Background: Although CB1, the most abundant neuronal receptors, and CB2 receptors are co-expressed in neurons, the CB1-CB2 relationship is unknown.
Results: CB1 and CB2 receptors form heteromers in neuronal cells and in the brain.
Conclusion: Activation of either receptor leads to negative modulation of the partner receptor via heteromers.
Significance: These heteromers may explain previous conflicting results and serve as therapeutic targets.
Keywords: Cannabinoid Receptors, Cannabinoids, G Protein-coupled Receptors (GPCRs), Membrane Proteins, Neurons, Heterodimers
Abstract
Exploring the role of cannabinoid CB2 receptors in the brain, we present evidence of CB2 receptor molecular and functional interaction with cannabinoid CB1 receptors. Using biophysical and biochemical approaches, we discovered that CB2 receptors can form heteromers with CB1 receptors in transfected neuronal cells and in rat brain pineal gland, nucleus accumbens, and globus pallidus. Within CB1-CB2 receptor heteromers expressed in a neuronal cell model, agonist co-activation of CB1 and CB2 receptors resulted in a negative cross-talk in Akt phosphorylation and neurite outgrowth. Moreover, one specific characteristic of CB1-CB2 receptor heteromers consists of both the ability of CB1 receptor antagonists to block the effect of CB2 receptor agonists and, conversely, the ability of CB2 receptor antagonists to block the effect of CB1 receptor agonists, showing a bidirectional cross-antagonism phenomenon. Taken together, these data illuminate the mechanism by which CB2 receptors can negatively modulate CB1 receptor function.
Introduction
The endocannabinoid system is known to have a broad impact on a variety of tissues. It has been shown to heavily influence cardiovascular and immune systems as well as to control progenitor cell proliferation (1–3). More recently, the endocannabinoid system has emerged as a major player in the complex web of neuromodulators, constituting a new intercellular communication network mainly involved in the control of neurotransmitter release (4–7). By acting as retrograde messengers at various synapses, these endogenous arachidonic acid derivatives participate in controlling processes, such as motor activity, memory and learning, appetite, emesis, nociception, and some motivational responses (8–14). There are two known cannabinoid receptors, CB1 and CB2, with strong evidence of a third in the form of GPR55, and all have been considered as therapeutic targets for basal ganglia disorders (15, 16). CB1 receptors mediate psychoactivity, whereas CB2 receptor-selective agonists lack psychoactivity but are implicated in the control of fundamental neural cell processes, such as proliferation and survival, and their pharmacological manipulation might be useful for both delaying the progression of neurodegenerative disorders and inhibiting the growth of glial tumors (17, 18). CB1 is the most abundant receptor in the central nervous system, and, besides its classical influence on mood and emotion, it has been demonstrated to play a role in the modulation of memory processing and in metabolism (19). The ubiquitous expression pattern of CB1 receptors reflects the complexity and the variety of functions the endocannabinoid system impacts in neuronal activity. CB1 receptors are often localized presynaptically, where their stimulation usually inhibits neurotransmitter release (4, 20, 21). In the striatal spine module, CB1 receptors are localized both pre- and postsynaptically (22). Presynaptically, CB1 receptors are localized in GABAergic terminals of interneurons or collaterals from medium spiny neurons and also in glutamatergic but not in dopaminergic terminals (23–27). Postsynaptically, CB1 receptors are localized in the somatodendritic area of medium spiny neurons (23–25), and both enkephalinergic and dynorphinergic medium spiny neurons express CB1 receptors (28). The related receptor, CB2, traditionally was thought to be expressed in peripheral tissue, where it can help control inflammation and various immunological responses (1), but recent reports have suggested that it too can be found in the brain (albeit at a lower expression level than CB1 receptors) (29) and can impact a variety of neuronal processes. Multiple studies have demonstrated the expression of CB2 receptors in different non-neuronal (30–33) and neuronal populations (31, 34–38) where CB2 receptors show a preferred postsynaptic localization (31, 35–37).
Both CB1 and CB2 receptors are members of the GPCR4 family and are Gi/Go-protein-coupled receptors (39). Agonist activation triggers inhibition of adenylyl cyclase and voltage-gated calcium channels, activation of potassium channels, mitogen-activated protein kinase (MAPK), and phosphoinositide-3 kinase (PI3K)/Akt signaling pathways (40, 41). Initially, CB1 and CB2 receptors, like other GPCRs, were thought to have acted as single signaling receptors, but studies in the last decade have convincingly shown that certain GPCRs in a variety of different tissues can also form homodimers and even heteromers (42). For the cannabinoid receptors, heteromers have been shown to exist between CB1 and the dopamine and adenosine receptors (43–47) as well as with angiotensin (AT1) (48), opioid μ1 (49), and orexin OX1 receptors (50). However, to date, no studies have examined the possible interactions between CB1 and CB2 receptors despite the fact that they have overlapping expression and that the two receptors have been shown to impact similar cellular processes. Here we report that CB1 and CB2 receptors can form functional heteromers in transfected cells and in a variety of brain tissues. Heteromer formation leads to a negative cross-talk between receptor agonists and antagonists, suggesting an additional level of molecular regulation between the two receptors.
MATERIALS AND METHODS
Fusion Proteins and Expression Vectors
The human cDNA for the CB1, CB2, and dopamine D4.4 receptors cloned in pcDNA3.1 were amplified without their stop codons using sense and antisense primers harboring either unique EcoRI and BamHI sites (CB1R, CB2R) or XhoI and EcoRI (D4.4R). The fragments were then subcloned to be in frame with Rluc into the EcoRI and BamHI restriction sites of an Rluc-expressing vector (pRluc-N1, PerkinElmer) or with enhanced YFP into the EcoRI and BamHI (CB1R, CB2R) or XhoI and EcoRI restriction sites of an enhanced YFP-expressing vector (EYFP-N1; enhanced yellow variant of GFP; Clontech) to give the plasmids that express CB1, CB2, or D4.4 receptors fused to Rluc or YFP on the C-terminal end of the receptor (CB2R-Rluc, CB2R-YFP, CB1R-YFP, or D4R-YFP). Expression of constructs was tested by confocal microscopy, and the receptor functionality was tested by the ERK1/2 activation pathway (see “Results”).
Cell Line Cultures and Transfection
Human embryonic kidney (HEK-293T), human neuroblastoma SH-SY5Y, and neuroblastoma and glioma hybrid NG108-15 cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 units/ml penicillin/streptomycin and 10% (v/v) heat-inactivated fetal bovine serum (FBS). Other supplements were 2 mm l-glutamine for HEK-293T and SH-SY5Y cells, 1 mm sodium pyruvate for SH-SY5Y cells, and 100 μm hypoxanthine, 0.02 μm aminopterin, 16 μm thymidine (HAT supplement) for NG108-15 cells. The human neuroblastoma SK-N-MC cells were grown in minimum essential medium supplemented with 2 mm l-glutamine, 100 IU/ml penicillin/streptomycin, 1 mm sodium pyruvate, and 10% (v/v) heat-inactivated FBS. All supplements were from Invitrogen. Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and were passaged when they were 80–90% confluent (i.e. approximately twice a week).
HEK-293T or SH-SY5Y cells were transiently transfected with the corresponding fusion protein cDNA by the ramified PEI (Sigma) method. Cells were incubated (4 h) with the corresponding cDNA together with ramified PEI (5 ml of 10 mm PEI for each mg of cDNA) and 150 mm NaCl in a serum-starved medium. After 4 h, the medium was changed to a fresh complete culture medium. 72 h after transfection, cells were washed twice in quick succession in Hanks' balanced salt solution (137 mm NaCl, 5 mm KCl, 0.34 mm Na2HPO4·12H2O, 0.44 mm KH2PO4, 1.26 mm CaCl2·2H2O, 0.4 mm MgSO4·7H2O, 0.5 mm MgCl2, 10 mm HEPES, pH 7.4) supplemented with 0.1% glucose (w/v), detached by gently pipetting, and resuspended in the same buffer. To control the cell number, sample protein concentration was determined using a Bradford assay kit (Bio-Rad) using bovine serum albumin dilutions as standards.
Primary Cultures of Rat Pinealocytes
Male Sprague-Dawley rats (3 month old, ∼350 g), receiving water and food ad libitum, were obtained from the animal facility of the Faculty of Biology (University of Barcelona). 4% isoflurane (2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane)-anesthetized animals were killed by decapitation at 20:00 h (after the light period), and pineal glands were immediately dissected. All procedures were approved by the Catalan Ethical Committee for Animal Use (CEAA/DMAH 4049 and 5664). Pinealocytes were prepared from rat pineal glands as described previously by da Silveira Cruz-Machado et al. (51). Briefly, pinealocytes were obtained by trypsinization (0.25%, 37 °C, 15 min) followed by mechanical dispersion in the presence of fetal bovine serum. Cells were pelleted and resuspended in defined culture medium BGJb (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (heat-inactivated), 100 units/ml penicillin/streptomycin (pH 7.4). The total number of cells and fractional survival was estimated by trypan blue exclusion. Cells (200,000 cells/well) were plated on polylysine-coated 6-well chamber plates and maintained at 37 °C, 5% CO2 for 48 h prior to use.
Rat Brain Slice Preparation
Rats were decapitated with a guillotine, and the brains were rapidly removed and placed in ice-cold oxygenated (O2/CO2, 95%/5%) Krebs-HCO3− buffer (124 mm NaCl, 4 mm KCl, 1.25 mm NaH2PO4, 1.5 mm MgCl2, 1.5 mm CaCl2, 10 mm glucose, and 26 mm NaHCO3, pH 7.4). The brains were sliced at 4 °C in a brain matrix (Zivic Instruments, Pittsburgh, PA) into 0.5-mm coronal slices. Slices were kept at 4 °C in Krebs-HCO3− buffer during the dissection of the nucleus accumbens and the globus pallidus. For signaling experiments, each slice was transferred into an incubation tube containing 1 ml of ice-cold Krebs-HCO3− buffer, and the ERK1/2 phosphorylation was determined as described below. For proximity ligation assays, slices were fixed with 4% paraformaldehyde solution for 1 h at room temperature with gentle agitation. The slices were then washed in TBS (50 mm Tris-HCl, 0.9% NaCl, pH 7.8) and treated for 5 min with 1% Na2BH4 dissolved in TBS, followed by successive TBS washes until all Na2BH4 was eliminated. Finally, the slices were cryopreserved in a 30% sucrose solution overnight at 4 °C and stored at −20 °C until sectioning. 15-μm-thick slices were cut on a freezing cryostat (Leica Jung CM-3000) and mounted on slide glass. Slices were thawed at 4 °C, washed in TBS, and rocked with the blocking solution (Olink Bioscience, Uppsala, Sweden) for 1 h at 37 °C in a humidified atmosphere.
