Background: The kinase mTOR is a key regulator of cellular growth and differentiation.
Results: The deubiquitinase USP9X interacts with mTOR and modulates its activity in C2C12 cells.
Conclusion: USP9X negatively regulates mTOR activity and muscle differentiation.
Significance: Pharmacological inhibition of USP9X could promote mTOR activity and differentiation to reverse muscle atrophy.
Keywords: Mass Spectrometry (MS), mTOR, Protein Complexes, Protein-Protein Interactions, Proteomics, Muscle Differentiation, Tandem Affinity Purification, USP9X
Abstract
The mammalian target of rapamycin (mTOR) is an atypical serine/threonine kinase that responds to extracellular environment to regulate a number of cellular processes. These include cell growth, proliferation, and differentiation. Although both kinase-dependent and -independent functions of mTOR are known to be critical modulators of muscle cell differentiation and regeneration, the signaling mechanisms regulating mTOR activity during differentiation are still unclear. In this study we identify a novel mTOR interacting protein, the ubiquitin-specific protease USP9X, which acts as a negative regulator of mTOR activity and muscle differentiation. USP9X can co-immunoprecipitate mTOR with both Raptor and Rictor, components of mTOR complexes 1 and 2 (mTORC1 and -2), respectively, suggesting that it is present in both mTOR complexes. Knockdown of USP9X leads to increased mTORC1 activity in response to growth factor stimulation. Interestingly, upon initiation of differentiation of C2C12 mouse skeletal myoblasts, knockdown of USP9X increases mTORC2 activity. This increase in mTORC2 activity is accompanied by accelerated differentiation of myoblasts into myotubes. Taken together, our data describe the identification of the deubiquitinase USP9X as a novel mTORC1 and -2 binding partner that negatively regulates mTOR activity and skeletal muscle differentiation.
Introduction
Mammalian target of rapamycin (mTOR),4 a 300-kDa kinase, belongs to the family of phosphatidylinositol kinase-related kinases and is evolutionarily conserved from yeast to humans. In cells, mTOR exists in two distinct multiprotein complexes, mTOR complex 1 (mTORC1) and 2 (mTORC2), defined by the presence of unique binding partners, regulatory-associated protein of mTOR (Raptor) and rapamycin-insensitive companion of mTOR (Rictor), respectively (1, 2). Raptor and Rictor are scaffold proteins that facilitate mTOR binding with its regulatory interactors and downstream substrates. Well established functions of mTORC1 include the regulation of ribosomal biogenesis, translation, transcription, stress response, and autophagy (3). mTORC2 is involved in actin organization in cells (3, 4). Although mTORC1 is the primary target inhibited by the drug rapamycin, prolonged treatment of cells with the drug also results in the inhibition of mTORC2 (5). The inhibition of mTORC2 may be a secondary effect brought about by sequestration of mTOR in a complex with rapamycin-FKBP12, thereby limiting the pool of mTOR available to form mTORC2.
The mTOR signaling pathway is regulated by growth factors, nutrients, cellular energy, and stress (3, 6). In response to these various upstream inputs, mTOR exerts its functions through regulation of a number of downstream targets. Two of the most well characterized targets of mTORC1 in mammalian cells are ribosomal protein S6 kinase 1 (S6K1) and eIF4E-binding protein (7). Activated S6K1 phosphorylates the 40 S ribosomal subunit S6 and promotes mRNA translation. The mTOR substrate eIF4E-binding protein is critical for mTOR mediated regulation of cap-dependent translation (8). Upon activation, mTORC2 can phosphorylate a number of substrates including Akt. Akt also acts upstream of mTOR signaling, but mTORC2-mediated Akt phosphorylation at Ser-473 is required for full activation of Akt kinase functions (9).
Given its central role in integrating growth factor and nutrient status with cell growth and metabolism, it is not surprising that mTOR is a critical regulator of muscle cell differentiation and regeneration (10). Rapamycin has been shown to inhibit muscle hypertrophy and regrowth of myofibers after denervation (11, 12). Whether the rapamycin-induced reduction in hypertrophy is mediated by inhibition of mTORC1, mTORC2, or both remains controversial. Independent studies have shown that genetic ablation of mTORC1 or mTORC2 activity by deletion of Raptor or Rictor, respectively, reduces the differentiation potential of myoblasts (13, 14). Besides mTOR kinase-dependent activities, kinase-independent functions of mTOR may also be important for early steps of muscle differentiation (11, 15). In addition, mTOR has been shown to modulate myogenesis by transcriptionally regulating insulin-like growth factor II expression (16). However, despite the wealth of information available on mTOR signaling, we still do not have a complete understanding of the proteins regulating this pathway, particularly during skeletal muscle differentiation. Identification of novel mTOR interactors will provide further insight into the modulation of biological role of mTOR in muscle cell differentiation.
In this report we identify and characterize the interaction between mTOR and its novel binding protein, the deubiquitinase enzyme USP9X (also known as fat facet in mouse (FAM)). To discover mTOR binding partners, we employed tandem affinity purification to isolate mTOR complexes and used mass spectrometry to determine the identity of interacting proteins. Using this approach, we identified several known interactors of mTOR as well as we isolated USP9X as a novel mTOR interactor. USP9X plays an essential role in development and can regulate cell adhesion and polarity and promote cell proliferation by deubiquitinating and stabilizing its substrates (17–21). USP9X mRNA and protein levels are elevated in undifferentiated human and mouse stem cells, and modest increases in USP9X can promote the self-renewing ability of progenitor cells (22–24). We show that USP9X interacts with both mTORC1 and mTORC2 and negatively regulates mTOR activity and differentiation. Co-purification of MAGED1, a known regulator of muscle and neuronal differentiation, in a tandem affinity-purified USP9X protein complex further supports the role of USP9X in modulation of muscle differentiation. Although USP9X does not seem to modulate mTOR protein levels, reduction of USP9X levels promotes mTOR functions in C2C12 myoblasts. Under growth factor starvation and stimulation, USP9X knockdown increases mTORC1 activity. However, upon differentiation, reducing USP9X levels results in higher mTORC2 activity. The increase in mTORC2 activity observed upon low USP9X levels is accompanied by accelerated differentiation of C2C12 myoblasts into myotubes.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293T cells (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM, Cellgro) supplemented with 10% heat inactivated fetal bovine serum (FBS, Invitrogen). C2C12 myoblasts (ATCC) were grown in DMEM supplemented with 10% fetal bovine serum (FBS, Cellgro). Cells were grown under 5% CO2 at 37 °C.
