Background: Alp12 is a novel plasmid-encoded actin-like protein from Clostridium tetani.
Results: Alp12 forms dynamically unstable filaments with an open helical cylinder structure composed of four protofilaments.
Conclusion: Specialized prokaryotic filament systems have evolved to execute a single function in comparison with the general multitasking force provider, double-stranded F-actin.
Significance: Repetitive Alp12 polymerization cycles may be incorporated into nanomachines.
Keywords: Actin, Cytoskeleton, Electron Microscopy (EM), Kinetics, Microbial Pathogenesis, Protein Assembly, TRIF, Dynamic Instability, Filament Structure, Plasmid Segregation
Abstract
Eukaryotic F-actin is constructed from two protofilaments that gently wind around each other to form a helical polymer. Several bacterial actin-like proteins (Alps) are also known to form F-actin-like helical arrangements from two protofilaments, yet with varied helical geometries. Here, we report a unique filament architecture of Alp12 from Clostridium tetani that is constructed from four protofilaments. Through fitting of an Alp12 monomer homology model into the electron microscopy data, the filament was determined to be constructed from two antiparallel strands, each composed of two parallel protofilaments. These four protofilaments form an open helical cylinder separated by a wide cleft. The molecular interactions within single protofilaments are similar to F-actin, yet interactions between protofilaments differ from those in F-actin. The filament structure and assembly and disassembly kinetics suggest Alp12 to be a dynamically unstable force-generating motor involved in segregating the pE88 plasmid, which encodes the lethal tetanus toxin, and thus a potential target for drug design. Alp12 can be repeatedly cycled between states of polymerization and dissociation, making it a novel candidate for incorporation into fuel-propelled nanobiopolymer machines.
Introduction
The structures of two classes of bacterial actin-like proteins (Alps)3 have been determined previously. First, MreB, which is involved in maintaining cell shape and is found in most prokaryotes (1), forms linear protofilaments arranged in rafts (2, 3). Second, ParM-like systems, the polymerizing motors that ensure ordered plasmid segregation prior to cell division, form helical filaments constructed from two protofilaments that resemble F-actin, although they differ in helical parameters and in handedness (4). The par locus of the R1 drug resistance plasmid encodes three components: a centromere-like site in the DNA (parC), a DNA-binding protein (ParR), and ParM (an Alp bearing NTPase activity) (5). In vivo, these components can form a linear assembly of multiple filaments (6), which positions pairs of plasmids at opposite ends of the rod-shaped bacteria by a polymerization mechanism, ensuring equal distribution of the plasmids between daughter cells (5). Other plasmid segregation motor systems that have been structurally characterized are pSK41-ParM (7) and AlfA (8, 9). The currently known crystal structures of diverse Alp monomers are similar despite low sequence homology (∼20%) (7, 10). Recently, a range of potential Alps, with low level sequence homology, was identified, including Alp12 from Clostridium tetani (11). The genes for Alp12, the tetanus toxin, and its direct transcriptional regulator TetR are harbored on the pE88 plasmid (12). Tetanus toxins block the release of neurotransmitters from presynaptic membranes of inhibitory neurons in the spinal cords of mammals, leading to continuous muscle contractions and death. Here, we show by three-dimensional electron microscopy reconstructions that C. tetani Alp12 filaments have a unique polymer structure that is entirely different from F-actin and that Alp12 filaments display dynamic behavior similar to microtubules.
EXPERIMENTAL PROCEDURES
Chemicals
Nucleotides and chemicals were purchased from Sigma, and fluorophores were bought from Invitrogen.
Proteins
N-terminally His-tagged Alp12 (Q89A01), cloned into the pSY5 vector (13), was transfected into Escherichia coli BL21(DE3) cells, which were grown to A600 ∼ 1 and induced overnight at 15 °C with isopropyl β-d-thiogalactopyranoside (0.2 mm). Alp12 was purified on a HisTrap column (GE Healthcare), cleaved on-column with PreScission protease to remove the His tag, and purified by gel filtration (Superdex 75, GE Healthcare). During the entire expression and purification procedures, no nucleotides were used to isolate Alp12 in a monomeric state. Unlike actin, Alp12 is stable without any nucleotide. The protein was flash-frozen and stored at −80 °C in gel filtration buffer (150 mm KCl, 1 mm MgCl2, 1 mm DTT, and 50 mm Tris, pH 7.5). Proteins were thawed, dialyzed against the appropriate buffer, and briefly centrifuged at high speed before use. Protein concentrations were determined using a NanoDrop spectrophotometer using theoretical extinction coefficients. Physiological concentrations of Alp12 were assumed to be similar to those of ParM-R1, which are in the range of 12–14 μm (5). The following buffers were used: high salt buffer (300 mm KCl, 1 mm MgCl2, 0.5 mm DTT, and 30 mm Hepes, pH 7.5) and low salt buffer (50 mm KCl, 1 mm MgCl2, 0.5 mm DTT, and 30 mm Hepes, pH 7.5). Although Alp12 was able to polymerize in both buffers, the high salt buffer was preferentially used in this study because the KCl concentration (300 mm) is close to the physiological level found in bacteria (14). Polymerization was initiated by adding NTP. ParM-R1 from E. coli was expressed and purified as described previously (10), and assembly was initiated by the addition of nucleotide in buffers as used for Alp12.
Light Scattering, Phosphate Release, and Kinetic Modeling
Assembly and disassembly of Alp12 at 24 °C were followed by light scattering at 90° using either a PerkinElmer Life Sciences LS 55 spectrometer for long-time measurements (initial delay time due to mixing by hand of ∼10–15 s) or a BioLogic stopped-flow machine to observe the early polymerization phase (initial delay time of ∼3 ms), monitored at 600 nm. The release of Pi upon nucleotide hydrolysis during Alp12 polymerization and disassembly was measured at 24 °C using a phosphate assay kit (E-6646, Molecular Probes) based on a method described previously (15). The absorbance at 360 nm was measured using an Ultrospec 2100 pro spectrophotometer (Amersham Biosciences). The polymerization kinetics were modeled using DYNAFIT (16, 17). DYNAFIT takes the polymerization scheme and converts it to a set of differential equations according to the law of mass action, solves the equations numerically, and fits the kinetic constants to the progressive curve using the Levenberg-Marquardt algorithm.
