Background: Caveolin-1 regulates cellular antioxidant capacity, but mechanisms remain unknown.
Results: Caveolin-1 interacts with Nrf2 and suppresses its transcriptional activity and down-regulates cellular antioxidant enzymes.
Conclusion: Caveolin-1 regulates cellular antioxidant capacity through interaction with Nrf2.
Significance: Clarifying how caveolin-1 regulates cellular antioxidant capacity and identifying a novel target to establish the contribution of oxidative stress to human pathologies.
Keywords: Antioxidants, Caveolin, Nrf2, Oxidative Stress, Protein-Protein Interactions
Abstract
The Nrf2 (nuclear erythroid 2 p45-related factor-2) signaling pathway is known to play a pivotal role in a variety of oxidative stress-related human disorders. It has been reported recently that the plasma membrane resident protein caveolin-1 (Cav-1) can regulate expression of certain antioxidant enzymes and involves in the pathogenesis of oxidative lung injury, but the detailed molecular mechanisms remain incompletely understood. Here, we demonstrated that Cav-1 inhibited the expression of antioxidant enzymes through direct interaction with Nrf2 and subsequent suppression of its transcriptional activity in lung epithelial Beas-2B cells. Cav-1 deficiency cells exhibited higher levels of antioxidant enzymes and were more resistant to oxidative stress induced cytotoxicity, whereas overexpression of Cav-1 suppressed the induction of these enzymes and further augmented the oxidative cell death. Cav-1 constitutively interacted with Nrf2 in both cytosol and nucleus. Stimulation of 4-hydroxynonenol increased the Cav-1-Nrf2 interaction in cytosol but disrupted their association in the nucleus. Knockdown of Cav-1 also disassociated the interaction between Nrf2 and its cytoplasmic inhibitor Keap1 (Kelch-like ECH-associated protein 1) and increased the Nrf2 transcription activity. Mutation of the resembling Cav-1 binding motif on Nrf2 effectively attenuated their interaction, which exhibited higher transcription activity and induced higher levels of antioxidant enzymes relative to the wild-type control. Altogether, these studies clearly demonstrate that Cav-1 inhibits cellular antioxidant capacity through direct interaction with Nrf2 and subsequent suppression of its activity, thereby implicating in certain oxidative stress-related human pathologies.
Introduction
Oxidative stress has been known to play an important role in the pathogenesis of certain human diseases such as neurodegeneration, arthrosclerosis, and oxidative lung injury (1). Endogenous antioxidant systems consist of a number of proteins or enzymes, and small molecules (e.g. vitamin E and glutathione, GSH), which cope with oxidative stress and maintain the redox environment of the body (2). The biosynthesis of GSH is catalyzed by glutamate cysteine ligase (GCL),4 which is a heterodimeric protein composed of a rate-limiting catalytic (GCLC) and another modifier subunit (3, 4). Variability in GCL expression is associated with several oxidative stress-related diseases (5). Stress response proteins such as heme oxygenase (HO)-1 are also well known to be cytoprotective against various oxidant or pro-oxidant insults (6, 7).
The transcription factor Nrf2 (nuclear erythroid 2 p45-related factor-2) represents a major inducible cellular and tissue defense against oxidative stress (8, 9), which has been implicated as the central protein that interacts with the antioxidant response element to activate gene transcription constitutively or in response to an oxidative stress signal (10). Under homeostatic conditions, Keap1 (Kelch-like ECH-associated protein 1) binds to Nrf2 and facilitates the degradation of Nrf2 via the proteasome system (11). Upon stimulation, Nrf2 dissociates from its cytoplasmic inhibitor Keap1, translocates to the nucleus, and by heterodimerizing with a small Maf protein, transactivates the expression of antioxidant response element-dependent detoxifying genes, including HO-1, GCLC, and thioredoxin reductase 1 (12–14).
Caveolae are 50- to 100-nm omega-shaped invaginations of the cell surface plasma membrane that are enriched in glycosphingolipids and cholesterol (15–16). Caveolae occur in a variety of cell types, including epithelial, endothelial cells, fibroblasts, smooth muscle cells, and adipocytes. Caveolin-1 (Cav-1), a 21–24-kDa protein, is a major resident scaffolding protein constituent of caveolae that participates in vesicular trafficking and signal transduction events (15, 16). This molecule has been known to interact with various pathophysiologically important molecules such as inflammatory regulator Toll-like receptor 4 (17), apoptotic factors Fas and survivin (18, 19), and autophagy molecule LC3B (20, 21). It has been reported recently that deletion of Cav-1 protects against hyperoxia-induced oxidative lung injury via up-regulation of HO-1 (22). Expression of HO-1 was elevated markedly in lung tissue or fibroblasts from Cav-1(−/−) mice, and hyperoxia induced the physical interaction between cav-1 and HO-1 which resulted in attenuation of HO activity (22). Volonte and Galbiati (23) also have found that Cav-1 could interact with thioredoxin reductase 1 and suppress its activity, and deletion of Cav-1 by siRNA increased the dimeric expression of thioredoxin reductase 1. These studies suggest that Cav-1 plays an important role in regulation of cellular antioxidant capacity.
It is reasonable that Cav-1 physically interacts with the antioxidant proteins and therefore suppresses their enzyme activity; however, how Cav-1 regulates the expression of these proteins remains unclear. As we have found that both HO-1 and thioredoxin reductase 1 could be transcriptionally regulated by the same transcription factor Nrf2 (14), we hypothesized that Cav-1 might regulate the expression of these antioxidant enzymes through regulation of Nrf2 activity. In the present study, we indeed demonstrated that Cav-1 interacted with the Keap1-Nrf2 system and suppressed the transcription activity of Nrf2, thereby regulating the expression of cellular antioxidant enzymes such as GCLC and HO-1 and subsequent cellular antioxidant capacity.
EXPERIMENTAL PROCEDURES
Cell Culture and Cell Viability Assay
Beas-2B lung epithelial cells were purchased from American Type Culture Collection (ATCC, CRL-9609TM), and mouse lung fibroblasts were isolated from wild-type C57BL6 or Cav-1 knock-out mice (The Jackson Laboratory, 004585) as described previously (24, 25). All cells were maintained in DMEM containing 10% fetal bovine serum and antibiotics. Compound 4-hydroxynonenol (4-HNE) was obtained from Calbiochem (393204). For determination of cell viability, Beas-2B cells were seeded at a density of 1 × 104 cells per well into a 96-well culture dish, and the conventional 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (Bio-Basic, Inc., 298-93-1) reduction assay was used as reported previously (13, 14).