Antibodies and Reagents
mTOR, Raptor, Rictor, phospho-S6 (Ser-235/236), total-S6, phospho-Akt (Ser-473), and total-Akt antibodies were from Cell Signaling. Other antibodies were polyclonal anti-USP9X (Abcam), monoclonal anti-USP9X (Santa Cruz), HRP-conjugated anti-actin antibody (Sigma), anti-V5 (Invitrogen), HRP-conjugated secondary antibodies (GE Healthcare), and Alexa 488- and 555-conjugated antibodies (Molecular Probes). The myogenin (MyoG) antibody developed by Woodring E. Wrigth and the myosin heavy chain antibody developed by Donald A. Fischman were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA. For proteolysis, sequencing grade, modified trypsin (porcine) was purchased from Promega (Madison, WI). Control siRNAs and those targeting USP9X and mTOR were purchased from Dharmacon, now part of Thermo Fisher Scientific. Protein chemistry reagents including iodoacetamide and dithiothreitol (DTT) were obtained from Sigma. HPLC solvents were obtained from Burdick & Jackson (Muskegon, MI).
Constructs
Tandem affinity purification (TAP)-tag constructs were obtained from Euroscarf (25). Human mTOR cDNA was a kind gift from Michael Hall (University of Basel, Switzerland). mTOR was PCR-amplified and cloned into pENTR-DTOPO Gateway entry clone (Invitrogen) according to the manufacturer's instructions. mTOR was PCR-amplified using the long range amplification kit (Qiagen) with the following primers: forward primer, 5′-CACCATGCTTGGAACCGGACCTGC-3′; reverse primer, 5′-TTACCAGAAAGGGCACCAGC-3′. N-terminal TAP-tagged mTOR was generated by performing an attL × attR recombination (LR) reaction between the mTOR entry clone and TAP-tag destination vector as recommended by manufacturer (Invitrogen). USP9X cDNA clones were kindly provided by Stephan Wood (Griffith University, Australia). TAP-tagged USP9X was generated using the same protocol as mTOR, except the TAP-tag was fused to the C terminus of USP9X.
DNA and siRNA Transfection
HEK293T cells were grown to 80% confluency in antibiotic-free media and transfected with plasmid DNA using Lipofectamine 200 (Invitrogen) as per the manufacturer's protocol. Transfection of C2C12 cells with siRNA was done using Lipofectamine 200. A day before transfection, 800,000 low passage C2C12 cells were plated in a 10-cm dish in the absence of antibiotics. The transfection mix was made by combining 133 μl of 20 μm siRNA with 156 μl of Lipofectamine 200 in 2.5 ml of serum free media and incubated at room temperature for 20 min. The transfection mix was then added to the cells dropwise. Five hours after adding the transfection mix, the media on cells was replaced with fresh growth media (without penicillin/streptomycin). Cells were then used for mTOR cell-based assays as described below. For overexpression, 67 μg of either empty vector control or V5-USP9X cDNA vector and 168 μl of Lipofectamine 200 were used for transfection of C2C12 myoblasts in a 10-cm dish. Transfection was carried out as discussed for siRNA. 48 h after transfection, cells were plated in media containing 10 μg/ml blasticidin. The cells were drug selected for at least 10 days before using for further experiments.
Tandem Affinity Purification
HEK293T cells were transiently transfected with either TAP-tag or TAP-mTOR constructs in five 10-cm dishes each. 48 h after transfection, each plate was lysed in 1 ml of lysis buffer: 40 mm HEPES, 120 mm NaCl, 2 mm EDTA, 0.3% CHAPS, with protease and phosphatase inhibitors. Lysis was carried out for 30 min on ice with Dounce homogenization. Cell lysates were cleared of debris by high speed spin on a tabletop centrifuge for 30 min at 4 °C, and proteins were estimated using bicinchoninic acid (BCA) kit (Pierce). 15–17 mg of cell lysate was used for tandem affinity purification. The first round of purification was done on IgG beads (Sigma) equilibrated in lysis buffer for 2 h at 4 °C with gentle rotation. 25 μl of bead slurry was used per mg of protein. The beads were spun down and washed twice in lysis buffer followed by 2 washes in TEV protease reaction buffer: 40 mm HEPES, 120 mm NaCl, 2 mm EDTA, 0.1% CHAPS without protease and phosphatase inhibitor. The beads were then incubated with TEV protease (Invitrogen), and the reaction was carried out overnight at 4 °C. After TEV reaction, the beads were spun down, the supernatant was collected, and its volume was measured. The beads were then washed with the TEV protease reaction buffer using the 1× volume as the TEV supernatant. The wash and the TEV supernatant were pooled, and the second round of purification was done using streptavidin beads (Pierce), 15 μl of bead slurry/mg of starting protein concentration. Before use, streptavidin beads were equilibrated with TEV protease reaction buffer. Streptavidin pulldown was performed at 4 °C for 2 h with gentle rotation. The beads were spun down and washed four times with TEV protease reaction buffer, and the purified complexes were eluted twice using 5 mm d-biotin at 4 °C; first elution was done for 30 min, and the second elution was for 15 min. The eluted samples were pooled together and used for further analysis. A fraction of sample from each step of purification was saved to test the efficiency of tandem affinity purification. For TAP-USP9X purification, the conditions were similar to that used for TAP-mTOR, except mild lysis buffer (10 mm Tris, pH 7.5, 100 mm NaCl, 0.02% Triton X-100, with protease and phosphatase inhibitors) was used for cells lysis and during purification. In all cases cells expressing the TAP-tag alone served as a negative control to identify nonspecific binding proteins (supplemental Table S1B).
Cell-based Assay to Monitor in Vivo mTOR Activity
C2C12 cells were transfected with either control or USP9X or mTOR siRNA (Dharmacon) as described above. 24 h after transfection, cells were split into 24-well plates and allowed to recover for a day. After recovery, cell were washed twice with phosphate buffered saline (PBS) and growth factor-starved in media containing 0.1% FBS. After 24 h of starvation, cells were stimulated with media containing 10% FBS and 100 nm insulin for 5 or 15 min. Starved and stimulated cell samples were collected in LDS buffer (Invitrogen), DTT was added to a final concentration of 100 mm, and samples were boiled for 5 min. The samples were analyzed for mTOR activity by measuring the phosphorylation state of S6 and Akt using Western blotting.