Electron Microscopy, Total Internal Reflection Fluorescence Microscopy, and Fluorescence Microscopy
In this study, we used negative stain, as it requires much less data analysis due to the high signal-to-noise ratio compared with cryo-electron microscopy. This is usually the best way to initially characterize a previously unknown filament system. Negative stain has been shown to fix the structures of filament systems, F-actin, and F-actin-myosin complexes in <10 ms, entirely preserving their ultrastructure, as determined by comparison with cryo-electron microcopy at ∼20 Å resolution (the resolution limit for negative stain) (18). A drop of Alp12 solution was applied to carbon-coated copper grids, blotted, stained with 1% uranyl acetate, and visualized under a Hitachi H-7600 electron microscope operated at 100 keV and at a nominal magnification of ×40,000. Films were digitized with a Zeiss Z/I Imaging PhotoScan 2000 scanner in 7-μm steps. Fourier transforms, filtered images, and three-dimensional reconstructions were obtained using the EOS software package (19). Labeling of Alp12 with fluorophores was done similarly as described for ParM-R1 (20, 21). Total internal reflection fluorescence microscopy was carried out on an inverted Nikon TE200-E microscope equipped with autofocus assist system using similar protocols as described previously for ParM-R1 (20, 21). In general, crowding agents (∼0.5–1% methyl cellulose or ∼5–10% polyvinyl alcohol) have to be used in total internal reflection fluorescence microscopy to trap filaments in the vicinity of the evanescent field even for F-actin, which could be further conjugated near the surface by myosin (22). As in the case of ParM-R1 (20, 21), this can lead to some bundle formation of Alp12. Filaments within bundles are likely to be stabilized by electrostatic interactions as shown for ParM-R1 (23). However, Alp12 bundles disintegrated over time, indicative of microtubule-like dynamic instability, as observed previously for ParM-R1 bundles (20, 21).
Visualization of Alp12 in yeast was performed as described (24). In brief, Schizosaccharomyces pombe cells carrying GFP-Alp12 were grown in minimal medium without thiamine for 20–24 h to induce expression, and cells were imaged using a Zeiss 510 Meta confocal microscope.
Outline of EM Reconstruction
ParM filament images were extracted from the electron micrographs and straightened. The digitized images were corrected for the phase of the contrast transfer function. (The amplitude of the contrast transfer function was corrected after the refinement.) The contrast of the images was inverted to allow the adoption of procedures used for cryo-electron micrographs. Initially, an averaged power spectrum was calculated from nine Alp12 filaments (see Fig. 1B). This showed clear layer lines. From this, the selection rule was assigned as either l = −1n + 27m (“right-handed helix”) or l = 1n + 27m (“left-handed helix”). The true pitch was ∼141 nm. Note that the handedness was not determined yet at this stage. We initially assumed the selection rule as l = −1n + 27m (right-handed helix-like F-actin) and produced a three-dimensional structure by using helical reconstruction (25). The layer lines up to the 72nd order (∼19.5 Å) were included.
FIGURE 1.
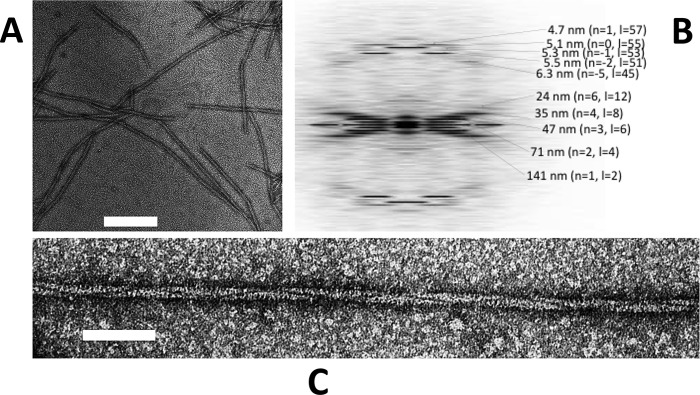
Typical electron micrographs of Alp12 filaments. A, individual filaments were observed to be relatively short. Scale bar = 200 nm. B, the calculated averaged Fourier transform (from nine Alp12 filaments) showed many layer lines typical for a helical filament. The first layer line at ∼141 nm classifies the helical repeat. C, closer view of an Alp12 filament. Scale bar = 50 nm.
Structure Refining
The refinement of the structure was carried out as described in more detail for ParM-R1 (see supplemental data in Ref. 20). The map from the helical reconstruction was used as an initial reference. Through using correlation maps, the polarity of each image was determined (26). Only filaments with a clear polarity and a clear correlation pattern in the correlation maps were selected. The Alp12-ATP filament structure was obtained from 91 filaments and 5582 particles, whereas, the Alp12-GMP-PNP envelope was generated from nine filaments and 422 particles. The contrast transfer function amplitude was corrected (26) after the iteration converged. The final resolution (19.7 Å) was evaluated by Fourier shell correlation (supplemental Fig. S11).
Constructing Atomic Model of Alp12 Filament
Alp12 homology models were docked into our final EM map by Situs (27). The fitting with the largest correlation value was selected. Then energy minimization was performed by NAMD (28) without any constraints from the EM map. Each non-hydrogen atom in the atomic model was replaced by a Gaussian density distribution with a radius of 4 Å. The 200% volume contour in the EM map was defined as zero, and the pixels with negative values were set to zero. Then, the Fourier shell correlation (29) was calculated between the EM map and the map from the homology model to evaluate the correctness (supplemental Fig. S11). The two maps were in agreement up to 24 Å resolution, which is less than the resolution of the EM map. This may be attributed to the Alp12 homology model not being entirely correct. Coordinates of the model were deposited into the EMDataBank (Protein Data Bank code 4APW; EMDB ID code for EM map, EMD-2068).