Transfection of siRNA and cDNA
Human Cav-1 siRNA was purchased from Dharmacon (L-003467-00), and the Cav1-mRFP (14434) and pcDNA3-EGFP-Nrf2 (21549) were obtained from Addgene. The mutant derivative of Nrf2 was performed using the QuikChange II site-directed mutagenesis kit (Stratagene, 200555), following the manufacturer's instructions. All siRNA and cDNAs were transfected into Beas-2B cells by using Lipofectamine 2000 (Invitrogen, 11668019).
Hydrogen Peroxide Assay
The consumption of H2O2 was examined using the hydrogen peroxide assay kit (Abcam). Beas-2B cells were plated at 3 × 104 cells per well into a 24-well culture dish, and after treatment of H2O2 for various time points, the culture supernatants were harvested for analysis of the H2O2 concentration following the supplied protocol.
Isolation of Cytoplasmic and Nuclear Fractions
A nuclear extract kit (Active Motif, 40010) was used for isolation of the cytoplasmic and nuclear fractions in Beas-2B cells, and the quality of the isolation was confirmed by Western blot analysis of specifically distributed proteins.
Immunoprecipitation and Immunoblotting (Western Blot) Analyses
Immunoprecipitation and immunoblotting were performed essentially as described previously (24, 25). Antibodies against Keap1 (sc-15246), Nrf2 (sc-722), GCLC (sc-28965), Lamin B (sc-373918), Hsp70 (sc-32239), and β-actin (sc-4778) were from Santa Cruz Biotechnology, Inc. The Cav-1 antibody was obtained from BD Transduction Laboratories (610406), and the HO-1 antibody was purchased from Stressgen (OSA-110). All blot images shown in the figures were representatives from at least three independent experiments.
Real-time Polymerase Chain Reaction (PCR)
Total RNA was extracted from cells using TRIzol (Invitrogen) according to the manufacturer's instructions and reverse-transcribed into cDNA using random hexamers and superscript II reverse transcriptase (Invitrogen). The relative levels of GCLC and HO-1 mRNA transcripts to control β-actin were determined by quantitative real-time PCR using the SYBR Green system (Takara) on a spectrofluorometric thermal cycler (iCycler; Bio-Rad).
Confocal Imaging Study
After treatment, cells were fixed with 4% paraformaldehyde and analyzed with the immunofluorescence staining protocol as described previously (20). Samples were viewed with an Olympus Fluoview 300 confocal laser scanning head with an Olympus IX70 inverted microscope.
Luciferase Reporter Assay
The antioxidant response reporter kit was purchased from Qiagen (CCS-5020L) and the Dual-Luciferase reporter assay system was obtained from Promega (E1910). The assay was performed following the manufacturer's instructions.
Statistics
All data of at least three independent experiments are expressed as the mean ± S.D. and analyzed by Student's t test. Values of p < 0.05 were considered to be statistically significant.
RESULTS
Effects of Cav-1 Depletion on Cellular Antioxidant Capacity
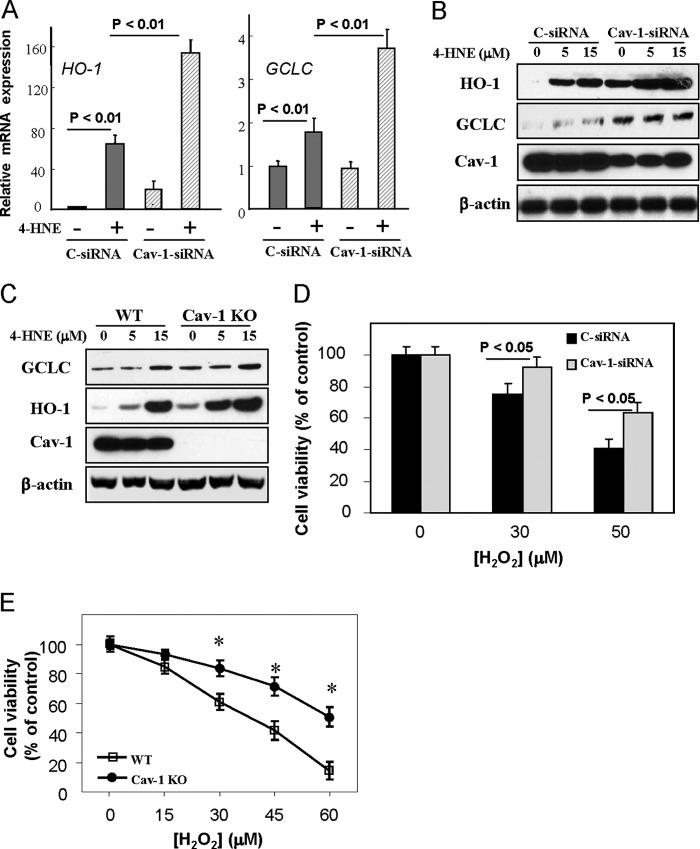
As an initial experiment toward determining the role of Cav-1 in regulation of cellular antioxidant capacity, human bronchial epithelial Beas-2B cells were transfected with Cav-1 siRNA and then treated with 15 μm of 4-HNE, an α,β-unsaturated aldehyde, which has been shown to effectively disrupt the Keap1-Nrf2 complex and to induce expression of antioxidant enzymes at low concentrations without any appreciable cytotoxicity (14, 26–28). In Beas-2B cells, 4-HNE at <15 μm also exhibited no considerable cytotoxicity (data not shown), but it significantly induced expressions of both mRNA transcripts (Fig. 1A) and protein levels (Fig. 1B) of HO-1 and GCLC, which were markedly higher in Cav-1 siRNA-treated cells relative to those in controls (Fig. 1, A and B). The Nrf2 expression showed no considerable changes in the Cav-1 siRNA-treated cells (data not shown). Similar findings were also observed in Cav-1 knock-out fibroblasts, in which 4-HNE treatment also induced higher protein level of HO-1 and GCLC (Fig. 1C). Consequently, the Cav-1 siRNA-treated Beas-2B cells and the Cav-1 knock-out fibroblasts were more resistant to H2O2-induced cell death (Fig. 1, D and E). These data clearly demonstrated an increased antioxidant capacity in Cav-1 deficiency cells.
FIGURE 1.
