping with other switches between constitutive and stress-responsive expression observed in yeasts (Tirosh et al. 2011), whose fitness advantages remain an area of active research.
Decades of work in genetic mapping have revealed suites of unlinked, often weakly acting loci to be the rule rather than the exception in explaining trait variation within species (Weiss 2008). The notion of genetic complexity has also become increasingly relevant in comparisons between species, as genomic methods enable studies of polygenic evolution over long timescales (Gasch et al. 2004; Ihmels et al. 2005; Ludwig et al. 2005; Tanay et al. 2005; Tsong et al. 2006; Borneman et al. 2007; Hinman et al. 2007; Hogues et al. 2008; Tuch et al. 2008; Perez and Groisman 2009; Lavoie et al. 2010; Piasecki et al. 2010). Understanding the evolutionary pressures at play in polygenic pathway evolution will require both observational molecular approaches and formal tests of natural selection, which together will continue to accelerate the dissection of the genetic basis of evolutionary novelties.
Supplementary Material
Supplementary figures S1 and S2 and tables 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Chris Hittinger for providing the S. kudriavzevii strain; Angela Kaczmarczyk, Ivy McDaniel, and Oh Kyu Yoon for strain handling and culture; Adam Boyko for providing PRFREQ software; Jeremiah Degenhardt, Dan Pollard, Eva Stukenbrock, and Oliver Zill for discussions; and Chris Ellison, Nicole King, Daniel Ortiz-Barrientos, Hua Tang, John Taylor, and two anonymous reviewers for helpful comments on the manuscript. This work was supported by a Burroughs-Wellcome Career Award at the Scientific Interface, an Ellison New Scholar Award in Aging, and NIH R01-GM087432 to R.B.B. and NIH R01-GM40282 to M. Slatkin.
Appendix
Consider two species with effective population sizes Ne(1) and Ne(2), which diverged t generations ago, and assume that they have the same generation time. Furthermore, assume that until t0 generations after their divergence, one of the species (say, species 1) underwent positive selection with selection coefficient s11, after which it underwent negative selection with selection coefficient s12, while species 2 has been under constant negative selection with coefficient s2.
In general, the expected number of substitutions along a given branch after time t is uip(si). Here, ui is the mutation rate in species i; p(si) is the probability of fixation of a new mutant and depends on the selection coefficient and the effective population size. Then, the expected difference in the number of substitutions D along the lineages to the two species is
The first term in the final result is positive by the assumption that s11 > s12; as such, the quantity D1 − D2 can be negative so long as s12 < s2 and t is sufficiently big compared with t0.
Thus, a lineage undergoing a short ancient period of positive selection followed by a long period of tight constraint can exhibit a reduced rate of fixed changes, relative to a lineage under constant and more modest negative selection.
References
- Arthur RK, Ruvinsky I. Evidence that purifying selection acts on promoter sequences. Genetics. 2011;189:1121–1126. doi: 10.1534/genetics.111.133637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CR, Tuch BB, Johnson AD. Extensive DNA-binding specificity divergence of a conserved transcription regulator. Proc Natl Acad Sci U S A. 2011;108:7493–7498. doi: 10.1073/pnas.1019177108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Bradley RK, Roberts A, Smoot M, Juvekar S, Do J, Dewey C, Holmes I, Pachter L. Fast statistical alignment. PLoS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Mostovoy Y, Dudoit S, Brem RB. Polygenic and directional regulatory evolution across pathways in Saccharomyces. Proc Natl Acad Sci U S A. 2010;107:5058–5063. doi: 10.1073/pnas.0912959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Wakeley J, Sawyer S, Hartl DL. Directional selection and the site-frequency spectrum. Genetics. 2001;159:1779–1788. doi: 10.1093/genetics/159.4.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, Thomas WK. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 2005;37:544–548. doi: 10.1038/ng1554. [DOI] [PubMed] [Google Scholar]
