Abstract
The 16S rRNA gene sequence diversity within the Phylum Actinobacteria was assessed from four sources: PCR-generated V6 sequence tags derived from seawater samples, metagenomic data from the Global Ocean Sampling (GOS) expedition, marine-derived sequences maintained in the Ribosomal Database Project (RDP), and select cultured strains for which sequence data is not yet available in the RDP. This meta-analysis revealed remarkable levels of phylogenetic diversity and confirms the existence of major, deeply rooted, and as of yet uncharacterized lineages within the phylum. A dramatic incongruence among cultured strains and those detected using culture-independent techniques was also revealed. Redundancy among the actinobacteria detected using culture-independent techniques suggests that greater sequence coverage or improved DNA extraction efficiencies may be required to detect the rare phylotypes that can be readily cultured from marine samples. Conversely, new strategies need to be developed for the cultivation of frequently observed but yet to be cultured marine actinobacteria.
Keywords: Marine actinobacteria, Bacterial diversity, Metagenomics, Actinomycetes
Introduction
The Phylum Actinobacteria represents a phylogenetically coherent lineage of high G + C Gram positive bacteria that is currently delineated into four Subclasses. The best known of these is the Subclass Actinobacteridae, which includes the Order Actinomycetales whose members are commonly referred to as actinomycetes. Actinomycetes are the most thoroughly studied of the actinobacteria and include a diverse assemblage of important agriculture and human health related taxa such as the plant symbionts Frankia spp., human pathogens such as Mycobacterium tuberculosis, and the most prolific source of microbial antibiotics discovered to date, the genus Streptomyces (Bérdy 2005). Recently, marine-derived actinomycetes have become recognized as a resource for biotechnology (Bull et al. 2000) thus creating incentive to expand upon the exploitable diversity within this group by sampling marine environments.
It has long been recognized that actinomycetes can be cultured from marine samples (Grein and Meyers 1958), yet it was not clear if these common soil bacteria should be considered terrestrial contaminants or an autochthonous component of the marine microbial community (Goodfellow and Haynes 1984). Early support for the existence of indigenous marine actinomycete populations came from the recovery of strains from deep-sea sediments (Weyland 1969), the description of the first marine species (Helmke and Weyland 1984), and the isolation of seawater dependent strains (Jensen et al. 1991). More recently, cultivation efforts have revealed considerable actinomycete diversity within marine samples (Gontang et al. 2007; Jensen et al. 2005; Magarvey et al. 2004; Maldonado et al. 2005b) and there is now evidence that actinomycetes are capable of growth in the marine environment (Mincer et al. 2005; Moran et al. 1995). The recent description of three marine genera (Han et al. 2003; Maldonado et al. 2005a; Yi et al. 2004) provides strong support for the concept of marine-specific actinomycetes, however some marine communities are dominated by taxa that have also been reported from land (Prieto-Davo et al. accepted) leaving questions about the extent to which the communities in these two habitats differ.
The application of molecular techniques to the field of marine microbial ecology has provided a new perspective on the diversity of actinobacteria in the sea (reviewed by Ward and Bora 2006). The first actinobacterial clones were simply identified as Gram-positive bacteria due to the low number of nucleotides sequenced (Fuhrman et al. 1993). Once longer sequences were obtained, it was possible to place these bacteria within the high G + C Gram positive lineage (Rappé et al. 1997) and ultimately within the Phylum Actinobacteria (Rappé et al. 1999). Today, actinobacteria are consistently observed when culture-independent techniques are applied to marine samples. They are recognized as a ubiquitous yet relatively small component of the marine bacterioplankton (Giovannoni and Stingl 2005) where they perform as of yet undefined ecological roles. It has also been revealed that marine actinobacteria include major new lineages in both the plankton (Rappé et al. 1999) and in association with sponges (Montalvo et al. 2005), however the phylogenetic diversity within these groups has generally not been analyzed in detail. Recent advances in DNA sequencing technology have led to the generation of large data sets that can now be mined for actinobacterial 16S rRNA gene sequences in an effort to provide a better understanding of the extant diversity of this Phylum as it occurs in the marine environment.
This paper presents a meta-analysis of the actinobacterial diversity detected in recent PCR-based (Sogin et al. 2006) and metagenomic (Rusch et al. 2007) surveys of marine bacterial diversity. The findings were then expanded to include a broader representation of culture-independent marine actinobacterial diversity by incorporating sequences retrieved from the Ribosomal Database Project (RDP). The diversity detected from these analyses was then contrasted with that identified using culture-dependent techniques based on additional RDP-derived sequences as well as select sequences obtained from strains maintained in a culture collection at the Scripps Institution of Oceanography. The results reveal high levels of diversity within the Phylum and a dramatic incongruence in the results obtained when culture-dependent and culture-independent methodologies are employed.
Methods
Compilation of V6 rRNA sequence tags
A total of 3693 Actinobacterial sequence tags were parsed from a previously published dataset that contains PCR-derived products spanning the V6 hypervariable region of cloned ribosomal RNA genes from four paired environmental DNA samples (Sogin et al. 2006). The paired samples consisted of seawater collected at two different depths from the same coordinates with three of the pairs (53R–55R, 112R–115R, and 137–138) originating from North Atlantic seawater and one (FS312–FS396) from diffuse hydrothermal vent fluids. The sequences were further parsed to include only those 2145 sequence tags with Basic Local Alignment Search Tool (BLAST) expectation scores ≤e–15 when queried against the V6RefDB database (Sogin et al. 2006). This value was chosen as it yielded best BLAST matches that displayed approximately 80% or greater query coverage. Although this criterion eliminated highly divergent sequences, it reduced the likelihood of making incorrect taxonomic assignments for sequence tags that were poorly represented in the database.