In Situ Proximity Ligation Assay (PLA)
Primary cultures of pinealocytes or SH-SY5Y cells transfected or not with 3 μg of cDNA corresponding to CB2R-HA (Missouri S&T Resource Center), were fixed in 4% paraformaldehyde for 15 min and washed with phosphate-buffered saline (PBS) containing 20 mm glycine to quench the aldehyde groups. After permeabilization with PBS-glycine containing 0.05% Triton X-100 for 5 min, cells were incubated for 1 h at room temperature with PBS containing 1% bovine serum albumin. Rat nucleus accumbens slices were obtained as described above. The receptor-receptor molecular interaction in these samples was detected using the Duolink II in situ PLA detection kit (Olink Bioscience). To detect heteromers in pinealocytes or in nucleus accumbens slices, the direct PLA-linked primary antibodies were used. The rabbit anti-CB1 receptor antibody (Thermo Scientific) was linked to a plus PLA probe, and the rabbit anti-CB2 receptor antibody (Cayman Chemical, Ann Arbor, MI) was linked to a minus PLA probe following the instructions of the supplier. After a 1-h incubation at 37 °C with the blocking solution in a preheated humidity chamber, pinealocytes or slices were incubated overnight with these PLA probe-linked antibodies (1:1000) at 4 °C. After washing with wash buffer at room temperature, samples were processed for ligation, amplification, and detection as described by the manufacturer. As negative controls, pinealocytes or slices were incubated overnight with the plus PLA probe-linked rabbit anti-CB1 receptor antibody and a goat anti-D4 receptor primary antibody (1:500; Santa Cruz Biotechnology) at 4 °C, followed by an incubation (2 h, 37 °C) in a preheated humidity chamber with Duolink II minus PLA probe anti-goat diluted in the antibody diluent. To detect heteromers in SH-SY5Y cells, transfected or non-transfected (as negative controls) cells were incubated (1 h, 37 °C) with the blocking solution in a preheated humidity chamber and then incubated overnight with the primary antibodies: rabbit anti-CB1 receptor antibody (1:1000; Thermo Scientific) and mouse monoclonal anti-HA tag antibody (1:1000; Abcam, Cambridge, UK) in the antibody diluent medium. SH-SY5Y cells were washed with washing buffer at room temperature and incubated (2 h, 37 °C) in a preheated humidity chamber with PLA probes detecting rabbit or mouse antibodies (Duolink II plus PLA probe anti-rabbit and Duolink II minus PLA probe anti-mouse) and processed as described above. Samples were mounted using the mounting medium with DAPI and observed in a Leica SP2 confocal microscope (Leica Microsystems, Mannheim, Germany).
Immunostaining
After 72 h of transfection, HEK-292T cells were fixed in 4% paraformaldehyde for 15 min and washed with PBS containing 20 mm glycine to quench the aldehyde groups. After permeabilization with PBS-glycine containing 0.05% Triton X-100 for 5 min, cells were incubated with PBS containing 1% bovine serum albumin. After 1 h at room temperature, protein-Rluc was labeled with the primary mouse monoclonal anti-Rluc antibody (1:100; Chemicon, Billerica, MA) for 1 h, washed, and stained with the secondary antibody Cy3 donkey anti-mouse (1:200; Jackson Immunoresearch Laboratories, West Grove, PA). Protein-YFP was detected by its fluorescence properties. The samples were rinsed several times and mounted with a medium suitable for immunofluorescence (30% Mowiol, Calbiochem). The samples were observed in a Leica SP2 confocal microscope.
RT-PCR
Total cellular RNA was isolated from HEK-293T, SH-SY5Y, SK-N-MC, or NG108-15 cells using the QuickPrep total RNA extraction kit (Amersham Biosciences) or from Macaca fascicularis spleen using TRIzol reagent (Invitrogen) as described previously (38). Total RNA (1 μg) was reverse-transcribed by random priming using Moloney murine leukemia virus reverse transcriptase, RNase H minus, and point mutant, following the protocol of two-step RT-PCR provided by Promega (Madison, WI). The resulting single-stranded cDNA was used to perform PCR amplification for the CB1 and CB2 receptors and GAPDH as an internal control of PCR technique using Taq DNA polymerase (Promega). Common primers to amplify human and rat cDNA were used. Primers to amplify CB1R were 5′-TGGGCAGCCTGTTCCTCAC-3′ (forward) and 5′-CATGCGGGCTTGGTC-3′ (reverse). Primers to amplify CB2R were 5′- CGTGGCTGTGCTCTATCTGA-3′ (forward) and 5′-AGCCAGCTCAGCAGGTAGTC-3′ (reverse). To amplify GAPDH, the primers used were 5′-CATCCTGCACCACCAACTGCTTAG-3′ (forward) and 5′-GCCTGCTTCACCACCTTCTTGATG-3′ (reverse). RNA without reverse transcriptions did not yield any amplicons, indicating that there was no genomic DNA contamination.
Bioluminescence Resonance Energy Transfer (BRET) Assays
HEK-293T cells were transiently co-transfected with the indicated amounts of plasmid cDNAs corresponding to the indicated fusion proteins (see Fig. 1 legend). To quantify receptor fluorescence expression, cells (20 μg of protein) were distributed in 96-well microplates (black plates with a transparent bottom; Porvair (King's Lynn, UK)), and fluorescence was read in a FluoStar Optima fluorimeter (BMG Labtechnologies, Offenburg, Germany) equipped with a high energy xenon flash lamp, using a 10-nm bandwidth excitation filter at a reading of 400 nm. Receptor fluorescence expression was determined as the fluorescence of the sample minus the fluorescence of cells expressing protein-Rluc alone. For BRET measurements, the equivalent of 20 μg of protein was distributed in 96-well microplates (Corning 3600, white plates; Sigma), and 5 μm coelenterazine H (Molecular Probes, Inc., Eugene, OR) was added. After 1 min of adding coelenterazine H, the readings were collected using a Mithras LB 940 (Berthold, Bad Wildbad, Germany), which allows the integration of the signals detected in the short wavelength filter at 485 nm (440–500 nm) and the long wavelength filter at 530 nm (510–590 nm). To quantify receptor-Rluc expression luminescence, readings were performed 10 min after the addition of 5 μm coelenterazine H. Cells expressing BRET donors alone were used to determine background. The net BRET is defined as (long wavelength emission/short wavelength emission) − Cf, where Cf corresponds to long wavelength emission/short wavelength emission for the Rluc construct expressed alone in the same experiment. BRET curves were fitted by using a non-linear regression equation, assuming a single phase with GraphPad Prism software (San Diego, CA)
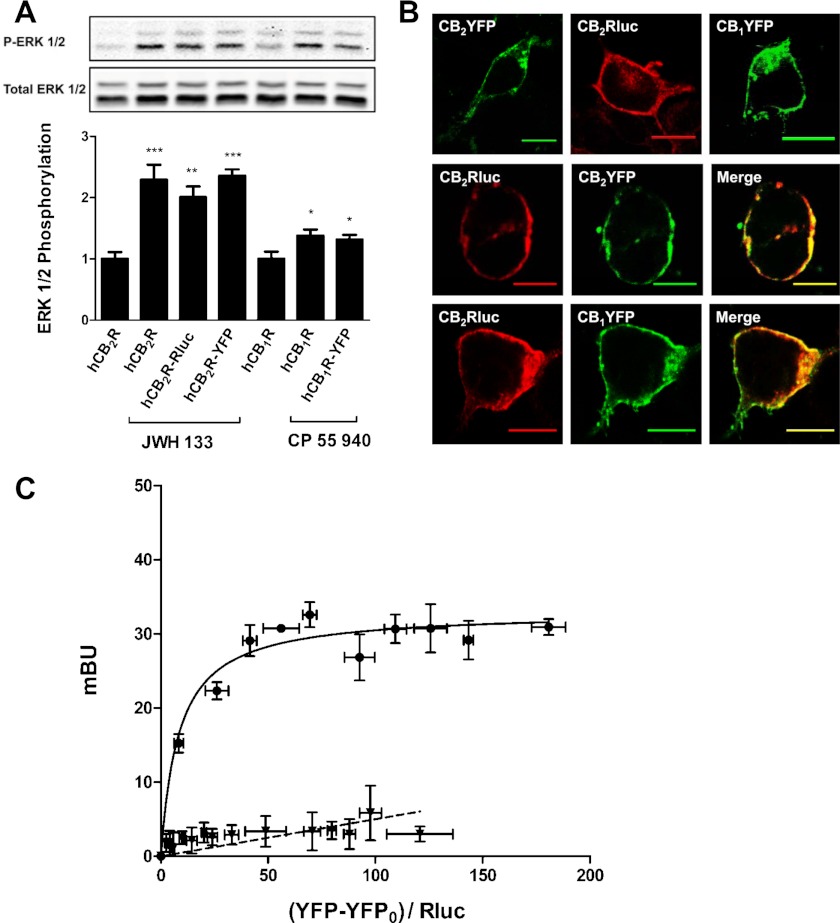
FIGURE 1.
CB2 receptors form heteromers with CB1 receptors in transfected cells. A, the functionality of fusion proteins in HEK-293T cells transfected with 1.5 μg of cDNA corresponding to CB1 receptor, CB2 receptor, CB2R-YFP, CB1R-YFP, or CB2R-Rluc. 72 h post-transfected cells expressing CB2R, CB2R-YFP, or CB2R-Rluc were treated for 7 min with vehicle (basal) or with JWH 133 (100 nm), and cells expressing CB1R or CB1R-YFP were treated for 7 min with vehicle (basal) or with CP 55940 (500 nm), and ERK1/2 phosphorylation was determined. Results (means ± S.E. (error bars) of four different experiments performed in duplicate) represent -fold over basal. Significant differences were analyzed by one-way ANOVA followed by Dunnett's multiple comparison post hoc test (*, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with basal). Above is a representative Western blot. B, confocal microscopy images of cells transfected with the plasmid corresponding to CB2R-YFP (1.5 μg), CB2R-Rluc (0.5 μg), or CB1R-YFP (1.5 μg) alone (top panels) or in combination (middle panels for CB2R-Rluc and CB2R-YFP and lower panels for CB2R-Rluc and CB1R-YFP). Proteins were identified by fluorescence or by immunocytochemistry as indicated under “Materials and Methods.” Colocalization is shown in yellow in merge panels. Scale bars, 10 μm. C, BRET saturation experiments showing CB1-CB2 receptor heteromerization were performed as described under “Materials and Methods” using cells transfected with 0.2 μg of cDNA corresponding to CB2R-Rluc and increasing amounts of cDNA (0–4 μg of cDNA) corresponding to CB1R-YFP (circles). As a negative control, cells were also transfected with cDNA corresponding to CB1R-Rluc (0.2 μg) and dopamine D4R-YFP (0–4 μg of cDNA) (triangles). Both fluorescence and luminescence for each sample were measured before every experiment to confirm similar donor expressions (∼100,000 bioluminescence units) while monitoring the increase in acceptor expression (100–20,000 net fluorescence units). The relative amount of BRET is given as the ratio between the net fluorescence of the acceptor (YFP-YFP0), and the luciferase activity of the donor (Rluc). BRET data are expressed as means ± S.E. of four different experiments grouped as a function of the amount of BRET acceptor. mBU, milli BRET units.
ERK1/2 and Akt/PKB Phosphorylation Assays
Each globus pallidus slice, obtained as described above, was transferred into an incubation tube containing 1 ml of ice-cold Krebs-HCO3− buffer, and the temperature was raised to 23 °C. After 30 min, the medium was replaced by 2 ml of Krebs-HCO3− buffer (23 °C) and was incubated under constant oxygenation (O2/CO2, 95%/5%) at 30 °C for 4–5 h in an Eppendorf Thermomixer (5 Prime, Inc., Boulder, CO). The medium was replaced by 200 μl of fresh Krebs-HCO3− buffer and incubated for 30 min before the addition of the desired concentrations of ligands. After the indicated incubation period, the solution was discarded, and slices were frozen on dry ice and stored at −80 °C until use. Transfected or non-transfected SH-SY5Y cells were cultured in serum-free medium for 16 h before the addition of any agent. Cells were treated or not with the indicated agonists for the indicated time. At the end of the incubation period, cells were rinsed with ice-cold phosphate-buffered saline. Cells or slices were lysed by the addition of 500 μl of ice-cold lysis buffer (50 mm Tris-HCl, pH 7.4, 50 mm NaF, 150 mm NaCl, 45 mm β-glycerophosphate, 1% Triton X-100, 20 μm phenyl-arsine oxide, 0.4 mm NaVO4, and protease inhibitor mixture). The cellular debris was removed by centrifugation at 13,000 × g for 5 min at 4 °C, and the protein was quantified by the bicinchoninic acid method using bovine serum albumin dilutions as a standard. Equivalent amounts of protein (10 μg) were separated by electrophoresis on a denaturing 7.5% SDS-polyacrylamide gel and transferred onto a PVDF-FL membrane. Odyssey blocking buffer (LI-COR Biosciences, Lincoln, NE) was then added, and the membrane was rocked for 90 min. The membrane was then incubated overnight with a mixture of a rabbit anti-phospho-Ser473 Akt antibody (1:2500; Signalway Antibody) to test the Akt phosphorylation or mouse anti-phospho-ERK1/2 antibody (1:2500; Sigma) to test ERK1/2 phosphorylation. As a control of the amount of protein loaded, rabbit anti-ERK1/2 antibody that recognizes both phosphorylated and non-phosphorylated ERK1/2 (1:40,000; Sigma) was used. Bands were visualized by the addition of IRDye 680 (anti-rabbit) antibody (1:10,000; Sigma) or IRDye 800 (anti-mouse) antibody (1:10,000; Sigma) or a mixture of both for 1 h and scanned by the Odyssey infrared scanner (LI-COR Biosciences). Band densities were quantified using the scanner software and exported to Excel (Microsoft, Redmond, WA). The level of phosphorylated proteins was normalized for differences in loading using the total ERK1/2 protein band intensities.