Effects of Cav-1 dificiency on cellular antioxidant capacity. A and B, Beas-2B cells were transfected with control (C-siRNA) or Cav-1 siRNA for 48 h and followed by a treatment of 4-HNE at 15 μm for an additional 6 h (A) or of 4-HNE at indicated concentrations for 24 h (B). Cells were then harvested for real-time PCR analysis of HO-1 and GCLC mRNA transcripts (A) or Western blot analysis of these proteins (B). C, WT or Cav-1 knock-out (Cav-1 KO) fibroblasts were treated with 4-HNE at indicated concentrations for 24 h and were subjected for Western blot analysis of GCLC and HO-1 expressions. D and E, cell viability assay for H2O2-induced cytotoxicity in Cav-1 siRNA treated Beas-2B cells (D) or Cav-1 KO fibroblasts (E). Beas-2B cells were treated with control or Cav-1 siRNA for 48 h and followed by the treatment of H2O2 at indicated concentrations for an additional 24 h (D). WT or Cav-1 KO fibroblasts were treated various concentrations of H2O2 for 24 h (E). *, significantly different from the corresponding value of control (p < 0.05).
Effects of Cav-1 Overexpression on Cellular Antioxidant Capacity
We next sought to determine whether increases on Cav-1 expression could exert an adverse effect in context of oxidative injury. Interestingly, 4-HNE induced mRNA expression of HO-1 (Fig. 2A) and GCLC (Fig. 2B) were attenuated effectively by Cav-1 mRFP overexpression in Beas-2B cells. As a result, these Cav-1 overexpressed cells were more susceptible to H2O2-induced cell death (Fig. 2C), which was in agreement with the results from Cav-1 deficiency cells shown in Fig. 1.
FIGURE 2.
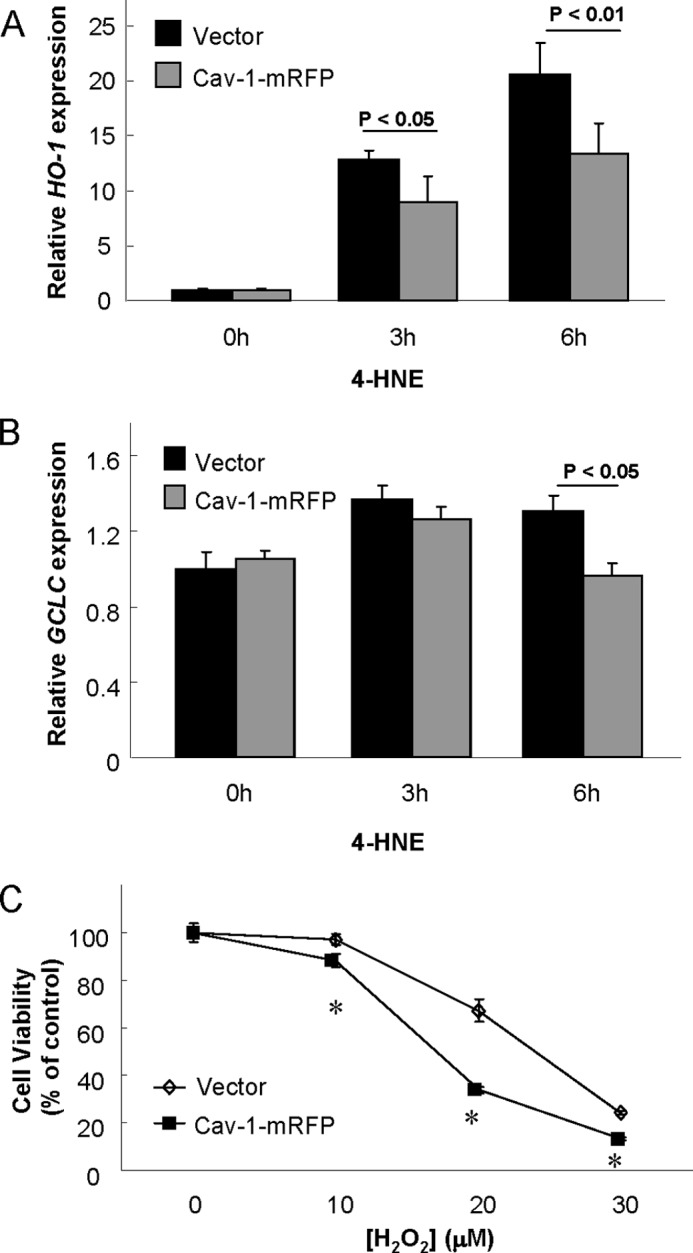
Effects of Cav-1 overexpression on cellular antioxidant capacity. Beas-2B cells were transfected with Cav-1 mRFP or its vector control for 48 h and followed by treatment of 15 μm of 4-HNE for indicated times (A and B), or various concentrations of H2O2 for 24 h (C). Cells were then subjected for real-time PCR analysis of HO-1 (A) or GCLC (B) mRNA expression, or 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay for cell viability (C). *, significantly different from the corresponding value of control (p < 0.05).
Kinetics of Cell Viability and H2O2 Consumption
The concentrations of H2O2 to induce cytotoxicity in Beas-2B cells appeared to be a little low, which led us further examined the detailed kinetics of cell viability and H2O2 consumption in these cells. H2O2 (50 μm) induced a rapid decline of cell viability to ∼50% within 1 h, which was further reduced to ∼30% at 3 h but remained at the same levels until 24 h (supplemental Fig. S1). The concentration of H2O2 was decreased promptly to less than half within 15 min and completely consumed by 1 h (supplemental Fig. S2), which was in agreement with the cell viability results (supplemental Fig. S1). These data suggested that Beas-2B cells contain normal levels of antioxidant defenses at basal conditions, which are able to rapidly degrade the H2O2. However, the cells survived from H2O2 appeared to be impaired on their proliferation capacity, with no considerable changes on the cell viability between 3 h and 24 h (supplemental Fig. S1).
