Background: The carbon catabolite repression 4 (Ccr4) family is implicated in RNA processing and modification.
Results: Ccr4d inhibits cell proliferation and induces G1 arrest by binding to p21 mRNA 3′-UTR and stabilizing p21 mRNA.
Conclusion: Ccr4d functions as an anti-proliferating protein through induction of cell cycle arrest via a p21-dependent and p53-independent pathway.
Significance: Ccr4d might have significant pathophysiological functions in carcinogenesis.
Keywords: Cell Biology, Cell Cycle, Cell Proliferation, Molecular Cell Biology, RNA-binding Protein, Ccr4d, mRNA Stability, p21
Abstract
Ccr4d is a new member of the Ccr4 (carbon catabolite repression 4) family of proteins that are implicated in the regulation of mRNA stability and translation through mRNA deadenylation. However, Ccr4d is not believed to be involved in mRNA deadenylation. Thus, its biological function and mechanistic activity remain to be determined. Here, we report that Ccr4d is broadly expressed in various normal tissues, and the expression of Ccr4d is markedly down-regulated during cell cycle progression. We showed that Ccr4d inhibits cell proliferation and induces cell cycle arrest at G1 phase. Our experiments further revealed that Ccr4d regulates the expression of p21 in a p53-independent manner. Mechanistic studies indicated that Ccr4d strongly bound to the 3′-UTR of p21 mRNA, leading to the stabilization of p21 mRNA. Interestingly, we found that the expression of Ccr4d is down-regulated in various tumor tissues. Collectively, our data indicate that Ccr4d functions as an anti-proliferating protein through the induction of cell cycle arrest via a p21-dependent and p53-independent pathway and suggest that Ccr4d might have an important role in carcinogenesis.
Introduction
The G1 phase of the cell cycle is a crucial integrator of internal and external cues, allowing cells to grow, process outside information, or repair damages before entering S phase (1). G1 phase progression is controlled by a set of cyclin-dependent kinases (CDKs),3 which are activated by associated cyclins but inhibited by two classes of CDK inhibitors. One group is the INK4 family, including p16INK4a, p15INK4b, p18INK4c, and p19INK4d, which specifically target CDK4 and CDK6 (2, 3). The other group is the CIP/KIP family, including p21Waf1/Cip1, p27Kip1, and p57Kip2, which inhibit a broad spectrum of CDKs (2, 3). p21Waf1/Cip1 (hereafter named p21) is a major mediator of the p53 signaling in controlling cell cycle transition from G1 to S and G2 to M (4). In addition to the transcriptional regulation by the p53 family, the level of p21 is found to be controlled by post-transcriptional mechanisms (5). Particularly, growing evidence suggests that p21 mRNA stability is regulated by a family of RNA-binding proteins that contain an RNA recognition motif and have an affinity to the 3′-untranslated region (3′-UTR) (6, 7). For example, HuD is the first RNA-binding protein that was found to regulate p21 mRNA stability (8). HuD belongs to the Elav (embryonic lethal, abnormal vision)-like protein family consisting of HuB, HuC, HuD, and HuR (9–11). All of them carry several RNA recognition motifs and are known to bind the AU-rich element-containing transcripts, including the p21 transcript (8). Apart from the Elav/Hu family of proteins, RNPC1 (also called RBM38), a RNA-binding protein with one RNA recognition motif domain, is shown to be required for the maintenance of the stability of the basal and stress-induced p21 transcript (12, 13). Interestingly, hnRNP K binds to the CU-rich elements in p21 3′-UTR and represses p21 translation (14), whereas AUF1 (hnRNP D) binds to p21 3′-UTR and decreases p21 mRNA stability (15). RBM42, a binding partner of hnRNP K, coordinates with hnRNP K in the maintenance of cellular ATP level under stress conditions by blocking p21 mRNA degradation (16). Moreover, NF90, a component of the spliceosome that contains double-stranded RNA binding motifs, is predominantly localized in the nucleus. Similar to HuR, NF90 is translocated from the nucleus to the cytoplasm and stabilizes p21 mRNA via binding to p21 3′-UTR (17).
Accurate gene expression requires a precise control of mRNA levels, which are determined by the relative rate of nuclear pre-mRNA synthesis and processing and cytoplasmic mRNA turnover. Deadenylation, the process of removing the poly(A) tail, is a rate-limiting step in mRNA turnover (18–20), which involves several deadenylases, including components of the Ccr4-Not complex. The Ccr4-Not complex controls mRNA metabolism through multiple mechanisms, including repression and activation of mRNA initiation, control of mRNA elongation as well as deadenylation, and subsequent degradation of mRNA (21–23). In yeast, the Ccr4-Not complex consists of nine core subunits: Ccr4 (carbon catabolite repression 4), Caf1 (CCR4-associated factor 1), Caf40, Caf130, and Not1 to -5. The deadenylation is catalyzed by the Ccr4 subunit in yeast but by the orthologs of both Ccr4 and Caf1 in other eukaryotes (19). These subunits can be divided into two subgroups: 1) proteins that contain DNase I-like domains of the exonuclease-endonuclease-phosphatase (EEP) superfamily, such as CNOT6 and CNOT6L, and 2) proteins that contain RNase D-like domains belonging to the DEDD family, such as CNOT7 and CNOT8 (19). The homologues Caf1a (CNOT7) and Caf1b (CNOT8) of Caf1, identified as components of the human Ccr4-Not complex, possess deadenylase activity mediated by DEDD nuclease domains and have overlapping roles in modulating cell proliferation (24), whereas a more distant homologue, hCaf1z (also named TOE1), forms a separate nuclear complex involved in mRNA metabolism (25).
Five Ccr4 homologues have been identified in human cells, including Ccr4a (CNOT6), Ccr4b (CNOT6L), Ccr4c (nocturnin, Ccn4L), Ccr4d (ANGEL2), and Ccr4e (ANGEL1). Of them, Ccr4a and Ccr4b share 74% amino acid identity with each other and contain leucine-rich repeats required for interactions with Caf1a/Caf1b (26). Both play a role in deadenylation and contribute to the prevention of cell death and senescence (27). Ccr4c is the ortholog of the deadenylase nocturnin, which has been characterized in mice and Xenopus laevis and is proposed to play a role in circadian function (28).
Ccr4d (ANGEL2) is a member of the EEP family based on sequence homology. It is more divergent from yeast Ccr4p and contains no leucine-rich repeat or leucine zipper. It is reported that Ccr4d, as a nucleocytoplasmic shuttling protein, mainly resides in the nucleus. Although exogenously expressed hCaf1z was shown to stimulate deadenylation and decay of reporter mRNAs, and Ccr4d interacts with hCaf1z, no significant effects on deadenylation and mRNA turnover of reporter mRNAs were observed upon exogenously expressed Ccr4d (25). Therefore, the cellular function and biological significance of Ccr4d remain to be determined.
In the current report, we found that Ccr4d is broadly expressed in various normal tissues, and its expression markedly decreased in cell cycle progression. Ectopic expression of Ccr4d resulted in the inhibition of cell proliferation and cell cycle arrest in G1 phase, and depletion of Ccr4d promoted cell growth and cell cycle progression. We further showed that depletion of Ccr4d led to a significant inhibition of p21 expression in a p53-independent manner. We demonstrated that Ccr4d bound to the 3′-UTR of p21 mRNA and showed that ectopic expression of Ccr4d resulted in an increase whereas Ccr4d knockdown led to a decrease in the activity of p21 3′-UTR luciferase reporter. Finally, we found that Ccr4d is down-regulated in a broad spectrum of tumor samples.
EXPERIMENTAL PROCEDURES
Bioinformatics
The open reading frame, conserved domains, and chromosomal location of Ccr4d were analyzed using the NCBI databases (available on the World Wide Web). RNA binding sites were predicted by the BindN program (29). The theoretical molecular weight and isoelectric point of Ccr4d were calculated using the Expacy Web site. Homologous alignment was analyzed by the ClustalW program (version 1.60) (30), and phylogenetic analysis was performed using the Jotun Hein method (31).
Northern Blotting
Human adult multiple tissue Northern blots (Clontech) or cancer profiling array (Clontech) were used, and a 500-bp probe of human ccr4d cDNA was labeled by random priming with [α-32P]dCTP (Amersham Biosciences). Hybridization was performed in ExpressHyb hybridization solution (Clontech) according to the manufacturer's instructions. The filter was exposed for autoradiography at −80 °C after stringent washing.