Evaluation of Neurite Outgrowth
SH-SY5Y cells seeded in 10-mm coated glass coverslips were transfected or not with 3 μg of cDNA corresponding to CB2R-YFP. 48 h post-transfection, cells were incubated for 24 h in serum-free growing medium in the absence or presence of 10 μm retinoic acid, 100 nm ACEA, or 50 nm JWH 133 (all from Tocris, Bristol, UK) alone or in combination. Cells were washed three times with PBS, fixed in 4% paraformaldehyde for 15 min, washed with PBS containing 20 mm glycine, permeabilized for 5 min with PBS-glycine buffer containing 0.05% Triton X-100, and blocked with PBS containing 1% BSA for 1 h at room temperature. Cells were labeled for 1 h with the primary mouse anti-MAP2 antibody (1:200; Calbiochem). Subsequently, cells were washed and stained with the secondary antibody, Cy3-conjugated affinity-purified donkey anti-mouse IgG (1:200 dilution; Jackson ImmunoResearch Laboratories), and nuclear staining was performed with Hoechst (1:1000, 1 mg/ml; Sigma). Coverslips were rinsed for 5 min in PBS containing 1% BSA and for 5 min in PBS-glycine buffer and mounted with Mowiol mounting medium. Confocal microscope observations were made with a Leica TCS SP2 microscope with a ×40 objective. Cell bodies and neurites present in 8–12 randomly selected fields were measured in each experiment using ImageJ software. Cells were considered to be differentiated if they had at least one process longer than the cell body, which would be regarded as a neurite. The results are expressed as the percentage of differentiated cells versus the total cell number in non-transfected cells or versus CB2R-YFP-expressing cells (detected by its own fluorescence) in transfected cells. At least three independent experiments were conducted for each treatment.
RESULTS
CB2 Receptors Form Heteromers with CB1 Receptors in Transfected Cells
A variety of GPCRs, including CB1 receptors, have been reported to be expressed as homomers and form heteromers with other GPCRs (43, 45, 49, 52), but it is not known whether CB2 and CB1 receptors can form heteromers. To test this, the BRET technique was used. The BRET technique requires the use of fusion proteins consisting of CB2R-Rluc, CB1R-YFP, and CB2R-YFP. Prior to the BRET experiments, we first confirmed that the fusion of Rluc or YFP to CB2 or CB1 receptors did not modify receptor function, as determined by ERK1/2 phosphorylation assays (Fig. 1A). In addition, we confirmed that the subcellular localization of the fusion proteins was indeed in the cell membrane, showing a high degree of colocalization when CB2R-Rluc and CB2R-YFP or CB2R-Rluc and CB1R-YFP were co-expressed (Fig. 1B). To test the ability of CB1 and CB2 receptors to form heteromers, BRET measurements were performed in transiently co-transfected HEK-293T cells using a constant amount of cDNA corresponding to CB2R-Rluc and increasing amounts of cDNA corresponding to CB1R-YFP. As can be seen in Fig. 1C, the BRET signal increased as a hyperbolic function of the amount of the CB1R-YFP expressed, reaching an asymptote. From the saturation curve, a BRETmax of 33 ± 1 milli BRET units (mBU) and a BRET50 of 8 ± 2 were calculated. The specificity of this interaction was demonstrated by comparing the BRET saturation curve with the low and linear BRET obtained for the negative control constituted by CB1R-Rluc and D4R-YFP (Fig. 1C). These results indicate that CB1 and CB2 receptors form heteromers in co-transfected cells.
CB1 and CB2 Receptors Form Heteromers in SH-SY5Y Neuroblastoma Cells
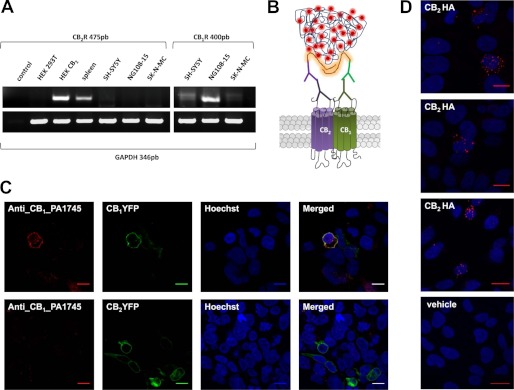
Knowing that CB1 and CB2 receptors form heteromers in HEK-293T transfected cells, we investigated whether they can form heteromers in a neuronal cell model. We first determined the endogenous receptor expression in different neuroblastoma cell lines. As shown in Fig. 2A, all neuroblastoma cells tested expressed the mRNA corresponding to CB1 receptor, but they did not express the mRNA corresponding to CB2 receptor. From these results, human SH-SY5Y neuroblastoma cells were selected. Non-transfected cells were used as CB1 receptor-expressing neuroblastoma cells, whereas SH-SY5Y cells transfected with HA tagged CB2 receptor were used as CB1 receptor- and CB2 receptor-expressing cells. The BRET technique is a powerful approach for looking at receptor interactions in co-transfected cells, but BRET cannot be easily applied in native cells endogenously expressing one or both receptors. To solve this, other direct and indirect methods can be used. Here we sought to determine if the endogenous expressed CB1 and transfected CB2 receptors could also form heteromers in a neuronal cell model. To do this, we employed the PLA, which is used to detect protein interactions. This direct method requires that both receptors be close enough to allow the two different antibody-DNA probes to be able to ligate (<17 nm) (53, 54). If the receptors are within sufficient proximity, a punctate fluorescent signal can be detected by confocal microscopy diagrammed in Fig. 2B. For these experiments, two different and specific primary antibodies directed against each of the two receptors were used. One was the well known commercial mouse anti-HA antibody to detect the HA-labeled CB2 receptor, and the other was a rabbit anti CB1 receptor antibody. The specificity of this last antibody was tested in CB1R-YFP- or CB2R-YFP-transfected HEK-293T cells (Fig. 2C). Colocalization of fluorescence due to YFP with the anti-CB1 receptor antibody staining was detected in CB1R-YFP-transfected cells but not in CB2R-YFP-transfected cells, and a lack of antibody-promoted staining was observed in non-transfected cells (cells that do not show fluorescence) (Fig. 2C). Using these antibodies in PLA experiments, the CB1-CB2 receptor heteromer expression in SH-SY5Y cells was demonstrated by punctuate fluorescent signal detected by confocal microscopy after excitation at 624 nm. This pattern was observed in CB2 receptor-expressing cells, whereas no signal was detected in non-transfected cells used as a negative control (Fig. 2D).
FIGURE 2.
CB1-CB2 receptor heteromers in neuroblastoma cells. A, CB1 and CB2 receptor mRNA expression in different neuroblastoma cells was analyzed by RT-PCR using total RNA from SH-SY5Y, SK-N-MC, or NG108-15 neuroblastoma cells and specific common primers for the human and rat CB2 and CB1 receptors or GAPDH as internal control of mRNA expression. As positive controls for the CB2 receptor expression, total RNAs from human spleen or HEK-293T cells stably expressing human CB2 receptors (HEK CB2) were used, and RNAs from HEK-293T cells (HEK-293T) or primers without RNA (control) were included as negative controls. B, a schematic representation of the PLA technology is shown. Receptors were recognized by primary antibodies and secondary antibodies linked to different DNA chains, one plus and one minus. If the two receptors are close enough, the two different antibody-DNA probes are able to ligate. Following an amplification process, the presence of fluorescently tagged nucleotides allows detection of a punctuate fluorescent signal by confocal microscopy. C, to test the specificity of rabbit anti-CB1 receptor primary antibody, HEK-293T cells were transfected with CB1R-YFP (1.5 μg of plasmid) (top panels) or CB2R-YFP (1.5 μg of plasmid) (bottom panels), and immunocytochemistry was performed with rabbit anti-CB1R, as indicated under “Materials and Methods.” CB1 receptor- or CB2 receptor-expressing cells were identified by their own fluorescence, and cell nuclei were stained with Hoechst (blue). Colocalization in merged images is shown in yellow. Scale bars, 10 μm. D, PLA in SH-SY5Y cells transfected with CB2R-HA (3 μg of plasmid) (top panels) or without transfection (vehicle; bottom panel). Red spots in three different fields from independent experiments with CB2 receptor-expressing cells (top panels) but not in non-transfected cells (bottom panel) indicate the CB1-CB2 receptor heteromer expression. Scale bars, 20 μm.
CB1-CB2 Receptor Heteromers Are Expressed in Rat Brain
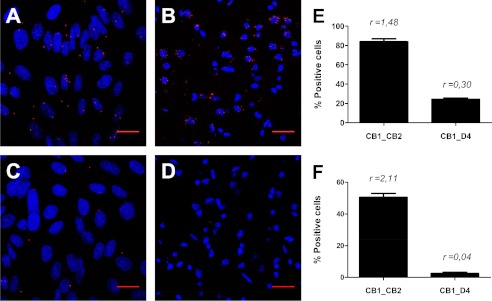
To gain more insight about the physiological relevance of CB1-CB2 receptor heteromers, we next investigated the expression of these heteromers in the rat brain taking advantage of the PLA approach used above. CB1 is the most abundant GPCR in the brain (55). Although CB2 receptor expression is much lower, sometimes even undetectable, it is also known to be present in different brain areas, impacting endocannabinoid signaling and colocalizing with CB1 receptors (32, 34, 36, 37, 56–59). Recently, it has been shown that both CB1 and CB2 receptors are co-expressed in the pineal gland, where they may be involved in the control of pineal physiology (57), and in the nucleus accumbens, where CB2 receptor controls cocaine intake (59). Despite evidence of their co-expression, nothing is known about CB1 and CB2 receptor molecular interactions in the brain. Here we determined the expression of CB1-CB2 receptor heteromers in rat pinealocytes and nucleus accumbens by PLA. We used the rabbit anti-CB1 receptor antibody described above and a rabbit anti-CB2 receptor antibody that is specific for the CB2 receptor, as previously demonstrated (57). Both antibodies were used directly linked to the PLA DNA probes, as described under “Materials and Methods.” PLA experiments were performed with pinealocytes obtained from rat pineal glands extracted from a rat sacrificed at 20 h (after the light period), when the expression of CB1 and CB2 receptors is more equilibrated (57), and with rat nucleus accumbens slices obtained as indicated under “Materials and Methods.” CB1-CB2 receptor heteromers in the primary cultures or in the slices were visualized as red spots in pinealocytes (Fig. 3A) or neurons (Fig. 3B) stained with Hoechst. Because CB1 and dopamine D4 receptors do not form heteromers in BRET experiments (see Fig. 1C) we considered this pair as the negative control and performed the same assay using the PLA DNA probe-linked anti-CB1 receptor antibody described above and a goat antibody against dopamine D4 receptor plus a goat secondary antibody linked to the corresponding PLA DNA probe (Fig. 3, C and D). A quantification of cells containing one or more red spots versus total cells (blue nucleus) and the ratio r (number of red spots/total cells) were determined in eight fields from three different experiments. 84% of pinealocytes expressed CB1-CB2 receptor heteromers with r = 1.48, in contrast with the 24% of positive cells with r = 0.3 detected in the negative control (Fig. 3E). Analogously 51% of nucleus accumbens cells (r = 2.11) expressed CB1-CB2 receptor heteromers, in contrast with 3% cells (r = 0.04) detected for the negative control (Fig. 3F). These results indicate that CB1-CB2 receptor heteromers are expressed in rat pinealocytes and in rat nucleus accumbens.