For each of the eight samples, the selected actinobacterial sequence tags were aligned using ClustalX (Thompson et al. 1997) and binned into groups of related sequences using the web based tool Clusterer (http://www.bugaco.com/mioritic/clusterer_jlp.php) with a distance parameter setting =10. Each singleton and cluster was defined as an Operational Taxonomic Unit (OTU) and the maximum number of sequences per cluster recorded for each sample (Table 1). Given that the length of the aligned sequence tags ranged from 79 to 90 nucleotides, this distance setting created sequence clusters that shared at least 87–90% identity in the V6 region. The sequence tags from each singleton and one representative from each cluster were then converted into longer sequences (≥1100 nucleotides) by retrieving the sequence associated with the best BLAST match to each query of the V6RefDB database (Sogin et al. 2006). These longer sequences were used for phylogenetic analyses as described below. In cases where the same BLAST match was obtained for different singletons or clusters within the same sample, the sequences were pooled to create one cluster.
Table 1.
Actinobacterial diversity within the V6 (Sogin et al. 2006) dataset
| Sample | Depth (m) | Actinobactial sequences (% total) | OTU singletons | OTU clusters | Max seq/cluster (% total) | OTUs total |
|---|---|---|---|---|---|---|
| 53R | 1400 | 97 (2.8) | 4 | 3 | 89 (91.8) | 7 |
| 55R | 500 | 198 (1.9) | 9 | 9 | 139 (70.2) | 18 |
| 112R | 4121 | 692 (14.5) | 6 | 6 | 637 (92.1) | 12 |
| 115R | 550 | 380 (4.9) | 5 | 8 | 351 (92.4) | 13 |
| 137 | 1710 | 354 (3.5) | 9 | 20 | 221 (62.4) | 29 |
| 138 | 710 | 347 (3.3) | 11 | 11 | 254 (73.32) | 22 |
| FS312 | 1529 | 17 (0.5) | 3 | 3 | 9 (52.9) | 6 |
| FS396 | 1537 | 60 (0.5) | 6 | 4 | 48 (80) | 10 |
| Total | 2145 | 53 | 64 | 117 |
Sequence tags were delineated into OTUs using the program Clusterer and the number of OTUs represented by single sequences (singletons) and groups of sequences (clusters) indicated along with the maximum number of sequences in any one cluster
Phylogenetic analyses of V6 sequence tags
The nearly full-length 16S rRNA gene sequences representing each actinobacterial OTU from all eight samples were compiled, aligned using ClustalX, and imported into MacClade (ver. 4.07, Sinauer Assoc., Sunderland, MA) for manual alignment and masking. Also included for reference in the multiple alignment was one sequence each from the Subclasses Rubrobacteridae, Acidimicrobidae, and Coriobacteridae. For the Subclass Actinobacteridae, one sequence was selected to represent the Order Bifidobacteriales and one sequence was selected to represent each of the 11 Suborders within the Order Actinomycetales. A neighbor-joining distance tree was created using PAUP (Swofford 2002). This tree was used to determine the taxonomic affiliations and site-specific distributions of the representative V6 actinobacterial sequence tags.
GOS sequence data compilation and analysis
A total of 4125 scaffolds that contained a 16S rRNA gene sequence were identified as part of a recent GOS publication (Rusch et al. 2007). These scaffolds originated from 41 aquatic, largely marine samples collected at 200 mile intervals along the eastern North American coast through the Gulf of Mexico and into the equatorial Pacific. Scaffolds <400 nucleotides in length were considered too short to be phylogenetically informative and eliminated from the pool. Of the remaining scaffolds, 150 were identified as members of the Phylum Actinobacteria based on BLAST analyses against the Ribosomal Database Project (RDP, http://rdp.cme.msu.edu/) and an additional 89 by comparison to The European Ribosomal RNA Database (http://bioinformatics.psb.ugent.be/webtools/rRNA/) for a total of 239 scaffolds. Since the scaffolds covered different regions of the 16S rRNA gene, the best BLAST matches (≥1100 nucleotides) for the 239 consensus sequences were used for phylogenetic analyses. These sequences were aligned using ClustalX and analyzed as above using Clusterer with a distance parameter of 120, which generated clusters of sequences that shared ca. 90% or greater 16S sequence identity. The sequence associated with each singleton and a representative from each cluster was used for phylogenetic analyses and to determine the taxonomic affiliations of the actinobacteria. The number of sequences associated with each scaffold (read depth) ranged from 1 to 19 (mean = 1.7) and was used to calculate sequence abundance. For sequence clusters, the total number of reads for all sequences in each cluster was calculated.
RDP sequence compilation and analysis
Two sets of sequences, distinguished based on the selection of cultured or non-cultured bacteria, were compiled using the Ribosomal Database Project (RDP, http://rdp.cme.msu.edu/) hierarchy browser (release 9) on 27 December 2006. Selection criteria for both data sets included good quality actinobacterial sequences >1200 nucleotides in length, both type and non-type strains, the taxonomy setting “nomenclatural” and the search string “marine”. The cultured search yielded 617 sequences to which an additional 33 sequences (GenBank accession numbers EU214912–214944, EF581384) were added from strains maintained at the Scripps Institution of Oceanography. The non-cultured search yielded 154 sequences to which the sequence from clone OM1 (U70710) was added, as it was not recovered with the search parameters employed yet represents an important reference strain (Rappé et al. 1999). These aligned datasets were analyzed as above using the program Clusterer with a distance parameter of 120, which generated clusters of sequences that share ≥90–92% identity for sequence lengths of 1200–1400 nucleotides. Each singleton and one representative of each sequence cluster were used to define the OTUs associated with both data sets. Neighbor-joining distance trees were created using PAUP to determine the taxonomic positions of the OTUs relative to reference actinobacterial sequences as previously described.
