Abstract
The shape of dose response of ionizing radiation (IR) induced cancer at low dose region, either linear non-threshold or J-shaped, has been a debate for a long time. This dose response relationship can be influenced by built-in capabilities of cells that minimize the fixation of IR-mediated DNA damage as pro-carcinogenic mutations. Key capabilities include sensing of damage, activation of cell cycle checkpoint arrests that provide time needed for repair of the damage as well as apoptosis. Here we describe computational modeling of the signaling pathways that link sensing of DNA damage and checkpoint arrest activation/apoptosis to investigate how these molecular-level interactions influence the dose response relationship for IR induced cancer. The model provides qualitatively accurate descriptions of the IR-mediated activation of cell cycle checkpoints and the apoptotic pathway, and of time-course activities and dose response of relevant regulatory proteins (e.g. p53 and p21). Linking to a two-stage clonal growth cancer model, the model described here successfully captured a monotonically increasing to a J-shaped dose response curve and identified one potential mechanism leading to the J-shape: the cell cycle checkpoint arrest time saturates with the increase of the dose.
Keywords: ionizing radiation, mammalian cell cycle, checkpoint arrest, apoptosis, nonlinear dynamical system
INTRODUCTION
Ionizing radiation (IR) is a special environmental stressor and causes DNA damage that, if not adequately repaired, results in pro-carcinogenic mutations and eventually in cancer (Little, 2003). A linear no threshold (LNT) dose-response model for IR-induced cancer is assumed as a default in the absence of information to the contrary (Upton, 2002). Recent Biological Effects of Ionizing Radiation (BEIR) VII reports concluded that the risk of cancer proceeds in a linear fashion at lower doses without a threshold (BEIR, 2006). The underlying assumption that supports LNT model is that a single DNA damage is capable of inducing a tumor. However, laboratory results of IR indicate a J-shaped or hormetic dose response relationship in the low dose region (Cohen 1995; Redpath et al. 2001; Boreham et al. 2006; Sakai et al. 2006; Day et al. 2007; Mitchel 2007a, b; Tubiana et al. 2006, 2009). To investigate the conflict between the experimental observations and the LNT model is important because the difference of the regulation costs based on LNT or J-shaped model are dramatic.
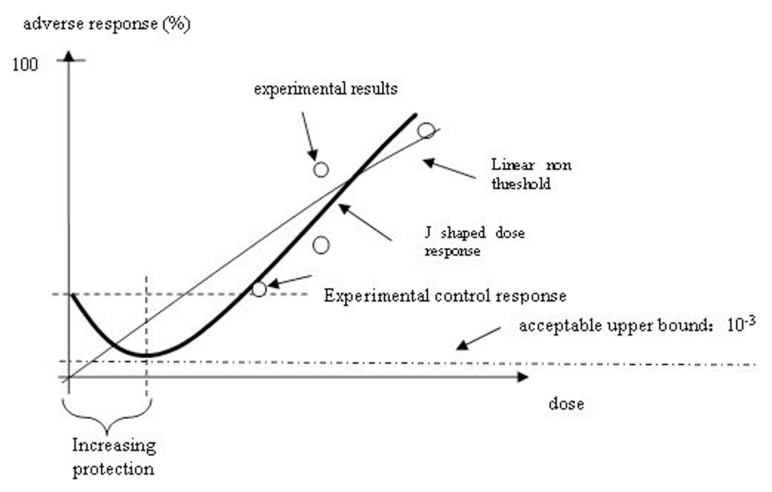
Figure 1 indicates the difference between the predicted adverse responses in the low dose region based on LNT model and J-shaped model, respectively (Calabrese and Ricci, 2010). It is seen that the regulatory concept of one in a million cancer risk becomes irrelevant when the J-shaped dose response model is correct. Thus we could spend billions of dollars to establish a regulation, which could just be an overprotection. Therefore more mechanism-based dose response model for IR needs to be developed in order to explore if the fundamental biological mechanism support a J-shaped dose response relationship as indicated in the experiments.
FIGURE 1.
Biphasic (hormetic) Dose-response Model for Cancer Incidence (the percent response in the controls must be non-zero). Protection is “optimized” because it is greatest at a dose range furthest away from a non-zero percentage response in the controls. The black dots identify exposure-response points that are – or should be – included in any complete analysis otherwise the empirical relationship (based on the white dots) cannot be estimated and thus the default appears to be sound when it is not.
The intent of the current manuscript is to introduce new conceptual approach to the analysis of dose-responses for IR-induced adverse health effects and to provide a preliminary example. Our concept is that computational models of biochemical signaling pathways can be developed and used to refine predictions of dose-response. For development of quantitatively accurate, predictive models it will be necessary to describe tissues consisting of multiple cell types where the different types each contribute in their own way to the overall function of the tissue. Such a model will probably need to incorporate not only cell type-specific data but also spatial information on the architecture of the tissue and on intercellular signaling. The scope of the current model is, however, more limited. Data obtained in a number of different biological systems are synthesized to describe a chimeric, “average” cell. Biochemical signaling pathways involved in sensing of DNA damage and in the activation of cell cycle checkpoint controls and the apoptotic pathway are described. As with any computational modeling effort, it is necessary to develop such initial descriptions (models) that can be iteratively refined. Our preliminary model thus defines a starting point which, with time, can evolve to a level of refinement where large amounts of detailed biological information are synthesized and a capability for robust predictions of dose- and time-response behaviors is obtained. We expect that our preliminary effort will be of interest not only to computational modelers interested in dose-response but also to radiation biologists who will (hopefully) develop the datasets needed to refine the model.
For IR-induced DNA damage, checkpoint arrest and apoptosis serve, among other things, as defensive responses. These responses lead ultimately to cell death or cell survival (Fei and El-Deiry, 2003). Both responses contribute to bystander effects (Wang et al. 2004; Azzam et al. 2000) and may also be involved in nonmonotonic dose-response (Conolly and Lutz, 2004). Moreover, checkpoint arrest and apoptosis have significant implications for cancer dose response. Checkpoint-induced delays in cell cycle increase cell generation time and thus decrease cell proliferation rate, which is an essential parameter in clonal growth models of tumor incidence (Moolgavkar and Knudson, 1981). Cell loss due to apoptosis leads to compensatory regenerative proliferation, which is a risk factor for cancer (Tan et al. 2003). The signaling networks that mediate these responses are sufficiently complex that computational models are useful (and perhaps even essential) adjuncts to laboratory studies of pathway structure and dynamic behavior (Bhalla et al. 2002; Qu et al. 2003a; Qu et al. 2003b; Novak et al. 2001).
MODEL DEVELOPMENT
A modular approach was used to construct the overall computational model linking IR-mediated DNA damage with activation of cell cycle checkpoint controls and apoptosis. Module 1 describes an empirical linkage of IR with DNA damage; Module 2 provides a molecular level description of the signaling pathways linking DNA damage with target proteins that induce checkpoint arrest and apoptosis; and Module 3 describes the molecular mechanisms of the G1/S and G2/M cell cycle checkpoint controls. Modules 2 and 3 are the main focus of this report. In future work the overall model could be extended by adding additional modules describing bystander effects and tumor incidence. Future work could also involve iterations of the current model structure and parameterization to reflect improvements in our understanding of the relevant biology.
Module 1
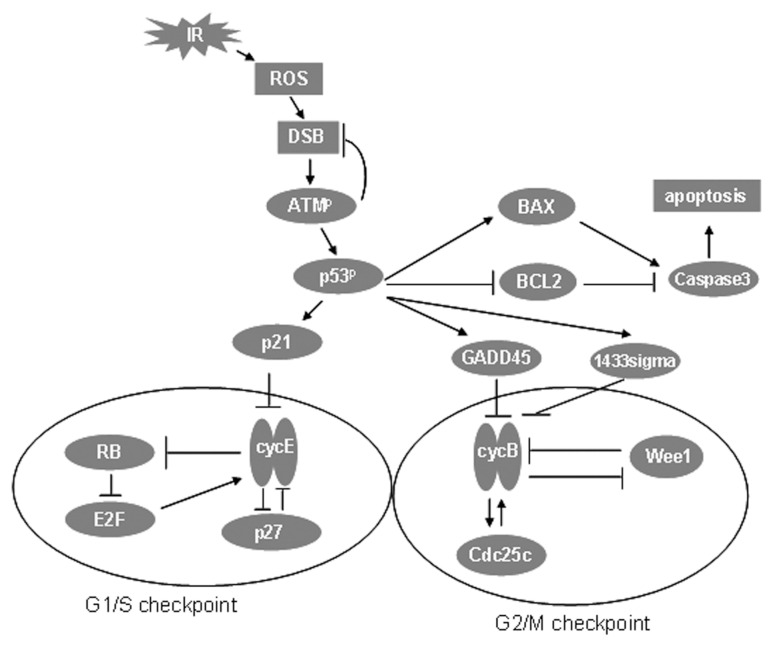
IR causes DNA damage, with double strand breaks (DSBs) being the most significant lesions (Criswell et al. 2003). DSB may arise through direct hits by IR or as secondary damage from reactive oxygen species (ROS) (Feinendegen et al. 1999; Feinendegen, 2002; Slupphaug et al. 2003). In Module 1 IR generates ROS and ROS cause DSB (Fig. 2). Direct formation of DSB by IR is not described. Repair of DSB is included in the model but we have not attempted to describe the detailed molecular mechanism of this process.
FIGURE 2.