FIGURE 3.
CB1-CB2 receptor heteromers in rat brain. The PLA was performed using primary cultures of rat pinealocytes (A and C) or rat nucleus accumbens slices (B and D). Pinealocytes (A) or slices (B) were treated with rabbit CB1 receptor and CB2 receptor primary antibodies directly linked to plus and minus PLA probes (see “Materials and Methods”). CB1-CB2 receptor heteromers were visualized as red spots around Hoechst-stained nuclei (A and B). Negative controls were performed with pinealocytes (C) or slices (D) treated with plus PLA probe-linked rabbit anti-CB1 receptor and with primary goat anti-D4 receptor detected with a minus PLA probe-linked secondary goat antibody. Scale bars, 20 μm. The percentage of pinealocytes (E) or nucleus accumbens neurons (F) containing one or more red spots versus total cells (blue nucleus) is given as well as r (number of red spots/total cells) values (E and F) from eight fields in three different experiments. Error bars, S.E.
Characterization of CB1-CB2 Receptor Heteromer Signaling
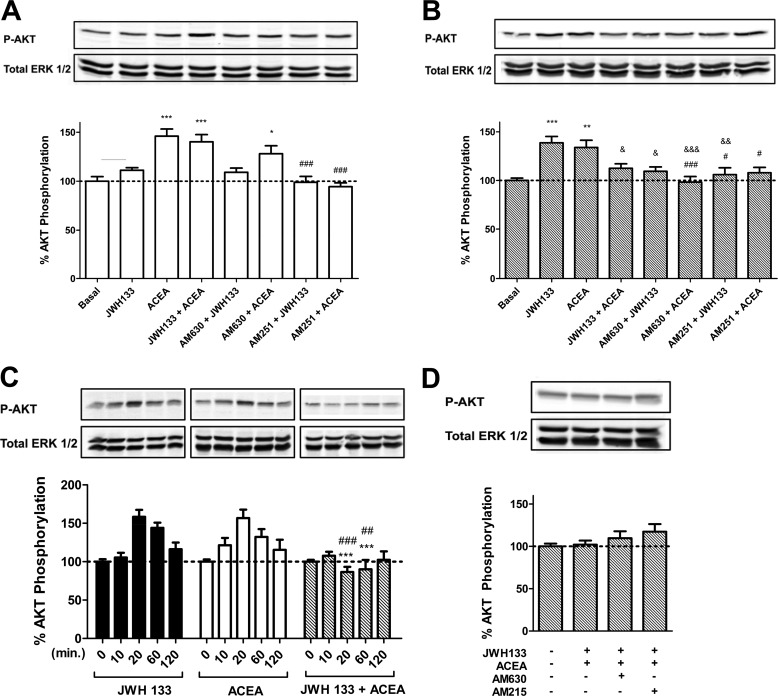
A common and often essential attribute of receptor heteromers is the ability to modify the downstream signaling versus the single constituent receptors. This type of receptor-receptor interaction has been observed for several receptor heteromers (60, 61). Because cannabinoid receptors have been previously described to be coupled to Akt/PKB protein activation in different cell types (40, 41, 62), we first investigated whether there were changes in Akt/PKB (Ser-473 Akt phosphorylation) signaling when heteromers were co-stimulated with both agonists or blocked with antagonists in SH-SY5Y neuronal cells. Non-transfected cells were used as CB1 receptor-expressing neuroblastoma cells, and cells transfected with CB2R-YFP were used as CB1- and CB2 receptor-expressing cells. Treatment with 100 nm CB1 receptor agonist ACEA induced Akt/PKB phosphorylation in both non-transfected and transfected cells (Fig. 4, A and B), and the CB2 receptor agonist JWH 133 induced Akt/PKB phosphorylation only in transfected cells (Fig. 4, A and B). Furthermore, JWH 133-induced Akt/PKB signaling was significantly diminished when cells expressing both receptors were co-stimulated with ACEA and JWH 133 (Fig. 4, B and D). These results indicate that a negative cross-talk exists between CB1 and CB2 receptors in Akt/PKB phosphorylation signaling. These results are not due to a change in the time in which the signaling peaks because differences were not observed in time-response curves when cells were activated with one or both agonists (Fig. 4C). It is important to point out that all ligands were first chosen based on their KI value (KI = 1.4 nm for ACEA ∼1400-fold selective for CB1 receptor; KI = 3.4 nm for JWH 133, ∼200-fold selective for CB2 receptor) and following a dose-response curve for Akt activation, where the lowest concentration that provided specific receptor signaling was chosen as the working concentration.
FIGURE 4.
Agonist and antagonist interactions between CB1 and CB2 receptors on Akt/PKB phosphorylation (P-AKT) in SH-SY5Y neuroblastoma cells. SH-SY5Y cells (A) or SH-SY5Y cells transfected with 3 μg of cDNA corresponding to CB2R-YFP (B–D) were used. In A and B, cells were treated for 20 min. with the agonists ACEA (100 nm) or JWH 133 (50 nm) alone or in combination or pretreated for 30 min with the antagonist AM630 (500 nm) or AM251 (200 nm) prior to agonist treatment. Akt/PKB phosphorylation was measured as indicated under “Materials and Methods.” Results (means ± S.E. (error bars) of four different experiments performed in duplicate) are expressed as -fold over basal (non-stimulated cells). Significant differences were analyzed by a one-way ANOVA followed by post-hoc Bonferroni's tests (*, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with basal. #, p < 0.05; ##, p < 0.01; ###, p < 0.001 compared with treated with ACEA alone. &, p < 0.05; &, p < 0.01; &&&, p < 0.001 compared with treated with JWH 133 alone). C, time-response curves of SH-SY5Y cells treated for the indicated times with 100 nm ACEA (white bars) or 50 nm JWH 133 (black bars) alone or in combination (gray bars). Results (means ± S.E. of four different experiments performed in duplicate) are expressed as -fold over basal (non-stimulated cells). Significant differences were analyzed by a one-way ANOVA followed by post hoc Bonferroni's tests (***, p < 0.001 compared with treated only with JWH 133. ##, p < 0.01; ###, p < 0.001 compared with treated only with ACEA alone). D, effect of 30 min 500 nm AM630 or 200 nm AM251 antagonist pretreatment in SH-SY5Y cells co-stimulated for 20 min with 100 nm ACEA and 50 nm JWH 133. Results (means ± S.E. of four different experiments performed in duplicate) are expressed as -fold over basal (non-stimulated cells). No significant differences were detected in D.
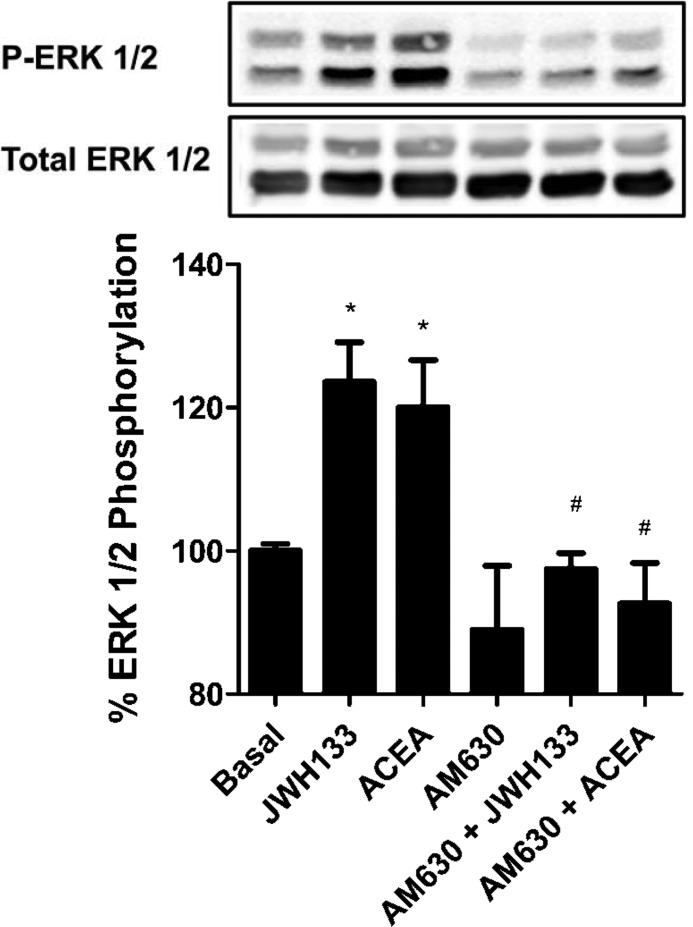
Looking at the effect of CB1 and CB2 receptor antagonists, the 100 nm ACEA-induced Akt/PKB phosphorylation was not significantly modified when non-transfected cells were pretreated with 500 nm AM630, a 165-fold selective CB2 receptor antagonist over CB1 receptor (63) (Fig. 4A), but it was completely counteracted when non-transfected and transfected cells were pretreated with 200 nm AM251, a 306-fold selective CB1 receptor antagonist over CB2 receptor (64), as expected for specific CB1-agonistic interaction (Fig. 4, A and B). However, interestingly, ACEA-induced Akt/PKB phosphorylation in cells expressing both receptors was also significantly prevented by pretreatment with AM630 (Fig. 4B). Moreover, treatment with 50 nm JWH 133, a 200-fold selective CB2 receptor agonist over CB1 receptor (65), did not induce Akt/PKB phosphorylation in non-transfected SH-SY5Y cells due to the lack of CB2 receptor expression in these cells (Fig. 4A) but was able to induce Akt/PKB phosphorylation in cells expressing CB1 and CB2 receptors (Fig. 4B). JWH 133-induced phosphorylation was blocked when cells expressing both cannabinoid receptors were pretreated with AM630, as expected for a specific CB2 receptor agonist, but was also prevented when cells expressing both receptors were pretreated with the CB1 receptor antagonist AM251. These results indicate that a bidirectional cross-antagonism exists between CB1 and CB2 receptors in Akt/PKB phosphorylation signaling. We used this heteromer characteristic to further test for the expression of CB1-CB2 receptor heteromers in rat brains. We selected globus pallidus slices for these experiments because it has been described that globus pallidus expresses a high amount of CB2 receptor (38). The Akt/PKB and ERK1/2 phosphorylation was determined in slices as indicated under “Materials and Methods.” CB1 and CB2 receptors were poorly coupled to Akt/PKB phosphorylation in the globus pallidus (results not shown), but the activation of both receptors increased ERK1/2 phosphorylation (Fig. 5). The JWH 133 agonist-induced ERK1/2 phosphorylation was blocked by AM630, as expected for a specific CB2 receptor antagonist, but this antagonist was also able to block the ERK1/2 phosphorylation induced by the CB1 receptor agonist ACEA, showing a cross-antagonism. These results strongly suggest that CB1-CB2 receptor heteromers may likely be expressed in the globus pallidus.
FIGURE 5.