Results
V6 sequence tag distribution
Members of the Phylum Actinobacteria were represented in all eight of the samples analyzed by Sogin et al. (2006). The deepest sample (112R, 4121 m) had the highest number (692) and percentage (14.5%) of actinobacteria, which were distributed among 12 OTUs (six clusters and six singletons, Table 1). The vast majority of the actinobacteria in this sample (92.1%) fell within one cluster represented by a previously cloned sequence (DQ513067) obtained from a deep-sea sediment fluid (Huber et al. 2006). This sequence also shows a high degree of identity (99%) to a sequence (DQ396300) cloned from a deep-sea octacoral (Penn et al. 2006). The paired Labrador seawater samples (137 and 138) were the most diverse, having the largest numbers of OTUs. The two samples obtained from hydrothermal vents (FS312 and FS396) had the lowest numbers and percentages of actinobacterial sequences, and these were among the least diverse possibly due to the elevated temperatures at these sites. The majority of the actinobacterial sequence tags (52.9–92.4%) analyzed from each of the eight samples fell within one OTU indicating that one group of related strains dominated each site. In many cases, the same OTUs were observed in multiple samples (see below). There was no clear relationship between actinobacterial abundance and depth (linear regression r2 = 0.3257).
V6 sequence tag taxonomy
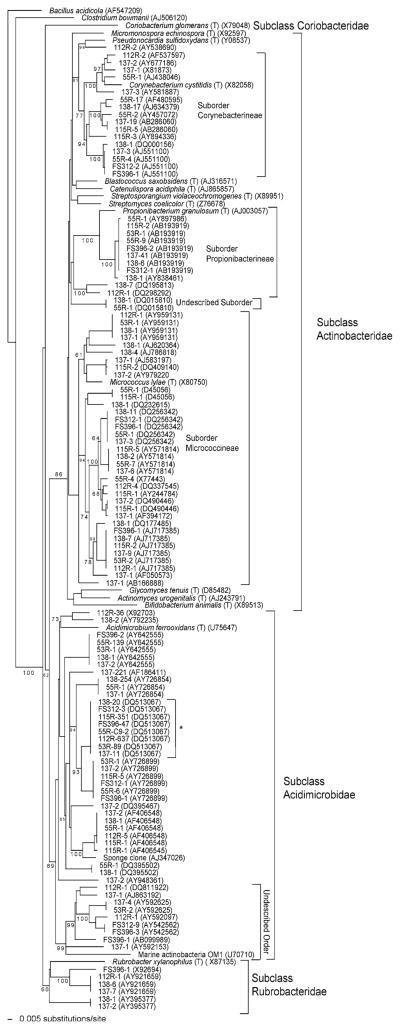
The taxonomic diversity of the actinobacteria identified from the V6 dataset was assessed using nearly full-length 16S rRNA gene sequences corresponding to the best BLAST match in the V6RefDB database for each OTU. This analysis revealed that members of three of the four currently described Subclasses within the Phylum had been detected (Fig. 1). The vast majority (1876 of 2145 or 87%) of the sequences however fall within the Subclass Acidimicrobidae (Table 2). Of these, only 38 sequences representing two OTUs clade with Acidimicrobium ferrooxidans (Fig. 1), the only described species within this Subclass as listed on the NCBI taxonomy browser (http://www.ncbi.nlm.nih.gov/sites/entrez?db=taxonomy). The Acidimicrobidae OTUs display considerable phylogenetic diversity and bifurcate into two deeply rooted lineages one of which includes OM1 (U70710), the marine actinobacterial sequence originally reported by Rappé et al. (1997). As previously suggested, these bacteria appear to represent a new Order within the Subclass (Rappé et al. 1997).
Fig. 1.
NJ phylogenetic dendogram created from nearly full-length (1200 informative nucleotides after masking) 16S rRNA gene sequences representing the actinobacterial sequence tags detected in the V6 dataset and reference strains. Sequence codes refer to sample number, number of sequences (>1 for sequences that represent clusters), and (accession number). * refers to the most abundant and frequently observed sequence
Table 2.