IR-mediated DNA damage, sensing of the damage by ATM, p53-dependent G1/S and G2/M cell cycle checkpoints and apoptosis. ( and —| imply activation and repression, respectively. Note the central role of p53 in the signaling pathways. The large ovals at the bottom left and right indicate proteins and interactions in the G1/S and G2/M checkpoints, respectively.
Module 2
Module 2 is the signal transduction module. The transducer and effector phases of molecular mechanisms that are activated in response to IR are described (Fig. 2). DSB activate ataxia telangiectasia-mutated kinase (ATM) by autophosphorylation (Criswell, et al. 2003; Bakkenist and Kastan, 2003; Bartek and Lukas, 2001; Mikkelsen and Wardman, 2003). ATM plays a role in DSB repair by binding to the broken ends of the DNA strands created by DSB. DSB repair is defective in ATM-deficient cells (Yang et al. 2004). We thus assumed that the rate of DSB repair (Module 1) is proportional to the activity of phosphorylated ATM, and to the amount of DSBs.
Notwithstanding its role in DNA repair, the primary function of ATM is to phosphorylate and activate the transcription factor p53 (Adams and Carpenter, 2006) (Fig. 2). p53 is a critical mediator of cellular responses to DNA damage in IR-exposed mammalian cells (Criswell et al. 2003; Mikkelsen and Wardman, 2003; Pietenpol and Stewart, 2002; Agarwal et al. 1998). p53 signals to the G1/S checkpoint through p21 and to the G2/M checkpoint through 14-3-3σ and GADD45 (Fei and El-Deiry, 2003; Criswell et al. 2003; Taylor and Stark, 2001).
Cyclin E and its dependent kinase (Cdk2) form a heterodimer (CycE) that stimulates progression of the cell cycle from G1 to S (Fig. 2). p21 inhibits CycE, and p53 mediated transcriptional activation of p21 thereby invokes the G1/S checkpoint arrest (Taylor and Stark, 2001; Sekiguchi and Huter, 1998). The role of CycE in the G1/S transition is covered in the description of Module 3.
Cyclin B and its dependent kinase (Cdk1) form a heterodimer (CycB) that stimulates progression of the cell from G2 into mitosis (Fig. 2). 14-3-3σ anchors CycB in the cytoplasm, thereby blocking mitosis (Taylor and Stark, 2001). GADD45 dissociates the CycB heterodimer and therefore also blocks mitosis (Taylor and Stark, 2001). p53 mediated activation of 14-3-3σ and GADD45 thus invokes the G2/M checkpoint arrest. The role of CycB in the G2/M transition is covered in the description of Module 3.
p53-dependent signaling also mediates entry into the apoptotic pathway through effects on Bax and Bcl-2 (Fig. 2) (Fei and El-Deiry, 2003). Bax is pro-apoptotic and is transcriptionally activated by p53 (Levine, 1997; Chao et al. 1995; Thomas et al. 2000). Bcl-2 is anti-apoptotic and is transcriptionally repressed by p53 (Fussenegger et al. 2000). Both Bax and Bcl-2 signal to caspase 3, which are proteases at the core of the apoptotic machinery (Fussenegger et al. 2000).
In addition to this relatively simple description of the p53-dependent checkpoint arrest and apoptosis pathways, two more complex versions of Module 2 are described in the Appendix that illustrate how pathway architecture determines the robustness (Kitano, 2004) of the system. Here, robustness refers to the ability to maintain a significant level of functionality when pathway components are missing or inoperative due to mutations. Redundancy in signaling pathways can also serve to enhance the degree of biochemical response to an input such as DNA damage. These pathways include Chk2-dependent checkpoint arrest (Fig. A-1) and c-Abl - p73 dependent apoptosis (Fig. A-2) (Levrero et al. 2000).
Module 3
Module 3 describes key aspects of the molecular mechanisms of the G1/S and G2/M cell cycle checkpoints. The G1/S checkpoint controls entry into S phase, ensuring that: (1) the cell is large enough to warrant a new round of DNA synthesis, (2) any damage suffered by the DNA has been repaired, and (3) external conditions are favorable for mitotic cell division (Elledge, 1996). The G2/M checkpoint guides entry into mitosis, confirming that (1) DNA is fully replicated, (2) any new damage sustained by the DNA has been repaired, and (3) the cell is large enough to divide (Gardner and Burke, 2000). Therefore, both G1/S and G2/M are important checkpoints relevant to DNA damage.
The term “DNA damage checkpoints” is sometimes used to refer to functional points in the cell cycle where the cell monitors for the presence of DNA damage and, if damage is found, initiates a cascade of events leading to cell cycle arrest until the damage is repaired (Laiho and Latonen, 2003). Therefore, the G1/S and G2/M checkpoints in the cell cycle are DNA damage checkpoints.
Module 3 was developed to describe the G1/S and G2/M checkpoints. Our approach was based on that used by Tyson and co-workers, where checkpoints are activated by inhibitory phosphorylation of key proteins, and cell growth (i.e., the increase in cell size or cell mass with time) drives progression around the cycle (Kapuy et al. 2009; Tyson and Novak, 2001; Tyson et al. 2001). It is assumed the key reactions which drive checkpoint transitions take place in nucleus. The concentrations of the effective key regulators accumulate when cell grows and the effective concentration should equal to the product of cell mass and actual concentration of the key regulators. Our description of the G1/S checkpoint is a synthesis of information taken from literature review. The description of G2/M checkpoint control is a modified version of the model developed by Tyson et al. (2001).
The key proteins controlling the G1 to S transition in a mammalian cell cycle are the CycEs (Koepp et al. 2001; Moberg et al. 2001), which are thought to initiate DNA replication (Iliakis et al. 2003). To keep our model as simple as possible, we only considered the heterodimer - cyclin E and Cdk2. No description of the interactions between CycE monomers was included. Since Cdk is only active when associated with its cyclin partner (Koepp et al. 2001), we considered it acceptable to ignore its monomer form in this initial model of the system.
E2F, Rb, and p27 all interact with cyclin E (Sherr and Roberts, 2004; Leng et al. 1997; Russo et al. 1996). E2F is a transcription factor for cyclin E and thus promotes G1/S progression (Bartek and Lukas, 2001; Yamasaki, 2003; Ohtani, 1999). Rb is thought to regulate progression through the G1 phase of the cell cycle (Ohtani, 1999; Gala et al. 2001; Morris and Dyson, 2001) by inhibiting the activity of E2F through formation of Rb-E2F complexes (Morris and Dyson, 2001; Wells et al. 2003; Weintraub et al. 1992; Arroyo and Raychaudhuri et al. 1992).
Formation of the Rb-E2F complex not only determines the activity of cyclin E but also is regulated by CycE. That is, Rb can be phosphorylated by CycE, inhibiting Rb activity and enabling release of active E2F to promote entry into S phase (Leng et al. 1997). CycE can phosphorylate both Rb and Rb-E2F complexes in the model, as experimental observations show (Qin et al. 1992). The net effect of the CycE-Rb-E2F-CycE regulatory loop is to form a molecular switch that synergizes CycE activation when CycE levels are rising (Halaban, 1999). These are the main controlling factors for Rb (Qu et al. 2003a; Degregori, 2004) and the total Rb activity is assumed to be constant and not regulated by other factors in the model.
Activity of CycE is also stimulated by interactions between CycE and p27. CycE is inactive when complexed with p27. Ubiquitylation of p27, however, causes dissociation of the CycE-p27 complex (Akli et al. 2004; Furukawa, 2002; Lane et al. 2001). The trigger for p27 ubquitylation is phosphorylation of p27 by CycE (Sherr, 2000; Li et al. 2004). As indicated by experimental observations, CycE is assumed in the model to phosphorylate both p27 and p27-cycE complexes (Sherr, 2000; Yang et al. 2004). The result of the CycE-p27-CycE regulatory loop is the same as that of the CycE-Rb-E2F-CycE loop, i.e., to synergize CycE activation as CycE levels are rising. The relative importance of these two loops in increasing CycE levels is not clear, although most of the literature to date has focused on the loop containing Rb. P27 activity is assumed to have a zero order synthesis and a first order degradation.
Our depiction of the G2/M checkpoint is based on the antagonism of CycB and Wee1 as well as the positive feedback of CycB and Cdc25c (Tyson et al. 2001; Tyson et al. 2002; Novak et al. 2007). Entry of the cell into mitosis is promoted by CycB, which is thus the key complex of the G2/M checkpoint (Kaufmann, 1998; Bahassi et al. 2004). The mutual phosphorylation between CycB and Wee1 inhibits each other. Cdc25c activates CycB by dephosphorylation; CycB activates Cdc25c by phosphorylation. These two feedbacks induce and then synergize the switch-like behavior of CycB in the G2 to M transition. The G2 state corresponds to an inactive CycB, an inactive Cdc25c and an active Wee1 activity. The mitotic state corresponds to an active CycB, an active Cdc25c and an inactive Wee1 activity.
Mathematical Methods and Software
The mathematical model is composed of a set of ordinary differential equations (ODEs). The equations and their derivations are described in the on-line auxiliary materials. The dynamical system contains 33 ODEs and 127 parameters. When available, specific parameter values were taken from previously published sources. For example, for the equations modified based on Tyson’s model of the cell cycle, we used the parameter values as published (Tyson et al. 2001). Whenever possible, the nature of the biological process was considered when assigning values to the kinetic constants. For example, rate constants for transcriptional reactions were assigned relatively small values, as transcription is a relatively slow process. When such information was not available, parameter values were assumed to generate the expected qualitative behaviors of the model. This approach to the assignment of parameter values has been used by others (Qu et al. 2003a; Qu et al. 2003b) and is reasonable for the first-generation model we have developed. A simple sensitivity study about the parameter values based on Monte Carlo simulation was conducted when comparing the simulation results with some of the experimental data during the model verification process. During the process, the parameter values are assumed to fit uniform distribution in the range of ±20% of the assigned parameter values.