CB1-CB2 receptor heteromer fingerprint was found in rat globus pallidus. Globus pallidus slices were obtained as described under “Materials and Methods” and treated for 10 min with 500 nm JWH 133 or 1 μm ACEA or pretreated for 20 min with 5 μm AM630 antagonist prior to agonist treatment. ERK1/2 phosphorylation (P-ERK 1/2) was determined as described under “Materials and Methods.” Results (means ± S.E. (error bars) of two different experiments performed in triplicate) are expressed as -fold over basal (non-stimulated cells). Significant differences were analyzed by a one-way ANOVA followed by post hoc Bonferroni's tests (*, p < 0.05 compared with basal; #, p < 0.05 compared with the respective agonist-treated cells). Above is a representative Western blot.
Functional Characterization of CB1-CB2 Receptor Heteromers on Neurite Outgrowth
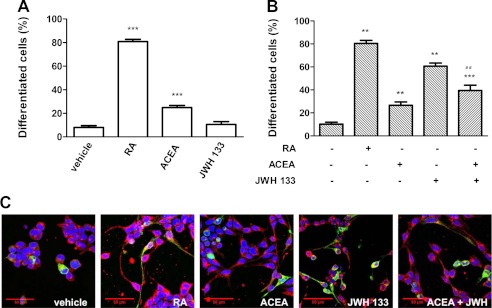
Because the endocannabinoid signaling pathway is involved in brain development and neural cell differentiation (66–68) and because activation of PI3K/Akt signaling is involved in neural differentiation of SH-SY5Y cells (69), we investigated the role of CB1-CB2 receptor heteromers on neuritogenesis in our SH-SY5Y neuronal cell model. Again, non-transfected cells were used as CB1 receptor-expressing neuroblastoma cells, and cells transfected with CB2R-YFP were used as CB1 and CB2 receptor-expressing cells. The human neuroblastoma cell line SH-SY5Y is a well characterized model system to study neuronal differentiation in vitro. These cells develop long extensions and express several neuronal markers when treated with different agents, including retinoic acid or phorbol esters (70). SH-SY5Y cells reduce their rate of growth and initialize differentiation, adopting a neuronal phenotype when exposed to 10 μm retinoic acid (71, 72). This reagent was therefore used as a control treatment for neuritogenesis (Fig. 6). The effect of CB1 and CB2 receptor agonists on neuritogenesis was analyzed by confocal microscopy, by quantifying neuritic processes in cells stained with an antibody against the microtubule-associated protein MAP-2 (anti-MAP-2). Treatment with ACEA induced, in both non-transfected (Fig. 6A) and transfected (Fig. 6B) SH-SY5Y cells, the appearance of neurites to a moderate extent compared with the effect exerted by retinoic acid. Treatment with JWH 133, a selective CB2 receptor agonist, did not induce neuritogenesis in non-transfected SH-SY5Y cells because they do not express CB2 receptor (Fig. 6A), but it induced neuritogenesis in cells expressing CB1 and CB2 receptors (Fig. 6B). When cells expressing CB1 and CB2 receptors were co-stimulated with JWH 133 and ACEA, the JWH 133-induced neuritogenesis was diminished (Fig. 6, B and C), indicating a negative cross-talk between CB1 and CB2 receptors in neuroblastoma cell differentiation, a phenomenon that is similar to the negative cross-talk observed at the Akt/PKB signaling.
FIGURE 6.
Agonist interactions between CB1 and CB2 receptors on neurite outgrowth. SH-SY5Y neuroblastoma cells (A) or SH-SY5Y cells transfected with 3 μg of cDNA corresponding to CB2R-YFP (B and C) were treated for 24 h with 10 μm retinoic acid (RA), 100 nm ACEA, or 50 nm JWH 133 alone or in combination. Neurite outgrowth was measured as indicated under “Materials and Methods.” Results are expressed as the percentage of differentiated cells versus the total cell number in non-transfected cells or versus CB2R-YFP expressing cells (detected by their own fluorescence) in transfected cells. Results are expressed as means ± S.E. (error bars) of 8–12 fields in three independent experiments. Significant differences between treated and non-treated cells (**, p < 0.01; ***, p < 0.001) or between co-activated cells compared with cells activated with JWH-133 (##, p < 0.01) were determined by a one-way ANOVA followed by post hoc Bonferroni's tests. In C, a representative image of cells expressing CB2R-YFP (green) treated as in B is shown. Treated cells were fixed, permeabilized, and processed for nuclear staining with Hoechst (blue) and for immunocytochemistry with anti-MAP2 antibody (red).
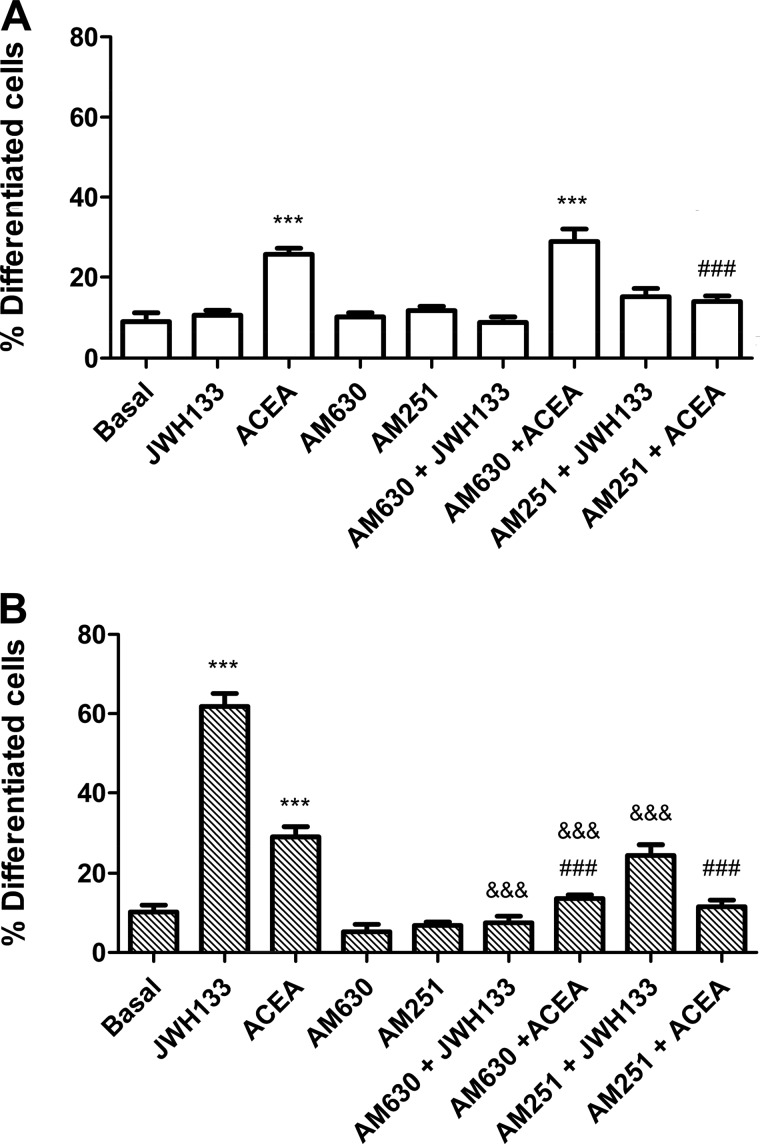
Finally, we evaluated whether the bidirectional cross-antagonism observed between CB1 and CB2 receptors in signaling can also be observed in the cannabinoid receptor-mediated neuritogenesis. As expected, the CB1 receptor antagonist AM251, but not the CB2 receptor antagonist AM630, completely blocked the CB1 receptor agonist ACEA-induced neuritogenesis in non-transfected SH-SY5Y cells (Fig. 7A). Interestingly, both AM251 and AM630 were able to prevent the ACEA-induced or the CB2 receptor agonist JWH 133-induced neuritogenesis in CB2 receptor-expressing SH-SY5Y cells (Fig. 7B). These results demonstrate the existence of a bidirectional cross-antagonism in the differentiation process that matches that observed in the Akt/PKB signaling described above. This indicates that the antagonist binding to one receptor in the CB1-CB2 receptor heteromer blocks the functionality of the entire heteromer.
FIGURE 7.
Antagonist interactions between CB1 and CB2 receptors on neurite outgrowth. SH-SY5Y neuroblastoma cells (A) or SH-SY5Y cell transfected with 3 μg of cDNA corresponding to CB2R-YFP (B) were treated for 24 h with 10 μm retinoic acid (RA), 100 nm ACEA, or 50 nm JWH 133 or pretreated for 30 min with 500 nm AM630 or AM251 antagonist prior to agonist treatment. Neurite outgrowth was measured as indicated under “Materials and Methods.” Results are expressed as the percentage of differentiated cells versus the total cell number in non-transfected cells or versus CB2R-YFP-expressing cells (detected by their own fluorescence) in transfected cells. Results are expressed as means ± S.E. (error bars) of 8–12 fields in three independent experiments. Significant differences between treated and non-treated cells (***, p < 0.001) or between antagonized cells compared with cells activated with JWH 133 (###, p < 0.001) or ACEA (&&&, p < 0.001) were determined by a one-way ANOVA followed by post hoc Bonferroni's tests.
DISCUSSION
The cannabinoid receptors CB1 and CB2 are increasingly becoming an important subject for investigation in a variety of neurological and immunological processes (1, 10–13). Although it has been described that both receptors can be co-expressed in the same brain areas (38, 57–59), the relationship between both receptors at the molecular level is not known. Among GPCRs, more and more evidence points to an important role of heteromer formation between GPCRs on receptor function modulation (42, 48, 60, 73). However, determination of the heteromer functional characteristics and, mainly, identification of heteromers in tissue is often a challenge. Here, we present evidence of CB2 receptor molecular and functional interaction with CB1 receptors. Several conclusions can be drawn from the data. First, CB1 and CB2 receptors can form heteromers in transfected cells and in a variety of brain tissues, including pineal gland, nucleus accumbens, and globus pallidus. Second, one specific characteristic of CB1-CB2 receptor heteromers is that of bidirectional cross-antagonism (i.e. the ability of CB1 receptor antagonists to block the effect of CB2 receptor agonists and, conversely, the ability of CB2 receptor antagonists to block the effect of CB1 receptor agonists). Third, agonist co-activation of CB1-CB2 receptor heteromers results in a negative cross-talk in Akt phosphorylation and neurite outgrowth.
Although it was known that CB1 receptors are highly expressed in the central nervous system, it was believed for a long time that CB2 receptors were restricted to the peripheral tissues (74). Recently, several studies have shown the expression of CB2 receptors in brain regions where CB1 receptors are also expressed (32, 34, 36, 37, 56–59), including the pineal gland (57), the nucleus accumbens (59), and the globus pallidus (38). Because the physiological role of the expression of two different receptors for the same endogenous ligands in the same brain regions is not obvious, we explored if both receptors are modulating each other and, for this reason, we first examined the possibility of direct receptor-receptor interaction by heteromerization. The identification of CB1-CB2 receptor heteromers was first performed via BRET, a biophysical technique, in co-transfected cells and by immunofluorescence using the PLA in a neuronal cell model. The definition of a receptor heteromer is that the heteromer is a macromolecular complex composed of at least two functional receptor units with biochemical properties that are demonstrably different from those of its individual receptors (42). Thus, we focused on the determination of the functional characteristics of CB1-CB2 receptor heteromers expressed in neuroblastoma cells. Both cannabinoid receptors have been shown to signal through the MAPK and Akt/PKB pathways (41, 62); therefore, we explored the implication of heteromer formation on these pathways. We first investigated whether there were changes in Akt/PKB (Ser-473 Akt phosphorylation) signaling when heteromers were both co-stimulated with agonists or blocked with antagonists. When neuroblastoma cells were co-stimulated with both receptor agonists, a negative cross-talk was observed between CB1 and CB2 receptors in Akt/PKB phosphorylation signaling. It has been described that activation of PI3K/Akt signaling is involved in neural differentiation of SH-SY5Y cells (69); according to this, we observed that CB1 and CB2 receptor agonists promoted neuritogenesis in our SH-SY5Y neuronal cell model, and, interestingly, we also observed a negative cross-talk in neuritogenesis when cells were co-stimulated with both receptor agonists. Thus, both CB1 and CB2 receptors might be negatively modulating each other in signaling pathways where endocannabinoids are involved, such as brain development and neural cell differentiation (66–68), Although these data, coupled with the BRET and PLA experiments, implicate heteromers in this receptor modulation, they could also be explained via simple signaling cross-talk rather than via physical interaction between receptors. However, the results obtained with the antagonists clearly show that CB1-CB2 receptor heteromers are the signaling units. Antagonists, by definition, do not signal; thus, the fact that a CB1 receptor-specific antagonist can block CB2 receptor signaling strongly argues against a cross-talk at the intracellular signaling level. A more likely explanation is that binding of the antagonist to CB1-CB2 receptor heteromers leads to a conformational change that reduces CB2 receptor-induced Akt/PKB phosphorylation. We found this cross-antagonism to be bidirectional in Akt/PKB phosphorylation signaling and in neurite growth.