Actinobacterial taxonomic distribution, abundance (number of sequences), and diversity (number of OTUs given in parentheses) in the V6, GOS, and RDP data sets
| Taxon | V6 | GOS | RDP uncultured | RDP cultured |
|---|---|---|---|---|
| Phylum/Class | ||||
| Actinobacteria | 2145 (60) | 239 (33), 407 | 155 (50) | 650 (48) |
| Subclass | ||||
| Rubrobacteridae | 18 (3) | 2 (2), 2 | 0 | 4 (2) |
| Coriobacteridae | 0 | 0 | 3 (2) | 0 |
| Undescribed Subclass | 0 | 0 | 4 (1) | 1 |
| Acidimicrobidae | 1876 (19) | 125 (17), 201 | 56 (34) | 0 |
| Order | ||||
| Acidimicrobiales | 1853 (12) | 36 (15), 49 | 39 (23) | 0 |
| Undescribed Order | 23 (7) | 89 (2), 152 | 17 (11) | 0 |
| Subclass | ||||
| Actinobacteridae | 251 (38) | 112 (14), 204 | 92 (13) | 645 (45) |
| Order | ||||
| Bifidobacteriales | 0 | 0 | 0 | 0 |
| Actinomycetales | 251 (38) | 112 (14), 204 | 92 (13) | 645 (45) |
| Suborder | ||||
| Actinomycineae | 0 | 0 | 0 | 1 |
| Catenulisporineae | 0 | 0 | 0 | 0 |
| Corynebacterineae | 83 (12) | 0 | 4 (3) | 9 (6) |
| Frankineae | 0 | 0 | 0 | 0 |
| Glycomycineae | 0 | 0 | 0 | 1 |
| Micrococcineae | 92 (19) | 22 (2), 74 | 55 (6) | 135 (15) |
| Micromonosporineae | 0 | 0 | 0 | 202 (1) |
| Propionibacterineae | 72 (5) | 0 | 4 (1) | 17 (6) |
| Pseudonocardineae | 2 (1) | 0 | 0 | 2 (2) |
| Streptomycineae | 0 | 0 | 1 | 250 (6) |
| Streptosporangineae | 0 | 0 | 0 | 24 (4) |
| Undescribed Suborder | 2 (1) | 90 (12), 130 | 28 (2) | 4 (3) |
Sequences labeled “undescribed” Subclass or Suborder fall outside of currently described taxa. GOS scaffolds were multiplied by read depth to estimate sequence abundance (italicized)
Many of the OTUs within the Subclass Acidimicrobidae were observed in more than one of the samples analyzed by Sogin et al. (2006). The most prominent of these is represented by DQ513067, which was the only OTU observed in all eight samples and accounted for 1160 (62.6%) of the total actinobacteria analyzed from the V6 data set (Fig. 1). The second most abundant OTU (AF186411) represented 221 sequence tags and was observed in only one sample (137) revealing an uneven distribution for some abundant phylotypes even among samples collected at the same location but from different depths. Although it only represented 10 sequences, AF406548 was observed in five samples indicating that some low abundance phylotypes were widely distributed. This OTU was also closely related to an actinobacterial sequence (AJ347026) previously reported from a marine sponge (Hentschel et al. 2002). Despite the numerical dominance of some OTUs, the 1876 sequences belonging to the Subclass Acidimicrobidae represent 19 distinct OTUs that differ by ca. 10% or greater in the V6 region.
The second most populated Subclass in the V6 data set was the Actinobacteridae, which accounted for 12% (251) of the actinobacterial sequences analyzed. All of these claded within the Order Actinomycetales, i.e., no members of the Order Bifidobacteriales were detected (Fig. 1). All of the Actinomycetales, commonly known as actinomycetes, fell within the Suborders Corynebacterineae, Propionibacterineae, and Micrococcineae. One OTU comprised of two sequences appears to represent a new Suborder that is most closely related to the Suborder Propionibacterineae. Although seven times fewer actinomycete sequences were detected relative to members of the Subclass Acidimicrobidae, twice as many OTUs (38) were identified within this Subclass. As within the Subclass Acidimicrobiae, some actinomycete OTUs were observed in multiple samples, with AB193919 (Suborder Propionibacterineae) being the most widely distributed (seven samples) and abundant (62 of 251 sequences). Outside of the Subclass Actinobacteridae, three OTUs representing 18 sequences claded with the Subclass Rubrobacteridae while no members of the Subclass Coriobacteridae were observed.
GOS dataset
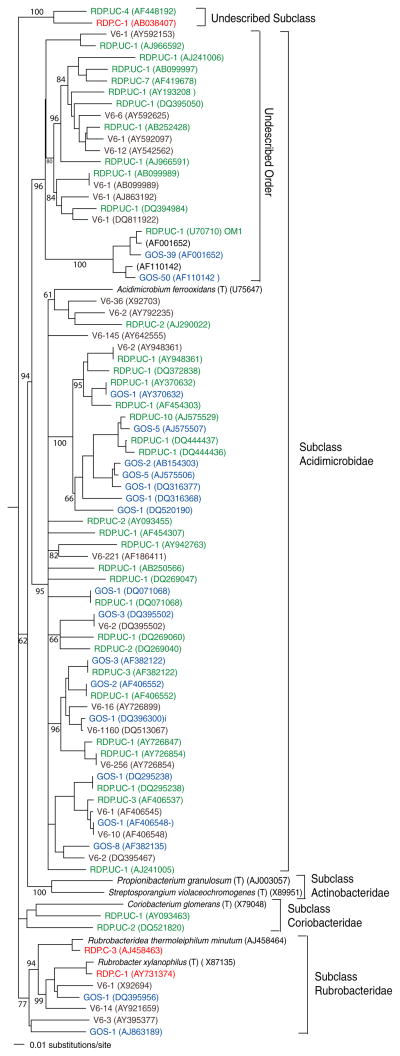
An analysis of the 16S rRNA gene sequences in the GOS data set (Rusch et al. 2007) revealed a total of 239 actinobacterial scaffolds of ≥400 nucleotides in length (Table 2). These scaffolds had best BLAST matches to 70 distinct 16S rRNA gene sequences. Detailed, sample-specific analyses were not performed on these sequences however 50% (119) originated from four samples collected from the Delaware Bay (GS11), the Chesapeake Bay (GS12), Lake Gatun, Panama (GS20), and a hypersaline lagoon in Ecuador (GS33). These sites appear to be highly influenced by terrestrial input relative to the majority of samples, which were collected from the open ocean. Cluster analysis of the 239 scaffolds places them into 33 OTUs. Phylogenetic analysis of these OTUs reveals they are evenly distributed between the Subclasses Acidimicrobidae and Actinobacteridae (Table 2). As in the V6 data set, the majority of the OTUs within the Subclass Acidimicrobidae (17) were widely distributed among diverse and uncultured lineages that are only distantly related to A. ferrooxidans (Fig. 2). Eighty-nine of the Acidimicrobidae scaffolds fell within two highly populated OTUs that account for 79% of the sequences and belong to the same undescribed Order that was also observed in the V6 data. These GOS scaffolds were only identified as members of the Phylum Actinobacteria by comparison with The European Ribosomal RNA Database and claded with OM1 (U70710), while the V6 sequences in this undescribed Order formed a separate lineage (Fig. 2). The remaining GOS OTUs were populated by relatively few sequences.