RNA Interference
siRNAs were synthesized by Shanghai Genepharma Co., Ltd. The sequences were as follows: non-silencing siRNA, UUCUCCGAACGUGUCACGU; ccr4d siRNA-1, GGACCCUGAAUACUCUGAUTT; ccr4d siRNA-2, AAACCUGAUGGCUGUGCUAUUTT; ccr4a siRNA, GGACCCUGAAUACUCUGAUTT; ccr4b siRNA, GGCCAUGGAUUACAUUAAATT; ccr4c siRNA, GGAGCCCAUUGAUCCUAAATT. siRNAs were transfected into cells using LipofectamineTM RNAiMAX (Invitrogen) according to the manufacturer's instructions.
Western Blotting
Western blotting was performed as described previously (32, 33). Antibodies used in this experiment were as follows: αβ-actin, α-tubulin, αCyclin D1, and αFLAG from Sigma-Aldrich; αE2F1, αCdk1, αCdk2, αCdk4, αCdc25a, αCyclin A, αCyclin B1, αCyclin E, and αp16 from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); αp53 (DO-1), αp21, and αp27 from MBL; and αPUMA from Cell Signaling Technology. Polyclonal antibody against Ccr4d was raised using synthetic peptide in rabbits.
Colony Formation Assay
MCF-7 cells transfected with control vector or FLAG-Ccr4d construct were maintained in culture medium supplemented with 1 mg/ml G418 for 14 days. The cells were stained with crystal violet for colony counting.
Cell Proliferation Assays
For the MTT assay, MCF-7 cells were treated with control siRNA or Ccr4d siRNA and seeded into 96-well plates. On the day of harvest, 5 mg/ml MTT was added into every well, and plates were incubated at 37 °C for 4 h. Cell viability was determined by measuring the absorbance of converted dye at a wavelength of 490 nm.
For analysis of BrdU incorporation, cells were transfected with FLAG-Ccr4d or Ccr4d siRNA for 48 or 72 h. 10 μm BrdU was incorporated for 1 h before cell fixation. DNA was denatured with 2 n HCl, 0.5% Triton X-100 for 20 min, and BrdU signal was detected with anti-BrdU antibody and Alexa Fluoro 488 goat anti-mouse IgG. Cells (n = 10,000) were collected using FACSCalibur (BD Biosciences) and analyzed with ModFit LT 3.0 (Verity Software House Inc., Topsham, ME).
Cell Growth Assay
U2OS cells were seeded into 24-well plates and treated with control siRNA or Ccr4d siRNA. Cultures in the well plates were measured at different time points by the CloneSelectTM Imager system (Genetix Ltd., New Milton, UK) and returned to the incubator.
Cell Synchronization and Flow Cytometry
MCF-7 cells were synchronized at the G0/G1 phase by serum starvation. Briefly, after cells were transfected with FLAG-Ccr4d constructs or siRNA for 24 h, cells were switched to conditioned medium without serum for 48 h. The synchronized cells were then cultured in medium containing 10% FBS for 24–27 h and were collected for cell cycle profile analysis by flow cytometry. Flow cytometry was performed as previously described (34). Briefly, cells were trypsinized and fixed in 70% ethanol at 4 °C overnight. After being washed with phosphate-buffered saline (PBS), cells were incubated with RNase A in PBS for 30 min at 37 °C and then stained with 50 mg/ml propidium iodide. Cell cycle data were collected using FACSCalibur (BD Biosciences) and analyzed with ModFit LT 3.0 (Verity Software House Inc.).
Real-time Reverse Transcription (RT)-PCR
Power SYBR Green PCR Master Mix and the ABI PRISM 7300 sequence detection system (Applied Biosystems, Foster City, CA) were used for real-time RT-PCR experiments. The primer pairs used were as follows: ccr4d forward primer (GCCAGACATCTTTGATTCATCTC) and reverse primer (TTCCCAATTCCGCTTTATCA); p21 forward primer (TGAGCCGCGACTGTGATG) and reverse primer (GTCTCGGTGACAAAGTCGAAGTT); GAPDH forward primer (GAAGGTGAAGGTCGGAGTC) and reverse primer (GAAGATGGTGATGGGATTC); ccr4a forward primer (CTTTGTGATAAATATGCGACCC) and reverse primer (CGTTTCAACCTCCTGAAGAC); ccr4b forward primer (CAATCTCGCAGTTCATCCAG) and reverse primer (ACATAACCGTGAATGATGCT); ccr4c forward primer (GAGGACGGATTGCCCTAGTA) and reverse primer (GTAGGCCAGGATTTCTTCCA).
Luciferase Reporter Assay
Luciferase activity was measured using a dual luciferase kit (Promega, Madison, WI) according to the manufacturer's protocol. Reporters used in this assay were the full-length p21 3′-UTR-Luc construct from Dr. Bryan R. Cullen (Duke University) and the pWWP-Luc (p21-Luc) construct from Dr. Bert Vogelstein (Johns Hopkins University).
RNA-binding Pull-down Assays
cDNA was used as a template for PCR amplification of different p21 UTRs and coding regions (CR). All forward primers contained the T7 promoter sequence (T7), CAGAGATGCATAATACGACTCACTATAGGGAGA. To prepare 5′-UTR and CR-A transcript (positions 1–570), forward primer (T7)GCCGAAGTCAGTTCCTTGT and reverse primer TTAGGGCTTCCTCTTGGAG were used. To prepare the 3′-UTR-B transcript (positions 571–1118), forward primer (T7)TCCGCCCACAGGAAGC and reverse primer ACCCTGCCCAACCTTAGA were used. To prepare 3′-UTR-C transcript (positions 1119–2109), forward primer (T7)GACCCTGAAGTGAGCACAG and TTTTTTTAAAGTCACTAAGAATCATT were used. For the biotin pull-down assay, PCR-amplified DNA was used as a template to transcribe biotinylated RNA by using T7 RNA polymerase in the presence of biotin-UTP and was purified as described previously (35). 2 μg of biotinylated transcripts were incubated with 60 μg of total cell lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway), and pull-down material was analyzed by Western blotting.
RNA-binding Protein Immunoprecipitation (RIP) Assay
RIP assay was performed according to the Magna RIPTM manufacturer's protocol (Millipore). Briefly, H1299 cells were transfected with FLAG-Ccr4d for 48 h, and total cell lysates were used for immunoprecipitation in the presence of excess anti-FLAG antibody or mouse normal IgG. The precipitated Ccr4d-RNA complexes were treated with RNase-free DNase to remove trapped genomic DNAs. The precipitated RNAs were purified by TRIzol reagent for cDNA synthesis, followed by RT-PCR amplification using sequence-specific primers. RNA in immunoprecipitated material was used in RT-PCRs to detect p21 mRNA by using different primers. The primer pairs used were 5′-UTR forward primer (GCCGAAGTCAGTTCCTTGTG) and reverse primer (GGCGCCTCCTCTGAGTGC), 3′-UTR-1 forward primer (TGTACCCTTGTGCCTCGC) and reverse primer (GAGAAGATCAGCCGGCGT), and 3′-UTR-2 forward primer (CAAAGGCCCGCTCTACATC) and reverse primer (GCCCAGCACTCTTAGGAAC).