Western Blotting and Immunoprecipitation (IP)
Protein samples were boiled in 1× LDS sample buffer (Invitrogen), and samples were run on 4–12% NuPAGE gradient gels (Invitrogen) using MOPS running buffer (Invitrogen). The proteins were then transferred to nitrocellulose membrane (Pall Life Sciences), blocked in 5% nonfat dry milk made up in TBST (Tris-buffered saline containing 0.1% of Tween 20). Western blot analysis for various proteins was performed according to manufacturer's recommendations. For co-IP, HA-mTOR or V5-USP9X was transiently expressed in HEK293T cells. 48 h after transfection, cells were lysed using mild lysis buffer: 10 mm Tris, pH 7.5, 100 mm NaCl, 0.02% Triton X-100, with protease and phosphatase inhibitors. Cell lysate was cleared of debris by spinning the sample at 16,000 × g for 15 min at 4 °C, and protein estimation was performed using BCA assay kit (Pierce). Equivalent amounts of protein were precleared using equilibrated protein G-agarose beads (GE Healthcare) for 1 h at 4 °C with rotation; 50 μl of bead slurry was used per mg of protein lysate. The beads were spun down, and the supernatant was used for IP experiments. For IP, cleared protein lysates were incubated with antibody against the protein being immunoprecipitated for 2 h at 4 °C with rotation, after which equilibrated protein G-agarose was added, and IP was continued for an additional 1 h. Finally, the beads were spun down and washed four times using the lysis buffer. USP9X IPs from C2C12 cells was performed using polyclonal USP9X antibody under the same IP conditions as that used for V5-USP9X IP. For USP9X IPs under differentiation, cells were either undifferentiated or differentiated for 3 or 7 days as discussed below, and USP9X IPs were performed. Immunoprecipitated samples were run on gels and proteins detected as described.
Immunofluorescence
C2C12 cells were plated in an 8-well chamber slide at 5000 cells per well. A day after plating, cells were fixed in 4% paraformaldehyde for 15 min at room temperature, washed 3 times with PBS, and permeabilized using 0.2% Triton X-100 in PBS for 15 min at room temperature. Cells were washed 4 times with PBS, blocked with 5% normal goat serum in PBS for 30 min at room temperature, and then incubated with primary antibodies overnight at 4 °C. For co-localization of USP9X and mTOR, mouse monoclonal USP9X antibody (Santa Cruz) at a 1–50 dilution and rabbit monoclonal mTOR antibody (Cell Signaling) at a 1–200 dilution in blocking buffer was used. After primary antibody incubation, cells were washed 4 times with PBS and incubated with goat anti-mouse Alexa 488 and goat anti-rabbit Alexa 555 antibodies at 1–200 dilution in blocking buffer. Secondary incubation was done at room temperature for 1 h followed by 4 PBS washes. Finally, cells were mounted using ProLong Gold antifade reagent with DAPI (Invitrogen) and 0.16–0.19-mm-thick coverslips (VWR). Images were taken using confocal microscope Zeiss LSM 510 with a 60× objective under oil immersion. The images were further processed using Bitplane Imaris 7.0 software and then compiled using Adobe Illustrator CS4. For MyoG and myosin heavy chain (MHC) immunofluorescence, C2C12 cells were plated in an 8-well chamber slide at 20,000 cells per well 24 h after transfection with siRNA and differentiated as described below. MyoG and MHC immunofluorescence were done similar to USP9X, except the experiment was done using donkey serum and the primary antibodies were used at 1–100 dilution. Donkey secondary anti-mouse Alexa 488 was used at a 1–500 dilution. The experiment was done in triplicate, and three images per replicate (a total of nine images per condition) were counted for analysis. MyoG and total nuclei was spot counted with Bitplane Imaris 7.0 software. Nuclei in MHC-positive cells were identified manually, and the count was automated using Bitplane Imaris 7.0 software.
C2C12 Cell Differentiation
C2C12 myoblasts were transfected with control or USP9X siRNA (Dharmacon) as described above and split into 24-well plates a day after transfection. Cells were allowed to recover for a day and then changed to differentiation media, which was DMEM supplemented with 2% heat inactivated horse serum. Cell samples for both conditions were collected from the start of differentiation in 24-h intervals using radio immunoprecipitation lysis buffer: 50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS containing protease and phosphatase inhibitors. Before lysis, cells were washed with PBS and then scraped in radio immunoprecipitation assay buffer and saved at −80 °C. At the end of the differentiation experiment, radio immunoprecipitation assay lysates were thawed on ice and lysed additionally for 30 min at 4 °C with gentle rotation. Cell debris was removed by spinning at 16,000 × g for 15 min at 4 °C, and protein concentration was determined using a BCA kit (Pierce). Equivalent protein amounts were run on SDS-PAGE gel and Western-blotted. Protein band intensity was quantitated using ImageQuant.
In-gel Digestion and Mass Spectrometry Analysis
Protein samples were boiled in 1× LDS sample buffer (Invitrogen), and samples were separated by one-dimensional SDS-PAGE on 4–12% NuPAGE gradient gels (Invitrogen) using MOPS running buffer (Invitrogen). The gel was stained with Sypro Ruby (Invitrogen), and gel bands were manually excised and trypsin-digested as follows. Gel spots were destained and dehydrated with acetonitrile. Proteins were reduced with 10 mm DTT at 56 °C for 60 min, alkylated with 55 mm iodoacetamide (37 °C, 45 min), and then incubated with 250 ng of sequencing grade trypsin (Promega) at 37 °C overnight. The resulting tryptic peptides were extracted from the gel with 50% acetonitrile, 5% formic acid. Samples were concentrated under vacuum and resuspended in 5% acetonitrile, 5% formic acid. Peptide mixtures were separated using an Eksigent nano-LC 2D HPLC system (Eksigent, Dublin, CA) and analyzed by electrospray ionization with a quadrupole time-of-flight mass spectrometer (QSTAR Elite, ABSCIEX, Concord, Canada) operating in the positive ion mode. Briefly, peptides were applied to a guard column (C18 Acclaim PepMap100, 300-μm inner diameter × 5 mm, 5-μm particle size, 100 Å pore size, Dionex, Sunnyvale, CA) and washed with the aqueous loading solvent (2% solvent B in A, flow rate: 20 μl/min) for 10 min. Subsequently, samples were transferred onto the analytical C18-nanocapillary HPLC column (C18 Acclaim PepMap100, 75-μm inner diameter × 15 cm, 3-μm particle size, 100 Å pore size, Dionex) and eluted at a flow rate of 300 nl/min either using Gradient 1 (2–40% solvent B in A (from 0 to 35 min), 40–80% solvent B in A (from 35 to 45 min), and at 80% solvent B in A (from 45 to 55 min), with a total runtime of 85 min (including mobile phase equilibration)) or using a slightly longer Gradient 2 (2–40% solvent B in A (from 0 to 45 min), 40–80% solvent B in A (from 45 to 60 min), and at 80% solvent B in A (from 60 to 70 min), with a total runtime of 105 min (including column equilibration)). Solvents were prepared as follows: mobile phase A, 2% acetonitrile, 98% of 0.1% formic acid (v/v) in water; mobile phase B, 98% acetonitrile, 2% of 0.1% formic acid (v/v) in water. Mass spectra and tandem mass spectra (MS/MS) were recorded in positive-ion mode with a resolution of 12,000–15,000 full width at half maximum. The nanospray needle voltage was typically 2300 V in HPLC-MS mode. Spectra were calibrated in static nanospray mode using MS/MS fragment ions of a Glu-fibrinogen B peptide standard (y1 fragment ion with m/z at 175.1195 and y11 fragment ion with m/z at 1285.5444). For collision-induced dissociation tandem mass spectrometry, the mass window for precursor ion selection of the quadrupole mass analyzer was set to ±1 m/z. The precursor ions were fragmented in a collision cell using nitrogen. Advanced information-dependent acquisition was employed for MS/MS data collection using the QSTAR Elite (Analyst QS 2.0)-specific features, including “Smart Collision” and “Smart Exit” (either with the fragment intensity multiplier set to 4 and maximum accumulation time at 2.5 s or for weaker samples with the fragment intensity multiplier set to 8 and maximum accumulation time at 3 s) to obtain MS/MS spectra for the six most abundant precursor ions after each survey scan. Dynamic exclusion features, based on value M, were set to a width of 50 mDa and exclusion duration of 120 s.