Fig. 2.
Neighbor-joining phylogenetic tree created from all non-Actinobacteridae 16S rRNA gene sequences (1120 informative nucleotides after masking) from the V6 (black), GOS (blue), RDP cultured (red), and RDP uncultured (green) datasets. Sequence codes refer to dataset, number of sequences, and (accession number). The tree was rooted with Bacillus acidicola (AF547209, not shown)
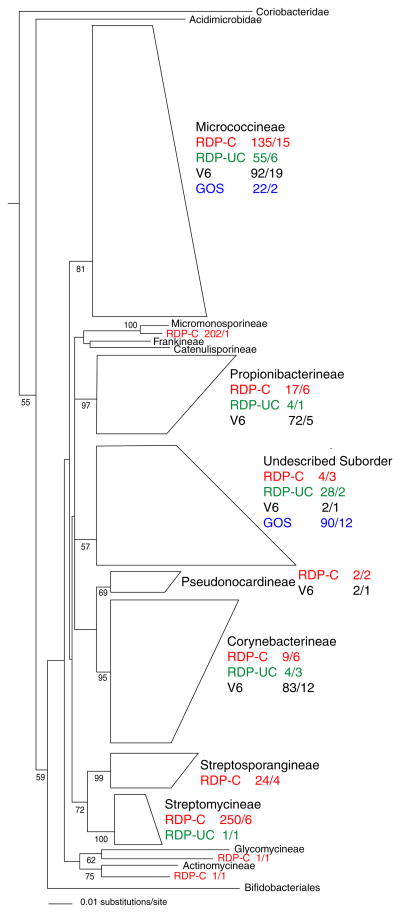
Members of the Subclass Actinobacteridae comprised 112 (47%) of the GOS actinobacterial sequences relative to 12% in the V6 dataset (Table 2). All of these sequences could be placed within the Order Actinomycetales with the vast majority (80%) falling into 12 OTUs within an undescribed Suborder that was also detected among the V6 data (Fig. 3). The remaining members of the Subclass Actinobacteridae fell within two OTUs in the Suborder Micrococcineae. This suborder was also well represented in the V6 data. In total, only one of the 11 recognized Suborders in the Order Actinomycetales was detected among the GOS data, three fewer than were observed in the V6 data. In contrast to that reported by Rusch et al. (2007), none of the GOS actinobacterial sequences fell within the Suborder Frankineae based on our phylogenetic analyses, which instead placed most of these sequences (e.g., AB021325) within an undescribed Suborder. This difference in taxonomic assignment may in part be due to differences in the length and nucleotide composition of the BLAST matches used in our analyses relative to the actual scaffolds, however weak bootstrap support for this new lineage warrants further phylogenetic study.
Fig. 3.
Neighbor-joining phylogenetic wedge tree created from all Actinobacteridae 16S rRNA gene sequences (998 informative nucleotides after masking) from the V6 (black), GOS (blue), RDP cultured (red), and RDP uncultured (green) datasets. Sequence codes refer to dataset, number of sequences/number of OTUs. The tree was rooted with Bacillus acidicola (AF547209, not shown)
RDP sequences (uncultured)
To assess marine actinobacterial diversity in a broader context, datasets comprised of cultured and uncultured members of the Phylum Actinobacteria were compiled from the RDP. A total of 155 uncultured actinobacterial sequences were identified from this search and grouped into 50 OTUs (Table 2). This represents a considerably greater OTU to sequence ratio than was detected from the V6 and GOS surveys. A small proportion of these sequences were identified as members of the Subclass Acidimicrobidae relative to the GOS and V6 datasets, however they represented the majority of the OTUs delineated from the RDP uncultured data including 11 within the previously discussed undescribed Suborder (Fig. 2). The uncultured RDP sequences also included the only sequences identified within the Subclass Coriobacteriales, both of which were derived from marine sediments. Although the majority of the actinobacterial sequences detected in the uncultured RDP dataset belonged to the Order Actinomycetales, 90% of these were identified as members of either the Suborder Micrococcineae or an undescribed Suborder. These results are largely consistent with the V6 and GOS culture-independent studies, which did not detect members of most Suborders within the Order Actinomycetales.