RESULTS
Cloning and Characterization of Ccr4d
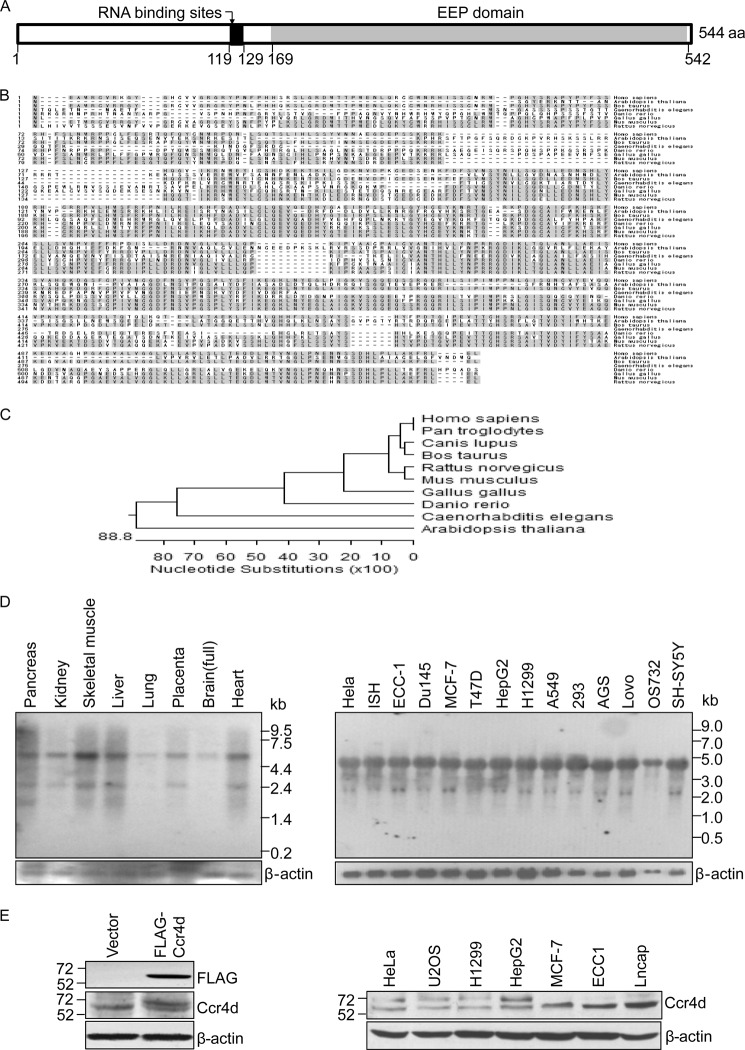
The molecular mechanism of tumorigenesis has been one of the primary research focuses in our laboratory (36–40). In an effort to identify more cell cycle regulators, we cloned a gene, ccr4d in GenBankTM (ID NM_144567), from a mammary cDNA library (Clontech) through functional screening. The cDNA of ccr4d is 4,668 bp in length and contains an open reading frame encoding for a protein of 544 amino acids. Bioinformatics analysis indicates that Ccr4d harbors an EEP domain (amino acids 169–542), thus belonging to the EEP family (Fig. 1A, EEP). This family of proteins includes magnesium-dependent endonucleases and a large number of phosphatases involved in intracellular signaling. BindN analysis indicates that Ccr4d harbors putative RNA binding sites (amino acids 119–129) (29). Amino acid sequence alignment revealed that human Ccr4d shares 100.0% identity with its Pan troglodytes homologue. The similarity of the amino acid sequence of Ccr4d to homologues in other organisms is 93.8% in Canis lupus, 91.7% in Bos taurus, 87.2% in Rattus norvegicus, 85.5% in Mus musculus, 69.2% in Gallus gallus, 58.0% in Danio rerio, 39.2% in Caenorhabditis elegans, and 35.7% in Arabidopsis thaliana (Fig. 1B). Phylogenetic analysis also indicates that Ccr4d is evolutionarily well conserved (Fig. 1C).
FIGURE 1.
Cloning, characterization, and expression of Ccr4d. A, a schematic representation of the structure of Ccr4d. A conserved domain (EEP domain) and putative RNA binding sites are shown. aa, amino acids. B, amino acid sequence alignment of Ccr4d from different species. Shaded residues represent conserved regions. C, phylogenetic analysis of evolutionary relationships among homologues of Ccr4d proteins from different species. D, Northern blotting analysis of ccr4d mRNA expression in normal human tissues and in various tumor cell lines. β-actin was used as a loading control. E, Western blotting analysis of Ccr4d protein expression. Left, MCF-7 cells were transfected with empty vector or FLAG-Ccr4d constructs. Cellular proteins were prepared, and Western blotting was performed with anti-FLAG or anti-Ccr4d antibody. Right, Ccr4d expression in a collection of tumor cell lines was analyzed using anti-Ccr4d antibody. β-actin was used as a loading control.
Northern blotting with human multiple tissue blots and mRNA extracted from different cell lines demonstrated that the ccr4d gene primarily transcribes a ∼4.6-kb message in various tissues (Fig. 1D, left) and cell lines (Fig. 1D, right), although an additional transcript was detected in some tissues, such as pancreas and skeletal muscle (Fig. 1D, left). We focused on the ∼4.6-kb transcript because it is the transcript that we initially cloned, and it is the transcript that exhibits a broad tissue distribution and expresses at a high level.
Western blotting analysis in MCF-7 breast carcinoma cells that were transfected with FLAG-tagged Ccr4d expression construct (FLAG-Ccr4d) using a monoclonal antibody against FLAG and in a collection of cell lines using polyclonal antibodies against Ccr4d, which we generated with synthetic Ccr4d peptide, indicated that Ccr4d is expressed as a protein of ∼65 kDa (Fig. 1E, left) and that endogenous Ccr4d is broadly expressed in a collection of cell lines (Fig. 1E, right).
Ccr4d Inhibits Cell Proliferation
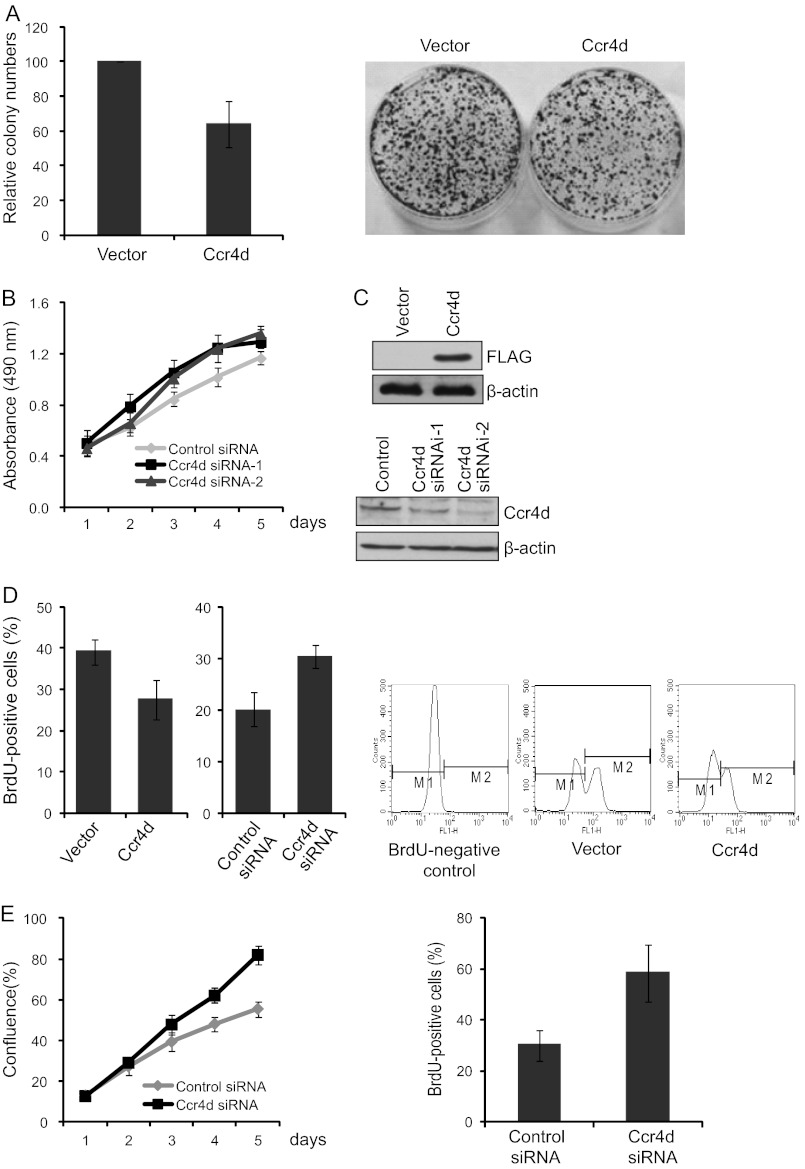
To gain insights into the biological function of the Ccr4d protein, we first examined the effect of Ccr4d on the proliferation of MCF-7 breast carcinoma cells. To this end, we created MCF-7 cell clones that stably express FLAG-Ccr4d. These clones were expanded, and their growth was examined by colony formation assays. It was demonstrated that ectopic expression of Ccr4d was associated with a significant inhibition of colony formation (∼35% decrease, p < 0.05; Fig. 2A, left). A presented profile is shown in Fig. 2A (right). On the other hand, knockdown of Ccr4d resulted in an increase in cell proliferation as measured by MTT assays (Fig. 2B). The overexpression and knockdown of Ccr4d were confirmed by Western blotting (Fig. 2C).
FIGURE 2.