To study the predictions of cell cycle behavior from the mathematical model, mathematical methods from nonlinear dynamics were applied. The results are displayed graphically, and illustrated by time course plots and phase plane plots for protein activity.
For a dynamical system with two variables x and y where dx/dt = f(x,y) and dy/dt = g(x,y), phase plane plots are used to study how x and y change relative to each other and to determine if the system has one or more steady states, i.e., f(x,y) = 0 and g(x,y) = 0. In the phase plane plot, a point on the x nullcline is identified by fixing the value of y and running the computational model to identify the corresponding steady state value of x. The entire x nullcline is developed by repeating this steady state analysis over the range of values of y that are of interest. The x nullcline is thus the set defined by f(x,y) = 0, or in other words, the set of values of x at which the system is at steady state for a range of values of y. Similarly, the y nullcline denotes steady state values of y for a range of values of x, g(x,y) = 0.
Intersections of the x and y nullclines identify steady states of the entire dynamical system. In a system with more than one steady state, each steady state is associated with a unique intersection of the nullclines. When the dynamical system has more than two state variables, the variables not represented in the 2-dimensional plot are assumed to be in a “pseudo-steady state”. That is, they change quickly with the variables of interest. In this manner, a system with more than two variables can also use phase plane plot to do analysis (Angeli et al. 2004).
MATLAB programs were developed to predict time course and draw phase plane plots (The Mathworks, Natick, MA, USA).
Data Transformation
Model predictions were qualitatively compared to experimental time course and dose response data taken from the literature. The experimental data are obtained in vitro and for specific types of IR. In order to compare model predictions with experimental data to see if the model prediction can capture the trend of experimental time course and dose response data, the experimental data were first linearly transformed and scaled. That is, the x and y coordinates of the transformed data were obtained by multiplying a constant to the x and y coordinates of the original data. For example, if the original data are represented by (X1o, Y1o), (X2o, Y2o), ...(Xno, Yno) and the transformed data is represented by (X1t, Y1t), (X2t, Y2t), ...(Xnt, Ynt), the original data and transformed data have the following relationship: X1t/X1o = X2t/X2o = ... Xnt/Xno = a and Y1t/Y1o = Y2t/Y2o = ... Ynt/Yno = b. Where, a and b are constants. In this manner, the consistency between model predictions and transformed data actually evaluates the ability of the model to qualitatively describe the time courses of protein activities and the shapes of dose response curves after exposure to IR.
RESULTS
Model Predictions Compared with Experimental Data
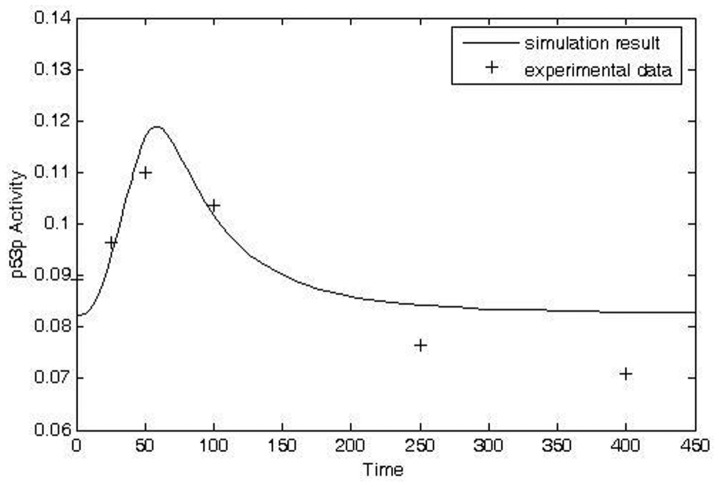
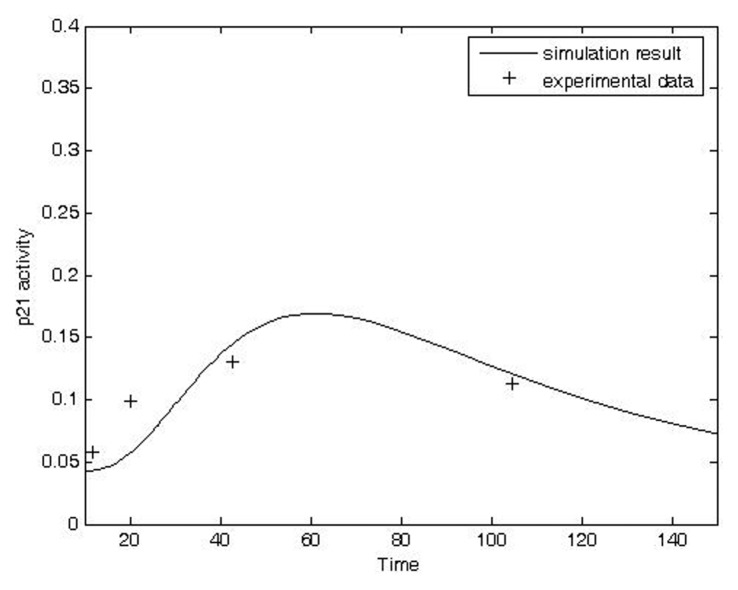
The model can be used to predict (simulate) post-irradiation effects of IR on protein activities. Total exposure to IR (IRD) is the product of the absorbed dose rate (IRR) and the exposure time (). Some post-irradiation experimental data for protein time courses are available, such as phosphorylated p53 and p21 in liver and confluent fibroblast cultures (Reynolds et al. 2004; Fournier et al. 2004). Qualitatively accurate simulations of phosphorylated p53 post irradiation activity and p21 post-irradiation activity were obtained for data from carbon-ion-irradiated confluent fibroblast cultures (Figs. 3 and 4, respectively; Fournier et al. 2004).
FIGURE 3.
Comparison of model prediction and transformed experimental data for time course of p53p activity in carbon ion-irradiated confluent fibroblasts. Original data from Fournier et al. (2004). (Simulation condition: IRR = 0.18, t = 40 unit; Experimental condition: dose: 0.5 Gy; measuring time: 0, 1.5, 3, 6, 12, 24 hours after radiation, respectively).
FIGURE 4.
Comparison of model prediction and transformed experimental data time course of p21 activity in carbon ion-irradiated confluent fibroblasts. Original data from Fournier et al. (2004). (Simulation condition: IRR = 0.5, t = 10 unit; Experimental condition: dose: 0.5 Gy; exposure duration: 0, 1.5, 3, 6, 12, 24 hours after radiation, respectively).
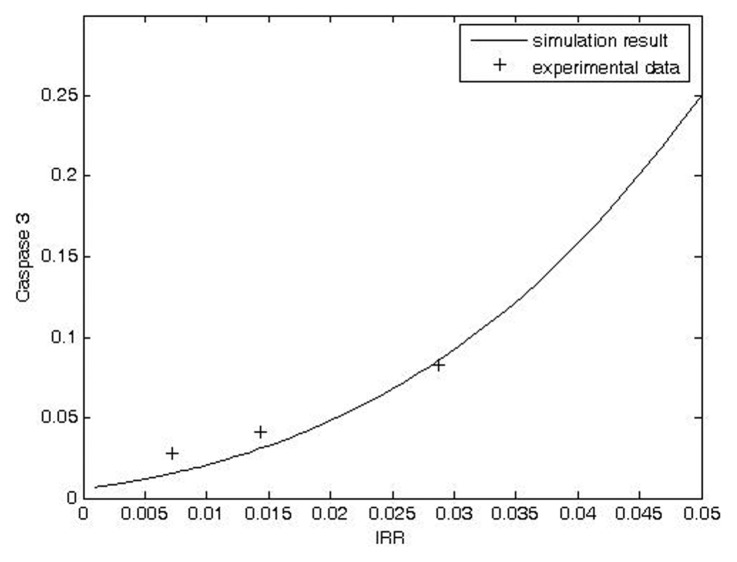
The model can also be used to predict the shapes of the dose response curves for protein activity versus IRR, DSB, or other upstream protein activity. Our simulation of a dose response curve for caspase 3 activity versus IRR provided a good description of transformed experimental data generated in a normal lymphoblastoid cell line by Fernet et al. (2003) (Fig. 5). Because Δt is a constant in the simulation, IRD is proportional to IRR in this case.
FIGURE 5.
Comparison of model prediction and transformed experimental data for dose response curve of caspase 3 activity versus 137Cs-irradiated lymphoblastoid cells. Original data from Fernet et al. (2003) (Experimental condition: dose: 5, 10 and 20 Gy).
The parameter values used for the simulations in Figs 3, 4 and 5 were held constant while the exposure values varied for different cases.
During these comparisons, a sensitivity study for the parameter values was conducted. That is, the parameter values were first assumed to fit uniform distributions with minimum value equal to 80% and maximum value equal to 120% of the assigned values. Then Monte Carlo simulation is used to randomly draw parameter values from these uniform distributions. In 18 repetitions, the consistency between the simulation results and the experimental data are still found (Fig. A-3).