- Dowell RD. Transcription factor binding variation in the evolution of gene regulation. Trends Genet. 2010;26:468–475. doi: 10.1016/j.tig.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Elyashiv E, Bullaughey K, Sattath S, Rinott Y, Przeworski M, Sella G. Shifts in the intensity of purifying selection: an analysis of genome-wide polymorphism data from two closely related yeast species. Genome Res. 2010;20:1558–1573. doi: 10.1101/gr.108993.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Babak T, Tsang J, Zhou Y, Zhang B, Mehrabian M, Schadt EE. Systematic detection of polygenic cis-regulatory evolution. PLoS Genet. 2011;7:e1002023. doi: 10.1371/journal.pgen.1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Moses AM, Schadt EE. Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc Natl Acad Sci U S A. 2010;107:2977–2982. doi: 10.1073/pnas.0912245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He BZ, Holloway AK, Maerkl SJ, Kreitman M. Does positive selection drive transcription factor binding site turnover? A test with Drosophila cis-regulatory modules. PLoS Genet. 2011;7:e1002053. doi: 10.1371/journal.pgen.1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Nguyen A, Davidson EH. Caught in the evolutionary act: precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev Biol. 2007;312:584–595. doi: 10.1016/j.ydbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Hittinger CT, Goncalves P, Sampaio JP, Dover J, Johnston M, Rokas A. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54–58. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins-Davis A, Townsend JP. Evolving gene expression: from G to E to GxE. Trends Ecol Evol. 2009;24:649–658. doi: 10.1016/j.tree.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogues H, Lavoie H, Sellam A, Mangos M, Roemer T, Purisima E, Nantel A, Whiteway M. Transcription factor substitution during the evolution of fungal ribosome regulation. Mol Cell. 2008;29:552–562. doi: 10.1016/j.molcel.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Kohn MH, Fang S, Wu CI. Inference of positive and negative selection on the 5' regulatory regions of Drosophila genes. Mol Biol Evol. 2004;21:374–383. doi: 10.1093/molbev/msh026. [DOI] [PubMed] [Google Scholar]
- Kuo D, Licon K, Bandyopadhyay S, Chuang R, Luo C, Catalana J, Ravasi T, Tan K, Ideker T. Coevolution within a transcriptional network by compensatory trans and cis mutations. Genome Res. 2010;20:1672–1678. doi: 10.1101/gr.111765.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, et al. (26 co-authors) Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics. 1998;149:2099–2104. doi: 10.1093/genetics/149.4.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JC, Groisman EA. Transcription factor function and promoter architecture govern the evolution of bacterial regulons. Proc Natl Acad Sci U S A. 2009;106:4319–4324. doi: 10.1073/pnas.0810343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki BP, Burghoorn J, Swoboda P. Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc Natl Acad Sci U S A. 2010;107:12969–12974. doi: 10.1073/pnas.0914241107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3. 2011;1:11–25. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi KR, Matsuo T, Takano-Shimizu-Kouno T. Two types of cis-trans compensation in the evolution of transcriptional regulation. Proc Natl Acad Sci U S A. 2011;108:15276–15281. doi: 10.1073/pnas.1105814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci U S A. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat Genet. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Wong KH, Barkai N, Struhl K. Extensive divergence of yeast stress responses through transitions between induced and constitutive activation. Proc Natl Acad Sci U S A. 2011;108:16693–16698. doi: 10.1073/pnas.1113718108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson DG, Boyko AR, Hernandez RD, et al. (11 co-authors) Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence. PLoS Genet. 2009;5:e1000592. doi: 10.1371/journal.pgen.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 2008;6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishnoi A, Sethupathy P, Simola D, Plotkin JB, Hannenhalli S. Genome-wide survey of natural selection on functional, structural, and network properties of polymorphic sites in Saccharomyces paradoxus. Mol Biol Evol. 2011;28:2615–2627. doi: 10.1093/molbev/msr085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss KM. Tilting at quixotic trait loci (QTL): an evolutionary perspective on genetic causation. Genetics. 2008;179:1741–1756. doi: 10.1534/genetics.108.094128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MD, Odom DT. Evolution of transcriptional control in mammals. Curr Opin Genet Dev. 2009;19:579–585. doi: 10.1016/j.gde.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.