Inhibition of cell proliferation induced by Ccr4d. A, colony formation assays. Left, MCF-7 cells stably expressing Ccr4d were maintained in culture medium for 14 days in the presence of 1 mg/ml G418 and stained with crystal violet. The number of colonies in each condition was counted and expressed as mean ± S.D. from triplicate experiments. A presented profile is shown on the right. B, induction of cell proliferation upon Ccr4d knockdown. MCF-7 cells were transfected with control siRNA or Ccr4d siRNA. The growth curve of the cells was measured by MTT assays. Bars, mean ± S.D. (error bars) for triplicate experiments. C, confirmation of Ccr4d overexpression and knockdown used in colony formation assays and MTT assays by Western blotting. β-actin was used as a loading control. D, BrdU incorporation assays. Two left panels, MCF-7 cells were transfected with FLAG-Ccr4d or Ccr4d siRNA for 48 or 72 h, and then BrdU was incorporated for 1 h. Cells (n = 10,000) were collected, and the percentage of BrdU-positive cells was measured by flow cytometry. Bars, mean ± S.D. (error bars) for triplicate experiments. A representative profile is shown in the three right panels. The cells without BrdU incorporation were used as a negative control for flow cytometry analysis. E, induction of cell proliferation of U2OS cells upon Ccr4d knockdown. Left, U2OS cells were transfected with control siRNA or Ccr4d siRNA. The percentage of cell confluence was measured every 24 h by the CloneSelectTM Imager system. Bars, mean ± S.D. (error bars) for triplicate experiments. Right, BrdU incorporation assays. U2OS cells were transfected with control siRNA or Ccr4d siRNA for 72 h, and then BrdU was incorporated for 1 h. Cells (n = 10,000) were collected, and the percentage of BrdU-positive cells was measured by flow cytometry. Bars, mean ± S.D. (error bars) for triplicate experiments.
To further determine the function of Ccr4d in cell proliferation, we employed a BrdU incorporation assay, a more sensitive and specific method. It was shown that the number of BrdU-positive cells was decreased by about 12% (p < 0.05) in MCF-7 cells upon ectopic expression of Ccr4d, and conversely, the number of BrdU-positive cells was increased by about 1.5-fold (p < 0.05) in Ccr4d-deficient MCF-7 cells (Fig. 2D).
Next, human osteosarcoma U2OS cells were used to test the inhibitory effect of Ccr4d on cell proliferation. We found that knockdown of Ccr4d resulted in an increase in cell proliferation of U2OS cells (Fig. 2E, left), as measured by the CloneSelectTM Imager system, an image-based visualization system for cell growth assessment. Further, it was shown that the number of BrdU-positive cells was increased by about 2.3-fold (p < 0.05) in Ccr4d-deficient U2OS cells (Fig. 2E, right) by a BrdU incorporation assay. Collectively, these results support the notion that Ccr4d has an inhibitory activity on cell proliferation.
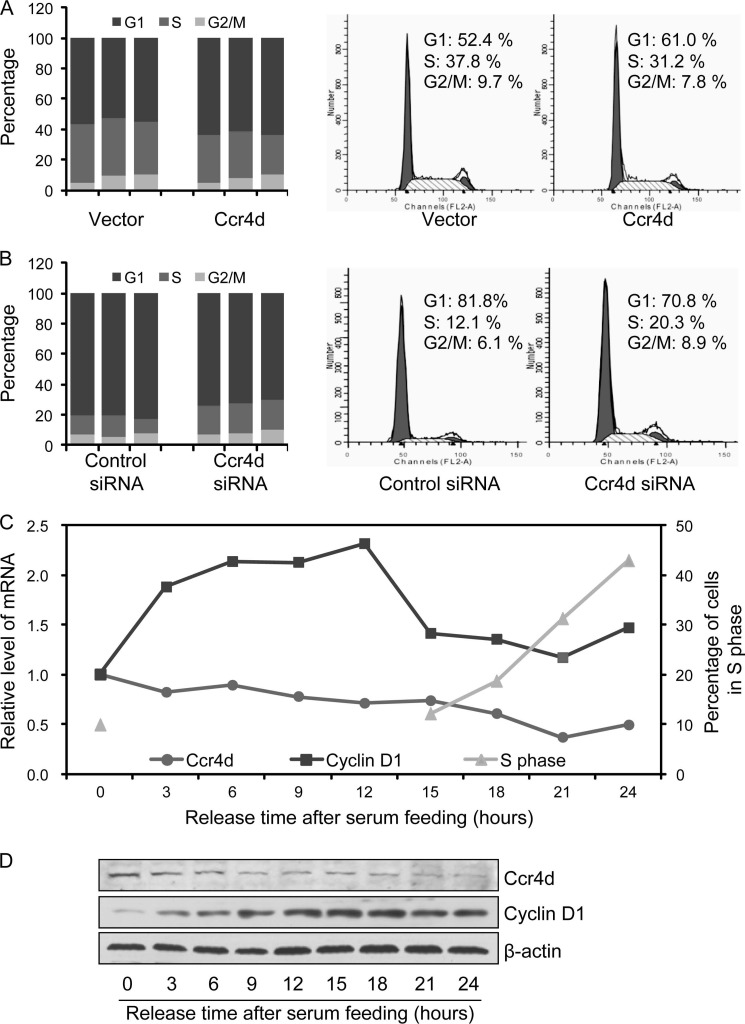
To determine whether the growth-inhibitory effect of Ccr4d represented a general inhibition of growth or a cell cycle phase-specific inhibition, synchronized MCF-7 cells that were transfected with FLAG-Ccr4d or with Ccr4d siRNA were released by refeeding serum. Cell cycle profiling was analyzed using propidium iodide staining and flow cytometry. We found a significant decrease (from 37.1 ± 2.1 to 29.7 ± 2.8%) in the population of cells in the S phase upon overexpression of Ccr4d (Fig. 3A), with a concomitant accumulation (from 54.7 ± 2.1 to 62.8 ± 1.6%) of the cell population in G1 phase; the cell population in G2/M phases was essentially unaffected (Fig. 3A). On the other hand, cell population increased in S phase from 12.0 ± 2.2 to 19.5 ± 1.4% and decreased in G1 phase from 81.0 ± 1.1 to 72.2 ± 2.0% upon Ccr4d knockdown (Fig. 3B). Therefore, it appeared that Ccr4d inhibits the proliferation of MCF-7 cells by blocking the G1-to-S transition of the cell cycle.
FIGURE 3.
Ccr4d induces cell cycle arrest in G1 phase. A, inhibition of cell cycle progression by Ccr4d. After the cells were transfected with vector or FLAG-Ccr4d construct for 24 h, MCF-7 cells were switched to conditioned medium without serum for 24 h. The cells were then cultured in medium containing 10% FBS for 24–27 h and were collected for cell cycle analysis by flow cytometry. The data represent three independent experiments in the left panel. A representative profile is shown in the right panel. B, Ccr4d knockdown promotes cell cycle progression. After MCF-7 cells were transfected with control siRNA or Ccr4d siRNA for 24 h, the cells were switched to conditioned medium without serum for 48 h. The cells were then cultured in medium containing 10% FBS for 24–27 h and were collected for cell cycle analysis by flow cytometry. The data represent three independent experiments in the left panel. A representative profile is shown in the right panel. C, the expression of ccr4d mRNA changed with cell cycle progression. MCF-7 cells were grown in normal medium to 60% confluence and switched to serum-free medium for 48 h followed by growth in medium replenished with serum for different times. Cells were then collected for real-time RT-PCR analysis. A representative profile is shown. D, the expression of Ccr4d protein changed with cell cycle progression. MCF-7 cells were grown in normal medium to a 60% confluence and switched to serum-free medium for 48 h, followed by growth in medium replenished with serum for different times. Cellular lysates were prepared for Western blotting analysis with antibodies against the indicated proteins. β-actin was used as a loading control.
In order to further support this argument, we next measured the expression of Ccr4d during cell cycle progression. For this purpose, MCF-7 cells were first cultured in serum-free medium for 48 h and were then fed with fresh medium containing 10% serum. Collection of cells at different time points and measurement of the expression of ccr4d mRNA by real-time RT-PCR showed that the expression of endogenous Ccr4d gradually declined with the cell cycle progression, reaching the lowest point after serum feeding for 21 h (Fig. 3C). The expression of cyclin D1 was also measured as a control for cell cycle progression. The cell cycle progression was also validated at the corresponding time by propidium iodide staining and flow cytometry (Fig. 3C). Further, collection of cells at different time points and measurement of the expression of Ccr4d protein by Western blotting showed that the protein expression of endogenous Ccr4d also gradually declined with the cell cycle progression (Fig. 3D). The expression of cyclin D1 was measured as a control for cell cycle progression. The expression profiling of Ccr4d during cell cycle progression supports the argument that Ccr4d exerts its growth-inhibitory effect by arresting cells in G1 phase.