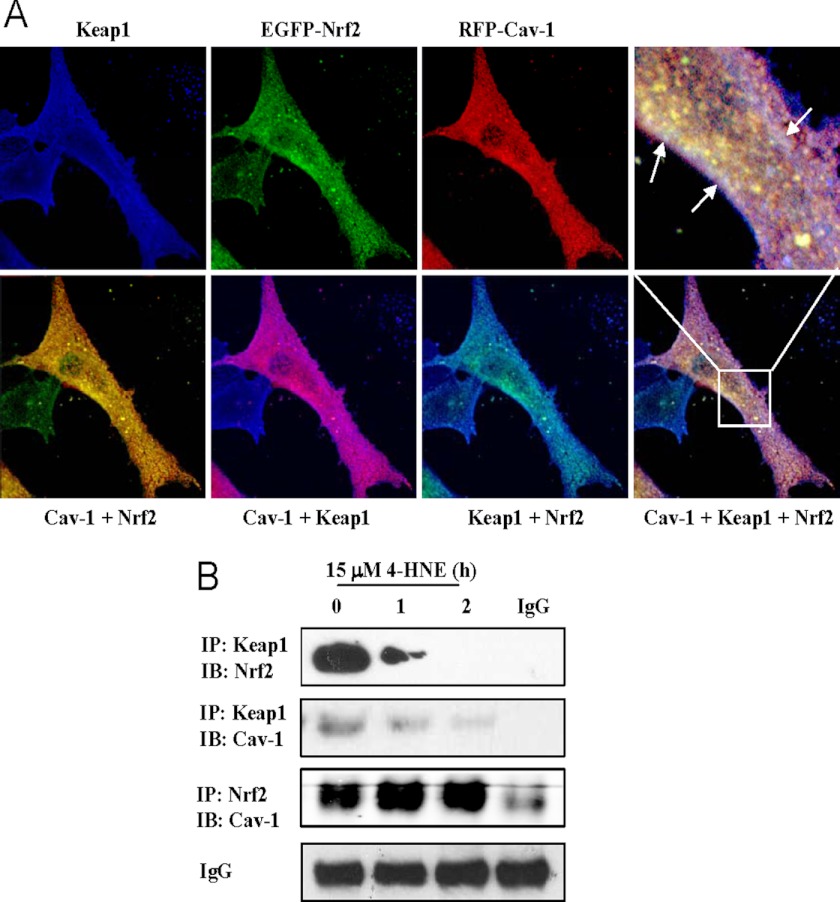
Interactions between Cav-1 and Keap1-Nrf2 System
As the Keap1-Nrf2 system plays a central role in regulation of the levels of cellular antioxidants such as HO-1 and GCLC (12–14), we then explored the possible cross-talk between Cav-1 and the Keap1-Nrf2 system. Confocal imaging analysis demonstrated that these three proteins constitutively interacted with each other at basal conditions (Fig. 3A), and these physical interactions were further confirmed by immunoprecipitation experiments (Fig. 3B). Under stimulation of 4-HNE, the Keap1-Nrf2 complex was disrupted rapidly, as well as did the Cav-1-Keap1 interaction (Fig. 3B). Interestingly, the association between Cav-1 and Nrf2 was enhanced slightly, rather than being disrupted by 4-HNE treatment (Fig. 3B), suggestive of that when Nrf2 was liberated from Keap1 by 4-HNE stimulation, Cav-1 subsequently could capture certain amount of free Nrf2 to suppress its activity.
FIGURE 3.
Interactions between Cav-1 and the Keap1-Nrf2 system. A, confocal image analysis of interactions between Cav-1 and the Keap1-Nrf2 system in Beas-2B cells. Cells were co-transfected with pcDNA3-EGFP-Nrf2 and Cav-1 mRFP for 48 h. Cells were then fixed and stained with Keap1 primary antibody and followed by incubation with a Dylight 405-conjugated second antibody. White arrows indicate overlapping areas of three colors. B, Beas-2B cells were treated with 15 μm of 4-HNE for indicated times, and harvested samples were immunoprecipitated (IP) and immunoblotted (IB) with indicated antibodies, respectively. Nonspecific IgG serves as input control.
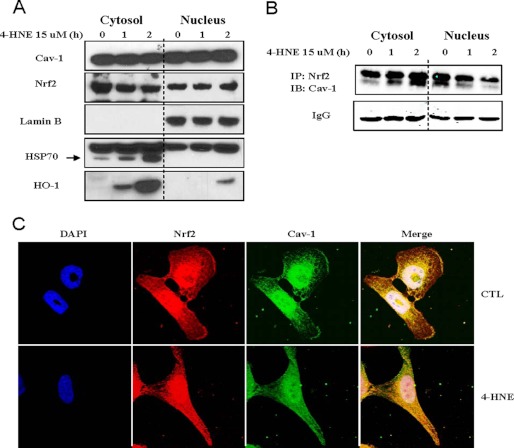
Kinetics of Cav-1-Nrf2 Interaction in Cytosol and Nucleus
The intriguing kinetics between Cav-1 and Nrf2 in whole cell lysate led us to further investigate their interaction in subcellular fractions. In general, the localization of Cav-1 is associated with the lipid rafts on plasma membrane, but in our experience, the cellular distribution of Cav-1 might be different in various cells. For example, we have observed that in human airway epithelial A549 cells, Cav-1 appeared to be localized predominantly on the plasma membrane, as the Cav-1 immunofluorescence formed a bright cycle on the cell surface but with less density in other areas (data not shown). However, in Beas-2B cells, as shown in Fig. 3A, Cav-1 was distributed not only on the plasma membrane but throughout the cells. Emerging evidence also has suggested that Cav-1 is localized in the nucleus in certain cell types (29–32). We were therefore interested in studying whether Cav-1 could interact with Nrf2 in nucleus of Beas-2B cells. To our surprise, we found that in Beas-2B cells almost half of Cav-1 was localized in nucleus (Fig. 4A), which was not likely due to experimental contamination of nucleus isolation, as the distributions of lamin B, heat shock protein 70 (HSP70), and HO-1 were distinct in either cytosol or nucleus (Fig. 4A). More interestingly, 4-HNE treatment significantly increased the Cav-1-Nrf2 association in cytosol, but this interaction was time-dependently decreased in nucleus (Fig. 4B). The double Cav-1 bands in Fig. 4B might appear because Caveolin-1 has two isoforms, of which the β-isoform contains residues 32–178, resulting in a protein 3 kDa smaller in size (15, 33). Confocal imaging studies further confirmed the complex kinetics of Cav-1-Nrf2 interaction (Fig. 4C). The yellow color formed by Cav-1 and Nrf2 was increased in cytosol after 4-HNE treatment, whereas the white color around the nucleus area appeared to be deceased slightly by 4-HNE stimulation (Fig. 4C). All of these findings strongly suggested that Cav-1 exhibited a suppressive effect on Nrf2 in both the cytosol and nucleus.
FIGURE 4.
Kinetics of Cav-1-Nrf2 interaction in cytosol and nucleus. A and B, Beas-2B cells were treated with 15 μm of 4-HNE for indicated times, and the cytoplasmic and nuclear fractions were isolated. Both fractions were subjected for Western blot analysis of indicated proteins (A), or immunoprecipitation (IP) with Nrf2 and immunoblotting (IB) with Cav-1 (B). Nonspecific IgG serves as input control. C, confocal image analysis of the Cav-1-Nrf2 interaction in the cytosol and nucleus. After treatment with or without (CTL, control) 15 μm of 4-HNE for 2 h, cells were subjected for immunofluorescence staining and confocal image analysis of Cav-1, Nrf2, and nuclei (DAPI).