Analysis of Checkpoint Arrest with Phase Plane Plots
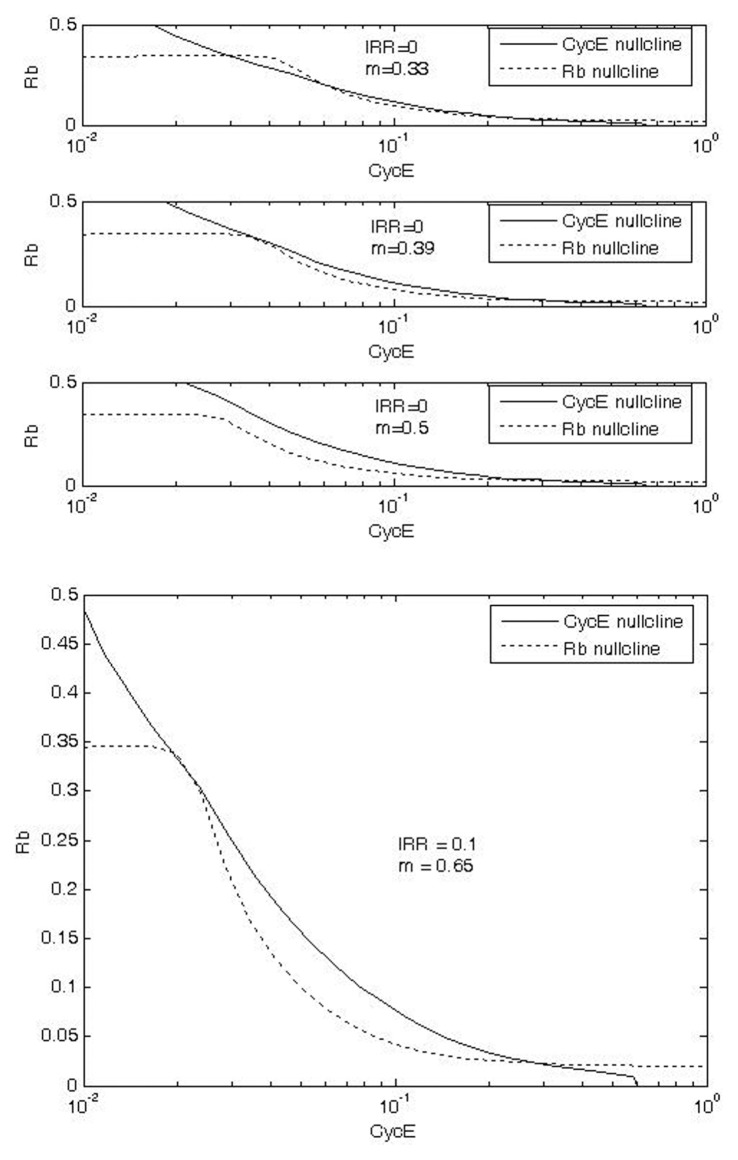
Phase plane plots were used to analyze predictions of checkpoint arrest following exposure to IR (Fig. 6). Phase plane comparisons of Rb and CycE nullclines in the G1/S checkpoint were developed for controls and IR-exposed conditions (Figs. 6a and 6b, respectively). As m (cell mass) increases from 0.33 to 0.39 to 0.5, the Rb nullcline moves to the left while CycE nullcline changes only for small values of Rb (Fig. 6a). At m = 0.33, the CycE and Rb nullclines intersect 3 times. These intersections define two stable steady states separated by an unstable steady state (the unstable steady state is also called a saddle point). The two stable steady states correspond to G1 and S. G1 is defined by high Rb and low CycE activities, while S is defined by low Rb and high CycE activities.
FIGURE 6.
Phase plane plots of CycE and Rb nullclines in G1/S checkpoint without (a) and with (b) exposure to IR. The three panels for (a) show that, as cell mass (m) increases (0.33 ‡ 0.39 ‡ 0.5), the Rb nullcline moves to the left relative to the CycE nullcline and the stable steady state at low CycE disappears, triggering the switch to S phase. Under exposure to IR, the switch is activated only at higher m, thus allowing extra time for DNA repair.
As m increases from 0.33 to 0.39, the intersection of nullclines defining G1 state converges with the saddle point. As m increases to 0.5, the intersection of nullclines that defines G1 disappears and the cell switches into the S steady state. Thus, in the absence of exposure to IR, 0.39 is a critical value of m. The G1/S transition does not occur until the cell grows to this critical size. For this initial version of our model, the key point here is not the particular value of m per se, but rather the realization that as the cell increases in size a critical point is reached at which the cell switches from G1 to S.
The critical cell size associated with the transition from G1 to S increases in the presence of DNA damage under IR exposure (Fig. 6b). In this case, with IRR = 0.1, the critical value of m is 0.65. Recall that the critical value for the control case is 0.39. Activation of the G1/S checkpoint is thus evidenced by the extra time spent in G1 as the cell grows to this larger size (m = 0.65 vs. m = 0.39). This extra time provides time for DNA repair.
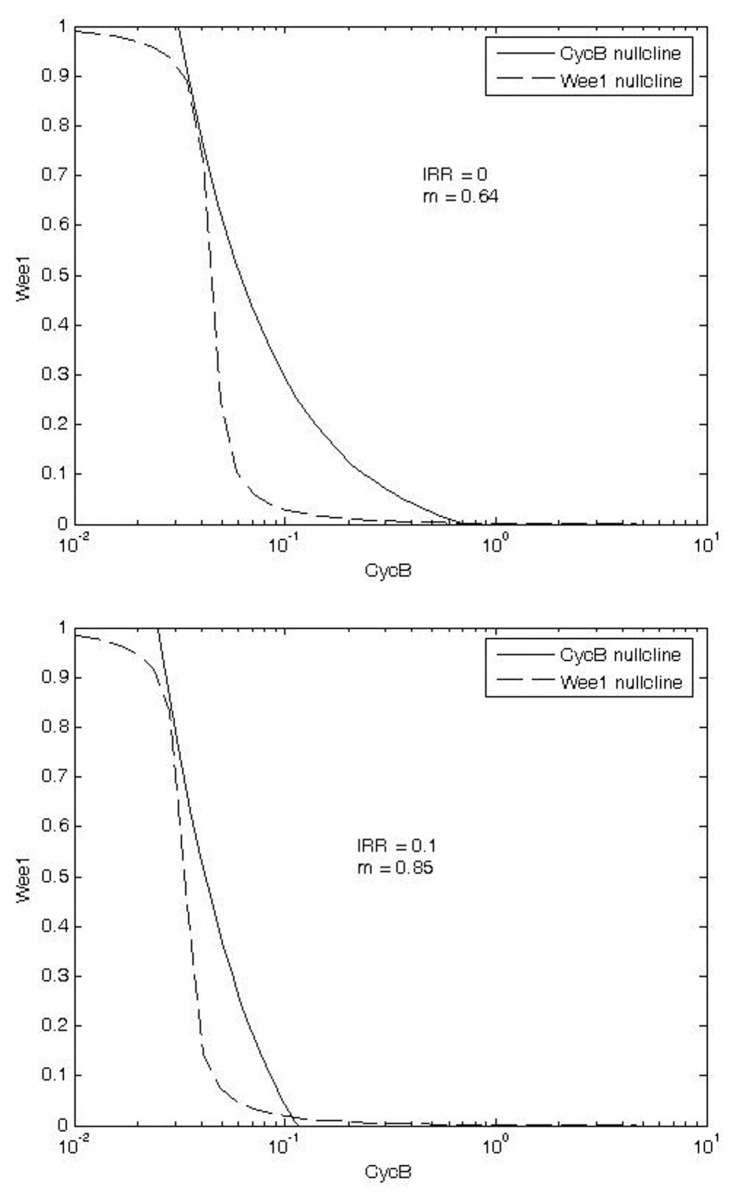
Phase plane plots comparing the CycB and Wee1 nullclines in the G1/S checkpoint also indicate that exposure to IR increases the critical value of m associated with the G2/M transition (Fig. 7).
FIGURE 7.
Phase plane plots of CycB and Wee1 nullclines in G2/M checkpoint without (a) and with (b) exposure to IR. These plots show that larger cell mass (m) is required (0.85 vs. 0.64) for switching out of G2/M in the presence of DNA damage under exposure to IR.
Analysis of the choice of cell survival or death given various doses of IR
IR exposure and its consequent DNA damage lead ultimately to cell death or cell survival. Here, we examine the biochemical mechanism by which this decision is made at the G1/S checkpoint. As CycE and Rb have switch-like behaviors at the G1 to S transition, a comparison of the IR-induced and basal activities of CycE and Rb was used to identify how the checkpoint is activated (Fig. 8). High caspase activity, which is associated with entry into the apoptotic pathway, was used as a surrogate measure of cell death.