RESULTS
Filament Assembly and Dynamics
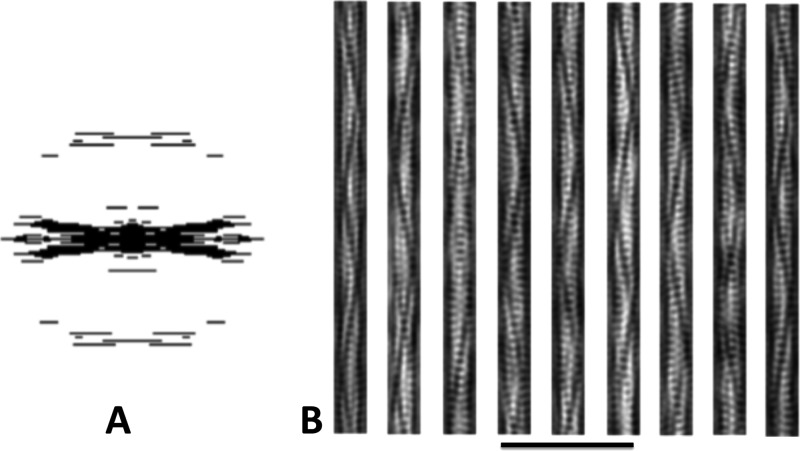
Initially, GFP-labeled Alp12 was expressed in yeast and was observed to form bundles of filaments by light microscopy (supplemental Fig. S1). Subsequently, Alp12 expressed in E. coli was purified in its monomeric form, and the protein was induced to polymerize by the addition of nucleotides into mainly single filaments. Filaments initiated by adding ATP, GTP, or non-hydrolyzable nucleotides (AMP-PNP or GMP-PNP) had similar appearances under the electron microscope (Fig. 1 and supplemental Fig. S2). Filaments were typically ∼0.5–3 μm in length. A unique feature of these filaments is a dark line spiraling along the filament length (Fig. 1C), which became very clear in filtered images (Fig. 2). We later show that this line can be attributed to a large cleft between antiparallel filament strands. The calculated Fourier transform showed layer lines typical for a helical filament, which could be indexed by the selection rule l = −2n + 55m (Fig. 1B). The addition of ADP or GDP did not facilitate polymer formation as revealed by both electron microscopy and light scattering (supplemental Fig. S3).
FIGURE 2.
Filtered images of Alp12. A, applied mask in Fourier space. B, examples of filtered projection structures using the back Fourier transformation in A. The trench and strands become clearer.
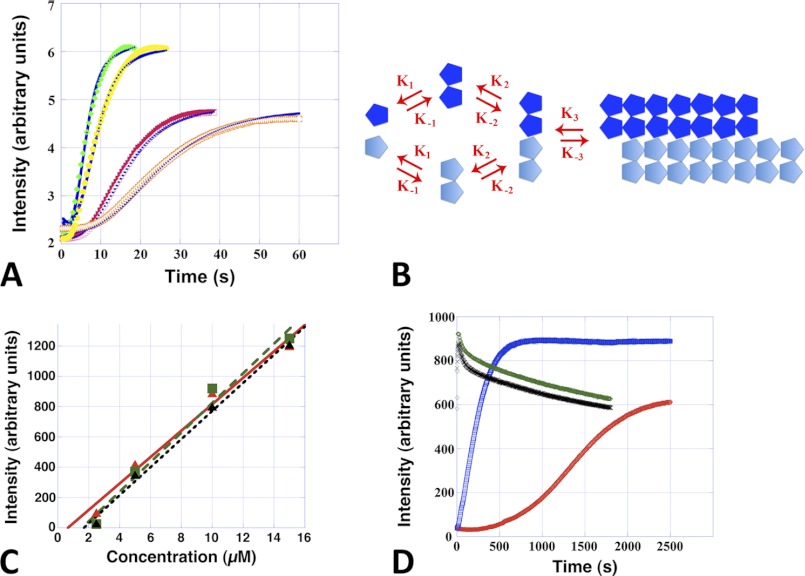
Alp12 filaments, like microtubules and F-actin, assemble via a nucleation-condensation reaction (30). Characteristics of this mechanism were the existence of a critical concentration, which was determined to be ∼2–3 μm for ATP and GTP and slightly less for non-hydrolyzable nucleotides (Fig. 3C), values that are similar to those for ParM-R1 (31); and a time lag in spontaneous polymerization, during which stable nuclei assembled (Fig. 3A and supplemental Fig. S4A). Polymerization was substantially faster than for F-actin but slower than for ParM-R1 (20), reaching a concentration-dependent maximum ∼15–20 s after the addition of ATP or GTP (Fig. 3A and supplemental Fig. S4A). Polymerization induced by non-hydrolyzable nucleotides was substantially slower (Fig. 3D). ATP appeared to be the more efficient nucleotide because the amount of polymer formed was often higher than with GTP at low NTP concentrations (0.1 mm); however, the levels were similar at physiological bacterial levels (∼1 mm NTP) (Fig. 3, A and D). The number of monomers required to form a nucleus was determined by plotting the log of the maximum rate of polymerization versus the log of the protein concentration (supplemental Fig. S4B) (32), suggesting that Alp12 filaments elongate from a nucleus composed of three to four monomers. To further clarify the polymerization kinetics, we used DYNAFIT to fit the experimental curves (16, 17). Several polymerization mechanisms were tested, but only one fitted well to the experimental light scattering data. This mechanism involves dimer formation, followed by dimer association into tetramers and subsequent elongation (Fig. 3, A and B). An initial conformational change due to NTP binding was not required. The kinetic parameters were almost identical for ATP and GTP (Table 1).
FIGURE 3.
Alp12 assembly kinetics in near-physiological high salt buffer. A, Alp12 polymerization followed by light scattering using a stopped-flow machine. Orange, 7.5 μm Alp12 and 1 mm GTP; red, 7.5 μm Alp12 and 1 mm ATP; yellow, 15 μm Alp12 and 1 mm GTP; green, 15 μm Alp12 and 1 mm ATP. Blue curves are the best fits obtained by DYNAFIT. B, the kinetic scheme used for the fit: monomers associate into dimers and then tetramers and then elongate. C, steady-state light scattering intensities plotted as a function of Alp12 concentration. Data points are averaged over three individual experiments. Green, Alp12-ATP; black, Alp12-GTP; red, Alp12-AMP-PNP. The intersection of the linearly approximated curves with the x axis defines the critical concentration. D, long-term light scattering observations. Due to mixing by hand, the initial 10–15 s could not be resolved, and the Alp12-ATP and Alp12-GTP curves start almost at the plateau of polymerization. Protein concentration was 8 μm. Green, 1 mm ATP; black, 1 mm GTP; blue, 1 mm AMP-PNP; red, 1 mm GMP-PNP. Note the slower polymerization in the presence of non-hydrolyzable nucleotides.
TABLE 1.