Bioinformatics Data Base Searches and Analysis of Mass Spectrometric Data
Protein Pilot 4.1 beta software, Version 4.1.46 (revision 460) with the Paragon Algorithm 4.0.0.0.459 developed by AB Sciex (26), was used to perform mass spectrometric data base searches. Peaklists for the QSTAR Elite LC-MS/MS data sets were generated using Protein Pilot 4.1.46 (revision 460). For Protein Pilot searches the following search parameters were used: trypsin enzyme specificity, carbamidomethyl (Cys) as a fixed modification, “thorough search effort setting” allowing for “biological modifications.” In contrast to other search engines, the “thorough” mode of Paragon Algorithm determines modifications using feature probabilities based on the specified settings (26). The mass tolerance for the QSTAR Elite data was generally considered <50 ppm on the precursor and the fragment ion levels. All data were searched using the publicly available SwissProt data base (SwissProt Homo sapiens, total sequence entries 20,239, “UniProt release 2010_05,” retrieved May, 2011). To assess false discovery rates and restrict the rates of false positive peptide/protein identifications, we used the Proteomics System Performance Evaluation Pipeline (PSPEP) tool available in ProteinPilot 4.1. This tool automatically creates a concatenated forward and reverse decoy data base and provides an Excel output of the experimentally determined false discovery rate at the spectral, peptide, and protein levels with standard statistical errors (26). The discriminating variable for the Paragon algorithm is the peptide confidence value, which is a 0–99-scaled real number (26). For all mTOR affinity experiments and corresponding data base searches, a cut-off peptide confidence value of 96 was chosen for which the Protein Pilot false discovery rate analysis tool (PSPEP) algorithm (26) provided a global false discovery rate of 1% and a local false discovery rate at 5% in all cases. Typically, a minimum of two detected peptides was required for the identification of a protein (see supplemental Tables S1, A, B, and D). In the few cases where only one unique peptide per protein was identified, the MS/MS spectrum was inspected “manually” (see supplemental Fig. S9) based on an adaptation of previously published criteria (27). Proteins with only one observed peptide were manually validated and added to the list only if the following criteria were satisfied: (a) a correct precursor ion charge state, (b) abundant fragment ions assigned (not more than 1 unassigned fragment ion over 50% of base peak intensity), (c) good signal to noise (signal to noise ratio of >3 for the majority of fragment ions), (d) a minimum of three consecutive y- or b-ions, (e) any y-ion with an N-terminal proline must be intense, and (f) an immonium ion for Arg (m/z 175.12) or Lys (m/z 147.11) present that is consistent with the expected C-terminal tryptic site.
RESULTS
Tandem Affinity Purification of in Vivo mTOR Complexes
A TAP-tag comprising protein G and streptavidin-binding peptide epitopes separated by the TEV protease cleavage site was fused to the N terminus of mTOR cDNA. Because epitope fusion to protein can lead to disruption of its function, we tested the TAP-mTOR protein for its kinase activity toward its substrate, S6K1. TAP-mTOR or the TAP-tag alone was transiently expressed in HEK293T cells, and purification was performed using IgG beads that bound to the protein G epitope on the TAP-tag. Purified complexes were then used for in vitro kinase activity on mTOR substrate S6K1. As shown in Fig. 1A, the TAP-mTOR fusion protein has kinase activity.
FIGURE 1.
Tandem affinity purification of TAP-mTOR complexes. A, kinase activity of TAP-mTOR protein is shown. TAP-tag and TAP-mTOR were immunoprecipitated using IgG beads, and in vitro kinase activity was measured using S6K1 as a substrate. B, Western blot (WB) analysis of TAP-mTOR purification is shown. TAP-tag (left panel) or TAP-mTOR (right panel) was transiently expressed in HEK293T cells, and tandem purification was performed using the protocol as described under “Experimental Procedures.” Samples were analyzed after TEV protease treatment (1st Step) and after biotin elution (2nd Step) to determine the efficiency of purification. Western blot analysis using anti-mTOR antibody (upper panel) and anti-raptor antibody (lower panel) were performed to monitor the purification of mTOR complexes. Relative loading of various fractions are shown by the numbers below the blot. C, tandem purification of TAP-mTOR complexes is shown. TAP-tag and TAP-mTOR complexes were purified as in B, and the final elutes were run on SDS-PAGE gels and analyzed using SYPRO Ruby staining. TAP-mTOR, USP9X, Raptor, Rictor, and TTI1 bands, as indicated by arrows, were identified using mass spectrometry analysis of protein bands after in-gel trypsin digestion. * indicates a nonspecific protein band.
It has been shown that the integrity of mTOR complexes is sensitive to the detergents used during extraction/purification (28). Therefore, as a first step, we established a tandem purification condition using CHAPs detergent for lysis. Under this condition we were able to retain mTOR complexes intact, at least with respect to its known interactor, Raptor (Fig. 1B). The purification of TAP-mTOR complexes was specific, as TAP-tag samples did not show mTOR or Raptor after both the first and second round of purification. Using this condition we then performed a large-scale purification of TAP-mTOR complexes from transiently transfected cells, separated the proteins present in the complex by one-dimensional-SDS-PAGE, and visualized using SYPRO Ruby staining (Fig. 1C). The identity of proteins present in the complex was determined by mass spectrometry. As seen in Fig. 1C TAP-mTOR lane, the most-abundant band detected was that of TAP-mTOR itself, which displayed high sequence coverage (supplemental Fig. S1A). This band was absent in the TAP-tag sample. Raptor and Rictor, components of mTORC1 and mTORC2, respectively, were detected in most of the purifications. We also detected RuvBL1 and RuvBL2, members of ATPases associated with diverse cellular functions (AAA+) and TTI1, a Tel2 interacting protein 1 homolog that interact with mTOR and regulates its levels and complex stability (29, 30). The identification of known interacting proteins in TAP-mTOR purified sample demonstrates the specificity of tandem purification for isolation of in vivo mTOR interactors. In addition, a 300-kDa deubiquitinase enzyme, USP9X, was detected as a novel mTOR interactor in our affinity purification. Tandem mass spectra of USP9X peptides are shown in supplemental Fig. S1, B and C. The specific mTOR interacting proteins that were identified by mass spectrometry are summarized in Table 1 (additional MS data are summarized in supplemental Tables S1, A and B).