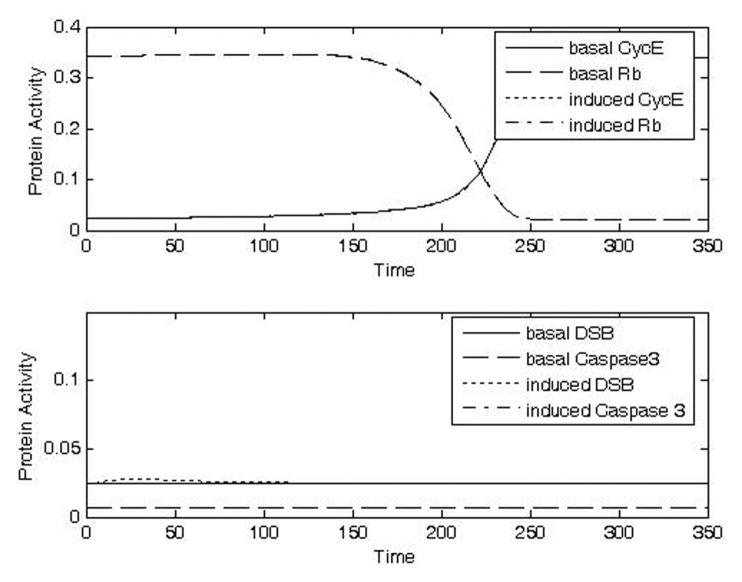
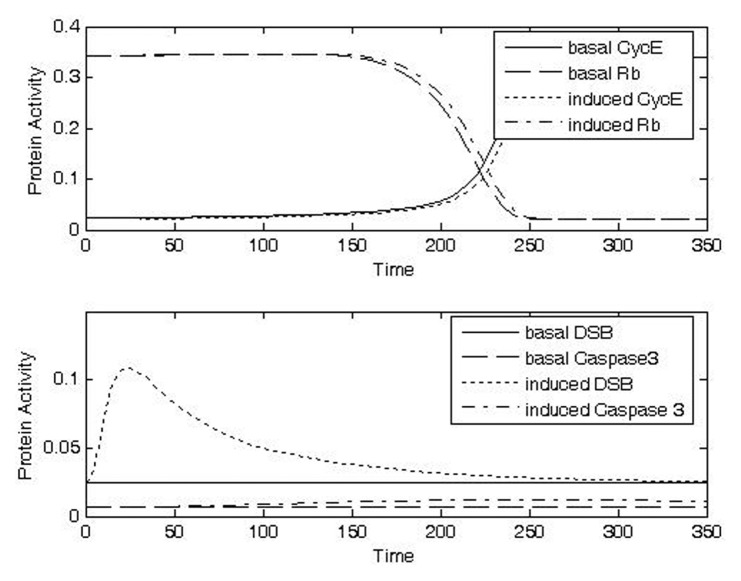
FIGURE 8A.
Analysis of protein activities at G1/S checkpoint for different doses of IR for CycE, Rb, DSB, caspase 3. Increasing doses of IR lead to increased levels of DSB (a–d, bottom panels), increased durations of checkpoint arrest as indicated by switching of CycE and Rb (a–d, top panels), and increased probability of apoptosis as indicated by increased levels of caspase3 (a–d, bottom panels). a) Case1: IRR = 0.01, Δt = 10; b) Case 2: IRR = 0.3, Δt = 10; c) Case 3: IRR = 0.5, Δt = 20; d) Case 4: IRR = 1, Δt = 20.
Exposure to a low dose of IR for a short duration (Case 1, IRR = 0.01, Δt = 10) was predicted to cause only small, transient changes in the amount of DSB (Fig. 8a, bottom panel), while no conspicuously visible effect was predicted for the activities of CycE and Rb (Fig. 8a, top panel) or caspase (Fig. 8a, bottom panel). These behaviors are consistent with rapid repair of a low level of DSB and with a consequent lack of significant propagation of signal to the cell cycle checkpoint.
Exposure to a higher dose of IR (Case 2, IRR = 0.3, Δt = 10) was predicted to delay the switch-like transitions in the activities of CycE and Rb (Fig. 8b, top panel), indicating arrest at the G1/S checkpoint. In this case the level of DNA damage (Fig. 8b, bottom panel) is sufficient to activate the signal transduction pathway and alter the time-course activities of the key regulatory activities at the checkpoint, providing additional time for DNA repair. This dose of IR was predicted to cause only a small increase caspase activity (Fig. 8b, bottom panel).
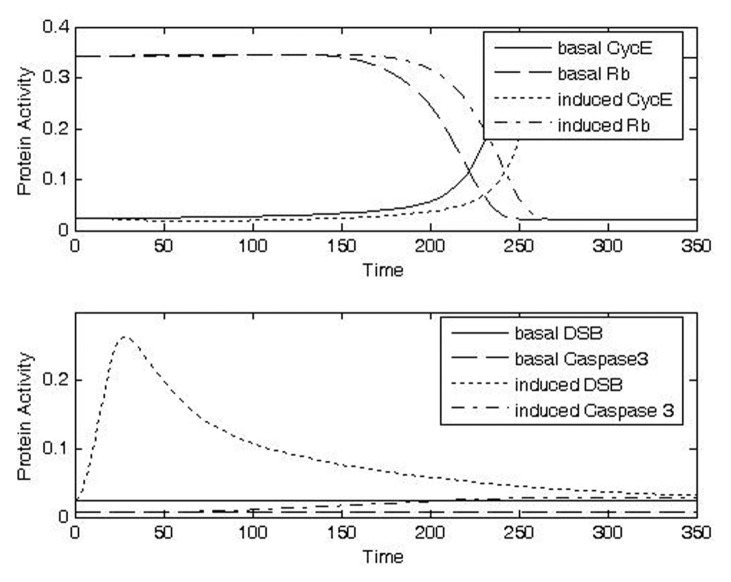
FIGURE 8B.
Exposure to a still higher dose of IR (Case 3, IRR = 0.5, Δt = 20) was predicted to trigger a correspondingly longer checkpoint arrest (Fig. 8c, top panel). The duration of the checkpoint arrest was predicted to be sufficient in this case to allow caspase activity to increase to a level associated with an apparently improved possibility of entry into the apoptotic pathway (Fig. 8c, bottom panel). With a sufficiently long checkpoint arrest, it is reasonable to think that some of the cells go into apoptosis during the arrest period; thus this prediction is consistent with data showing that cell survival is reduced in association with p53-dependent G1 arrest (Gupta et al. 1996).
FIGURE 8C.
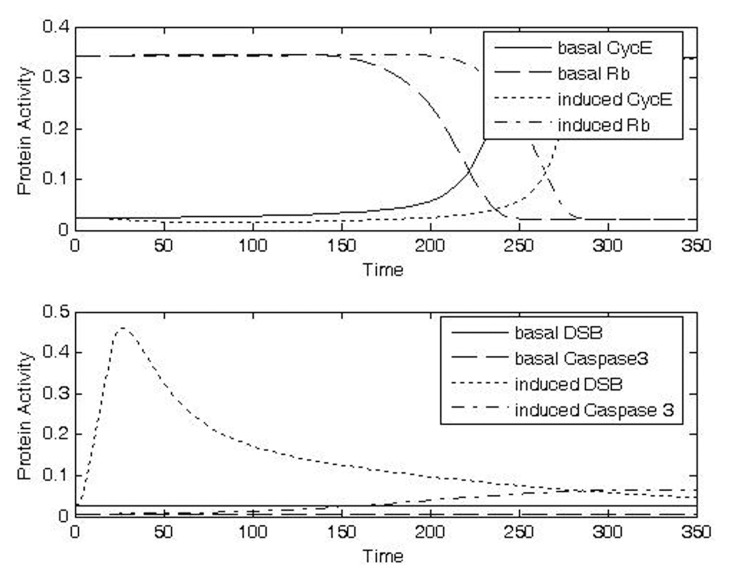
At the highest dose evaluated (Case 4, IRR = 1, Δt = 20), caspase activity was predicted to increase the likelihood for apoptosis dramatically (Fig. 8d, bottom panel). An increased likelihood of apoptosis in the presence of high levels of DNA damage has been seen experimentally (Sun et al. 2005).
FIGURE 8D.
Dose response curve for checkpoint arrest and caspase 3 activities
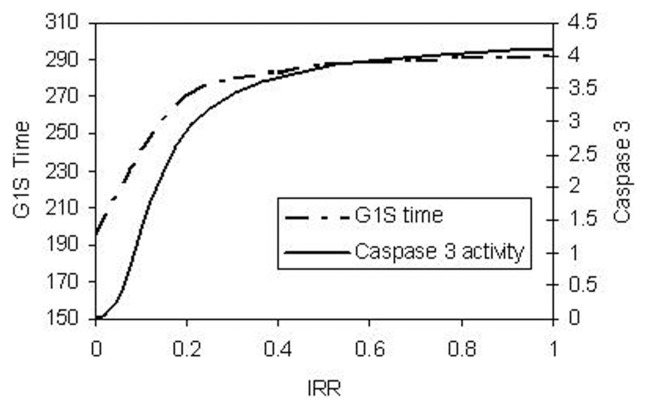
The length of time spent at the G1/S checkpoint and caspase 3 activities are functions of the dose rate IRR (Fig. 9). Both the time in the G1/S checkpoint and caspase 3 activity initially increase with IR dose and then saturate.
FIGURE 9.
G1/S time and caspase 3 activities as a function of IRR.
Robustness analysis
The robustness of checkpoint activation in response to DNA damage was analyzed by comparing the relative duration of checkpoint arrest when mediated only by the p53 dependent pathway (Fig. 2) with the duration resulting from the combination of p53- and Chk2-mediated signaling (Fig. A-1). The robustness of activation of the apoptotic pathway was analyzed by comparing predicted caspase 3 activity based only on p53 dependent signaling with combined signaling through p53 and p73 (Fig. A-2). A detailed description of these results is given in Appendix.
DISCUSSION
Our long-range goal is to understand how the relevant signaling pathways and adaptive capabilities of cells exposed to IR influence the shapes of dose-response curves for IR-mediated cancers. The present work, a first step towards this goal, describes an initial (i.e., 1st generation) computational model of signaling pathways linking IR-mediated DNA damage with cell cycle checkpoint arrest and apoptosis. In this model, Module 1 (Fig. 2) links IR exposure to DNA damage but does not contain detailed information about specific kinds of IR, doses and dose-rates, the production of ROS, the kinds of DNA damage produced, and the repair of the damage. While these are all important components of the IR-exposure-health effect continuum, detailed descriptions were beyond the scope of the current investigation. Our description of the G2/M checkpoint in Module 3 (Fig. 2) followed the work of Tyson et al. (2001). We therefore focused our efforts largely on the development of Module 2 (Fig. 2) and on the G1/S checkpoint component of Module 3 (Fig. 2) with the goal of producing a reasonably detailed though admittedly not comprehensive description of the pathway that senses DNA damage and transmits signals to cell cycle checkpoints and the apoptotic pathway.