Calculated rate constants for the kinetic scheme in Fig. 3B
K values are expressed in μm−1 s−1.
| Alp12 concentration | Nucleotide concentration | K1 | K−1 | K2 | K−2 | Ke | K−e |
|---|---|---|---|---|---|---|---|
| 7.5 μm | 1 mm ATP | 6.71 | 0.3 | 43.82 | 0.0001 | 9.65 | 0.15 |
| 7.5 μm | 1 mm GTP | 6.71 | 0.3 | 43.82 | 0.0001 | 9.65 | 0.09 |
| 15 μm | 1 mm ATP | 6.71 | 0.3 | 43.82 | 0.0001 | 9.65 | 0.47 |
| 15 μm | 1 mm GTP | 6.71 | 0.3 | 43.82 | 0.0001 | 9.65 | 0.3 |
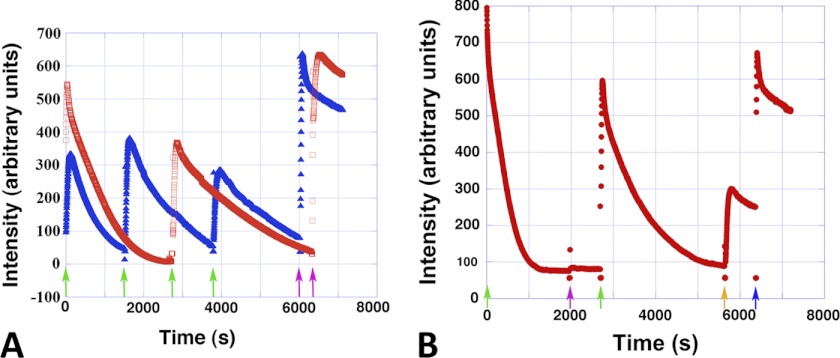
Light scattering experiments indicated that the Alp12 filaments that were polymerized in the presence of non-hydrolyzable nucleotides were stable (Fig. 3D), whereas filaments assembled by either ATP or GTP were dynamically unstable. However, Alp12 filament disassembly was slower than for ParM-R1 and produced a different curve shape (supplemental Fig. S5A). Filament assembly could be induced to undergo repeated cycles of polymerization and depolymerization by the addition of new ATP or GTP after the filaments had disassembled (Fig. 4A).
FIGURE 4.
Repeated polymerization-depolymerization cycles. A, Alp12 (8 μm) could be cycled repeatedly through polymerization and depolymerization. At each green arrow, 100 μm NTP was added. At the purple arrows, 1 mm NTP was added. Red, ATP; blue, GTP. B, ATP and GTP were not completely exchangeable. The amount of nucleotide added 100 μm ATP (green arrows), 100 μm GTP (purple arrow), 1 mm GTP (orange arrow), and 1 mm ATP (blue arrow). Note that 100 μm GTP was not sufficient to repolymerize Alp12. The Alp12 concentration was 20 μm.
Nevertheless, ATP and GTP were not completely interchangeable. When a polymerization-depolymerization cycle was initiated by ATP and an equal amount of GTP was added after the filaments had dissociated, Alp12 did not repolymerize (Fig. 4B), yet further addition of ATP facilitated repolymerization (Fig. 4B). In contrast, repolymerization occurred when a polymerization-depolymerization cycle was initiated by GTP, followed by the addition of ATP after the initial filament dissociation (supplemental Fig. S5B). Therefore, it appears likely that Alp12 operates mainly with ATP within the bacterial cell.
The rate of disassembly was dependent on the initial amount of NTP added (Fig. 5C and supplemental Fig. S6). Disassembly appeared to be biphasic, consisting of a very fast initial decline, followed by a long and steady slower decline, which tapered out to reach the base line. The initial fast decline seemed to be similar for all NTP concentrations, whereas the second phase showed an exponential decline of depolymerization with increasing NTP concentrations (supplemental Fig. S6, C and D). Real-time total internal reflection fluorescence microscopy confirmed that filaments disintegrated over time and repolymerized after the addition of fresh NTP (supplemental Fig. S7 and Movies 1 and 2).
FIGURE 5.
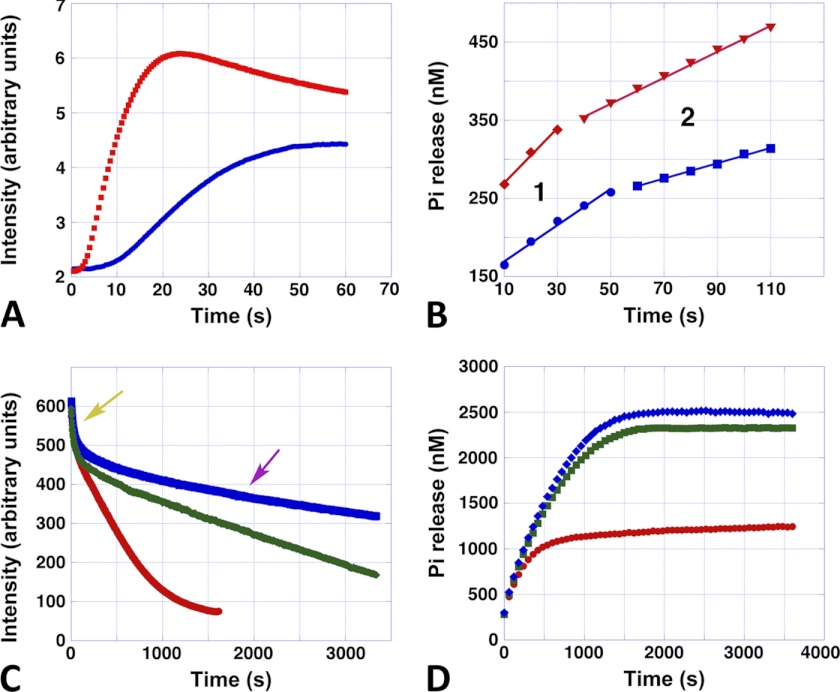
Assembly, disassembly, and phosphate release. A, typical stopped-flow polymerization curves of Alp12 polymerized by 1 mm ATP. Blue, 7.5 μm Alp12; red, 15 μm Alp12. B, corresponding Pi release. Blue, 7.5 μm Alp12; red, 15 μm Alp12. The first ∼10 s could not be followed due to mixing by hand. Note that Pi release consisted of two phases (phases 1 and 2). C, disassembly monitored by light scattering. The Alp12 concentration was 15 μm. The ATP concentrations initially added were 100 μm (red), 500 μm (green), and 1000 μm (blue). Note the fast decreasing phase (yellow arrow), which was similar for all ATP concentrations. The second disassembly phase (purple arrow) was slower and depended on ATP concentration. D, corresponding Pi release. The Alp12 concentration was 15 μm. The ATP concentrations were 100 μm (red), 500 μm (green), and 1000 μm (blue). Pi release increased until it reached a plateau. The time to reach the plateau took substantially longer at 500 μm ATP than at 100 μm ATP, yet the difference between 500 and 1000 μm ATP was small. Similar behavior was observed for the total amount of Pi released (plateau height).