Regulation of Keap1-Nrf2 Interaction and Nrf2 Transcription Activity by Cav-1
To further investigate the mechanisms of Cav-1 in regulation of cellular antioxidant capacity, we next sought to examine the role of Cav-1 in Keap1-Nrf2 interaction and subsequent Nrf2 transcription activity. Interestingly, siRNA-dependent knockdown of Cav-1 eventually decreased the Keap1-Nrf2 interaction at basal level (Fig. 5A, left panels), whereas overexpression of Cav-1 exhibited no appreciable effects on the Keap1-Nrf2 association (Fig. 5A, right panels), indicating that Cav-1 at least in part mediates the Keap1-Nrf2 interaction at basic conditions. The antioxidant response element luciferase reporter assay revealed that the Nrf2 transcription activity was increased notably in Cav-1 siRNA-treated cells but was decreased significantly in cells overexpressed with Cav-1 mRFP (Fig. 5B), which was completely in agreement with the results of the antioxidant gene expressions shown in Figs. 1 and 2.
FIGURE 5.
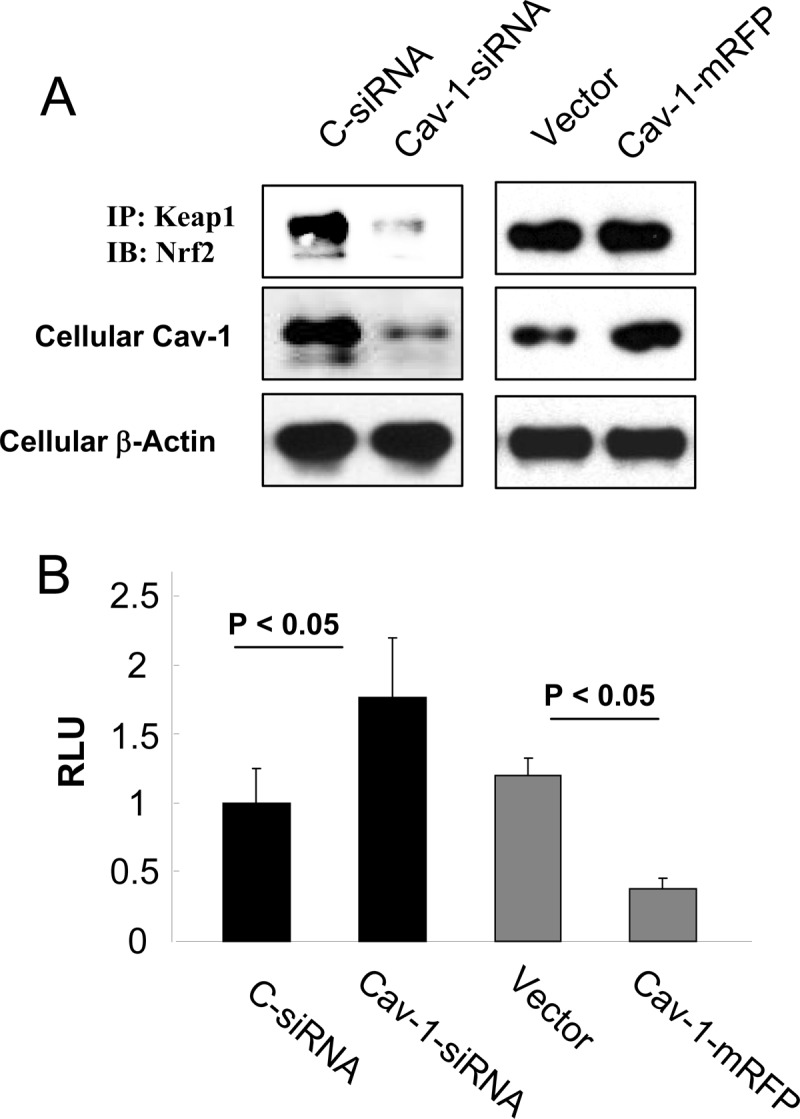
Role of Cav-1 in regulation Keap1-Nrf2 interaction and Nrf2 transcription activity. A, Beas-2B cells were transfected with control (C-siRNA) or Cav-1 siRNA, or Cav-1 mRFP or its vector control for 48 h and were harvested for immunoprecipitation (IP) or Western blot (IB) analysis as indicated. B, Beas-2B cells were co-transfected with the antioxidant response element reporter plasmid and the indicated siRNA or cDNA for 48 h and were harvested for luciferase assay. RLU, relative luciferase unit.
Mutation of the Cav-1-binding Motif on Nrf2 Influences Their Physical Interaction and Subsequent Cellular Antioxidant Capacity
Proteins that bind to Cav-1 typically contain canonical Cav-1-binding motifs (CBMs), ΦXΦXXXXΦ or ΦXXXXΦXXΦ, where Φ refers to an aromatic amino acid (Trp, Phe, or Tyr), and X is any nonaromatic amino acid, although proteins without such motifs also are capable of binding to Cav-1 (15, 20). The primary structure of Nrf2 contains a sequence 281FGDEFYSAFI290 resembling a consensus CBM. To examine whether the CBM sequence on Nrf2 mediates the Nrf2-Cav-1 interaction, we generated amino acid substitution mutants of Nrf2 at position Tyr-286. Interestingly, the wild-type Nrf2 overexpression increased levels of Nrf2-Cav-1 were almost abolished by Y286A mutation (Fig. 6A), an aromatic-to-nonaromatic amino acid substitution. Transfection of wild-type Nrf2 also increased the Keap1-Nrf2 interaction, but intriguingly, the mutation of CBM on Nrf2 further slightly increased the Keap1-Nrf2 association (Fig. 6A). Furthermore, without Cav-1 binding, the Y286A mutation led to an increased transcription activity of Nrf2 relative to its wild-type control (Fig. 6B), and subsequently, the cellular levels of antioxidants such as HO-1 were further enhanced in the Y286A-transfected cells (Fig. 6C), and these cells were more resistant to H2O2-induced cytotoxicity (Fig. 6D).
FIGURE 6.