Our main objective in developing the 1st-generation model was to describe the relevant biology only at the level of detail necessary to generate qualitatively accurate representations of checkpoint activation and of the choice between checkpoint activation and apoptosis. The biochemical detail in these modules, including signaling pathways and regulatory switches at the checkpoints, is sufficient for this purpose: DNA damage activates checkpoints; more severe damage leads to extended delays at checkpoints and greater probabilities that cells will apoptose. The current model does successfully reproduce measured time-courses of key proteins (e.g., p53, p21, and caspase 3, Figs. 3, 4 and 5, respectively). These are activities expected to underlie nonlinear dose-response behaviors in cells and tissues exposed to IR. However, since measured values are lacking for many of the parameters of the model (Table A-3), its predictive capabilities are best categorized as qualitative but not quantitative. Development of a quantitatively accurate version of the model would require (1) for each parameter either a data-derived value or a complete sensitivity analysis (e.g. considering the correlations between the parameter values) showing that model behaviors are not sensitive to its value and (2) additional dose-response and time course datasets for the activities of key signaling and regulatory proteins.
Our model contains less molecular-level details than others more concerned with the mechanistic details of cell cycle control and programmed cell death (e.g., Albeck et al. 2008; Chen et al. 2007; Csikasz-Nagy et al. 2009; Cui et al. 2008; Eissing et al. 2004; Han et al. 2005; Li et al. 2009; Qu et al. 2004; Zamborszky et al. 2007; Zhang et al. 2009). The recent model of apoptosis developed by Tyson and co-workers for example, (Zhang et al. 2009), contains a detailed description of the apoptotic pathway reflecting a new, more detailed understanding of the underlying signaling and regulatory motifs (e.g. BH3, cytochrome c, SMAC and XIAP, feedbacks between BH3&BCL and caspase 9 & caspase 3). Further development of the present model, where both qualitative and quantitative accuracy would be desired, are likely to incorporate the kinds of mechanistic insights being obtained by these more mechanistically advanced modeling efforts.
The data used to develop the model were obtained from a variety of test systems. Ideally, data would be obtained under uniform experimental conditions using a single biological model. At the level of the biology it describes, the computational model is thus a chimera. Much of the biology in the model is evolutionarily conserved, leading us to think that the model structure is likely to be robust across cell types and species. The chimerical nature of the model should, however, be viewed as a source of uncertainty in the evaluation of its predictions.
It is interesting to note that Albert and Othmer (2003) found that the spatial and temporal pattern of expression of the segment polarity genes in Drosophila melanogaster is determined by the topology of the gene network and by whether components are absent or present. The behavior of the network is relatively insensitive to the absolute levels of the mRNAs and proteins and the functional details of their interactions. There is thus some hope that insight into key features of the behavior of the signaling pathways and regulatory activities that coordinate DNA repair, cell cycle checkpoints and apoptosis will not require exhaustive enumeration of protein concentrations and reaction rate constants. Additional work will be required to tell whether or not this is the case.
Different versions of Module 2 incorporating parallel signaling pathways (e.g., Figs. A-1 and A-2) provided a capability for examination of robustness, an essential feature of complex biological systems (Aderem and Smith, 2004; Kitano, 2004). For example, parallel signaling was predicted to increase the duration of the delay at the G1/S checkpoint for a given exposure to IR (Fig. A-4), allowing more time for both DNA repair and for caspases activation leading to apoptosis. That is, a second protective response from the parallel signaling pathway will synergize the response from the initial pathway and makes the system more robust.
The current model is effectively a model of average behavior of cell population. In future versions the model could be extended to describe a virtual tissue, - a heterogeneous population of cells - thereby providing a capability for simultaneously describing cells in different phases of the cell cycle as well as the cell-to-cell communication involved in bystander effects (Matsumoto et al. 2004). While several models for bystander effects have been described (Brenner et al. 2001; Nikjoo and Khvostunov, 2003; Little et al. 2005), none of them contains such a level of biological detail.
Entry into the apoptotic pathway is determined by the activity of biochemical signaling pathways while necrosis is due to relatively large amounts of damage associated with generalized disruption of signaling and of cellular architecture. In developing the initial model, we were primarily interested in describing intracellular signaling pathways linking detection of DNA damage with activation of cell cycle checkpoint controls and with the apoptotic pathway. These signaling-mediated events are likely to be the important determinants of the shape of the dose-response curves at low doses. At somewhat higher doses, necrosis will become more important and a description of necrosis could be included in subsequent versions of the model.
Since our long-range goal is to understand the dose-responses for IR-mediated cancers, linkage to a multistage, clonal growth model for cancer (Moolgavkar and Knudson, 1981; Moolgavkar et al. 1988) is another desirable extension of the current model. Initial results show that the model developed here, when linked to the two-stage clonal growth cancer model, can describe either a monotonic increasing or a J-shaped dose response curve, depending on the parameter values used (Zhao and Ricci, 2010). Saturation of checkpoint arrest time, as shown in Fig. 9, is the underlying mechanism of the J-shape (Zhao and Ricci, 2010).
While the current model was motivated by an interest in the health effects of IR, its structure is relevant to other environmental stressors that generate DNA DSB, since the signaling pathways from DSB to the cell cycle and to apoptosis are presumably independent of the character of the stressor inducing the DSB. Furthermore, other protein activities and signaling pathways could be incorporated as relevant new data become available or to allow the model to address new questions. For instance, the description of DNA repair in the current version of the model is superficial. More detailed descriptions could be developed to describe, for example, the actual biochemical mechanisms of repair of DSB.
In conclusion, we have developed an initial example of a conceptual approach that can improve our ability to predict dose- and time-response curves for adverse health effects related to IR exposure. Future research leading to refinements in model structure and to laboratory measurements of key parameter values could lead to more mature models with a robust ability to predict health risks on the basis of relevant biological mechanisms. Such a model could serve to either confirm the suitability of current policy- or default-based approaches to risk assessments for IR or, potentially, to suggest that something other than low-dose linearity is more appropriate.
ACKNOWLEDGEMENT
The authors thank Qiang Zhang for his stimulating discussion and contribution to review. The research was originally supported by grant to the CIIT Centers for Health Research (now Hamner Institutes for Health Sciences) from Department of Energy BER Grant No. DE-FG02-03ER63669. Yuchao Zhao also thanks the funding from State Key Joint Laboratory of Environment Simulation and Pollution Control (Beijing Normal University) 10Y05ESPCN and the Research Fund for the Doctoral Program of Higher Education (SRFDP) for recent support.
ABBREVIATIONS
- CycE
Total Cyclin E/Cdk 2 dimer
- CycEdp
Dephosphorylated form of CycE
- E2F
E2F transcription factor
- Rb
Dephosphorylated (p−) Retinoblastoma
- Rbp
Phosphorylated (p+) Rb
- RbE2F
Rb_E2F complex
- RbpE2F
Rb_E2F complex with Rb p+
- p27
p27 p−
- p27p
p27p+
- p27CycE
p27_Cyclin E_Cdk trimer
- CycEp21
P21_Cyclin E_Cdk trimer
- cAbl
cAblp−
- cAblp
cAblp+
- p73p
p73p+
- RbcAbl
Rb_cAbl complex
- m
Cell mass
- CycB
Total Cyclin B/Cdk1 dimer
- CycBdp
Dephosphorylated form of CycB
- Cdc20T
Total Cdc20
- Cdc20A
Activated Cdc20
- IEP
Hypothetical “Intermediary enzyme”
- Dissociator
The sum of GADD45 and 1433 sigma
Footnotes
DISCLAIMER
This research was supported by the Office of Science (BER), U.S. Department of Energy, Grant No. DE-FG02-03ER63669 to the Hamner Institutes for Health Sciences.
This research may not necessarily reflect the views of the U.S. Environmental Protection Agency and no official endorsement should be inferred.