Interestingly, the bulk assembly and disassembly kinetics of Alp12, which displayed an initial peak in polymer concentration, followed by a sharp dip and a slower decreasing phase, especially noticeable at high protein concentrations (Fig. 5C), are reminiscent of ParM-R1 kinetics (31). The rise and initial fall in Alp12 polymer appear to represent a population of rapidly nucleated filaments that elongate together and undergo somewhat synchronous catastrophe. Microtubules undergo similar synchronous behavior under conditions in which nucleation is fast and/or nucleotide dissociation is slow (33, 34).
Buildup of ADP or GDP did not appear to have a major effect on repeated polymerization-depolymerization cycles (Fig. 4A), yet the addition of NDP to Alp12 polymers could cause fast depolymerization, as observed by light scattering (supplemental Fig. S8). Whereas the addition of an equal amount of NDP to Alp12-NTP caused only a small decrease in scattering intensity, the addition of 10-fold excess NDP over NTP led to very rapid depolymerization (supplemental Fig. S8, A and B). Therefore, NDP, especially if added in excess, may be able to exchange with the bound NTP (or NTP-Pi) in the filament, leading to increased dynamic instability, as outlined schematically in supplemental Fig. S9. The addition of new NTP to a sample previously destabilized by NDP again led to filament formation (supplemental Fig. S8, C and D, and Fig. S9).
To learn more about the state of nucleotide within filament over time, we measured the release of Pi. Release of Pi appeared to be triphasic (Fig. 5, A and B). Directly after the addition of NTP, Pi release was fastest. This phase appeared to be coupled to the initial polymerization phase of Alp 12, lasting ∼20–40 s depending on the protein concentration. At the maximum of polymer formation, only ∼3.5% of all monomers within filaments had released their phosphate (Fig. 5B). When filaments switched to depolymerization mode, Pi release slowed, tapering out at a plateau (Fig. 5, B and D, and supplemental Fig. S10, A and B). The length of the second slower phase of Pi release, for a given Alp12 concentration, was to some extent coupled to the amount of NTP used for polymerization and therefore was related to the depolymerization rates at different NTP concentrations (Fig. 5, C and D, and supplemental Fig. S10D). For a given Alp12 concentration, phase 2 was about twice as long at higher NTP concentrations (∼500–1000 μm) than at low NTP concentrations (100 μm) (Fig. 5D). The rates of phases 1 and 2, as well as the height of the plateau of Pi release, were proportional to the concentration of Alp12 at a given NTP concentration (Fig. 5B and supplemental Fig. S10, A–C). The total amount of Pi released during a polymerization-depolymerization cycle at low NTP concentrations was only about half of that at higher NTP concentrations (Fig. 5D and supplemental Fig. S10D). The plateau appeared to be saturating at NTP levels of ∼1 mm, levels that are close to physiological in bacterial cells. Nevertheless, the release of Pi by Alp12 was surprisingly low. Even at high NTP concentrations, the percentage of monomers that had released their Pi was only ∼17% at the end of a polymerization-depolymerization cycle (Fig. 5D).
Filament Structure
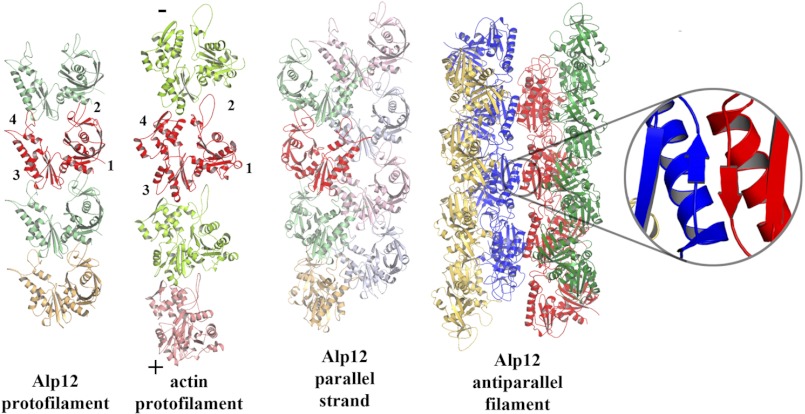
EM reconstructions revealed that the Alp12 filament consists of two antiparallel strands (Fig. 6 and supplemental Figs. S11 and S12). Homology models of the Alp12 monomer were fitted into the electron density map. All models were also mirrored, as the handedness of Alp12 is presently unknown. Only one model corresponding to the homology model based on pSK41-ParM could be accurately placed (Fig. 5F and supplemental Figs. S11 and Fig. S13) as a right-handed helix (l = −1n + 27m; Δφ = ±13.38°). The constructed model and the EM map (19.7 Å resolution) were well correlated up to 24 Å resolution as measured by the Fourier shell correlation method (supplemental Fig. S14). In total, the Alp12 filament is constructed from four protofilaments (Fig. 7), with each antiparallel strand of the Alp12 filament composed of two very similar parallel protofilaments (supplemental Fig. S15). Subunits within a protofilament associate through interactions that are globally similar to actin (Figs. 7 and 8). However, two protofilaments associate into a strand through a completely different interface from that observed for actin (Figs. 7 and 8, C and D). The two antiparallel strands are aligned with a gentle twist to form a helix with a repeat of ∼141 nm (Fig. 1B) through an interface that may involve a common β-sheet (residues 200–202) (Fig. 7 and supplemental Fig. S16). The dark line in the original EM images reflects the fact that the four protofilaments do not form a complete cylinder; rather, they give the appearance of an incomplete cylinder with a large trench (Fig. 9). Larger structures, consisting of more than two strands, are prevented from forming by the incompatibility of strand geometry with filament twist at larger radii (supplemental Fig. S16, B and C).
FIGURE 6.
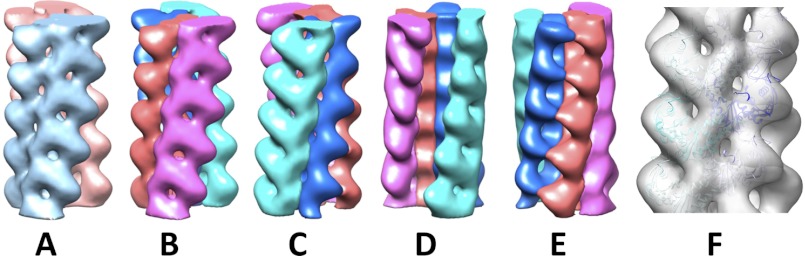
Electron density map. A, the two antiparallel strands are colored cyan and pink. B–E, each strand consists of two parallel protofilaments individually presented in different colors. The cyan strand in A is the red and purple protofilaments. The pink strand in A is the cyan and green protofilaments. B, strand 1 face; C, strand 2 face; D, trench face; E, back face. A comparison of B with C highlights the antiparallel nature of the two strands in the Alp12 filament. F, fitting of the model into the electron density map. The three-dimensional reconstruction was obtained from 91 filaments and 5582 particles, and the resolution was 19.7 Å. See supplemental Fig. S12 for an enlarged view.