Ccr4d Inhibits Cell Cycle through Regulation of Cell Cycle Inhibitor p21 in p53-independent Manner
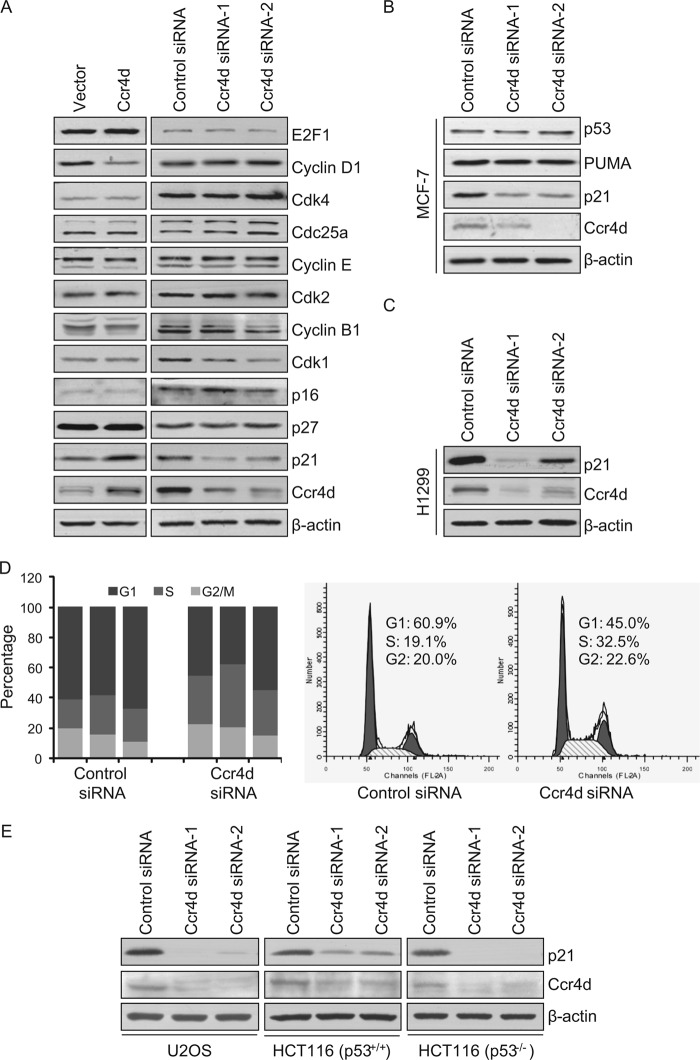
As stated earlier, although a unified mechanistic model is still lacking, the Ccr4 proteins are believed to be involved in proper processing of target mRNAs. To explore the molecular mechanism underlying Ccr4d-induced G1 arrest of MCF-7 cells and to identify the potential mRNA targets for Ccr4d, the expression of a collection of proteins, including E2F1, cyclin D1, Cdk4, Cdc25a, cyclin E, Cdk2, cyclin B1, Cdk1, p16, p21, and p27, whose functions are closely related to cell cycle control was first examined by Western blotting upon overexpression and knockdown of Ccr4d. Our results indicated that the expression of none of these proteins was significantly affected by overexpression or knockdown of Ccr4d, except for p21, whose protein level was significantly up-regulated upon overexpression of Ccr4d and down-regulated upon Ccr4d knockdown (Fig. 4A).
FIGURE 4.
Identification of p21 as a target for Ccr4d. A, the effect of Ccr4d on the expression of cell cycle-related proteins. MCF-7 cells were transfected with FLAG-Ccr4d construct or Ccr4d siRNA, and cellular lysates were prepared for Western blotting analysis with antibodies against the indicated proteins. B, expression of p53 and PUMA upon Ccr4d knockdown in MCF-7 cells. Cells were transfected with control siRNA and Ccr4d siRNA for 72 h, and cellular lysates were prepared for Western blotting analysis with antibodies against the indicated proteins. β-actin was used as a loading control. C, the expression of p21 upon Ccr4d knockdown in H1299 cells. Cells were transfected with control siRNA and Ccr4d siRNA for 72 h, and cellular lysates were prepared for Western blotting analysis with antibodies against the indicated proteins. β-actin was used as a loading control. D, Ccr4d knockdown promotes H1299 cell cycle progression. After cells were transfected with control siRNA or Ccr4d siRNA for 24 h, the cells were switched to conditioned medium without serum for another 24 h. The cells were then cultured in medium containing 10% FBS for 27 h and were collected for cell cycle analysis by flow cytometry. The data represent three independent experiments in the left panel. A representative profile is shown in the right panel. E, expression of p21 upon knockdown of Ccr4d in p53+/+ and p53−/− isogenic cell lines. Cells were transfected with control siRNA and Ccr4d siRNA for 72 h, and cellular lysates were prepared for Western blotting analysis with antibodies against the indicated proteins. β-actin was used as a loading control.
Because p53 is the major regulator of p21, we tested whether the expression of p53 and its other target proteins was altered under similar conditions. Interestingly, the expression of p53 and PUMA, another downstream target of p53 (41), was not changed upon Ccr4d knockdown (Fig. 4B). Furthermore, in p53-negative H1299 lung cancer cells, knockdown of Ccr4d was also associated with a decreased expression of p21, as measured by Western blotting (Fig. 4C). Moreover, knockdown of Ccr4d in H1299 cells also resulted in a decrease in cell population in G1 phase from 61.9 ± 4.4 to 45.7 ± 8.6% and an increase in cell population in the S phase from 22.3 ± 3.3 to 34.9 ± 6.5%, whereas the cell population in G2/M phases was essentially unaffected (Fig. 4D).
To strengthen the argument that Ccr4d regulates p21 in a p53-independent manner, various p53+/+ and p53−/− isogenic cell lines, including human osteosarcoma U2OS cells (p53 wild type), human colon cancer HCT116 (p53+/+) cells, and HCT116 (p53−/−) cells were used to test the expression of p21 upon Ccr4d knockdown. It was demonstrated that knockdown of Ccr4d was also associated with a deceased expression of p21, as measured by Western blotting (Fig. 4E). These data suggest that the regulation of p21 by Ccr4d is independent of p53.
Ccr4d Up-regulates p21 Expression through Binding to 3′-UTR of p21 mRNA and Stabilizing p21 mRNA
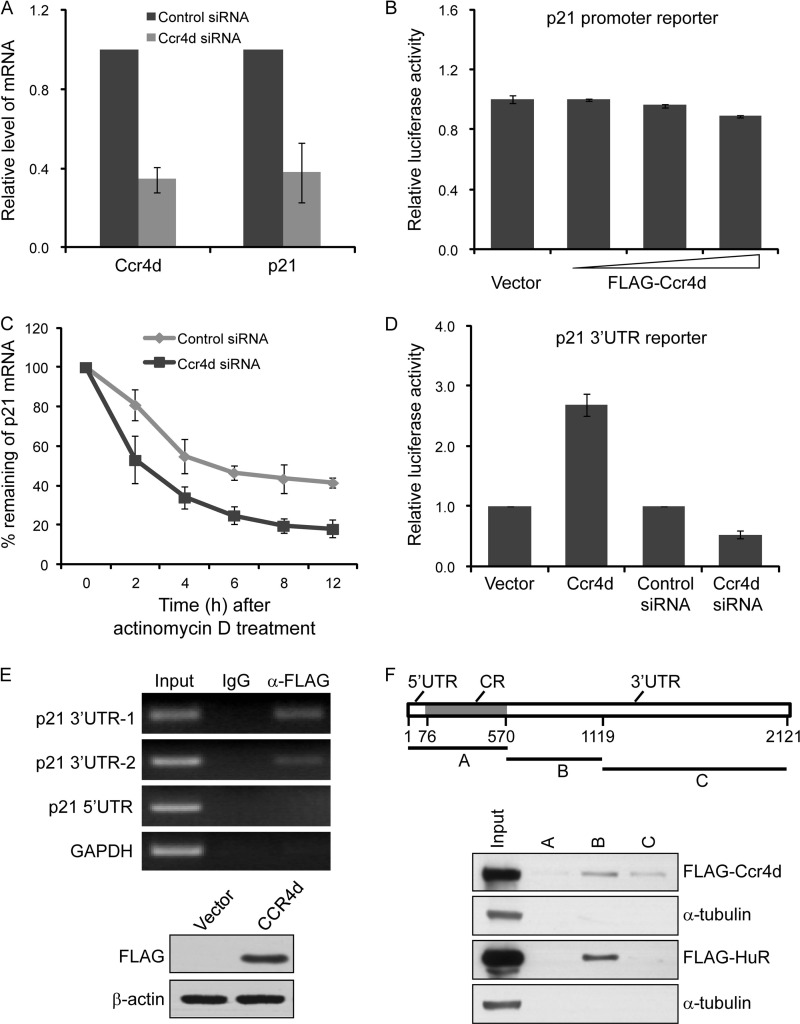
To further explore the molecular mechanism governing the regulation of p21 by Ccr4d, MCF-7 cells were transfected with Ccr4d siRNA, and the expression of p21 mRNA was measured by real-time RT-PCR. The results indicated that knockdown of Ccr4d led to a significant down-regulation of p21 mRNA (Fig. 5A). However, subsequent reporter assays in H1299 cells with p21 promoter-driven luciferase revealed that p21 transcription was not affected by Ccr4d overexpression, suggesting that the regulation of p21 by Ccr4d did not occur at the transcriptional level (Fig. 5B). Therefore, we next measured the rate of p21 mRNA clearance by real-time RT-PCR in MCF-7 cells that were treated with Ccr4d siRNA in the presence of actinomycin D, a transcription inhibitor. These experiments revealed that Ccr4d depletion led to a marked decrease in the stability of p21 mRNA, with a half-life of ∼9.1 h in untreated cells and ∼4.9 h in treated cells (Fig. 5C).