REFERENCES
- Adams M, Carpenter PB. Tying the loose ends together in DNA double strand break repair with 53BP1. Cell Div. 2006;1:1–19. doi: 10.1186/1747-1028-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aderem A, Smith KD. A systems approach to dissecting immunity and inflammation. Semin Immunol. 2004;16:55–67. doi: 10.1016/j.smim.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Agarwal ML, Taylor WR, Chernov MV, Chernova OB, Stark GR. The p53 network. J Biol Chem. 1998;27:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- Akli S, Zheng PJ, Multani AS, Wingate HF, Pathak S, Zhang N, et al. Tumor-specific low molecular weight forms of cyclin E induce genomic instability and resistance to p21, p27, and antiestrogens in breast cancer. Cancer Res. 2004;64:3198–3208. doi: 10.1158/0008-5472.can-03-3672. [DOI] [PubMed] [Google Scholar]
- Albeck JB, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitiative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30(1):11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert R, Othmer HG. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J Theor Biol. 2003;223:1–18. doi: 10.1016/s0022-5193(03)00035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeli D, Ferrell JE, Sontag ED. Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc Natl Acad Sci USA. 2004;101(7):1822–1827. doi: 10.1073/pnas.0308265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo M, Raychaudhuri P. Retinoblastoma-repression of E2F-dependent transcription depends on the ability of the retinoblastoma protein to interact with E2F and is abrogated by the adenovirus E1A oncoprotein. Nucleic Acids Res. 1992;20:5947–5954. doi: 10.1093/nar/20.22.5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam EI, de Toledo SM, Waker AJ, Little JB. High and low fluences of alpha-particles induce a G1 checkpoint in human diploid fibroblasts. Cancer Res. 2000;60:2623–2631. [PubMed] [Google Scholar]
- Bahassi el M, Hennigan RF, Myer DL, Stambrook PJ. Cdc25C phosphorylation on serine 191 by Plk3 promotes its nuclear translocation. Oncogene. 2004;23:2658–2663. doi: 10.1038/sj.onc.1207425. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. Pathways governing G1/S transition and their response to DNA damage. FEBS Lett. 2001;490:117–122. doi: 10.1016/s0014-5793(01)02114-7. [DOI] [PubMed] [Google Scholar]
- BEIR. National Academy of Sciences. Washiongton, DC: National Academies Press; 2006. Health risks from exposure to low levels of ionizing radiation: BEIR VII Phase 2 Committee to Assess Health Risks from Exposure to Low Levels of Ionzing Radiation Board of Radiation Effects Research Division of Earth and Life Studeis National Research Council of the National Academies. [Google Scholar]
- Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. [DOI] [PubMed] [Google Scholar]
- Boreham D, Dolling J, Somers C, Quinn J, Mitchel REJ. The adaptive response and protection against heritable mutation and fetal malformations. Dose Response. 2006;4(4):317–326. doi: 10.2203/dose-response.06-104.Boreham. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner DJ, Little JB, Sachs RK. The bystander effect in radiation oncogenesis: II. A quantitative model. Radiat Res. 2001;155:402–408. doi: 10.1667/0033-7587(2001)155[0402:tbeiro]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Ricci PF. Encyclopedia of Environmental Health. Elsevier Publishers; 2010. Hormesis in Environmental Health. (In press) [Google Scholar]
- Chen C, Cui J, Lu R, Wang S, Zhang S. Modeling of the role of a Bax-activation switch in the mitochondrial apoptosis decision. Biophys J. 2007;92:4303–4315. doi: 10.1529/biophysj.106.099606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DT, Linette GP, Boise LH, White LS, Thompson CB, Korsmeyer SJ. Bcl-XL and Bcl-2 repress a common pathway of cell death. J Exp Med. 1995;182:821–828. doi: 10.1084/jem.182.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BL. Test of the linear-no threshold theory of radiation carcinogenesis for inhaled radon decay products. Health Phys. 1995;68(2):157–174. doi: 10.1097/00004032-199502000-00002. [DOI] [PubMed] [Google Scholar]
- Conolly RB, Lutz WK. Nonmonotonic dose-response relationships: mechanistic basis, kinetic modeling, and implications for risk assessment. Toxicol Sci. 2004;77:151–157. doi: 10.1093/toxsci/kfh007. [DOI] [PubMed] [Google Scholar]
- Criswell T, Leskov K, Miyamoto S, Luo G, Boothman DA. Transcription factors activated in mammalian cells after clinically relevant doses of ionizing radiation. Oncogene. 2003;22:5813–5827. doi: 10.1038/sj.onc.1206680. [DOI] [PubMed] [Google Scholar]
- Csikasz-Nagy A, Kapuy O, Toth A, Pal C, Jensen LJ, Uhlmann F, Tyson JJ, Novak B. Cell cycle regulation by feed-forward loops coupling transcription and phosphorylation. Mol Syst Biol. 2009;5:236. doi: 10.1038/msb.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J, Chen H, Lu H, Sun T, Shen P. Two independent positive feedbacks and bistability in the bcl-2 apoptotic switch. PloS ONE. 2008;3:e1469. doi: 10.1371/journal.pone.0001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T, Zeng G, Hooker A, Bhat M, Scott BR, Turner DR, Sykes PJ. Adaptive response for chromosomal inversions in pKZ1 mouse prostate induced by low doses of X radiation delivered after a high dose. Radiat Res. 2007;167:682–692. doi: 10.1667/RR0764.1. [DOI] [PubMed] [Google Scholar]
- DeGregori J. The Rb network. J Cell Sci. 2004;117:3411–3413. doi: 10.1242/jcs.01189. [DOI] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Eissing T, Conzelmann H, Gilles ED, Allgower F, Bullinger E, Scheurich P. Bistability analyses of a caspase activation model for receptor-induced apoptosis. 2004;279(35):36892–36897. doi: 10.1074/jbc.M404893200. [DOI] [PubMed] [Google Scholar]
- Fei P, El-Deiry WS. P53 and radiation responses. Oncogene. 2003;22:5774–5783. doi: 10.1038/sj.onc.1206677. [DOI] [PubMed] [Google Scholar]
- Feinendegen LE, Bond VP, Sondhaus CA, Altman KI. Cellular signal adaptation with damage control at low doses versus the predominance of DNA damage at high doses. C R Acad Sci III. 1999;322:245–251. doi: 10.1016/s0764-4469(99)80051-1. [DOI] [PubMed] [Google Scholar]
- Feinendegen LE. Reactive oxygen species in cell responses to toxic agents. Hum Exp Toxicol. 2002;21:85–90. doi: 10.1191/0960327102ht216oa. [DOI] [PubMed] [Google Scholar]
- Fernet M, Angele S, Dork T, Hall J. Variation in radiation-induced apoptosis in ataxia telangiectasia lymphoblastoid cell lines. Int J Radiat Biol. 2003;79:193–202. doi: 10.1080/0955300031000085731. [DOI] [PubMed] [Google Scholar]
- Fournier C, Wiese C, Taucher-Scholz G. Accumulation of the cell cycle regulators TP53 and CDKN1A (p21) in human fibroblasts after exposure to low- and high-LET radiation. Radiat Res. 2004;161:675–684. doi: 10.1667/rr3182. [DOI] [PubMed] [Google Scholar]
- Furukawa Y. Cell cycle control genes and hematopoietic cell differentiation. Leuk Lymphoma. 2002;43:225–231. doi: 10.1080/10428190290005973. [DOI] [PubMed] [Google Scholar]
- Fussenegger M, Bailey JE, Varner J. A mathematical model of caspase function in apoptosis. Nat Biotechnol. 2000;18:768–774. doi: 10.1038/77589. [DOI] [PubMed] [Google Scholar]
- Gala S, Marreiros A, Stewart GJ, Williamson P. Overexpression of E2F-1 leads to cytokine-independent proliferation and survival in the hematopoietic cell line BaF-B03. Blood. 2001;97:227–234. doi: 10.1182/blood.v97.1.227. [DOI] [PubMed] [Google Scholar]
- Gardner RD, Burke DJ. The spindle checkpoint: two transitions, two pathways. Trends Cell Biol. 2000;10:154–158. doi: 10.1016/s0962-8924(00)01727-x. [DOI] [PubMed] [Google Scholar]
- Gupta N, Vij R, Haas-Kogan DA, Israel MA, Deen DF, Morgan WF. Cytogenetic damage and the radiation-induced G1-phase checkpoint. Radiat Res. 1996;145:289–298. [PubMed] [Google Scholar]
- Halaban R. Melanoma cell autonomous growth: the Rb/E2F pathway. Cancer Metastasis Rev. 1999;18:333–343. doi: 10.1023/a:1006396104073. [DOI] [PubMed] [Google Scholar]
- Han Z, Yang L, MacLellan WR, Weiss JN, Qu Z. Hysteresis and cell cycle transition: How crutial is it? Biophys J. 2005;88:1626–1634. doi: 10.1529/biophysj.104.053066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliakis G, Wang Y, Guan J, Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- Kapuy O, He E, Lopez-Aviles S, Uhlmann F, Tyson JJ, Novak B. System-level feedbacks control cell cycle progression. FEBS Lett. 2009;22 doi: 10.1016/j.febslet.2009.08.023. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WK. Human topoisomerase II function, tyrosine phosphorylation and cell cycle checkpoints. Proc Soc Exp Biol Med. 1998;217:327–334. doi: 10.3181/00379727-217-44240. [DOI] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nature Reviews Genetics. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Laiho M, Latonen L. Cell cycle control, DNA damage checkpoints and cancer. Ann Med. 2003;35:391–397. doi: 10.1080/07853890310014605. [DOI] [PubMed] [Google Scholar]
- Lane HA, Motoyama AB, Beuvink I, Hynes NE. Modulation of p27/Cdk2 complex formation through 4D5-mediated inhibition of HER2 receptor signaling. Ann Oncol. 2001;12(Suppl 1):S21–22. doi: 10.1093/annonc/12.suppl_1.s21. [DOI] [PubMed] [Google Scholar]