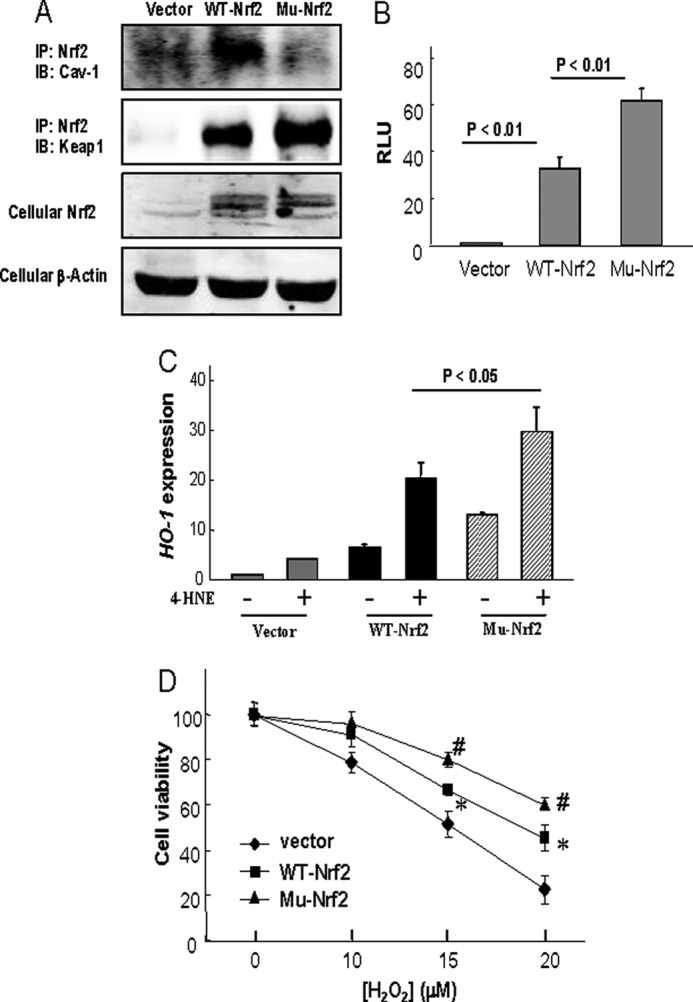
Effects of Nrf2 Y286A mutation on Cav-1-Nrf2 reaction and subsequent cellular antioxidant capacity. A, Beas-2B cells were transfected with control vector, pcDNA3-EGFP-Nrf2, or Y286A mutated Nrf2 (Mu-Nrf2) for 48 h and were harvested for immunoprecipitation (IP) or Western blot (IB) analysis as indicated. B, Beas-2B cells were co-transfected with the antioxidant response element reporter plasmid and the indicated cDNA for 48 h and were harvested for luciferase assay. RLU, relative luciferase unit. C and D, Beas-2B cells were transfected with control vector, pcDNA3-EGFP-Nrf2 (WT-Nrf2), or Y286A mutated Nrf2 (Mu-Nrf2) for 48 h and followed by 15 μm of 4-HNE treatment for 6 h for HO-1 mRNA expression (C) or various concentrations of H2O2 for 24 h for cell viability assay (D). *, significantly different from the corresponding value of vector control (p < 0.05); #, significantly different from the corresponding value of WT-Nrf2 (p < 0.05).
DISCUSSION
Cav-1 is the major structural component of caveolae, which exerts a variety of biological functions, including the regulation of cholesterol homeostasis, vesicular transport, proliferation, and apoptosis in a diversity of cell types (15, 16). Recently, this molecule also has been implicated as a modulator of oxidative stress. Jin and co-workers have reported that Cav-1 knock-out mice exhibit higher resistance to hyperoxia-induced acute lung injury and oxidative cell death, through regulation of a variety of signaling molecules such as survivin (19), Cyr61 (34), Fas and Bid (18), and up-regulation of HO-1 (22). Cav-1 also interacts with thioredoxin reductase 1 and suppresses its activity, and deletion of Cav-1 increases the dimeric expression of thioredoxin reductase 1 (23). However, how Cav-1 regulates the expression of these antioxidant proteins remains unclear. Here, we clearly demonstrate that Cav-1 negatively regulates the expression of antioxidant enzymes through direct interaction with Nrf2 and subsequent suppression of its transcription activity in lung epithelial Beas-2B cells.
The typical Cav-1-protein interaction occurs through the scaffolding domain in Cav-1 and the CBM in target proteins (15), such as endothelial nitric oxide synthase (35), Toll-like receptor 4 (17), and activin receptor-like kinase 1 (36). We have observed recently that the autophagy protein LC3B contain an amino acid sequence very similar to CBM, and this resembling structure mediates the Cav-1-LC3B interaction (20). In the present study, we also have found that Nrf2 contains such a similar CBM, which also mediates its interaction with Cav-1, as mutation of the CBM-like structure on Nrf2 effectively disrupts their association and subsequently enhances the cellular antioxidant capacity. Thus, activating Nrf2 pathways through its CDM mutation may represent an effective genetic approach against oxidative stress-induced damage.
The kinetics of Cav-1-Nrf2 interaction appears to be complicated in Beas-2B cells, as 4-HNE treatment time-dependently increased the Cav-1-Nrf2 interaction in cytosol but decreased their association in nucleus (Fig. 4B). We have observed previously that Cav-1 basically interacts with autophagic proteins LC3B and apoptotic factor Fas, and their interactions are disrupted under stimulation (20), whereas interaction between Toll-like receptor 4 and Cav-1 is further enhanced by LPS treatment in macrophage (17). Additional studies of subcellular trafficking of Cav-1 and Nrf2 in context of oxidative stress might be warranted. Nevertheless, the present study clearly demonstrates a unique and heretofore uncharacterized interaction of Cav-1 with Nrf2, with functional implications as one of the most apical regulatory events in oxidative stress signaling with respect to the downstream production of antioxidant enzymes.
It may be noteworthy that the interactions among Cav-1-Nrf2-Keap1 complex are various depending on the sources of Cav-1 and Nrf2. Cav-1 siRNA-treated Beas-2B cells displayed a markedly decreased Keap1-Nrf2 association at basic conditions (Fig. 5A), suggestive of that endogenous Cav-1 may mediate, at least in part, the constitutive Keap1-Nrf2 interaction. However, overexpression of exogenous Cav-1 caused no considerable changes on the Keap1-Nrf2 association (Fig. 5A) but further suppressed the Nrf2 transcription activity (Fig. 5B) and down-regulated the antioxidant gene expression (Fig. 2), indicating that exogenous Cav-1 could also suppress endogenous Nrf2 activity without affecting the Keap1-Nrf2 association. Overexpression of wild-type Nrf2 significantly increased its interactions with both endogenous Cav-1 and Keap1, and mutation of CBM resembling structure on Nrf2 effectively prevented its interaction with Cav-1 but further enhanced its association with Keap1 (Fig. 6A). These findings suggest that both Cav-1 and Keap1 function as important regulators that spontaneously interact with Nrf2 and try to suppress its activity. Furthermore, these data also indicate that the interaction between endogenous Keap1 and exogenous Nrf2 is no more mediated by endogenous Cav-1, although endogenous Keap1 also can effectively react with both exogenous Cav-1 and Nrf2 as demonstrated by the immunofluorescence staining (Fig. 3A).