FIGURE 7.
Model of Alp12 filament. The interactions between monomers in the Alp12 protofilament are grossly similar to those in the actin protofilament. The red monomers have been aligned, and the subdomains labeled are the barbed (+) and pointed (−) ends of actin. The Alp12 strand is composed of two parallel protofilaments in an arrangement that is not similar to actin. The Alp12 filament is composed of two antiparallel strands connected through subdomain 3, possibly through a β-sheet interaction (inset).
FIGURE 8.
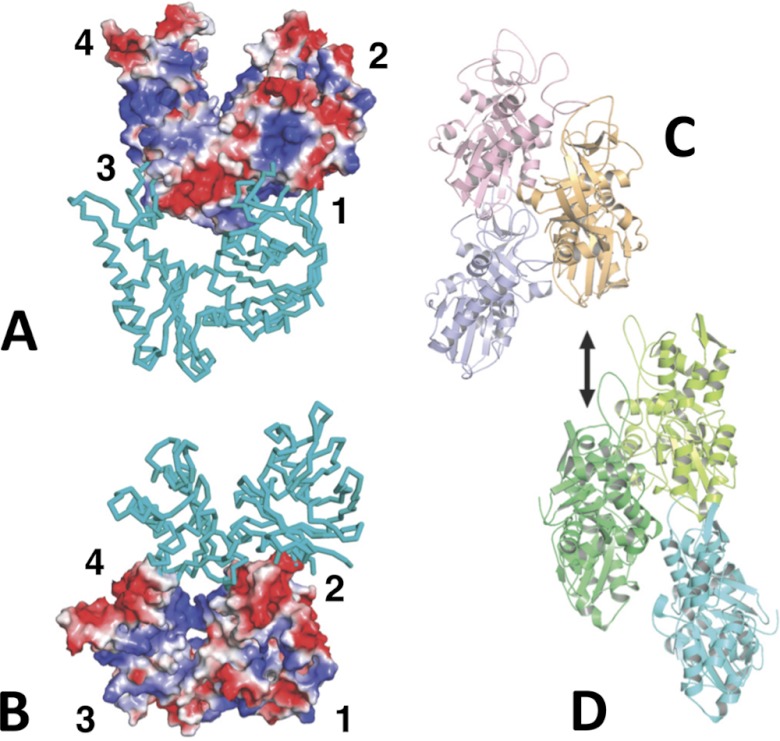
Alp12 interactions. A and B, charged interaction surfaces within the Alp12 protofilament. The two views are tilted relative to each other to see the electrostatic interactions at the interface. In particular, subdomain 2 of the lower monomer offers an acidic surface to a basic region on subdomain 1 of the upper monomer. C, Alp12 arrangement of three monomers within a parallel strand. D, three monomers within F-actin whereby the dark green monomer is in the same orientation as the orange Alp12 monomer in C. Alp12 uses different surfaces to form a strand from two protofilaments.
FIGURE 9.
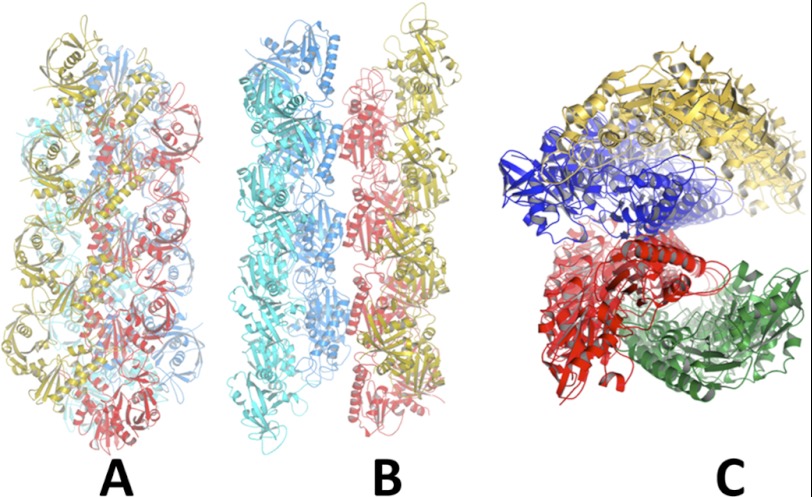
Three views of Alp12 filament. A, focusing on a parallel strand (yellow and red) with the second parallel strand (blue and cyan) related by an antiparallel manner. B, focusing on the trench. C, filament viewed end-on.
Monomers within each protofilament are in a more “open” configuration than F-actin (Fig. 6). Recently, the crystal structure of ParM-R1 revealed two different conformations: without associated nucleotide, the two large domains were in an open conformation that closed by ∼30° upon nucleotide binding (10).
The structure of the ParM-R1 filament has previously been determined to be mostly in a “closed” monomer conformation, and it is presumed to switch into an open conformation when NDP is released to disassemble (10, 20). Here, Alp12 filaments are composed of open monomers from the beginning. Because Pi release from the Alp12 filament is slow (Fig. 5), most monomers can be expected to be in NTP- or NTP-Pi-bound states in the first 1–2 min after polymerization, which was the time frame for the EM fixation. Hence, if the ParM-R1 precedent holds, namely that the open conformation leads to dynamic instability, Alp12 may polymerize in a form that is primed for dissociation. This is consistent with the fact that Alp12 polymerized by the non-hydrolyzable nucleotide GMP-PNP appears to have a very similar filament structure (supplemental Fig. S17).