A question arising from our findings is the following. Are the CB1-CB2 receptor heteromers indeed expressed in the brain? We explored this possibility using different approaches. The cross-antagonism discussed above can be considered as a heteromer fingerprint and can be exploited to demonstrate the expression of CB1-CB2 receptor heteromers in globus pallidus, a brain region that expresses a high amount of CB2 receptors (38). CB1 and CB2 receptors were poorly coupled to Akt/PKB phosphorylation in the rat globus pallidus, but the activation of both receptors increased ERK1/2 phosphorylation in rat globus pallidus slices. A clear cross-antagonism was observed for this signaling pathway, indicating that CB1-CB2 receptor heteromers are expressed in the globus pallidus. A second question arising from our findings is the following. Can CB1-CB2 receptor heteromers explain some of the reported results in the brain? Recently it has been described that both CB1 and CB2 receptors are co-expressed in the pineal gland, where they may be involved in the control of pineal physiology (57). In fact, CB1 and CB2 receptors and the enzymes catalyzing endocannabinoid biosynthesis and degradation were expressed in pinealocytes, and immunosignals for the CB2 receptor did not vary under a 12-h light/12-h dark cycle, whereas CB1 receptor immunoreaction was significantly reduced at the end of the light phase, when the expression of both receptors is more balanced (57). We isolated pinealocytes from rat pineal glands extracted at the end of the light period, and by taking advantage of the PLA, we demonstrated the expression of CB1-CB2 receptor heteromers in pinealocytes. In the pineal gland, the rhythm in melatonin biosynthesis is under control of norepinephrine-mediated regulation of arylalkylamine N-acetyltransferase, the penultimate enzyme of melatonin biosynthesis (75), and it has been described that phytocannabinoids like tetrahydrocannabinol reduce arylalkylamine N-acetyltransferase activity and attenuate melatonin biosynthesis in rat pineal glands (76). Thus, our results favor the hypothesis that through the negative cross-talk in CB1-CB2 receptor heteromers, CB2 receptor, mainly at the end of the light period, can negatively modulate the CB1 receptor-mediated tetrahydrocannabinol effect on arylalkylamine N-acetyltransferase activity and thus modulate melatonin synthesis. Another brain region where the CB2 receptor expression was reported is the nucleus accumbens, where CB2 receptor may be directly involved in many of the neurochemical and motivational properties of cocaine that are responsible for addiction (59). In fact, Xi et al. (59) found that CB2 receptor agonists directly infused into the nucleus accumbens decreased the cocaine intake, CB2 receptors regulate the psychomotor stimulant properties of cocaine, and CB2 receptors are endogenously activated by endocannabinoids in nucleus accumbens, where they control locomotor activity. Because CB1 receptors are also expressed in the striatum (24, 28) and are reported to be involved in the locomotor activity control (28), we determined the expression of CB1-CB2 receptor heteromers in rat nucleus accumbens slices by PLA. We found CB1-CB2 receptor heteromer expression in this brain region. Although the CB2 receptor-mediated effects on locomotor activity are also seen in CB1−/− mice (59), our results provide a new perspective by which CB1 and CB2 receptors might be modulating each other to control locomotion through CB1-CB2 receptor heteromers, using one partner as a “brake” for the other partner's action when both are co-expressed in the same neuron. CB1 receptors are extremely abundant, but CB2 receptors, at least in neurons, are thought to be much less abundant. Altering the expression of CB2 receptors may provide a possible level of regulation of CB1 receptor function by changing the amount of CB1-CB2 receptor heteromers. Further studies will have to be performed to investigate this possibility.
Finally, our data may provide explanations for several previously controversial points concerning CB1 and CB2 receptors. A confounding problem with the cannabinoid receptors has been the expression levels of the two receptors in the brain. Specifically, there have been varying reports as to the amount of CB2 receptor in the brain and whether those levels change in pathological settings as well as speculation on the role of the neuronal CB2 receptor (18, 31, 34, 35, 37, 38, 77–82). Our results suggest that, even at low expression levels, CB2 receptors could have a significant effect on signaling from CB1 receptor by reducing the cellular response through CB1 receptor. This modulation could be up-regulated upon injury or disease, supporting the thesis proposed recently by Onaivi (83) that CB1 and CB2 receptors can work independently and/or cooperatively in differing neuronal populations. This interdependence in neurons expressing both receptors could be through heteromers. At the ligand level, low and high levels of cannabinoid receptor ligands have given different results (84, 85), which were interpreted as affecting different populations of neurons, or by the presence of another cannabinoid ligand-mediated receptor, such as TRPV1 or GPR55, in the same neurons, but an alternative explanation could also be the presence of receptor heteromers. Conflicting results have also been observed on the serotonin system at high and low doses of the nonspecific cannabinoid receptor agonist WIN55212,2. At low doses, there was an increase in neuronal excitation that decreased at higher concentrations. The authors argued these effects must be through CB1 receptor because they could be blocked by the CB1 receptor-specific antagonist, rimonabant. However, we show here that a CB1 receptor antagonist could also block CB2 receptor-mediated signaling via receptor heteromers. It would be interesting to revisit these experiments using a CB2 receptor-specific antagonist as well. Heteromers could also help explain seemingly opposite effects seen with the ligands 2-arachidonoylglycerol and anandamide, which have been reported to have seemingly opposite effects on striatal spiny neurons and sensory neurons of the dorsal root ganglion (reviewed by di Marzo (86)). Part of these effects could also be through differential signaling via CB1-CB2 receptor heteromers. More studies will be required to elucidate how these heteromers behave in different tissues at different concentrations of these two endogenous agonists. Another complication has been the discovery that multiple isoforms of the CB2 receptors are expressed. Perhaps the different isoforms of the CB2 receptor can differentially modulate CB1 receptor and/or vice versa, depending on the tissue environment. Further studies may provide a clue as to how, at the mechanistic level, CB2 receptor is altering CB1 receptor signaling or vice versa and help clarify if any differences in isoforms exist. Finally, a third family of the cannabinoid receptors has recently been proposed, the GPR55 receptor (87, 88), with which a functional interaction with CB2 receptor has also been described (89, 90). It will be interesting to further characterize its role, if any, in the brain and whether it too can form heteromers with the other members of the family. In conclusion, we report the presence of CB1-CB2 receptor heteromers in a variety of brain regions. These heteromers may have a profound impact on CNS function in a variety of neurological and immunological systems, and our data suggest that these heteromers must be taken into account when designing therapeutic approaches toward alterations involving the endocannabinoid system.
Acknowledgment
We thank Jasmina Jiménez (University of Barcelona) for technical help.
Note Added in Proof
An article by Kleyer et al. (91) showing similar findings has recently been published.
This work was supported by Spanish Ministerio de Economia and Competitividad Grants SAF2010-18472, SAF2008-03229-E, and SAF2008-03118-E within the framework of the Era-NET Neuron program) and a grant for collaborative projects (Grant PI2011/02-7) from the Centro de Investigación Biomédica en Red sobre Enfermedades Neurodegenerativas (CIBERNED).
- GPCR
- G protein-coupled receptor
- PLA
- proximity ligation assay(s)
- BRET
- bioluminescence resonance energy transfer
- ANOVA
- analysis of variance
- ACEA
- arachidonyl-2-chloroethylamide.