TABLE 1.
Specific proteins identified in mTOR affinity experiments
| Protein scorea | Seq. Cov.b | SwissProt accession no. | Gene name | Protein name | Unique peptidesc | Unique precursorsc |
|---|---|---|---|---|---|---|
| % | ||||||
| 163.4 | 35.8 | P42345 | MTOR | Serine/threonine-protein kinase mTOR | 83 | 100 |
| 24.1 | 13.2 | Q8N122 | RPTOR | Regulatory-associated protein of mTOR | 14 | 17 |
| 8.01 | 4.0 | O43156 | TTI1 | TEL2-interacting protein 1 homolog | 4 | 4 |
| 4.91 | 7.5 | Q9Y265 | RUVB1 | RuvB-like 1 | 3 | 3 |
| 4.75 | 0.9 | Q93008 | USP9X | Ubiquitin carboxyl-term. hydrolase FAF-X/USP9X | 2 | 2 |
| 2.24 | 4.3 | Q9Y230 | RUVB2 | RuvB-like 2 | 2 | 2 |
| 0.99 | 0.5 | Q6R327 | RICTR | Rapamycin-insensitive companion of mTOR | 1 | 1 |
a Protein Pilot 4.1 database search score according to Shilov et al. (26).
b Amino acid sequence coverage (Seq. Cov.) of proteins in %.
c A peptide confidence score cutoff of 96 was applied.
USP9X Co-immunoprecipitates mTOR, Raptor, and Rictor
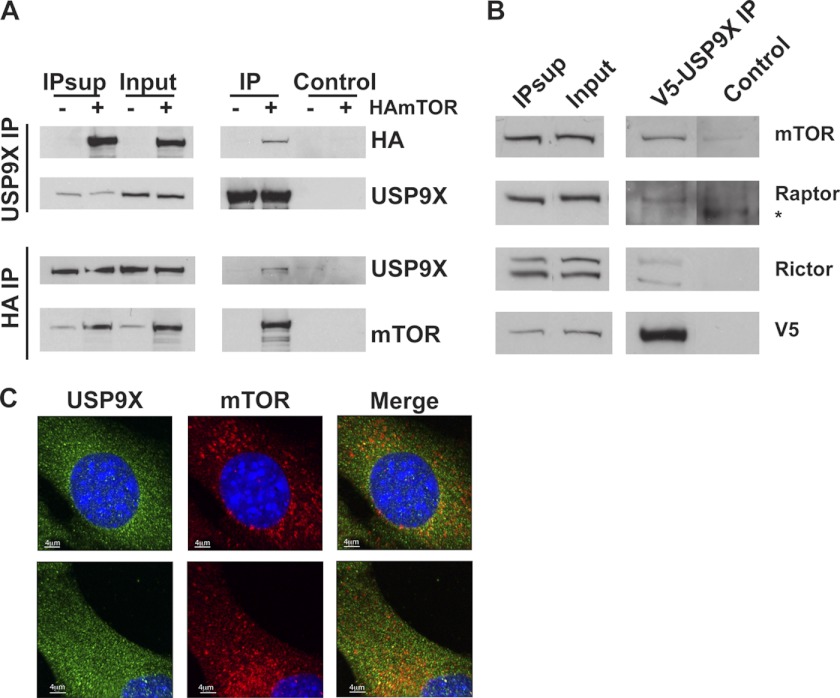
To further verify the interaction of USP9X and mTOR, endogenous USP9X IP from HEK293T cells overexpressing HA-tagged mTOR was performed. As shown in Fig. 2, A, top panels, USP9X can co-IP HAmTOR. The reverse, i.e. co-IP of endogenous USP9X with HAmTOR, was also observed (Fig. 2A, bottom panels). To determine whether USP9X can interact with mTORC1 or mTORC2 or both, V5-tagged USP9X was transiently expressed in HEK293T cells, and V5 IP was performed. V5-USP9X was able to co-IP endogenous mTOR, Raptor, and Rictor, suggesting that USP9X can interact with mTORC1 and mTORC2, respectively (Fig. 2B). However, the interaction of USP9X with endogenous mTOR and its interactor was relatively weak, as only a small fraction of mTOR/Raptor/Rictor was co-immunoprecipitated with V5-USP9X. Consistent with this, only a small percentage of cellular mTOR co-localized with USP9X in perinuclear regions (Fig. 2C).
FIGURE 2.
USP9X interacts with mTOR, Raptor, and Rictor. A, co-immunoprecipitation of mTOR and USP9X is shown. Hemagglutinin (HA)-tagged mTOR was transiently expressed in HEK293T cells, and USP9X (top panels) and HAmTOR (bottom panels) were immunoprecipitated (IP) followed by HA, USP9X, and mTOR Western blot analysis as shown. B, shown is co-immunoprecipitation of mTOR, Raptor, and Rictor with USP9X. V5-tagged USP9X was transiently expressed in HEK293T cells, and V5 IP was performed as in A. V5 IP was then analyzed using antibodies against mTOR, Raptor, Rictor, and V5 antibodies. Control lanes in A and B show pulldown using Protein G beads only. 5% of input and IP supernatants (IP Sup) and 100% of IPs and control were used for Western blot analyses. * indicates a nonspecific band recognized by anti-Raptor antibody. C, Co-localization of mTOR and USP9X is shown. Exponentially growing C2C12 cells were fixed with paraformaldehyde and permeabilized with Triton X-100. Immunofluorescence was performed using primary antibodies against endogenous USP9X and mTOR and fluorescently labeled secondary antibodies. Two different representative images are shown (top and bottom panel).