FIGURE 5.
Ccr4d induced p21 expression by enhancement of p21 mRNA stability. A, regulation of p21 expression by Ccr4d. MCF-7 cells were transfected with control siRNAs or Ccr4d siRNAs, and total RNAs were prepared and analyzed for p21 mRNA expression by real-time RT-PCR. Upon normalization to GAPDH transcript levels, the percentage of p21 transcript levels compared with untreated cells was plotted as mean ± S.D. from triplicate measurements. B, effect of Ccr4d on p21 promoter-driven luciferase reporter activity. H1299 cells were co-transfected with p21 promoter-driven luciferase constructs and different amounts of FLAG-Ccr4d constructs, and then cells were collected, and luciferase activity was measured and normalized to that of Renilla. Bars, mean ± S.D. for triplicate measurements. C, assay of p21 mRNA stability. MCF-7 cells were transfected with Ccr4d siRNA and were treated with 5 μm actinomycin D for different times. The data were normalized against the expression of GAPDH. Each point represents the mean ± S.D. for triplicate measurements. D, effect of Ccr4d on p21 3′-UTR-driven luciferase reporter activity. H1299 cells were co-transfected with p21 3′-UTR-driven luciferase constructs and FLAG-Ccr4d constructs or Ccr4d siRNA, and then cells were collected, and luciferase activity was measured and normalized to that of Renilla. Bars, mean ± S.D. for triplicate measurements. E, binding of Ccr4d to p21 mRNA 3′-UTR. Top, RIP assay was performed using total protein extracts from H1299 cells that were transiently transfected with FLAG-Ccr4d constructs for 48 h, and endogenous target transcripts of the corresponding immunoprecipitated materials were detected by an RT-PCR assay using p21 sequence-specific primers. PCR products corresponding to p21 mRNA were visualized on agarose gel. The PCR product of GAPDH served as a negative control. Bottom, Ccr4d overexpression used in RIP assays was confirmed by Western blotting. F, binding assay of Ccr4d to p21 mRNA 3′-UTR by RNA pull-down. Top, schematic drawing of the full-length p21 cDNA and various transcripts A, B, and C derived from the 5′-UTR and CR and the 3′-UTR used in this study. Bottom, RNA pull-down assay was performed using biotinylated transcripts of p21, and proteins of RNA pull-down were detected by Western blotting with anti-FLAG antibody. Binding of HuR to p21 transcript was taken as a positive control, and α-tubulin was used as a negative control.
3′-UTR is a key regulatory element involved in the regulation of mRNA stability (42). Indeed, it was reported that several RNA-binding proteins regulate mRNA stability by directly binding to the 3′-UTR of p21 mRNA (13, 43). Therefore, to characterize Ccr4d-regulated p21 expression, we next investigated whether Ccr4d could be functionally and physically associated with p21 mRNA 3′-UTR. For this purpose, H1299 cells that were overexpressed with Ccr4d or treated with Ccr4d siRNA were transfected with a p21 3′-UTR-driven luciferase reporter construct. Forty-eight hours after transfection, cellular lysates were prepared and analyzed for luciferase activity. As shown in Fig. 5D, overexpression of Ccr4d was associated with a marked elevation of the p21 mRNA 3′-UTR-driven luciferase reporter activity, and Ccr4d knockdown was accompanied by a drastic repression of the reporter activity.
We then examined whether or not Ccr4d could bind to p21 mRNA 3′-UTR by a RIP assay. In these experiments, H1299 cells were transiently transfected with the FLAG-Ccr4d construct. Total proteins were extracted and immunoprecipitated with an anti-FLAG antibody or normal mouse IgG. The precipitated Ccr4d-RNA complexes were treated with RNase-free DNase to remove trapped genomic DNAs, and the precipitated RNAs were purified by TRIzol reagent for cDNA synthesis, followed by RT-PCR amplification using sequence-specific primers. Ccr4d was found to be able to bind 3′-UTR but not 5′-UTR of p21 mRNA, and a significant amount of 3′-UTR PCR product was amplified from anti-FLAG-precipitated materials (Fig. 5E). RNA pull-down assays were also performed using various biotinylated transcripts of p21 from the 5′-UTR and CR or 3′-UTR. The results of the experiments showed that Ccr4d bound strongly to the 3′-UTR-B transcript and slightly weakly to 3′-UTR-C transcript (Fig. 5F). RNA-binding protein HuR was used as a positive control, which only bound to the B transcript of p21 3′-UTR. α-Tubulin was used as a negative control. Taken together, these data support an argument that Ccr4d up-regulates p21 expression through its binding to p21 mRNA 3′-UTR and stabilizing p21 mRNA.
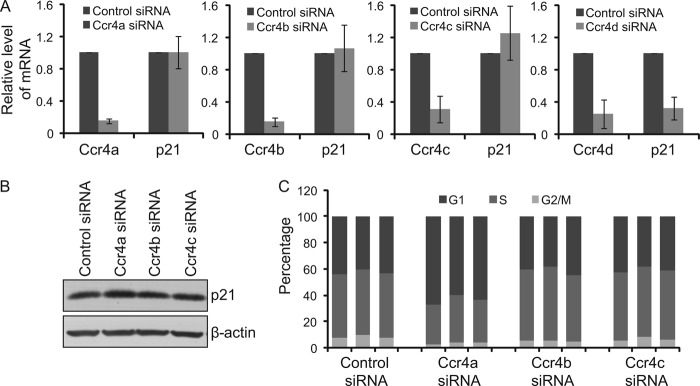
We carried out a number of experiments to address whether other members of Ccr4 could also regulate p21 expression and affect cell cycle progression. First, we analyzed the effect of Ccr4 family members on p21 mRNA expression in MCF-7 cells by real-time RT-PCR and Western blotting. We found that, different from Ccr4d, other members of Ccr4 (e.g. Ccr4a, -b, and -c) have no significant effect on expression of p21 mRNA (Fig. 6A), although the protein level of p21 was increased upon Ccr4a knockdown but not upon Ccr4b knockdown or Ccr4c knockdown (Fig. 6B). Next, the effect of Ccr4 family members on the cell cycle of MCF-7 cells were tested by flow cytometry assay. It was shown that cell cycle arrest was induced at G1 phase upon Ccr4a knockdown, and no significant changes were observed in the cell cycle of MCF-7 cells upon Ccr4b or Ccr4c knockdown compared with control (Fig. 6C).
FIGURE 6.
Specificity of Ccr4d on p21 expression. A, the mRNA expression of p21 upon knockdown of Ccr4 family members in MCF-7 cells. Cells were transfected with control siRNA and Ccr4 siRNA for 72 h, and total RNAs were prepared and analyzed for p21 mRNA expression by real-time RT-PCR. Upon normalization to GAPDH transcript levels, the percentage of p21 transcript levels compared with untreated cells was plotted as mean ± S.D. (error bars) from triplicate measurements. B, protein expression of p21 upon knockdown of Ccr4 family members in MCF-7 cells. Cells were transfected with control siRNA and Ccr4a, -b, or -c siRNA for 72 h, and cellular lysates were prepared for Western blotting analysis with antibodies against the indicated proteins. β-actin was used as a loading control. C, effect of knockdown of Ccr4 family members on cell cycle progression. After MCF-7 cells were transfected with control siRNA or Ccr4a, -b, or -c siRNA for 24 h, the cells were switched to conditioned medium without serum for 48 h. The cells were then cultured in medium containing 10% FBS for 27 h and were collected for cell cycle analysis by flow cytometry.