DISCUSSION
The cell shape-determining protein MreB from various bacterial species has been shown to assemble as single-stranded protofilaments (2, 3, 35). In contrast, all force-generating motors known to be involved in segregating plasmids (ParM-R1, pSK41-ParM, and AlfA) were found to be helical polymers composed of two protofilaments gently winding around each other (2, 7–9, 36). A double helical design has one major disadvantage, namely in its polar design, which can bind a plasmid only at one end. Therefore, helical motor filaments like ParM-R1 rely on the crowdedness inside the bacterial cell to form randomly oriented bundles, which can capture plasmids at both ends (6). Here, we have characterized a novel actin-like filament system from C. tetani that can act as a polymerizing motor, displays microtubule-like dynamics at steady state similar to ParM-R1, and has an actin-like protofilament structure. However, the structure of the filament differs dramatically from F-actin both in the interaction interface that brings pairs of protofilaments together to form a strand and in the antiparallel association between two strands. This four-protofilament, two-stranded antiparallel design has advantages in ensuring equal binding and distribution of plasmids for segregation (Fig. 10) and is considerably stiffer than a double-stranded helix, which is of importance when moving heavy loads like DNA in a directed manner. As monomers in the Alp12 filament were found to be in a open conformation primed for disassembly, stabilization of the filament may be expected through interaction with yet to be described C. tetani pE88 components corresponding to the ParR-parC complex (Fig. 6). This Alp12-ATP filament structure is the first of a bacterial actin that has been solved in its dynamic unstable form; the structures of other actins like ParM-R1 have been determined only in their stable conformation with non-hydrolyzable nucleotides like GMP-PNP (20). It is now obvious that many different designs of filamentous polymerizing motors have been probed during evolution (4). The most common form seems to be helical polymer formed from two intertwining protofilaments. Alp12 construction features have not been observed before, and it will be interesting to see if similar actin designs have been adopted in other bacterial species.
FIGURE 10.
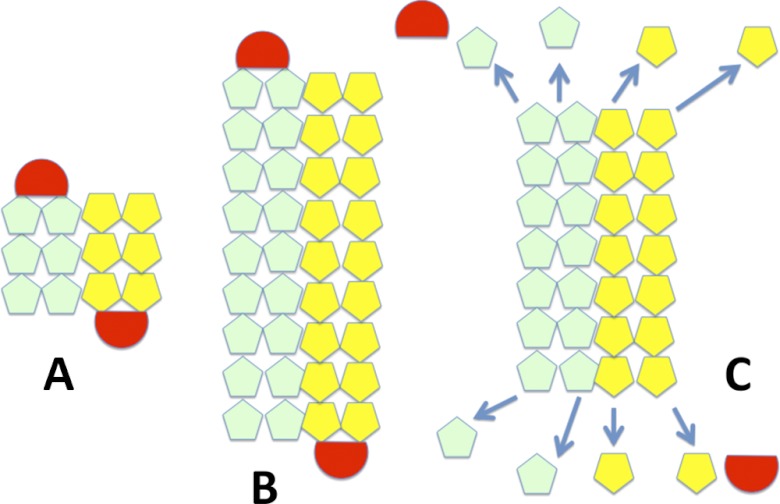
Model for plasmid segregation of Alp12. A, initially, a short filament of antiparallel strands, each consisting of two parallel protofilaments, forms (light green and yellow), which can capture a plasmid complex at each end (red). B, Alp12 and other bacterial plasmid segregation proteins (ParM-R1, pSK41-ParM, and AlfA) are polymerizing motors that push plasmids to opposite ends of the bacteria via a polymerization mechanism. The bound plasmid (similar to the ParR-parC complex in ParM-R1) prevents immediate disassembly of the filament, C, filaments that fail to capture plasmids or, after successful plasmid segregation and plasmid dissociation, the unbound filament ends are vulnerable to dynamic instability and filament dissociation (arrows). This scheme allows the Alp12 filament system to repetitively probe the cytoplasm to capture and segregate the pE88 plasmid.
The kinetics of Alp12 also appear to be specialized. The assembly kinetics differ from all previously investigated actins, which often required trimer formation prior to elongation. Alp12 forms dimers, which associate into tetramers before polymerizing, a nucleation process that appears logical considering its four-protofilament design. Alp12 is the second bacterial actin after ParM-R1 that displays dynamic instability. However, the depolymerization rates and the mechanisms by which instability is achieved may differ between the two systems. Higher resolution structural and further kinetic data are necessary to characterize the differences in more detail. Whereas ParM-R1 preferred GTP to ATP (20) as polymerization fuel, Alp12 appeared to be mainly an ATPase. Phosphate release from Alp12 was slow, and only ∼17% of all monomers had released their Pi at the end of a polymerization-depolymerization cycle, indicating that a disassembling unit may be substantially larger than a single monomer, believed to be the dissociating unit for ParM-R1 (20). For ParM-R1-GTP, phosphate release also appeared triphasic, with an initial burst followed by a slower release. Nevertheless, a plateau was reached significantly faster than with Alp12 (20). At present, the correlation between Pi release, kinetics, and structural state is not entirely obvious in the Alp12 system and may require cryo-electron microscopy for further elucidation. Another complication arises from the observation that bound NTP or NTP-Pi within an Alp12 filament can be readily replaced by NDP, leading to rapid filament dissociation. Likewise, NDP can be replaced by NTP, leading to rapid elongation. The addition of an ATP-regeneration system (37) had no major effects on the results, as one may expect considering the small amount of Pi released during a polymerization-depolymerization cycle. Repeated cycling of an actin-like protein between phases of filament formation after the addition of fuel in the form of NTP followed by spontaneous dissociation is, to our knowledge, a new phenomenon, which potentially may be incorporated into the design of fuel-propelled nanobiopolymer machines for industrial applications. Furthermore, because Alp12 is responsible for segregating the pE88 plasmid, which encodes the lethal tetanus toxin, a search for Alp12 polymerization cycle inhibitors may be an interesting strategy for drug development.
Acknowledgments
D. P. thanks J. Usukura and S. Minakata (Nagoya University) for kind support in using the Hitachi H-7600 electron microscope.
This work was supported by the Agency for Science, Technology, and Research (A*STAR) of Singapore (to D. P., L. J. L., B. X., and R. C. R.); Joint Council Office Grant Project 10/03/FG/06/04 (to D. P. and R. C. R.); and a grant-in-aid for scientific research and a grant-in-aid for young scientists from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Daiko Research Foundation (to A. N.).

This article contains supplemental Figs. S1–S17 and Movies 1 and 2.