- Leng X, Connell-Crowley L, Goodrich D, Harper JW. S-Phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr Biol. 1997;7:709–712. doi: 10.1016/s0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J Cell Sci. 2000;113(Pt 10):1661–1670. doi: 10.1242/jcs.113.10.1661. [DOI] [PubMed] [Google Scholar]
- Li P, Li C, Zhao X, Zhang X, Nicosia SV, Bai W. p27(Kip1) stabilization and G(1) arrest by 1,25-dihydroxyvitamin D(3) in ovarian cancer cells mediated through down-regulation of cyclin E/cyclin-dependent kinase 2 and Skp1-Cullin-F-box protein/Skp2 ubiquitin ligase. J Biol Chem. 2004;279:25260–25267. doi: 10.1074/jbc.M311052200. [DOI] [PubMed] [Google Scholar]
- Li S, Brazhnik P, Sobral B, Tyson JJ. Temporal controls of the asymmetric cell division cycle in Caulobacter crescentus. PLoS Comput Biol. 2009;5(8):e1000463. doi: 10.1371/journal.pcbi.1000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MP. Risks associated with ionizing radiation. Br Med Bull. 2003;68:259–275. doi: 10.1093/bmb/ldg031. [DOI] [PubMed] [Google Scholar]
- Little MP, Filipe JA, Prise KM, Folkard M, Belyakov OV. A model for radiation-induced bystander effects, with allowance for spatial position and the effects of cell turnover. J Theor Biol. 2005;232:329–338. doi: 10.1016/j.jtbi.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Takahashi A, Ohnishi T. Radiation-induced adaptive responses and bystander effects. Biol Sci Space. 2004;18:247–254. doi: 10.2187/bss.18.247. [DOI] [PubMed] [Google Scholar]
- Mikkelsen RB, Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 2003;22:5734–5754. doi: 10.1038/sj.onc.1206663. [DOI] [PubMed] [Google Scholar]
- Mitchel REJ. Cancer and low dose responses in vivo: implications for radiation protection. Dose–Response. 2007a;5:284–291. doi: 10.2203/dose-response.07-014.Mitchel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel REJ. Low doses of radiation reduce risk in vivo. Dose-Response. 2007b;5:1–10. doi: 10.2203/dose-response.06-109.Mitchel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg KH, Bell DW, Wahrer DC, Haber DA, Hariharan IK. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- Moolgavkar SH, Dewanji A, Venzon DJ. A stochastic two-stage model for cancer risk assessment. I. The hazard function and the probability of tumor. Risk Anal. 1988;8:383–392. doi: 10.1111/j.1539-6924.1988.tb00502.x. [DOI] [PubMed] [Google Scholar]
- Moolgavkar SH, Knudson AG., Jr Mutation and cancer: a model for human carcinogene-sis. J Natl Cancer Inst. 1981;66:1037–1052. doi: 10.1093/jnci/66.6.1037. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- Nikjoo H, Khvostunov IK. Biophysical model of the radiation-induced bystander effect. Int J Radiat Biol. 2003;79:43–52. [PubMed] [Google Scholar]
- Novak B, Pataki Z, Ciliberto A, Tyson JJ. Mathematical model of the cell division cycle of fission yeast. Chaos. 2001;11:277–286. doi: 10.1063/1.1345725. [DOI] [PubMed] [Google Scholar]
- Novak B, Tyson JJ, Gyorffy B, Csikasz-Nagy A. Irreversible cell-cycle transitons are due to systmes-level feedback. Nat Cell Biol. 2007;9:724–728. doi: 10.1038/ncb0707-724. [DOI] [PubMed] [Google Scholar]
- Ohtani K. Implication of transcription factor E2F in regulation of DNA replication. Front Biosci. 1999;4:D793–804. doi: 10.2741/ohtani. [DOI] [PubMed] [Google Scholar]
- Pietenpol JA, Stewart ZA. Cell cycle checkpoint signaling: cell cycle arrest versus apoptosis. Toxicology. 2002;181–182:475–481. doi: 10.1016/s0300-483x(02)00460-2. [DOI] [PubMed] [Google Scholar]
- Qin XQ, Chittenden T, Livingston DM, Kaelin WG., Jr Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- Qu Z, MacLellan WR, Weiss JN. Dynamics of the cell cycle: checkpoints, sizers, and timers. Biophys J. 2003a;85:3600–3611. doi: 10.1016/S0006-3495(03)74778-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z, Weiss JN, MacLellan WR. Regulation of the mammalian cell cycle: a model of the G1-to-S transition. Am J Physiol Cell Physiol. 2003b;284:C349–364. doi: 10.1152/ajpcell.00066.2002. [DOI] [PubMed] [Google Scholar]
- Qu Z, Weiss JN, MacLellan WR. Coordination of cell growth and cell division: a mathematical study. J Cell Sci. 2004;117:4199–4207. doi: 10.1242/jcs.01294. [DOI] [PubMed] [Google Scholar]
- Redpath J, Liang D, Taylor T, James C, Christie E, Elmore E. The shape of dose-response curve for radiation-induced neoplastic transformation in vitro: evidence for an adaptive response against neoplastic transformation at low doses of low-LET radiation. Radiat Res. 2001;156:700–707. doi: 10.1667/0033-7587(2001)156[0700:tsotdr]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Reynolds R, Witherspoon S, Fox T. The infant mouse as a in vivo model for the detection and study of DNA damage-induced changes in the liver. Mol Carcinog. 2004;40:62–72. doi: 10.1002/mc.20017. [DOI] [PubMed] [Google Scholar]
- Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- Sakai L, Nomura T, Ina Y. Enhancement of bio-protective functions by low dose/dose-rate radiation. Dose-Response. 2006;4:327–332. doi: 10.2203/dose-response.06-115.Sakai. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi T, Hunter T. Induction of growth arrest and cell death by overexpression of the cyclin-Cdk inhibitor p21 in hamster BHK21 cells. Oncogene. 1998;16:369–380. doi: 10.1038/sj.onc.1201539. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18(22):2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Sun F, Anantharam V, Latchoumycandane C, Kanthasamy A, Kanthasamy AG. Dieldrin Induces Ubiquitin-Proteasome Dysfunction in {alpha}-Synuclein Overexpressing Dopaminergic Neuronal Cells and Enhances Susceptibility to Apoptotic Cell Death. J Pharmacol Exp Ther. 2005;315:69–79. doi: 10.1124/jpet.105.084632. [DOI] [PubMed] [Google Scholar]
- Tan YM, Butterworth BE, Gargas ML, Conolly RB. Biologically motivated computational modeling of chloroform cytolethality and regenerative cellular proliferation. Toxicol Sci. 2003;75:192–200. doi: 10.1093/toxsci/kfg152. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- Thomas A, Giesler T, White E. p53 mediates bcl-2 phosphorylation and apoptosis via activation of the Cdc42/JNK1 pathway. Oncogene. 2000;19:5259–5269. doi: 10.1038/sj.onc.1203895. [DOI] [PubMed] [Google Scholar]
- Tubiana M, Aurengo A, Averbeck D, Masse R. Recent reports on the effect of low doses of ionizing radiation and its dose–effect relationship. Radiat Environ Biophys. 2006;44:245–251. doi: 10.1007/s00411-006-0032-9. [DOI] [PubMed] [Google Scholar]
- Tubiana M, Feinendegen LE, Yang Ch, Kaminski JM. The linear no–threshold relationship is inconsistent with radiation biologic and experimental data. Radiology. 2009;251:13–22. doi: 10.1148/radiol.2511080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Chen K, Novak B. Network dynamics and cell physiology. Nat Rev Mol Cell Biol. 2001;2:908–916. doi: 10.1038/35103078. [DOI] [PubMed] [Google Scholar]
- Tyson JJ, Novak B. Regulation of the eukaryotic cell cycle: molecular antagonism, hysteresis, and irreversible transitions. J Theor Biol. 2001;210:249–263. doi: 10.1006/jtbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- Tyson JJ, Csikasz-Nagy A, Novak B. The dynamics of cell cycle regulation. Bioessays. 2002;24:1095–1109. doi: 10.1002/bies.10191. [DOI] [PubMed] [Google Scholar]
- Upton AC. Carcinogenic effects of low-level ionizing radiation: problems and prospects. In Vivo. 2002;16:527–533. [PubMed] [Google Scholar]
- Wang B, Ohyama H, Shang Y, Fujita K, Tanaka K, Nakajima T, et al. Adaptive response in embryogenesis: IV. Protective and detrimental bystander effects induced by X radiation in cultured limb bud cells of fetal mice. Radiat Res. 2004;161:9–16. doi: 10.1667/rr3106. [DOI] [PubMed] [Google Scholar]
- Weintraub SJ, Prater CA, Dean DC. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- Wells J, Yan PS, Cechvala M, Huang T, Farnham PJ. Identification of novel pRb binding sites using CpG microarrays suggests that E2F recruits pRb to specific genomic sites during S phase. Oncogene. 2003;22:1445–1460. doi: 10.1038/sj.onc.1206264. [DOI] [PubMed] [Google Scholar]
- Yamasaki L. Role of the RB tumor suppressor in cancer. Cancer Treat Res. 2003;115:209–239. doi: 10.1007/0-306-48158-8_9. [DOI] [PubMed] [Google Scholar]
- Yang J, Xu ZP, Huang Y, Hamrick HE, Duerksen-Hughes PJ, Yu YN. ATM and ATR: sensing DNA damage. World J Gastroenterol. 2004;10:155–160. doi: 10.3748/wjg.v10.i2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborszky J, Hong CI, Csikasz NA. Computational analysis of mammalian cell division gated by a circadian clock: quantized cell cycle and cell size control. J Biol Rhythms. 2007;22(6):542–553. doi: 10.1177/0748730407307225. [DOI] [PubMed] [Google Scholar]
- Zhang T, Brazhnik P, Tyson JJ. Computational Analysis of Dynamical Responses to the Intrinsic Pathway of Programmed Cell Death. Biophys J. 2009;97:415–434. doi: 10.1016/j.bpj.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ricci PF. Modeling dose-response at low dose: a systems biology approach for ionizing radiation. Dose Response. 2010;8(4):456–477. doi: 10.2203/dose-response.09-054.Zhao. [DOI] [PMC free article] [PubMed] [Google Scholar]