It might be also of interest to note that half of Cav-1 in Beas-2B cells is constitutively localized in the nucleus (Fig. 4A). Cav-1 is often described as a plasma membrane protein; however, it already has been demonstrated that Cav-1 can be localized in the nucleus of ovarian carcinoma cells in association with nuclear matrix and chromatin (29), of endothelial cells in association with receptors internalized through caveolae-mediated endocytosis (30, 31), as well as of human diploid fibroblasts (32). The endocytosis mechanism may clearly explicate the stimulation-induced Cav-1 nucleus localization (30–31) but the constitutive expression of Cav-1 in nucleus is difficult to interpret. This cell type-specific constitutive Cav-1 nucleus localization might be due to some uncharacterized different localization signals, which requires further investigation. Moreover, though our present study has suggested that nucleus-localized Cav-1 is critically involved in regulation of certain gene transcription, further studies also are required to demonstrate the additional physiological functions of Cav-1 in nucleus and the underlying molecular mechanisms.
Cav-1 has been shown to play important roles in a number of human diseases; however, whether the role of Cav-1 in these diseases is due to its regulation of cellular antioxidant capacity through Nrf2 requires further investigations, probably depending on how importantly oxidative stress plays a role in the pathogenesis of the diseases. Apparently, Cav-1 knock-out mice being more resistant to hyperoxia-induced acute lung injury (18, 19, 22, 34) should due at least in part to the enhanced antioxidant capacity. It has also been reported that genetic loss of caveolin-1 confers dramatic protection against atherosclerosis (37), and this function of Cav-1 deficiency might also be linked to the increased antioxidant capacity, as antioxidants are well recognized to exert protective effects in atherosclerosis (1, 38).
In conclusion, the present study represents an initial effort to demonstrate that Cav-1 functions as a spontaneous inhibitor of Nrf2 in both cytosol and nucleus, thereby suppressing its transcription activity and attenuating subsequent cellular antioxidant capacity. These results also suggest that activation of the Nrf2 pathway through its CDM mutation may represent an effective genetic approach against oxidative stress-induced human diseases.
This work was supported in part by Young Investigator Award 30825019 by the National Natural Science Foundation of China (to H. H. S.), Distinguished Young Investigator Award R2110323 by the Natural Science Foundation of Zhejiang Province (to Z. H. C.), and General Projects of the National Natural Science Foundation of China Grants 31170859 (to Z. H. C.) and 81170037 (to W. L.).

This article contains supplemental Figs. S1 and S2.
- GCL
- glutamate cysteine ligase
- GCLC
- GCLC, GLC catalytic subunit
- Cav-1
- caveolin-1
- HO-1
- heme oxygenase-1
- Keap1
- Kelch-like ECH-associated protein 1
- Nrf2
- nuclear erythroid 2 p45-related factor–2
- 4-HNE
- 4-hydroxynonenol
- RLU
- relative luciferase unit
- EGFP
- enhanced GFP
- CBM
- Cav-1-binding motif.
REFERENCES
- 1. Halliwell B., Gutteridge J. M. (eds) (2007) Free Radicals in Biology and Medicine, 4th Ed., Clarendon Press, Oxford, UK [Google Scholar]
- 2. Papas A. M. Editor, (1999) Antioxidant Status, Diet, Nutrition, and Health, CRC Press, Boca Raton, FL [Google Scholar]
- 3. Rahman I., Biswas S. K., Jimenez L. A., Torres M., Forman H. J. (2005) Glutathione, stress responses, and redox signaling in lung inflammation. Antioxid. Redox Signal. 7, 42–59 [DOI] [PubMed] [Google Scholar]
- 4. Lu S. C. (2009) Regulation of glutathione synthesis. Mol. Aspects Med. 30, 42–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Franklin C. C., Backos D. S., Mohar I., White C. C., Forman H. J., Kavanagh T. J. (2009) Structure, function, and post-translational regulation of the catalytic and modifier subunits of glutamate cysteine ligase. Mol. Aspects Med. 30, 86–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morse D., Lin L., Choi A. M., Ryter S. W. (2009) Heme oxygenase-1, a critical arbitrator of cell death pathways in lung injury and disease. Free Radic. Biol. Med. 47, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ryter S. W., Alam J., Choi A. M. (2006) Heme oxygenase-1/carbon monoxide: From basic science to therapeutic applications. Physiol. Rev. 86, 583–650 [DOI] [PubMed] [Google Scholar]
- 8. Chan K., Kan Y. W. (1999) Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl. Acad. Sci. U.S.A. 96, 12731–12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dinkova-Kostova A. T., Holtzclaw W. D., Cole R. N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. (2002) Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. U.S.A. 99, 11908–11913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alam J., Stewart D., Touchard C., Boinapally S., Choi A. M., Cook J. L. (1999) Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274, 26071–26078 [DOI] [PubMed] [Google Scholar]
- 11. Ramos-Gomez M., Kwak M. K., Dolan P. M., Itoh K., Yamamoto M., Talalay P., Kensler T. W. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 98, 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Itoh K., Tong K. I., Yamamoto M. (2004) Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic. Biol. Med. 36, 1208–1213 [DOI] [PubMed] [Google Scholar]
- 13. Chen Z. H., Yoshida Y., Saito Y., Sekine A., Noguchi N., Niki E. (2006) Induction of adaptive response and enhancement of PC12 cell tolerance by 7-hydroxycholesterol and 15-deoxy-delta(12,14)-prostaglandin J2 through up-regulation of cellular glutathione via different mechanisms. J. Biol. Chem. 281, 14440–14445 [DOI] [PubMed] [Google Scholar]
- 14. Chen Z. H., Saito Y., Yoshida Y., Sekine A., Noguchi N., Niki E. (2005) 4-Hydroxynonenal induces adaptive response and enhances PC12 cell tolerance primarily through induction of thioredoxin reductase 1 via activation of Nrf2. J. Biol. Chem. 280, 41921–41927 [DOI] [PubMed] [Google Scholar]