Expression of Ccr4d Is Down-regulated in Human Tumor Samples
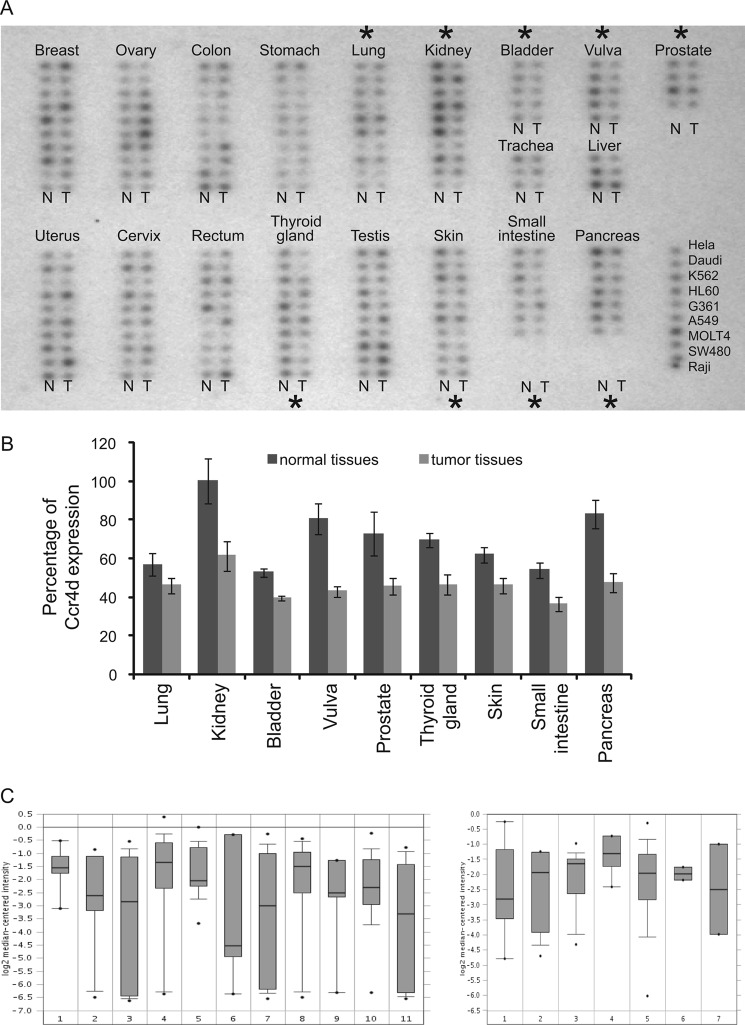
The positive regulation of p21 by Ccr4d and growth-inhibitory effect of Ccr4d suggest that Ccr4d could be implicated in carcinogenesis. In order to support this, we compared the expression profiles of Ccr4d in tumor samples versus normal tissues using a cancer profiling array (Clontech) by Northern blotting. The results indicated that the expression of ccr4d mRNA was down-regulated in cancers originating from a broad spectrum of tissue, including kidney, bladder, vulva, prostate, thyroid, skin, small intestine, and pancreas (Fig. 7A). The data from the cancer profiling array were quantified and shown in Fig. 7B. Further bioinformatics analysis of the cancer microarray ONCOMINE data base (Compendia Bioscience) (44–46) yielded a similar result (Fig. 7C). Although these analyses are still preliminary, together with our observations of the positive regulation of p21 by Ccr4d and the growth-inhibitory effect of Ccr4d, they nevertheless support a role of Ccr4d in carcinogenesis.
FIGURE 7.
Expression of ccr4d mRNA decreased in tumor tissues. A, expression profile of Ccr4d in human tumors. A cancer profiling array containing cDNA samples from 154 paired human tumors (T) and normal tissue (N) from individual patients was hybridized with 32P-labeled Ccr4d-specific probe. Human cancer cell lines are spotted on the right. Black asterisks, significant changes compared with normal tissues. B, data from the cancer profiling array were quantified using ImageQuant software, and the statistically significant changes are shown. Bars, mean ± S.E. (error bars). C, bioinformatics analysis of the cancer microarray ONCOMINE data base. The expression of Ccr4d is down-regulated in various carcinomas, including breast cancers. Left, the numbers 1–11 represent different types of tumor (45) as follows (numbers in parentheses represent the number of tumor tissues that were tested in this assay). 1, bladder cancer (8); 2, clear cell renal cell carcinoma (11); 3, colorectal adenocarcinoma (23); 4, ductal breast carcinoma (26); 5, gastroesophageal adenocarcinoma (12); 6, hepatocellular carcinoma (7); 7, lung adenocarcinoma (14); 8, ovarian serous surface papillary carcinoma (27); 9, pancreatic adenocarcinoma (6); 10, prostate adenocarcinoma (26); 11, squamous cell lung carcinoma (14). Right, numbers 1–7 represent different types of breast carcinoma (46) as follows (numbers in bracket represent the number of tumor tissues that were tested in this assay). 1, breast carcinoma (8); 2, invasive ductal and invasive lobular breast carcinoma (11); 3, invasive lobular breast carcinoma (13); 4, invasive tubular and lobular carcinoma (4); 5, invasive ductal breast carcinoma (158); 6, medullary breast carcinoma (2); 7, mucinous breast carcinoma (2).
DISCUSSION
In this study, we showed that ectopic expression of the endonuclease/exonuclease/phosphatase family protein Ccr4d resulted in inhibition of colony formation and that knockdown of this protein promoted cell growth in MCF-7 breast cancer cells. We demonstrated that ectopic expression of Ccr4d induced G1 arrest, whereas Ccr4d knockdown promoted cell cycle progression in both MCF-7 breast cancer cells and H1299 lung cancer cells. In agreement with these observations, we showed that mRNA level of endogenous Ccr4d markedly declined during cell cycle progression of MCF-7 cells. These results suggest that Ccr4d functions as an anti-proliferating factor by induction of cell cycle arrest in G1 phase and that suppression of Ccr4d expression might be a prerequisite for cells to reenter S phase.
Significantly, we showed that Ccr4d positively regulates the expression of p21, a direct mediator of cell cycle arrest at G1 phase. Although its expression is under p53-mediated transcriptional control, an increasing body of literature documents that p21 expression is also subject to post-transcriptional modulations. For example, the stability of p21 mRNA is regulated by HuR in response to UV light, epidermal growth factor, or prostaglandin A2 treatment (43, 47, 48). HuR is predominantly a nuclear protein. Upon these treatments, HuR is translocated from nucleus to cytosol, where it binds to the 3′-UTR of p21 transcript and enhances its stability (43, 47, 48). It was also shown that RNPC1a regulates the stability of basal and stress-induced p21 transcript through its binding to the 3′-UTR of the p21 transcript (12). Interestingly, RNPC1a is also capable of inducing cell cycle arrest in G1 (13). In this report, we showed that Ccr4d is yet another factor that binds to the 3′-UTR of p21 transcript and regulates the stability of p21 mRNA. Whether or not cooperative or antagonistic actions exist for different proteins in the regulation of the stability of p21 mRNA is currently unknown and will be an interesting subject for future investigations.
p21 expression increases following exposure to a wide variety of stress agents, including genotoxins, oxidants, and metabolic perturbations, and increased p21 expression is believed to be associated with growth arrest (5). In this regard, it is interesting to note that Ccr4d expression was also induced in response to DNA damage (49). In addition, testicular germ cell tumors responded well to cisplatin-based chemotherapy and showed a low incidence of acquired resistance compared with most somatic tumors (49). Microarray analysis has shown that the expression of ANGEL2 (Ccr4d) increased significantly in testicular germ cell lines after cisplatin treatment (49). These observations are consistent with our results that Ccr4d displays a growth-inhibitory activity and is functionally connected to p21.
mRNA stability is one of the key regulators in controlling the expression of many proteins. Dysregulation of mRNA stability has been associated with human diseases, including cancer, inflammatory disease, and Alzheimer disease (50). In this sense, our finding that Ccr4d positively regulates the stability of p21 mRNA indicates that Ccr4d might have significant pathophysiological functions. Indeed, we showed that the expression of Ccr4d is deregulated in human cancers from various tissue origins. Future investigations are warranted to further delineate the cellular activity of Ccr4d.
This work was supported by Natural Science Foundation of China Grants 81130048 and 30921062 (to Y. S.) and 30871252 (to X. Y.) and Ministry of Science and Technology of China 973 Program Grant 2011CB504204 (to Y. S.).
- CDK
- cyclin-dependent kinase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- EEP
- endonuclease-exonuclease-phosphatase
- RIP
- RNA-binding protein immunoprecipitation
- CR
- coding region(s).