RDP sequences (cultured)
A total of 650 cultured members of the Phylum Actinobacteria, including 33 not yet available in the RDP, were selected for analysis and delineated into 48 OTUs (Table 2). Phylogenetic analyses of these OTUs revealed that only two (representing four strains) were members of the Subclass Acidimicrobidae. This result is in stark contrast to those obtained from the culture-independent diversity assessments. Interestingly, a cultured representative (AB038407) of what appears to represent an undescribed Subclass was identified providing an opportunity for the taxonomic evaluation of this lineage. Unlike the culture-independent data sets, the cultured members of the Subclass Actinobacteridae were widely distributed throughout the Order Actinomycetales (Fig. 3) and included members of five Suborders that were not detected in either the V6 or GOS data sets. Two of these Suborders (the Micromonosporineae and Streptomycineae) accounted for 70% of the cultured actinomycete sequences. These sequences however, were delineated into only seven OTUs indicating that the cultured diversity within these groups is relatively low and limited to a narrow range of taxa.
Discussion
Culture-independent studies of marine samples have revealed that members of the Phylum Actinobacteria are frequently observed in the marine bacterial community (Giovannoni and Stingl 2005). Actinobacteria are also readily cultured from marine samples when selective isolation techniques are applied with recent efforts leading to the description of at least three new genera (Han et al. 2003; Maldonado et al. 2005a; Yi et al. 2004). In some cases, the results of culture-independent studies have proven useful as a guide to the choice of selective cultivation methods (e.g., Maldonado et al. 2005b), while in others, progress has been made in cultivating new diversity (e.g., Magarvey et al. 2004; Takami et al. 1997). Despite these advances, the relationship between cultured and culture-independent marine actinobacterial diversity is not well defined. In the present study, a meta-analysis was performed on culture-independent actinobacterial sequences compiled from a PCR-based study designed to access the rare ocean biosphere (Sogin et al. 2006), an extensive metagenomic dataset consisting largely of seawater samples from the Global Ocean Sampling (GOS) expedition (Rusch et al. 2007), and sequences derived from the Ribosomal Database Project (RDP, http://rdp.cme.msu.edu/). Comparisons of this diversity with that detected using culture-based methods confirm the existence of extensive, uncharacterized marine actinobacterial diversity (e.g., Rappé et al. 1999; Montalvo et al. 2005) and reveal a dramatic incongruence between the bacteria detected using the two approaches.
The PCR-based study by Sogin et al. (2006) employed 454 sequencing technology to generate ca. 118,000 sequence tags spanning the V6 variable region of the 16S rRNA gene. This study was designed to access the under-explored rare biosphere and revealed that bacterial communities were one to two orders of magnitude more complex than previously reported. Among the sequences analyzed from this study, those assigned to the Phylum Actinobacteria represented a variable and in some cases considerable (up to 14.5%) component of the V6 sequence tags in each sample. These actinobacteria were dominated by members of the Subclass Acidimicrobidae, a poorly known group comprised of only one formally described species. Although major new lineages within this Subclass were detected, a high level of redundancy was also observed, with as many as 637 sequences from one sample (112R) being assigned to the same OTU (AJ347026). Although the clustering criteria for this dataset grouped sequences into OTUs that shared as little as 90% sequence identity in the V6 region, it is none-the-less clear that a few highly populated taxa dominated the Acidimicrobidae analyzed in this study. It would be interesting to explore these taxa in more detail to get a better understanding of their species abundance and fine-scale phylogenetic architecture. Given that 1548 potential actinobacterial sequences were excluded from our analyses due to poor coverage when queried against the V6RefDB database, it would also be of interest to obtain nearly full-length sequences for these highly divergent lineages so that accurate taxonomic assignments can be made. The remaining Actinobacteria detected among the V6 sequence tags were actinomycetes (Order Actinomycetales). Although relatively few actinomycete sequences were observed, they were delineated into approximately three times as many OTUs as were identified among the Subclass Acidimicrobidae. Our analysis of the Phylum Actinobacteria in the V6 data set supports the suggestion that more sequencing may be required to detect the low abundance taxa that occur in the environments sampled (Huber et al. 2007).
A recent publication based on the GOS expedition reported 4125 16S rRNA gene sequences (Rusch et al. 2007) of which we identified 239 as members of the Phylum Actinobacteria. As in the V6 data set, the majority of these sequences could be placed in the Subclass Acidimicrobidae. The GOS Acidimicrobidae included two OTUs representing 89 sequences that were only identified as actinobacteria by comparison with The European Ribosomal RNA Database. These sequences belong to what appears to represent an undescribed Order that was also detected in the V6 and RDP uncultured data. The GOS sequences within this new lineage however were closely related to OM1, the planktonic actinobacterial sequence originally reported by Rappé et al. (1997) while the V6 sequences form a distinct lineage.
The 70 unique GOS actinobacterial BLAST matches represent singletons when a cluster analysis is performed using a distance parameter setting corresponding to 97% 16S rRNA sequence identity. This high level of species diversity, despite the conservative level of sequence identity applied, suggests that considerably more sequencing would be required to provide a comprehensive assessment of the actinobacterial diversity in the samples analyzed. Our phylogenetic analyses of the GOS sequences within the Subclass Actinobacteridae places the majority in an undescribed Suborder (Fig. 3), as opposed to the Suborder Frankineae (Rusch et al. 2007), suggesting that the taxonomic novelty of these sequences may be greater than previously recognized. Uncultured actinobacteria retrieved from the RDP generally fall within the same lineages as those detected from the V6 and GOS analyses indicating that these groups have been repeatedly and independently observed in marine samples.