USP9X Is Negative Regulator of mTORC1 Activity in Myoblasts
To analyze the effects of USP9X interaction on mTORC1 and mTORC2 activity, we assayed the phosphorylation status of S6K1 and Akt, respectively. As phosphorylation of S6K1 by mTOR leads to its activation and S6 phosphorylation at Ser-235/236, phospho-S6 was used to measure mTOR-mediated S6K1 activation. Because of their responsiveness to changes in external serum and insulin levels, we decided to use C2C12 myoblasts to assess the in vivo phosphorylation state of mTOR substrates. USP9X levels were knocked down in C2C12 myoblasts followed by serum starvation for 24 h to reduce the basal level of S6 and Akt phosphorylation. The cells were then stimulated for varying lengths of time, and phosphoprotein and total protein levels of S6 and Akt were analyzed. As shown in Fig. 3, A and B, knockdown of USP9X caused a robust increase in S6 phosphorylation. This increase in S6 phosphorylation was observed even under starved condition and remained elevated at all time points measured upon stimulation (Fig. 3B). This result suggests that USP9X is a negative regulator of mTORC1 activity. Interestingly, no significant change in Akt phosphorylation was detected upon USP9X knockdown (Fig. 3, C and D). siRNA-mediated knockdown of USP9X was efficient, as its protein levels remained low during the course of the experiment (Fig. 3E). Effects observed on mTOR activity upon USP9X knockdown using siRNA pool (Fig. 3) were reproduced using two individual duplexes targeting USP9X (supplemental Fig. S2). As prolonged starvation can initiate differentiation of C2C12 cells, we also analyzed the effects of USP9X knockdown with only a 5-h serum starvation followed by serum and insulin stimulation. Under this condition, the effects of knockdown on mTORC1 and mTORC2 activity were similar to that observed upon 24 h of starvation followed by stimulation (supplemental Fig. S3). Therefore, irrespective of the duration of starvation, the data indicate that although USP9X can interact with both mTORC1 and mTORC2, it appears to only regulate mTORC1 kinase activity under these conditions.
FIGURE 3.
USP9X knockdown increases S6 phosphorylation but not Akt phosphorylation. A, B, C, and D, USP9X levels were knocked down using small interfering RNA (siRNA) in mouse skeletal muscle C2C12 cells. To measure mTOR kinase activity toward its substrates S6K1 and Akt in vivo, transfected C2C12 cells were deprived of serum for 24 h followed by stimulation with media containing insulin and serum for the indicated times. As phosphorylation of S6K by mTOR leads to its activation and S6 phosphorylation at Ser-235/236, phospho-S6 was used to measure mTOR mediated S6K activation. Phospho-S6 Ser-235/236, total S6, phospho-Akt Ser-473, and total Akt levels were detected using Western blot analysis, and images were quantitated using ImageQuant. Phospho-S6 and Akt levels were normalized to total S6 and Akt levels, respectively. A single representative Western blot (top panel) and its quantitation (bottom panel) are shown in A and C. Quantitation of phospho-S6 and Akt levels for three biological replicates at various time points (0, 5, and 15 min) are shown in B and D, respectively. E, siRNA-mediated knockdown of USP9X relative to control siRNA was measured by USP9X Western blot. An actin Western blot is shown for relative loading of the samples. * represents p value < 0.05, ** represents p value < 0.005, and *** represents p value < 0.0005.
Tandem Affinity Purification of in Vivo USP9X Complexes
To gain greater insight into the functions of USP9X, we performed tandem affinity purification of USP9X complexes and identified the bound proteins using mass spectrometry. Purification of TAP-USP9X was specific, as the most abundant band in the sample was that of USP9X (sequence coverage shown in supplemental Fig. S1D). The USP9X band was absent in samples from cells expressing TAP-tag alone (Fig. 4). MARCH7, an ubiquitin ligase and a known USP9X-binding protein was identified with high confidence in the TAP-USP9X complex (Fig. 4 and supplemental Table S1, C and D) (31). The mTOR interactor RuvBL1 was also present in the USP9X pulldown. Interestingly, USP9X co-purified a protein complex comprised of MAGED1, a known regulator of muscle and neuronal differentiation, and its ubiquitin ligase Praja1 (32). Praja1 also interacts with USP9X in a yeast two-hybrid analysis.5 These observations suggested that the interaction of USP9X and mTOR might have roles in muscle differentiation.
FIGURE 4.
Tandem purification of TAP-USP9X complexes. TAP-tag and TAP-USP9X complexes were purified from HEK293T cells, and the final elutes were run on SDS-PAGE gels and analyzed using SYPRO Ruby staining. USP9X, MAGED4, MAGED1, Praja1, MARCH7, and RuvBL1 bands, as indicated by arrows, were identified by mass spectrometry (details of all the proteins identified by mass spectrometry are shown in supplemental Fig. S1E). * indicates nonspecific protein band; ** indicates TAP-tag.
USP9X Knockdown Accelerates Muscle Cell Differentiation
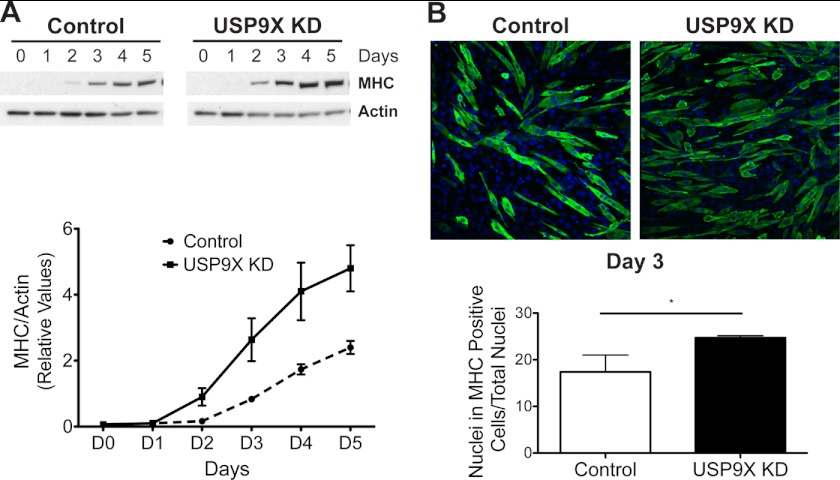
Interaction of USP9X with MAGED1, a protein involved in muscle differentiation, along with observed effects of USP9X knockdown on mTOR activity prompted us to examine the role of USP9X in modulation of cell differentiation. 48 h after siRNA transfection, C2C12 cells were transferred to differentiation media, and protein samples were collected for Western blot analysis every day for 5 days (outlined in supplemental Fig. S4). As seen in Fig. 5A levels of transcription factor MyoG, a marker for muscle differentiation, were detectable by Day1 in both control and USP9X siRNA-transfected cells. However, MyoG levels were higher in cells where USP9X levels had been reduced (Fig. 5A). We also monitored the nuclear localization of MyoG in differentiating cells by immunofluorescence (Fig. 5B). A higher percentage of cells showed nuclear staining for MyoG at day 2 of differentiation upon USP9X knockdown relative to control cells, and this increase was statistically significant (Fig. 5B). To further evaluate the effect of reducing USP9X levels on C2C12 differentiation, we performed Western blot and immunofluorescence analysis for another differentiation marker, MHC. MHC levels were detected by Western blot starting at day 2 of differentiation. Similar to MyoG, MHC levels were elevated in USP9X knocked down cells at all days during differentiation (Fig. 6A). In addition, MHC immunofluorescence at day 3 of differentiation showed that a higher number of cells stained positive for MHC after USP9X knockdown. Differentiation index, i.e. percentage of nuclei present in MHC-positive cells relative to total nuclei was calculated and showed a significant increase upon USP9X knockdown (Fig. 6B, bottom). It is important to point out that USP9X levels remained low in USP9X siRNA-transfected cells compared with the control for the entire duration of the experiment (supplemental Fig. S4).