REFERENCES
- 1. Klein T. W., Newton C., Larsen K., Lu L., Perkins I., Nong L., Friedman H. (2003) The cannabinoid system and immune modulation. J. Leukoc. Biol. 74, 486–496 [DOI] [PubMed] [Google Scholar]
- 2. Aguado T., Monory K., Palazuelos J., Stella N., Cravatt B., Lutz B., Marsicano G., Kokaia Z., Guzmán M., Galve-Roperh I. (2005) The endocannabinoid system drives neural progenitor proliferation. FASEB J. 19, 1704–1706 [DOI] [PubMed] [Google Scholar]
- 3. Mackie K., Stella N. (2006) Cannabinoid receptors and endocannabinoids. Evidence for new players. AAPS J. 8, E298–E306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piomelli D. (2003) The molecular logic of endocannabinoid signaling. Nat. Rev. Neurosci. 4, 873–884 [DOI] [PubMed] [Google Scholar]
- 5. Katona I., Urbán G. M., Wallace M., Ledent C., Jung K. M., Piomelli D., Mackie K., Freund T. F. (2006) Molecular composition of the endocannabinoid system at glutamatergic synapses. J. Neurosci. 26, 5628–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freund T. F., Katona I., Piomelli D. (2003) Role of endogenous cannabinoids in synaptic signaling. Physiol. Rev. 83, 1017–1066 [DOI] [PubMed] [Google Scholar]
- 7. Marsicano G., Goodenough S., Monory K., Hermann H., Eder M., Cannich A., Azad S. C., Cascio M. G., Gutiérrez S. O., van der Stelt M., López-Rodriguez M. L., Casanova E., Schütz G., Zieglgänsberger W., Di Marzo V., Behl C., Lutz B. (2003) CB1 cannabinoid receptors and on-demand defense against excitotoxicity. Science 302, 84–88 [DOI] [PubMed] [Google Scholar]
- 8. Ameri A. (1999) The effects of cannabinoids on the brain. Prog. Neurobiol. 58, 315–348 [DOI] [PubMed] [Google Scholar]
- 9. Howlett A. C. (2002) The cannabinoid receptors. Prostaglandins Other Lipid Mediat. 68, 619–631 [DOI] [PubMed] [Google Scholar]
- 10. Venance L., Maldonado R., Manzoni O. (2004) [Endocannabinoids in the central nervous system] Med. Sci. 20, 45–53 [DOI] [PubMed] [Google Scholar]
- 11. Schneider U., Seifert J., Karst M., Schlimme J., Cimander K., Müller-Vahl K. R. (2005) [The endogenous cannabinoid system. Therapeutic implications for neurologic and psychiatric disorders] Nervenarzt 76, 1062, 1065,–1066, 1068–1072 passim [DOI] [PubMed] [Google Scholar]
- 12. Ashton J. C. (2007) Cannabinoids for the treatment of inflammation. Curr. Opin. Investig. Drugs 8, 373–384 [PubMed] [Google Scholar]
- 13. Cabral G. A., Raborn E. S., Griffin L., Dennis J., Marciano-Cabral F. (2008) CB2 receptors in the brain. Role in central immune function. Br. J. Pharmacol. 153, 240–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Katona I., Freund T. F. (2008) Endocannabinoid signaling as a synaptic circuit breaker in neurological disease. Nat. Med. 14, 923–930 [DOI] [PubMed] [Google Scholar]
- 15. Romero J., Lastres-Becker I., de Miguel R., Berrendero F., Ramos J. A., Fernández-Ruiz J. (2002) The endogenous cannabinoid system and the basal ganglia. biochemical, pharmacological, and therapeutic aspects. Pharmacol. Ther. 95, 137–152 [DOI] [PubMed] [Google Scholar]
- 16. Mackie K. (2006) Cannabinoid receptors as therapeutic targets. Annu. Rev. Pharmacol. Toxicol. 46, 101–122 [DOI] [PubMed] [Google Scholar]
- 17. Palazuelos J., Aguado T., Egia A., Mechoulam R., Guzmán M., Galve-Roperh I. (2006) Non-psychoactive CB2 cannabinoid agonists stimulate neural progenitor proliferation. FASEB J. 20, 2405–2407 [DOI] [PubMed] [Google Scholar]
- 18. Fernández-Ruiz J., Romero J., Velasco G., Tolón R. M., Ramos J. A., Guzmán M. (2007) Cannabinoid CB2 receptor. A new target for controlling neural cell survival? Trends Pharmacol. Sci. 28, 39–45 [DOI] [PubMed] [Google Scholar]
- 19. Zanettini C., Panlilio L. V., Alicki M., Goldberg S. R., Haller J., Yasar S. (2011) Effects of endocannabinoid system modulation on cognitive and emotional behavior. Front. Behav. Neurosci. 5, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Di Marzo V., Melck D., Bisogno T., De Petrocellis L. (1998) Endocannabinoids. Endogenous cannabinoid receptor ligands with neuromodulatory action. Trends Neurosci. 21, 521–528 [DOI] [PubMed] [Google Scholar]
- 21. Maldonado R., Valverde O., Berrendero F. (2006) Involvement of the endocannabinoid system in drug addiction. Trends Neurosci. 29, 225–232 [DOI] [PubMed] [Google Scholar]
- 22. Ferré S., Goldberg S. R., Lluis C., Franco R. (2009) Looking for the role of cannabinoid receptor heteromers in striatal function. Neuropharmacology 56, Suppl. 1, 226–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pickel V. M., Chan J., Kash T. L., Rodríguez J. J., MacKie K. (2004) Compartment-specific localization of cannabinoid 1 (CB1) and μ-opioid receptors in rat nucleus accumbens. Neuroscience 127, 101–112 [DOI] [PubMed] [Google Scholar]
- 24. Pickel V. M., Chan J., Kearn C. S., Mackie K. (2006) Targeting dopamine D2 and cannabinoid-1 (CB1) receptors in rat nucleus accumbens. J. Comp. Neurol. 495, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Köfalvi A., Rodrigues R. J., Ledent C., Mackie K., Vizi E. S., Cunha R. A., Sperlágh B. (2005) Involvement of cannabinoid receptors in the regulation of neurotransmitter release in the rodent striatum. A combined immunochemical and pharmacological analysis. J. Neurosci. 25, 2874–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mátyás F., Yanovsky Y., Mackie K., Kelsch W., Misgeld U., Freund T. F. (2006) Subcellular localization of type 1 cannabinoid receptors in the rat basal ganglia. Neuroscience 137, 337–361 [DOI] [PubMed] [Google Scholar]
- 27. Uchigashima M., Narushima M., Fukaya M., Katona I., Kano M., Watanabe M. (2007) Subcellular arrangement of molecules for 2-arachidonoyl-glycerol-mediated retrograde signaling and its physiological contribution to synaptic modulation in the striatum. J. Neurosci. 27, 3663–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martín A. B., Fernandez-Espejo E., Ferrer B., Gorriti M. A., Bilbao A., Navarro M., Rodriguez de Fonseca F., Moratalla R. (2008) Expression and function of CB1 receptor in the rat striatum. Localization and effects on D1 and D2 dopamine receptor-mediated motor behaviors. Neuropsychopharmacology 33, 1667–1679 [DOI] [PubMed] [Google Scholar]
- 29. Svízenská I., Dubový P., Sulcová A. (2008) Cannabinoid receptors 1 and 2 (CB1 and CB2), their distribution, ligands, and functional involvement in nervous system structures. A short review. Pharmacol. Biochem. Behav. 90, 501–511 [DOI] [PubMed] [Google Scholar]
- 30. Ashton J. C., Friberg D., Darlington C. L., Smith P. F. (2006) Expression of the cannabinoid CB2 receptor in the rat cerebellum. An immunohistochemical study. Neurosci. Lett. 396, 113–116 [DOI] [PubMed] [Google Scholar]
- 31. Brusco A., Tagliaferro P. A., Saez T., Onaivi E. S. (2008) Ultrastructural localization of neuronal brain CB2 cannabinoid receptors. Ann. N.Y. Acad. Sci. 1139, 450–457 [DOI] [PubMed] [Google Scholar]
- 32. Golech S. A., McCarron R. M., Chen Y., Bembry J., Lenz F., Mechoulam R., Shohami E., Spatz M. (2004) Human brain endothelium. Coexpression and function of vanilloid and endocannabinoid receptors. Brain Res. Mol. Brain Res. 132, 87–92 [DOI] [PubMed] [Google Scholar]
- 33. Maresz K., Carrier E. J., Ponomarev E. D., Hillard C. J., Dittel B. N. (2005) Modulation of the cannabinoid CB2 receptor in microglial cells in response to inflammatory stimuli. J. Neurochem. 95, 437–445 [DOI] [PubMed] [Google Scholar]
- 34. Van Sickle M. D., Duncan M., Kingsley P. J., Mouihate A., Urbani P., Mackie K., Stella N., Makriyannis A., Piomelli D., Davison J. S., Marnett L. J., Di Marzo V., Pittman Q. J., Patel K. D., Sharkey K. A. (2005) Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science 310, 329–332 [DOI] [PubMed] [Google Scholar]
- 35. Gong J. P., Onaivi E. S., Ishiguro H., Liu Q. R., Tagliaferro P. A., Brusco A., Uhl G. R. (2006) Cannabinoid CB2 receptors. Immunohistochemical localization in rat brain. Brain Res. 1071, 10–23 [DOI] [PubMed] [Google Scholar]
- 36. Onaivi E. S., Ishiguro H., Gong J. P., Patel S., Perchuk A., Meozzi P. A., Myers L., Mora Z., Tagliaferro P., Gardner E., Brusco A., Akinshola B. E., Liu Q. R., Hope B., Iwasaki S., Arinami T., Teasenfitz L., Uhl G. R. (2006) Discovery of the presence and functional expression of cannabinoid CB2 receptors in brain. Ann. N.Y. Acad. Sci. 1074, 514–536 [DOI] [PubMed] [Google Scholar]
- 37. Brusco A., Tagliaferro P., Saez T., Onaivi E. S. (2008) Postsynaptic localization of CB2 cannabinoid receptors in the rat hippocampus. Synapse 62, 944–949 [DOI] [PubMed] [Google Scholar]
- 38. Lanciego J. L., Barroso-Chinea P., Rico A. J., Conte-Perales L., Callén L., Roda E., Gómez-Bautista V., López I. P., Lluis C., Labandeira-García J. L., Franco R. (2011) Expression of the mRNA coding the cannabinoid receptor 2 in the pallidal complex of Macaca fascicularis. J. Psychopharmacol. 25, 97–104 [DOI] [PubMed] [Google Scholar]
- 39. Pertwee R. G. (1997) Pharmacology of cannabinoid CB1 and CB2 receptors. Pharmacol. Ther. 74, 129–180 [DOI] [PubMed] [Google Scholar]
- 40. Gómez del Pulgar T., Velasco G., Guzmán M. (2000) The CB1 cannabinoid receptor is coupled to the activation of protein kinase B/Akt. Biochem. J. 347, 369–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Molina-Holgado E., Vela J. M., Arévalo-Martín A., Almazán G., Molina-Holgado F., Borrell J., Guaza C. (2002) Cannabinoids promote oligodendrocyte progenitor survival. Involvement of cannabinoid receptors and phosphatidylinositol-3 kinase/Akt signaling. J. Neurosci. 22, 9742–9753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferré S., Baler R., Bouvier M., Caron M. G., Devi L. A., Durroux T., Fuxe K., George S. R., Javitch J. A., Lohse M. J., Mackie K., Milligan G., Pfleger K. D., Pin J. P., Volkow N. D., Waldhoer M., Woods A. S., Franco R. (2009) Building a new conceptual framework for receptor heteromers. Nat. Chem. Biol. 5, 131–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carriba P., Ortiz O., Patkar K., Justinova Z., Stroik J., Themann A., Müller C., Woods A. S., Hope B. T., Ciruela F., Casadó V., Canela E. I., Lluis C., Goldberg S. R., Moratalla R., Franco R., Ferré S. (2007) Striatal adenosine A2A and cannabinoid CB1 receptors form functional heteromeric complexes that mediate the motor effects of cannabinoids. Neuropsychopharmacology 32, 2249–2259 [DOI] [PubMed] [Google Scholar]
- 44. Carriba P., Navarro G., Ciruela F., Ferré S., Casadó V., Agnati L., Cortés A., Mallol J., Fuxe K., Canela E. I., Lluís C., Franco R. (2008) Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nat. Methods 5, 727–733 [DOI] [PubMed] [Google Scholar]
- 45. Navarro G., Carriba P., Gandía J., Ciruela F., Casadó V., Cortés A., Mallol J., Canela E. I., Lluis C., Franco R. (2008) Detection of heteromers formed by cannabinoid CB1, dopamine D2, and adenosine A2A G-protein-coupled receptors by combining bimolecular fluorescence complementation and bioluminescence energy transfer. ScientificWorldJournal 8, 1088–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tebano M. T., Martire A., Chiodi V., Pepponi R., Ferrante A., Domenici M. R., Frank C., Chen J. F., Ledent C., Popoli P. (2009) Adenosine A2A receptors enable the synaptic effects of cannabinoid CB1 receptors in the rodent striatum. J. Neurochem. 110, 1921–1930 [DOI] [PubMed] [Google Scholar]