It is of interest to note that phylogenetically distinct, cultured marine representatives of the Subclass Actinobacteridae differ from non-marine-derived taxa largely at the genus and species level (data not shown) suggesting a relatively recent divergence from their terrestrial relatives. These marine-derived strains are scattered throughout the phylogenetic tree indicating that a deeply rooted marine actinomycete lineage has yet to be cultured, and that the ability to inhabit both environments appears to have evolved independently in multiple lineages. This may not be the case for members of the Subclass Acidimicrobidae, in which marine sequences form diverse and deeply rooted lineages within the Phylum Actinobacteria. It will be useful to determine the marine-specificity of these lineages and, once obtained in culture, if they display specific adaptations to life in the sea. The fact that 12% of the GOS 16S sequences were Gram-positive yet few Gram-positive specific protein domains were detected in the metagenomic analysis (Yooseph et al. 2007) suggests that the marine representatives of the Subclass Acidimicrobidae may possess unique structural and/or biochemical features. It is noteworthy that less than twice the number of actinobacterial OTUs was detected among the V6 data relative to the GOS study despite the fact that nearly five times as many actinobacterial sequences were analyzed. This is likely due to the fact that the GOS samples originated from a broader range of environments that included estuarine and at least one lake sample, which accounted for most to the actinobacterial sequences.
The discrepancy observed between cultured and culture-independent marine actinobacterial diversity could be accounted for in a number of ways. Given that the majority of the cultured actinobacteria fall within the Suborders Micromonosporineae and Streptomycineae, it is possible that these bacteria occur in the marine environment largely as spores, the detection of which is biased against due to relatively poor DNA extraction efficiencies when culture-independent methods are applied. It is also possible that these actinomycetes occur at abundances below the detection limits of most culture-independent methods (e.g., Mincer et al. 2005), yet can be readily observed using the highly selective methods required for the isolation of spore-forming actinomycetes. In support of this, marine actinomycetes belonging to the genus Salinispora are generally observed in marine sediments at abundances of ca. 103 colony-forming units (CFUs) per ml (Mincer et al. 2002). Assuming 108–109 total bacteria per ml sediment (Whitman et al. 1998), it is unlikely that these actinomycetes would be represented in the 4125 16S rRNA gene scaffolds detected in the GOS data set. Finally, most cultured marine actinomycetes tend to be recovered from benthic sources while most culture-independent studies have focused on seawater samples. Although it has been shown that culture-independent techniques can be used to detect members of the Suborders Micromonosporineae and Streptomycineae in marine sediments when appropriate PCR primers are used (Mincer et al. 2005; Stach et al. 2003), it remains unclear to what extent these bacteria also occur in seawater. Given that culturing efforts frequently target specific taxa, the bias towards members of the Suborders Micromonosporineae and Streptomycineae in the cultured RDP data is likely due to interest in these groups as a resource for biotechnology in addition to ineffective search parameters for identifying the environmental source of sequences in the RDP.
In conclusion, considerable actinobacterial diversity has been detected in recent PCR-based and metagenomic analyses of marine samples. This diversity includes what appears to represent a new Subclass within the Phylum Actinobacteria, a new Order within the Subclass Acidimicrobidae, and a new Suborder within the Order Actinomycetales. In contrast, the actinobacteria cultured from marine samples have largely been limited to actinomycetes with new taxa being recognized at the species and genus levels. The incongruence between the actinobacteria observed using the two methodologies is a clear indication of the limitations of these complimentary approaches. It is likely that the extensive actinobacterial diversity observed to date in the marine environment represents only a small subset of the extant diversity within this ubiquitous marine phylum. Future studies will undoubtedly continue to expand our understanding of marine actinobacterial diversity and hopefully reveal the growth requirements of currently uncultivated taxa so they can be evaluated from ecological and biotechnological perspectives.
Acknowledgments
We thank Saul A. Kravitz for his invaluable help in accessing sequence assemblies from the Global Ocean Sampling dataset and Douglas H. Bartlett for generously providing computational resources. P.R.J. acknowledges funding from the National Institutes of Health, International Cooperative Biodiversity Groups program (grant U01-TW007401-01) and NOAA Grant #NAO40AR4170038, California Sea Grant College Program Project #R/MP-98, through NOAA’S National Sea Grant College Program, U.S. Dept. of Commerce. The statements, findings, conclusions and recommendations are those of the author(s) and do necessarily reflect the views of California Sea Grant, NOAA or the U.S. Dept. of Commerce.