- 15. Cohen A. W., Hnasko R., Schubert W., Lisanti M. P. (2004) Role of caveolae and caveolins in health and disease. Physiol. Rev. 84, 1341–1379 [DOI] [PubMed] [Google Scholar]
- 16. Chidlow J. H., Jr., Sessa W. C. (2010) Caveolae, caveolins, and cavins: Complex control of cellular signaling and inflammation. Cardiovasc. Res. 86, 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang X. M., Kim H. P., Nakahira K., Ryter S. W., Choi A. M. (2009) The heme oxygenase-1/carbon monoxide pathway suppresses TLR4 signaling by regulating the interaction of TLR4 with caveolin-1. J. Immunol. 182, 3809–3818 [DOI] [PubMed] [Google Scholar]
- 18. Zhang M., Lee S. J., An C., Xu J. F., Joshi B., Nabi I. R., Choi A. M., Jin Y. (2011) Caveolin-1 mediates Fas-BID signaling in hyperoxia-induced apoptosis. Free Radic. Biol. Med. 50, 1252–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang M., Lin L., Lee S. J., Mo L., Cao J., Ifedigbo E., Jin Y. (2009) Deletion of caveolin-1 protects hyperoxia-induced apoptosis via survivin-mediated pathways. Am. J. Physiol. Lung Cell Mol. Physiol. 297, L945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Z. H., Lam H. C., Jin Y., Kim H. P., Cao J., Lee S. J., Ifedigbo E., Parameswaran H., Ryter S. W., Choi A. M. (2010) Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc. Natl. Acad. Sci. U.S.A. 107, 18880–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ryter S. W., Lam H. C., Chen Z. H., Choi A. M. (2011) Deadly triplex: Smoke, autophagy and apoptosis. Autophagy 7, 436–437 [DOI] [PubMed] [Google Scholar]
- 22. Jin Y., Kim H. P., Chi M., Ifedigbo E., Ryter S. W., Choi A. M. (2008) Deletion of caveolin-1 protects against oxidative lung injury via up-regulation of heme oxygenase-1. Am. J. Respir. Cell Mol. Biol. 39, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Volonte D., Galbiati F. (2009) Inhibition of thioredoxin reductase 1 by caveolin 1 promotes stress-induced premature senescence. EMBO Rep. 10, 1334–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X. M., Zhang Y., Kim H. P., Zhou Z., Feghali-Bostwick C. A., Liu F., Ifedigbo E., Xu X., Oury T. D., Kaminski N., Choi A. M. (2006) Caveolin-1: A critical regulator of lung fibrosis in idiopathic pulmonary fibrosis. J. Exp. Med. 203, 2895–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim H. P., Chen Z. H., Choi A. M., Ryter S. W. (2009) Analyzing autophagy in clinical tissues of lung and vascular diseases. Methods Enzymol. 453, 197–216 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y., Sano M., Shinmura K., Tamaki K., Katsumata Y., Matsuhashi T., Morizane S., Ito H., Hishiki T., Endo J., Zhou H., Yuasa S., Kaneda R., Suematsu M., Fukuda K. (2010) 4-hydroxy-2-nonenal protects against cardiac ischemia-reperfusion injury via the Nrf2-dependent pathway. J. Mol. Cell Cardiol. 49, 576–586 [DOI] [PubMed] [Google Scholar]
- 27. Chen Z. H., Yoshida Y., Saito Y., Noguchi N., Niki E. (2006) Adaptive response induced by lipid peroxidation products in cell cultures. FEBS Lett. 580, 479–483 [DOI] [PubMed] [Google Scholar]
- 28. Chen Z. H., Niki E. (2006) 4-Hydroxynonenal (4-HNE) has been widely accepted as an inducer of oxidative stress. Is this the whole truth about it or can 4-HNE also exert protective effects? IUBMB Life 58, 372–373 [DOI] [PubMed] [Google Scholar]
- 29. Sanna E., Miotti S., Mazzi M., De Santis G., Canevari S., Tomassetti A. (2007) Binding of nuclear caveolin-1 to promoter elements of growth-associated genes in ovarian carcinoma cells. Exp. Cell Res. 313, 1307–1317 [DOI] [PubMed] [Google Scholar]
- 30. Gobeil F., Jr., Bernier S. G., Vazquez-Tello A., Brault S., Beauchamp M. H., Quiniou C., Marrache A. M., Checchin D., Sennlaub F., Hou X., Nader M., Bkaily G., Ribeiro-da-Silva A., Goetzl E. J., Chemtob S. (2003) Modulation of pro-inflammatory gene expression by nuclear lysophosphatidic acid receptor type-1. J. Biol. Chem. 278, 38875–38883 [DOI] [PubMed] [Google Scholar]
- 31. Feng Y., Venema V. J., Venema R. C., Tsai N., Caldwell R. B. (1999) VEGF induces nuclear translocation of Flk-1/KDR, endothelial nitric oxide synthase, and caveolin-1 in vascular endothelial cells. Biochem. Biophys. Res. Commun. 256, 192–197 [DOI] [PubMed] [Google Scholar]
- 32. Chrétien A., Piront N., Delaive E., Demazy C., Ninane N., Toussaint O. (2008) Increased abundance of cytoplasmic and nuclear caveolin 1 in human diploid fibroblasts in H2O2-induced premature senescence and interplay with p38α(MAPK). FEBS Lett. 582, 1685–1692 [DOI] [PubMed] [Google Scholar]
- 33. Scherer P. E., Tang Z., Chun M., Sargiacomo M., Lodish H. F., Lisanti M. P. (1995) Caveolin isoforms differ in their N-terminal protein sequence and subcellular distribution. Identification and epitope mapping of an isoform-specific monoclonal antibody probe. J. Biol. Chem. 270, 16395–16401 [DOI] [PubMed] [Google Scholar]
- 34. Jin Y., Kim H. P., Cao J., Zhang M., Ifedigbo E., Choi A. M. (2009) Caveolin-1 regulates the secretion and cytoprotection of Cyr61 in hyperoxic cell death. FASEB J. 23, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ghosh S., Gachhui R., Crooks C., Wu C., Lisanti M. P., Stuehr D. J. (1998) Interaction between caveolin-1 and the reductase domain of endothelial nitric-oxide synthase. Consequences for catalysis. J. Biol. Chem. 273, 22267–22271 [DOI] [PubMed] [Google Scholar]
- 36. Santibanez J. F., Blanco F. J., Garrido-Martin E. M., Sanz-Rodriguez F., del Pozo M. A., Bernabeu C. (2008) Caveolin-1 interacts and cooperates with the transforming growth factor-β type I receptor ALK1 in endothelial caveolae. Cardiovasc. Res. 77, 791–799 [DOI] [PubMed] [Google Scholar]
- 37. Frank P. G., Lee H., Park D. S., Tandon N. N., Scherer P. E., Lisanti M. P. (2004) Genetic ablation of caveolin-1 confers protection against atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 24, 98–105 [DOI] [PubMed] [Google Scholar]
- 38. Niki E. (2004) Antioxidants and atherosclerosis. Biochem. Soc. Trans. 32, 156–159 [DOI] [PubMed] [Google Scholar]