REFERENCES
- 1. Massagué J. (2004) G1 cell cycle control and cancer. Nature 432, 298–306 [DOI] [PubMed] [Google Scholar]
- 2. Sherr C. J., Roberts J. M. (1995) Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 3. Sherr C. J. (1998) Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12, 2984–2991 [DOI] [PubMed] [Google Scholar]
- 4. Harper J. W., Elledge S. J., Keyomarsi K., Dynlacht B., Tsai L. H., Zhang P., Dobrowolski S., Bai C., Connell-Crowley L., Swindell E. (1995) Inhibition of cyclin-dependent kinases by p21. Molecular biology of the cell 6, 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jung Y. S., Qian Y., Chen X. (2010) Examination of the expanding pathways for the regulation of p21 expression and activity. Cell. Signal. 22, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abbas T., Dutta A. (2009) p21 in cancer. Intricate networks and multiple activities. Nat. Rev. Cancer 9, 400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. (2004) Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. U.S.A. 101, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Joseph B., Orlian M., Furneaux H. (1998) p21Waf1 mRNA contains a conserved element in its 3′-untranslated region that is bound by the Elav-like mRNA-stabilizing proteins. J. Biol. Chem. 273, 20511–20516 [DOI] [PubMed] [Google Scholar]
- 9. King P. H., Levine T. D., Fremeau R. T., Jr., Keene J. D. (1994) Mammalian homologs of Drosophila ELAV localized to a neuronal subset can bind in vitro to the 3′-UTR of mRNA encoding the Id transcriptional repressor. J. Neurosci. 14, 1943–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma W. J., Cheng S., Campbell C., Wright A., Furneaux H. (1996) Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 271, 8144–8151 [DOI] [PubMed] [Google Scholar]
- 11. Myer V. E., Fan X. C., Steitz J. A. (1997) Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 16, 2130–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shu L., Yan W., Chen X. (2006) RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 20, 2961–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho S. J., Zhang J., Chen X. (2010) RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 38, 2256–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yano M., Okano H. J., Okano H. (2005) Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J. Biol. Chem. 280, 12690–12699 [DOI] [PubMed] [Google Scholar]
- 15. Lal A., Mazan-Mamczarz K., Kawai T., Yang X., Martindale J. L., Gorospe M. (2004) Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 23, 3092–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fukuda T., Naiki T., Saito M., Irie K. (2009) hnRNP K interacts with RNA binding motif protein 42 and functions in the maintenance of cellular ATP level during stress conditions. Genes Cells 14, 113–128 [DOI] [PubMed] [Google Scholar]
- 17. Kao P. N., Chen L., Brock G., Ng J., Kenny J., Smith A. J., Corthésy B. (1994) Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells. NF45 and NF90. J. Biol. Chem. 269, 20691–20699 [PubMed] [Google Scholar]
- 18. Wilusz C. J., Wormington M., Peltz S. W. (2001) The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2, 237–246 [DOI] [PubMed] [Google Scholar]
- 19. Collart M. A., Panasenko O. O. (2012) The Ccr4-Not complex. Gene 492, 42–53 [DOI] [PubMed] [Google Scholar]
- 20. Kruk J. A., Dutta A., Fu J., Gilmour D. S., Reese J. C. (2011) The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 25, 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Denis C. L., Chen J. (2003) The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 73, 221–250 [DOI] [PubMed] [Google Scholar]
- 22. Collart M. A., Timmers H. T. (2004) The eukaryotic Ccr4-Not complex. A regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog. Nucleic Acid Res. Mol. Biol. 77, 289–322 [DOI] [PubMed] [Google Scholar]
- 23. Lau N. C., Kolkman A., van Schaik F. M., Mulder K. W., Pijnappel W. W., Heck A. J., Timmers H. T. (2009) Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem. J. 422, 443–453 [DOI] [PubMed] [Google Scholar]
- 24. Aslam A., Mittal S., Koch F., Andrau J. C., Winkler G. S. (2009) The Ccr4-NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol. Biol. Cell 20, 3840–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wagner E., Clement S. L., Lykke-Andersen J. (2007) An unconventional human Ccr4-Caf1 deadenylase complex in nuclear cajal bodies. Mol. Cell. Biol. 27, 1686–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dupressoir A., Morel A. P., Barbot W., Loireau M. P., Corbo L., Heidmann T. (2001) Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mittal S., Aslam A., Doidge R., Medica R., Winkler G. S. (2011) The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Mol. Biol. Cell 22, 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baggs J. E., Green C. B. (2003) Nocturnin, a deadenylase in Xenopus laevis retina. A mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol. 13, 189–198 [DOI] [PubMed] [Google Scholar]
- 29. Wang L., Brown S. J. (2006) BindN. A web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 34, W243–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thompson J. D., Higgins D. G., Gibson T. J. (1994) ClustalW. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hein J. (1990) Unified approach to alignment and phylogenies. Methods Enzymol. 183, 626–645 [DOI] [PubMed] [Google Scholar]
- 32. Zhang H., Yi X., Sun X., Yin N., Shi B., Wu H., Wang D., Wu G., Shang Y. (2004) Differential gene regulation by the SRC family of coactivators. Genes Dev. 18, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yi X., de Vries H. I., Siudeja K., Rana A., Lemstra W., Brunsting J. F., Kok R. M., Smulders Y. M., Schaefer M., Dijk F., Shang Y., Eggen B. J., Kampinga H. H., Sibon O. C. (2009) Stwl modifies chromatin compaction and is required to maintain DNA integrity in the presence of perturbed DNA replication. Mol. Biol. Cell 20, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yi X., Lemstra W., Vos M. J., Shang Y., Kampinga H. H., Su T. T., Sibon O. C. (2008) A long-term flow cytometry assay to analyze the role of specific genes of Drosophila melanogaster S2 cells in surviving genotoxic stress. Cytometry A 73, 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang W., Martindale J. L., Yang X., Chrest F. J., Gorospe M. (2005) Increased stability of the p16 mRNA with replicative senescence. EMBO Rep. 6, 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang X., Yu W., Shi L., Sun L., Liang J., Yi X., Li Q., Zhang Y., Yang F., Han X., Zhang D., Yang J., Yao Z., Shang Y. (2011) HAT4, a Golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Mol. Cell 44, 39–50 [DOI] [PubMed] [Google Scholar]
- 37. Shi L., Sun L., Li Q., Liang J., Yu W., Yi X., Yang X., Li Y., Han X., Zhang Y., Xuan C., Yao Z., Shang Y. (2011) Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 7541–7546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li R., Zhang H., Yu W., Chen Y., Gui B., Liang J., Wang Y., Sun L., Yang X., Zhang Y., Shi L., Li Y., Shang Y. (2009) ZIP, a novel transcription repressor, represses EGFR oncogene and suppresses breast carcinogenesis. EMBO J. 28, 2763–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Zhang H., Liang J., Yu W., Shang Y. (2007) EMBO J. 26, 2645–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu H., Chen Y., Liang J., Shi B., Wu G., Zhang Y., Wang D., Li R., Yi X., Zhang H., Sun L., Shang Y. (2005) Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature 438, 981–987 [DOI] [PubMed] [Google Scholar]
- 41. Nakano K., Vousden K. H. (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 42. Mignone F., Gissi C., Liuni S., Pesole G. (2002) Untranslated regions of mRNAs. Genome Biol. 3, REVIEWS0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang W., Furneaux H., Cheng H., Caldwell M. C., Hutter D., Liu Y., Holbrook N., Gorospe M. (2000) HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20, 760–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. (2004) ONCOMINE. A cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Su A. I., Welsh J. B., Sapinoso L. M., Kern S. G., Dimitrov P., Lapp H., Schultz P. G., Powell S. M., Moskaluk C. A., Frierson H. F., Jr., Hampton G. M. (2001) Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 61, 7388–7393 [PubMed] [Google Scholar]
- 46. Wang Y., Klijn J. G., Zhang Y., Sieuwerts A. M., Look M. P., Yang F., Talantov D., Timmermans M., Meijer-van Gelder M. E., Yu J., Jatkoe T., Berns E. M., Atkins D., Foekens J. A. (2005) Gene-expression profiles to predict distant metastasis of lymph node-negative primary breast cancer. Lancet 365, 671–679 [DOI] [PubMed] [Google Scholar]
- 47. Yang X., Wang W., Fan J., Lal A., Yang D., Cheng H., Gorospe M. (2004) Prostaglandin A2-mediated stabilization of p21 mRNA through an ERK-dependent pathway requiring the RNA-binding protein HuR. J. Biol. Chem. 279, 49298–49306 [DOI] [PubMed] [Google Scholar]
- 48. Giles K. M., Daly J. M., Beveridge D. J., Thomson A. M., Voon D. C., Furneaux H. M., Jazayeri J. A., Leedman P. J. (2003) The 3′-untranslated region of p21WAF1 mRNA is a composite cis-acting sequence bound by RNA-binding proteins from breast cancer cells, including HuR and poly(C)-binding protein. J. Biol. Chem. 278, 2937–2946 [DOI] [PubMed] [Google Scholar]
- 49. Duale N., Lindeman B., Komada M., Olsen A. K., Andreassen A., Soderlund E. J., Brunborg G. (2007) Molecular portrait of cisplatin induced response in human testis cancer cell lines based on gene expression profiles. Mol. Cancer 6, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hollams E. M., Giles K. M., Thomson A. M., Leedman P. J. (2002) MRNA stability and the control of gene expression. Implications for human disease. Neurochem. Res. 27, 957–980 [DOI] [PubMed] [Google Scholar]