FIGURE 5.
Knock down of USP9X elevates MyoG levels. Cells were transfected with either control or USP9X siRNA and 48 h later transferred to differentiation media (day 0). Cells were imaged, and protein samples were collected every 24 h starting at day 0 (days 0 to 5). A, differentiation marker MyoG expression in transfected cells was analyzed using Western blot analysis (top panel). Actin was used as the loading control. Changes in MyoG levels during differentiation of control or USP9X siRNA-transfected cells were quantitated and plotted (bottom). B, MyoG was visualized via immunofluorescence at day 2 of differentiation. Quantitation of MyoG-positive nuclei relative to total nuclei (stained with DAPI) is shown. MyoG is in green, and DAPI is in blue. ** represents p value < 0.005.
FIGURE 6.
Knock down of USP9X results in elevated levels of MHC. A, differentiation samples from Fig. 5 were analyzed for the expression of MHC marker using Western blot analysis (top panel). Actin was used as the loading control. Changes in MHC levels during differentiation of control or USP9X siRNA-transfected cells were quantitated and plotted (bottom). B, MHC was visualized via immunofluorescence at day 3 of differentiation. Differentiation index, percentage of nuclei in MHC-positive cells relative to total nuclei (stained with DAPI) is shown (bottom). MHC is in green, and DAPI is in blue. * represents p value < 0.05.
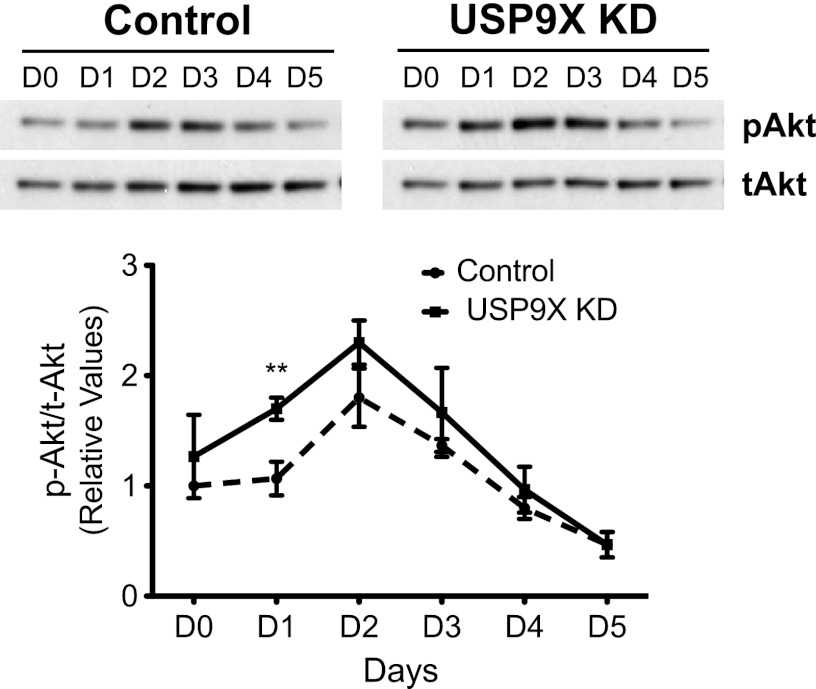
As mTOR activity is essential for muscle differentiation, we wanted to evaluate whether USP9X knockdown increased mTOR activity during muscle differentiation. Similar to the cell-based assay for measuring in vivo mTOR activity, we analyzed the phosphorylation of S6 and Akt as a measure of mTORC1 and mTORC2 activity, respectively. Interestingly, reduced USP9X levels resulted in elevated levels of Akt phosphorylation, particularly at day 1 (Fig. 7). An effect of USP9X knockdown on Akt phosphorylation during differentiation indicates that USP9X acts as a negative regulator of mTORC2 under these conditions. We did not observe any significant change in S6 phosphorylation during differentiation when comparing USP9X knockdown cells to control (supplemental Fig. S5). These results indicate that reduction in USP9X levels activates mTORC2 and accelerates muscle differentiation. We were unable to determine the effect of V5-tagged USP9X overexpression on mTOR-mediated differentiation of C2C12 myoblasts. Although V5-USP9X was detected in transfect cells using an antibody to the V5 tag, this did not result in increased total USP9X protein levels in these cells. This was true regardless of V5-USP9X expression levels in these cells (supplemental Fig. S6).
FIGURE 7.
USP9X knockdown increases Akt phosphorylation during differentiation. Protein samples collected during differentiation (Fig. 5) were analyzed for phospho-Akt and total-Akt levels using Western blot analysis, and images were quantitated using ImageQuant. A single representative Western blot is shown in the top panel. Phospho-Akt (Ser-473) levels were normalized to total-Akt levels, and data from three biological replicates are plotted in the bottom panel. ** represents p value < 0.005.
DISCUSSION
We report here the identification of the deubiquitinase USP9X as a novel mTOR interactor that negatively regulates mTOR activity and skeletal muscle differentiation. USP9X was originally cloned as the mammalian ortholog of the Drosophila fat facets gene that regulates polarity and cell fate in the retina and is essential for normal eye development and early embryonic viability (33, 34). Furthermore, overexpression of Drosophila fat facets in Drosophila neuronal cells leads to synaptic hypertrophy and disruption of synaptic functions (35). In mouse, USP9X is expressed in unfertilized oocytes and preimplantation embryos and is required for development (17). Unlike mTOR, whose hyperactivation can cause exhaustion of stem cell populations (36), USP9X has been shown to promote self-renewal of neural stem cells. Recent data show that a modest increase in USP9X levels dramatically enhanced the self-renewing properties of in vitro-derived neuronal progenitors (24). However, the molecular mechanisms underlying the “stemness”-promoting properties of USP9X are unclear. Our study using C2C12 cells, where “progenitor” myoblasts are differentiated into myotubes, indicates that the activity of USP9X inhibits myotube formation in part by modulating mTOR activity. Reduction of USP9X expression in C2C12 myoblasts increases mTOR activity that is accompanied by accelerated differentiation of cells upon transferring them to differentiation media.