Contributor Information
Paul R. Jensen, Email: pjensen@ucsd.edu, Scripps Institution of Oceanography, University of California San Diego, La Jolla, CA 92093, USA
Federico M. Lauro, Environmental Microbiology Initiative, School of Biotechnology and Biomolecular Sciences, The University of New South Wales, Sydney 2052, Australia
References
- Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Bull AT, Ward AC, Goodfellow M. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol Mol Biol Rev. 2000;64:573–606. doi: 10.1128/mmbr.64.3.573-606.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, McCallum K, Davis AA. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanonni SJ, Stingl U. Molecular diversity and ecology of microbial plankton. Nature. 2005;437:343–348. doi: 10.1038/nature04158. [DOI] [PubMed] [Google Scholar]
- Gontang EA, Fenical W, Jensen PR. Phylogenetic diversity of Gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol. 2007;73:3272–3282. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M, Haynes JA. Actinomycetes in marine sediments. In: Ortiz-Ortiz L, Bojalil LF, Yakoleff V, editors. Biological, biochemical, and biomedical aspects of actinomycetes. Academic Press, Inc; Orlando: 1984. pp. 453–472. [Google Scholar]
- Grein A, Meyers SP. Growth characteristics and antibiotic production of actinomycetes isolated from littoral sediments and materials suspended in sea water. J Bacteriol. 1958;76:457–463. doi: 10.1128/jb.76.5.457-463.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Nedashkovzkaya OI, Mikhailov VV, Kim SB, Bae KS. Salinibacterium amurkyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int J Syst Evol Microbiol. 2003;53:2061–2066. doi: 10.1099/ijs.0.02627-0. [DOI] [PubMed] [Google Scholar]
- Helmke E, Weyland H. Rhodococcus marinonascens sp. nov., an actinomycete from the sea. Int J Syst Bacteriol. 1984;34:127–138. [Google Scholar]
- Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J, Moore BS. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol. 2002;68:4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber JA, Johnson HP, Butterfield DA, Baross JA. Microbial life in ridge flank crustal fluids. Environ Microbiol. 2006;8:88–99. doi: 10.1111/j.1462-2920.2005.00872.x. [DOI] [PubMed] [Google Scholar]
- Huber JA, Welch DBM, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Jensen PR, Dwight R, Fenical W. Distribution of actinomycetes in near-shore tropical marine sediments. Appl Environ Microbiol. 1991;57:1102–1108. doi: 10.1128/aem.57.4.1102-1108.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Gontang E, Mafnas C, Mincer TJ, Fenical W. Culturable marine actinomycete diversity from tropical Pacific Ocean sediments. Environ Microbiol. 2005;7:1039–1048. doi: 10.1111/j.1462-2920.2005.00785.x. [DOI] [PubMed] [Google Scholar]
- Magarvey NA, Keller JM, Bernan V, Dworkin M, Sherman DH. Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl Environ Microbiol. 2004;70:7520–7529. doi: 10.1128/AEM.70.12.7520-7529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado L, Mincer TJ, Fenical W, Goodfellow M, Jensen PR, Ward AC. Salinispora gen nov., a home for obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol. 2005a;55:1759–1766. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- Maldonado LA, Stach JEM, Pathomaree W, Ward AC, Bull AT, Goodfellow M. Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie van Leeuwenhoek. 2005b;87:11–18. doi: 10.1007/s10482-004-6525-0. [DOI] [PubMed] [Google Scholar]
- Mincer TJ, Jensen PR, Kauffman CA, Fenical W. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments. Appl Environ Microbiol. 2002;68:5005–5011. doi: 10.1128/AEM.68.10.5005-5011.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincer TJ, Fenical W, Jensen PR. Cultured and culture-independent diversity within the obligate marine actinomycete genus Salinispora. Appl Environ Microbiol. 2005;71:7019–7028. doi: 10.1128/AEM.71.11.7019-7028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalvo NF, Mohamed NM, Enticknap JJ, Hill RT. Novel actinobacteria from marine sponges. Antonie van Leeuwenhoek. 2005;87:29–36. doi: 10.1007/s10482-004-6536-x. [DOI] [PubMed] [Google Scholar]
- Moran MA, Rutherford LT, Hodson RE. Evidence for indeginous Streptomyces populations in a marine environment determined with a 16S rRNA probe. Appl Environ Microbiol. 1995;61:3695–3700. doi: 10.1128/aem.61.10.3695-3700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn K, Wu D, Eisen JA, Ward N. Characterization of bacterial communities associated with deep-sea corals on the Gulf of Alaska seamounts. Appl Environ Microbiol. 2006;72:1680–1683. doi: 10.1128/AEM.72.2.1680-1683.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Davo A, Fenical W, Jensen PR. Actinomycete diversity in marine sediments. Aquat Microbial Ecol (accepted) [Google Scholar]
- Rappé MS, Kemp PF, Giovannoni SJ. Phylogenetic diversity of marine picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr. 1997;42:811–826. [Google Scholar]
- Rappé MS, Gordon DA, Vergin KL, Giovannoni SJ. Phylogeny of Actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst Appl Microbiol. 1999;22:106–112. [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, et al. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern tropical Pacific. PLOS Biol. 2007;5:398–431. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndi GJ. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci. 2006;32:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stach JE, Maldonado LA, Masson DG, Ward AC, Goodfellow M, Bull AT. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl Environ Microbiol. 2003;69:6189–6200. doi: 10.1128/AEM.69.10.6189-6200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates; Sunderland: 2002. paup*. [Google Scholar]
- Takami H, Inoue A, Fuji F, Horikoshi K. Microbial flora in the deepest sea mud of the Mariana T drench. FEMS Microbiol Lett. 1997;152:279–285. doi: 10.1111/j.1574-6968.1997.tb10440.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AC, Bora N. Diversity and biogeography of marine actinobacteria. Curr Opin Microbiol. 2006;9:279–286. doi: 10.1016/j.mib.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Weyland H. Actinomycetes in North Sea and Atlantic Ocean sediments. Nature. 1969;223:858. doi: 10.1038/223858a0. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Schuman P, Sohn K, Chun J. Serinicoccus marinus gen. nov., sp. nov., a novel actinomycete with L-ornithine and L-serine in the peptidoglycan. Int J Syst Evol Microbiol. 2004;54:1585–1589. doi: 10.1099/ijs.0.03036-0. [DOI] [PubMed] [Google Scholar]
- Yooseph S, Sutton G, Rusch DB, et al. The Sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLOS Biol. 2007;5:432–466. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]