- 47. Akirav I., Fattore L. (2011) Cannabinoid CB1 and dopamine D1 receptors partnership in the modulation of emotional neural processing. Front. Behav. Neurosci. 5, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rozenfeld R., Gupta A., Gagnidze K., Lim M. P., Gomes I., Lee-Ramos D., Nieto N., Devi L. A. (2011) AT1R-CBR heteromerization reveals a new mechanism for the pathogenic properties of angiotensin II. EMBO J. 30, 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hojo M., Sudo Y., Ando Y., Minami K., Takada M., Matsubara T., Kanaide M., Taniyama K., Sumikawa K., Uezono Y. (2008) μ-Opioid receptor forms a functional heterodimer with cannabinoid CB1 receptor. Electrophysiological and FRET assay analysis. J. Pharmacol. Sci. 108, 308–319 [DOI] [PubMed] [Google Scholar]
- 50. Ward R. J., Pediani J. D., Milligan G. (2011) Heteromultimerization of cannabinoid CB1 receptor and orexin OX1 receptor generates a unique complex in which both protomers are regulated by orexin A. J. Biol. Chem. 286, 37414–37428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. da Silveira Cruz-Machado S., Carvalho-Sousa C. E., Tamura E. K., Pinato L., Cecon E., Fernandes P. A., de Avellar M. C., Ferreira Z. S., Markus R. P. (2010) TLR4 and CD14 receptors expressed in rat pineal gland trigger NFKB pathway. J. Pineal Res. 49, 183–192 [DOI] [PubMed] [Google Scholar]
- 52. Mackie K. (2005) Cannabinoid receptor homo- and heterodimerization. Life Sci. 77, 1667–1673 [DOI] [PubMed] [Google Scholar]
- 53. Söderberg O., Leuchowius K. J., Gullberg M., Jarvius M., Weibrecht I., Larsson L. G., Landegren U. (2008) Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45, 227–232 [DOI] [PubMed] [Google Scholar]
- 54. Trifilieff P., Rives M. L., Urizar E., Piskorowski R. A., Vishwasrao H. D., Castrillon J., Schmauss C., Slättman M., Gullberg M., Javitch J. A. (2011) Detection of antigen interactions ex vivo by proximity ligation assay. Endogenous dopamine D2-adenosine A2A receptor complexes in the striatum. BioTechniques 51, 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Howlett A. C., Bidaut-Russell M., Devane W. A., Melvin L. S., Johnson M. R., Herkenham M. (1990) The cannabinoid receptor. Biochemical, anatomical, and behavioral characterization. Trends Neurosci. 13, 420–423 [DOI] [PubMed] [Google Scholar]
- 56. Núñez E., Benito C., Pazos M. R., Barbachano A., Fajardo O., González S., Tolón R. M., Romero J. (2004) Cannabinoid CB2 receptors are expressed by perivascular microglial cells in the human brain. An immunohistochemical study. Synapse 53, 208–213 [DOI] [PubMed] [Google Scholar]
- 57. Koch M., Habazettl I., Dehghani F., Korf H. W. (2008) The rat pineal gland comprises an endocannabinoid system. J. Pineal Res. 45, 351–360 [DOI] [PubMed] [Google Scholar]
- 58. Suárez J., Llorente R., Romero-Zerbo S. Y., Mateos B., Bermúdez-Silva F. J., de Fonseca F. R., Viveros M. P. (2009) Early maternal deprivation induces gender-dependent changes on the expression of hippocampal CB1 and CB2 cannabinoid receptors of neonatal rats. Hippocampus 19, 623–632 [DOI] [PubMed] [Google Scholar]
- 59. Xi Z. X., Peng X. Q., Li X., Song R., Zhang H. Y., Liu Q. R., Yang H. J., Bi G. H., Li J., Gardner E. L. (2011) Brain cannabinoid CB2 receptors modulate cocaine's actions in mice. Nat. Neurosci. 14, 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Moreno E., Vaz S. H., Cai N. S., Ferrada C., Quiroz C., Barodia S. K., Kabbani N., Canela E. I., McCormick P. J., Lluis C., Franco R., Ribeiro J. A., Sebastião A. M., Ferré S. (2011) Dopamine-galanin receptor heteromers modulate cholinergic neurotransmission in the rat ventral hippocampus. J. Neurosci. 31, 7412–7423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. González S., Rangel-Barajas C., Peper M., Lorenzo R., Moreno E., Ciruela F., Borycz J., Ortiz J., Lluís C., Franco R., McCormick P. J., Volkow N. D., Rubinstein M., Floran B., Ferré S. (2012) Dopamine D4 receptor, but not the ADHD-associated D4.7 variant, forms functional heteromers with the dopamine D2S receptor in the brain. Mol. Psychiatry 17, 650–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sánchez M. G., Ruiz-Llorente L., Sánchez A. M., Díaz-Laviada I. (2003) Activation of phosphoinositide 3-kinase/PKB pathway by CB1 and CB2 cannabinoid receptors expressed in prostate PC-3 cells. Involvement in Raf-1 stimulation and NGF induction. Cell. Signal. 15, 851–859 [DOI] [PubMed] [Google Scholar]
- 63. Hosohata Y., Quock R. M., Hosohata K., Makriyannis A., Consroe P., Roeske W. R., Yamamura H. I. (1997) AM630 antagonism of cannabinoid-stimulated [35S]GTPγS binding in the mouse brain. Eur. J. Pharmacol. 321, R1–R3 [DOI] [PubMed] [Google Scholar]
- 64. Pertwee R. G. (2005) Pharmacological actions of cannabinoids. Handb. Exp. Pharmacol. 168, 1–51 [DOI] [PubMed] [Google Scholar]
- 65. Huffman J. W., Liddle J., Yu S., Aung M. M., Abood M. E., Wiley J. L., Martin B. R. (1999) 3-(1′,1′-Dimethylbutyl)-1-deoxy-δ8-THC and related compounds. Synthesis of selective ligands for the CB2 receptor. Bioorg. Med. Chem. 7, 2905–2914 [DOI] [PubMed] [Google Scholar]
- 66. Fernández-Ruiz J., Gómez M., Hernández M., de Miguel R., Ramos J. A. (2004) Cannabinoids and gene expression during brain development. Neurotox. Res. 6, 389–401 [DOI] [PubMed] [Google Scholar]
- 67. Harkany T., Guzmán M., Galve-Roperh I., Berghuis P., Devi L. A., Mackie K. (2007) The emerging functions of endocannabinoid signaling during CNS development. Trends Pharmacol. Sci. 28, 83–92 [DOI] [PubMed] [Google Scholar]
- 68. Mulder J., Aguado T., Keimpema E., Barabás K., Ballester Rosado C. J., Nguyen L., Monory K., Marsicano G., Di Marzo V., Hurd Y. L., Guillemot F., Mackie K., Lutz B., Guzmán M., Lu H. C., Galve-Roperh I., Harkany T. (2008) Endocannabinoid signaling controls pyramidal cell specification and long-range axon patterning. Proc. Natl. Acad. Sci. U.S.A. 105, 8760–8765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. López-Carballo G., Moreno L., Masiá S., Pérez P., Barettino D. (2002) Activation of the phosphatidylinositol 3-kinase/Akt signaling pathway by retinoic acid is required for neural differentiation of SH-SY5Y human neuroblastoma cells. J. Biol. Chem. 277, 25297–25304 [DOI] [PubMed] [Google Scholar]
- 70. Påhlman S., Hoehner J. C., Nånberg E., Hedborg F., Fagerström S., Gestblom C., Johansson I., Larsson U., Lavenius E., Ortoft E. (1995) Differentiation and survival influences of growth factors in human neuroblastoma. Eur. J. Cancer 31A, 453–458 [DOI] [PubMed] [Google Scholar]
- 71. Påhlman S., Ruusala A. I., Abrahamsson L., Mattsson M. E., Esscher T. (1984) Retinoic acid-induced differentiation of cultured human neuroblastoma cells. A comparison with phorbolester-induced differentiation. Cell Differ. 14, 135–144 [DOI] [PubMed] [Google Scholar]
- 72. Encinas M., Iglesias M., Liu Y., Wang H., Muhaisen A., Ceña V., Gallego C., Comella J. X. (2000) Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J. Neurochem. 75, 991–1003 [DOI] [PubMed] [Google Scholar]
- 73. Fiorentini C., Busi C., Spano P., Missale C. (2010) Dimerization of dopamine D1 and D3 receptors in the regulation of striatal function. Curr. Opin. Pharmacol. 10, 87–92 [DOI] [PubMed] [Google Scholar]
- 74. Munro S., Thomas K. L., Abu-Shaar M. (1993) Molecular characterization of a peripheral receptor for cannabinoids. Nature 365, 61–65 [DOI] [PubMed] [Google Scholar]
- 75. Klein D. C. (2007) Arylalkylamine N-acetyltransferase. “The Timezyme”. J. Biol. Chem. 282, 4233–4237 [DOI] [PubMed] [Google Scholar]
- 76. Koch M., Dehghani F., Habazettl I., Schomerus C., Korf H. W. (2006) Cannabinoids attenuate norepinephrine-induced melatonin biosynthesis in the rat pineal gland by reducing arylalkylamine N-acetyltransferase activity without involvement of cannabinoid receptors. J. Neurochem. 98, 267–278 [DOI] [PubMed] [Google Scholar]
- 77. Benito C., Núñez E., Tolón R. M., Carrier E. J., Rábano A., Hillard C. J., Romero J. (2003) Cannabinoid CB2 receptors and fatty acid amide hydrolase are selectively overexpressed in neuritic plaque-associated glia in Alzheimer's disease brains. J. Neurosci. 23, 11136–11141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ehrhart J., Obregon D., Mori T., Hou H., Sun N., Bai Y., Klein T., Fernandez F., Tan J., Shytle R. D. (2005) Stimulation of cannabinoid receptor 2 (CB2) suppresses microglial activation. J. Neuroinflammation 2, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Onaivi E. S., Ishiguro H., Gong J. P., Patel S., Meozzi P. A., Myers L., Perchuk A., Mora Z., Tagliaferro P. A., Gardner E., Brusco A., Akinshola B. E., Liu Q. R., Chirwa S. S., Hope B., Lujilde J., Inada T., Iwasaki S., Macharia D., Teasenfitz L., Arinami T., Uhl G. R. (2008) Functional expression of brain neuronal CB2 cannabinoid receptors are involved in the effects of drugs of abuse and in depression. Ann. N.Y. Acad. Sci. 1139, 434–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sagredo O., González S., Aroyo I., Pazos M. R., Benito C., Lastres-Becker I., Romero J. P., Tolón R. M., Mechoulam R., Brouillet E., Romero J., Fernández-Ruiz J. (2009) Cannabinoid CB2 receptor agonists protect the striatum against malonate toxicity. Relevance for Huntington's disease. Glia 57, 1154–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. García-Gutiérrez M. S., Manzanares J. (2011) Overexpression of CB2 cannabinoid receptors decreased vulnerability to anxiety and impaired anxiolytic action of alprazolam in mice. J. Psychopharmacol. 25, 111–120 [DOI] [PubMed] [Google Scholar]
- 82. Onaivi E. S., Ishiguro H., Gu S., Liu Q. R. (2012) CNS effects of CB2 cannabinoid receptors. Beyond neuro-immuno-cannabinoid activity. J. Psychopharmacol. 26, 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Onaivi E. S. (2011) Commentary. Functional neuronal CB2 cannabinoid receptors in the CNS. Curr. Neuropharmacol. 9, 205–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bambico F. R., Katz N., Debonnel G., Gobbi G. (2007) Cannabinoids elicit antidepressant-like behavior and activate serotonergic neurons through the medial prefrontal cortex. J. Neurosci. 27, 11700–11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Bellocchio L., Lafenêtre P., Cannich A., Cota D., Puente N., Grandes P., Chaouloff F., Piazza P. V., Marsicano G. (2010) Bimodal control of stimulated food intake by the endocannabinoid system. Nat. Neurosci. 13, 281–283 [DOI] [PubMed] [Google Scholar]
- 86. Di Marzo V. (2011) Endocannabinoid signaling in the brain. Biosynthetic mechanisms in the limelight. Nat. Neurosci. 14, 9–15 [DOI] [PubMed] [Google Scholar]
- 87. Pertwee R. G. (2007) GPR55. A new member of the cannabinoid receptor clan? Br. J. Pharmacol. 152, 984–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ryberg E., Larsson N., Sjögren S., Hjorth S., Hermansson N. O., Leonova J., Elebring T., Nilsson K., Drmota T., Greasley P. J. (2007) The orphan receptor GPR55 is a novel cannabinoid receptor. Br. J. Pharmacol. 152, 1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Balenga N. A., Aflaki E., Kargl J., Platzer W., Schröder R., Blättermann S., Kostenis E., Brown A. J., Heinemann A., Waldhoer M. (2011) GPR55 regulates cannabinoid 2 receptor-mediated responses in human neutrophils. Cell Res. 21, 1452–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Irving A. (2011) New blood brothers. The GPR55 and CB2 partnership. Cell Res. 21, 1391–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kleyer J., Nicolussi S., Taylor P., Simonelli D., Furger E., Anderle P., Gertsch J. (2012) Cannabinoid receptor trafficking in peripheral cells is dynamically regulated by a binary biochemical switch. Biochem. Pharmacol. 83, 1393–1412 [DOI] [PubMed] [Google Scholar]