The atomic coordinates and structure factors (code 4APW) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Alp
- actin-like protein
- GMP-PNP
- guanosine 5′-(β,γ-imino)triphosphate
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
REFERENCES
- 1. Carballido-Lopez R., Errington J. (2003) A dynamic bacterial cytoskeleton. Trends Cell Biol. 11, 577–583 [DOI] [PubMed] [Google Scholar]
- 2. van den Ent F., Amos L. A., Löwe J. (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413, 39–44 [DOI] [PubMed] [Google Scholar]
- 3. Popp D., Narita A., Maeda K., Fujisawa T., Ghoshdastider U., Iwasa M., Maéda Y., Robinson R. C. (2010) Filament structure, organization, and dynamics in MreB sheets. J. Biol. Chem. 285, 15858–15865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Popp D., Robinson R. C. (2011) Many ways to build an actin filament. Mol. Microbiol. 80, 300–308 [DOI] [PubMed] [Google Scholar]
- 5. Jensen R. B., Gerdes K. (1999) Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 18, 4076–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salje J., Zuber B., Löwe J. (2009) Electron cryomicroscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science 323, 509–512 [DOI] [PubMed] [Google Scholar]
- 7. Popp D., Xu W., Narita A., Brzoska A. J., Skurray R. A., Firth N., Ghoshdastider U., Maéda Y., Robinson R. C., Schumacher M. A. (2010) Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation. J. Biol. Chem. 285, 10130–10140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Polka J. K., Kollman J. M., Agard D. A., Mullins R. D. (2009) The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J. Bacteriol. 191, 6219–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Popp D., Narita A., Ghoshdastider U., Maeda K., Maéda Y., Oda T., Fujisawa T., Onishi H., Ito K., Robinson R. C. (2010) Polymeric structures and dynamic properties of the bacterial actin AlfA. J. Mol. Biol. 397, 1031–1041 [DOI] [PubMed] [Google Scholar]
- 10. van den Ent F., Møller-Jensen J., Amos L. A., Gerdes K., Löwe J. (2002) F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 21, 6935–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Derman A. I., Becker E. C., Truong B. D., Fujioka A., Tucey T. M., Erb M. L., Patterson P. C., Pogliano J. (2009) Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability, and treadmilling in Alp7A. Mol. Microbiol. 73, 534–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brüggemann H., Bäumer S., Fricke W. F., Wiezer A., Liesegang H., Decker I., Herzberg C., Martinez-Arias R., Merkl R., Henne A., Gottschalk G. (2003) The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. U.S.A. 100, 1316–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chumnarnsilpa S., Lee W. L., Nag S., Kannan B., Larsson M., Burtnick L. D., Robinson R. C. (2009) The crystal structure of the C terminus of adseverin reveals the actin-binding interface. Proc. Natl. Acad. Sci. U.S.A. 106, 13719–13724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cayley S., Lewis B. A., Guttman H. J., Record M. T. (1991) Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 222, 281–300 [DOI] [PubMed] [Google Scholar]
- 15. Webb M. R. (1992) A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuzmic P. (1996) Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 17. Kuzmic P. (2009) DYNAFIT: a software package for enzymology. Methods Enzymol. 467, 247–280 [DOI] [PubMed] [Google Scholar]
- 18. Zhao F. Q., Craig R. (2003) Capturing time-resolved changes in molecular structure by negative staining. J. Struct. Biol. 141, 43–52 [DOI] [PubMed] [Google Scholar]
- 19. Yasunaga T., Wakabayashi T. (1996) Extensible and object-oriented system EOS supplies a new environment for image analysis of electron micrographs of macromolecules. J. Struct. Biol. 116, 155–160 [DOI] [PubMed] [Google Scholar]
- 20. Popp D., Narita A., Oda T., Fujisawa T., Matsuo H., Nitanai Y., Iwasa M., Maeda K., Onishi H., Maéda Y. (2008) Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 27, 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Popp D., Yamamoto A., Iwasa M., Narita A., Maeda K., Maéda Y. (2007) Concerning the dynamic instability of actin homolog ParM. Biochem. Biophys. Res. Comm. 353, 109–114 [DOI] [PubMed] [Google Scholar]
- 22. Kuhn J. R., Pollard T. D. (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Popp D., Narita A., Iwasa M., Maéda Y., Robinson R. C. (2010) Molecular mechanism of bundle formation by the bacterial actin ParM. Biochem. Biophys. Res. Comm. 391, 1598–1603 [DOI] [PubMed] [Google Scholar]
- 24. Srinivasan R., Mishra M., Wu L., Yin Z., Balasubramanian M. K. (2008) The bacterial cell division protein FtsZ assembles into cytoplasmic rings in fission yeast. Genes Dev. 22, 1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stewart M. (1988) Computer image processing of electron micrographs of biological structures with helical symmetry. J. Electron Microsc. Tech. 9, 325–358 [DOI] [PubMed] [Google Scholar]
- 26. Narita A., Maéda Y. (2007) Molecular determination by electron microscopy of the actin filament end structure. J. Mol. Biol. 365, 480–501 [DOI] [PubMed] [Google Scholar]
- 27. Wriggers W., Birmanns S. (2001) Using Situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol. 133, 193–202 [DOI] [PubMed] [Google Scholar]
- 28. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Heel M., Schatz M. (2005) Fourier shell correlation threshold criteria. J. Struct. Biol. 151, 250–262 [DOI] [PubMed] [Google Scholar]
- 30. Oosawa F., Asakura S. (1975) Thermodynamics of the Polymerization of Protein, Academic Press, London [Google Scholar]
- 31. Garner E. C., Campbell C. S., Mullins R. D. (2004) Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 32. Nishida E., Sakai H. (1983) Kinetic analysis of actin polymerization. J. Biochem. 93, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 33. Melki R., Carlier M. F., Pantaloni D. (1988) Oscillations in microtubule polymerization: the rate of GTP regeneration on tubulin controls the period. EMBO J. 7, 2653–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mandelkow E. M., Mandelkow E. (1992) Microtubule oscillations. Cell Motil. Cytoskeleton 22, 235–244 [DOI] [PubMed] [Google Scholar]
- 35. Bean G. J., Amann K. (2008) Polymerization properties of the Thermotoga maritima actin MreB: roles of temperature, nucleotides, and ions. Biochemistry 47, 826–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galkin V. E., Orlova A., Rivera C., Mullins R. D., Egelman E. H. (2009) Structural polymorphism of the ParM filament and dynamic instability. Structure 17, 1253–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He Z. H., Chillingworth R. K., Brune M., Corrie J. E., Trentham D. R., Webb M. R., Ferenczi M. A. (1997) ATPase kinetics on activation of rabbit and frog permeabilized isometric muscle fibers: a real-time phosphate assay. J. Physiol. 501, 125–148 [DOI] [PMC free article] [PubMed] [Google Scholar]