We employed tandem affinity purification to isolate mTOR complexes from cell extracts. The presence of a TAP-tag at the N terminus of mTOR did not disrupt its interaction with known interactors nor did it prevent mTOR kinase activity toward its substrate S6K1 (Fig. 1, A and B). Mass spectrometry analysis of purified complexes identified the known mTOR interactors, Raptor, Rictor, RuvBL1, RuvBL2, and TTI1 (29, 30). In addition to these known interactors, we also observed the deubiquitinase USP9X complexed with mTOR. We confidently identified USP9X by mass spectrometry in two independent TAP-mTOR purifications and confirmed the interaction by co-immunoprecipitation. A ubiquitin-specific protease, USP9X, can remove polyubiquitin chains from its substrates, such as AF-6, β-catenin, MCL1, ITCH, MARCH7, and ErbB2 leading to their stabilization and regulation of various cellular functions including cell adhesion, polarity, and cell proliferation (17–21, 31, 37, 38). USP9X can also remove polyubiquitin chains and mono-ubiquitin from its substrates, AMPK-related kinases and Smad4, respectively, causing changes in their activities without affecting their stability (39, 40). We did not detect any change in mTOR protein levels upon knocking down USP9X (supplemental Fig. S7). However, knockdown of USP9X increased mTORC1 activity in non-differentiating cells and mTORC2 in differentiating cells.
Reduction in USP9X levels by RNA interference increased mTORC1 activity in C2C12 myoblasts in a growth factor stimulation assay. No change in mTORC2 activity was observed in this assay (Fig. 3). The effect of USP9X knockdown using pooled siRNA on mTOR activity was not due to off-target effects. Two distinct USP9X siRNA duplexes showed similar effects on mTORC1/2 activity (supplemental Fig. S2). As USP9X interacts with Raptor and Rictor, components of mTORC1 and -2, respectively, it was surprising that only mTORC1 activity was affected by changes in USP9X levels in this assay. Several groups have reported that constitutively active mTORC1 substrate S6K1 can reduce mTORC2 activity. Activated S6K1 can inhibit input signaling from insulin and insulin-like growth factor 1 by phosphorylation and thereby degradation of insulin receptor substrate 1 (41–43). In addition, S6K1 can also phosphorylate Rictor and regulate signaling through mTORC2 (44, 45). Therefore, it is possible that we did not observe any changes in mTORC2 activity in myoblasts due to hyperactivation of mTORC1, particularly under serum-starved conditions, and initiation of negative feedback regulation. Alternatively, a kinase(s) other than mTOR that is not regulated by USP9X may be responsible for phosphorylating Akt in C2C12 cells (46–48). However, this is unlikely as reduction in mTOR levels in C2C12 cells results in lower Akt phosphorylation compared with control when analyzed in parallel to USP9X knockdown (supplemental Fig. S3B).
Using the same tandem affinity purification strategy employed for mTOR, we identified known and novel USP9X interacting proteins. In this pulldown assay we were able to detect MARCH7, a known USP9X interactor. We did not, however, observe mTOR in our purified USP9X complex. Although the absence of mTOR in USP9X pulldown suggests the transient or weak nature of mTOR/USP9X interaction, the presence of the mTOR interactor RuvBL1 indicates that at least some components of the mTOR/USP9X complex overlap with components of mTORC1 and mTORC2. In addition, we identified two melanoma antigen family members, MAGED1 and MAGED4, associated with USP9X. Interestingly, MAGED1 co-purified with its ubiquitin ligase Praja1 (32). MAGED1 is involved in a number of cellular processes including transcriptional regulation, apoptosis, cell cycle progression, and differentiation (49–51). A number of studies have shown that MAGED1 plays an important role in muscle and neuronal differentiation. In particular, overexpression of MAGED1 in C2C12 cells promoted differentiation by inhibiting the Msx2 homeodomain protein (52). Moreover, it has been reported that MAGED1 levels increase in differentiating C2C12 cells, and knockdown of MAGED1 impairs muscle differentiation in vivo (53). Together, interaction of MAGED1 with USP9X and the effects of USP9X knockdown on mTOR activity in myoblasts along with known functions of USP9X in promoting cellular stemness suggested a role of USP9X in muscle differentiation.
To assess the effects of USP9X on myogenesis, we differentiated C2C12 myoblasts after knocking down USP9X. The siRNA-mediated knockdown of USP9X was very efficient, and USP9X protein levels remained low during the entire course of the experiment (supplemental Fig. S4). Reduction in USP9X levels resulted in accelerated differentiation of myoblasts as monitored by the elevated protein levels of differentiation markers MyoG and MHC (Figs. 5 and 6). Although the magnitudes of increase for these two markers was different as measured by Western blot versus immunofluorescence, both techniques revealed significant increases upon USP9X knockdown. Initiation of differentiation in cells with decreased USP9X levels was also accompanied by a significant increase in Akt phosphorylation at Ser-473 at day 1 (Fig. 7). We did not observe any changes in mTORC1-mediated S6 phosphorylation (supplemental Fig. S5). Our data are consistent with published results showing that elevated mTORC2 and not mTORC1 activity can enhance muscle differentiation (14, 16). However, we cannot rule out the possibility that mTORC1 substrates other than S6K1 may have a role during myogenesis. Differential effects of USP9X knockdown on Akt phosphorylation observed in differentiating cells can be attributed to a lack of negative feedback resulting from elevated mTORC1 activity as seen in myoblasts. It is also possible that in differentiating cells, mTORC2 is more sensitive to USP9X levels than it is in myoblasts due to distinct growth conditions. The change in mTORC2 activity observed upon UPS9X knockdown is consistent with the interaction of USP9X with Rictor.
It will be of interest to further elucidate the precise mechanism by which USP9X regulates mTOR activity during differentiation. However, given the fact that USP9X is in a complex with mTOR and that their interaction is retained during differentiation (supplemental Fig. S8), the mechanism is likely to involve a physical interaction. We were unable to measure differentiation under conditions of USP9X overexpression. Expression of a V5-tagged USP9X in C2C12 cells did not result in changes in total USP9X. This indicates that C2C12 cells may be sensitive to increased levels of USP9X and down-regulate expression of the endogenous gene in response to overexpression (supplemental Fig. S6).
As knockdown of USP9X enhances the exit of C2C12 myoblasts from the cell cycle and undergo differentiation, our observations are consistent with the known functions of USP9X in promoting self-renewal of stem cells and inhibition of differentiation. Our study indicates that this established activity of USP9X operates in part through mTOR signaling. Surprisingly, we did not detect any change in USP9X protein levels upon initiation of differentiation in control cells (supplemental Fig. S4). It is possible that during differentiation of skeletal muscle, USP9X functions are modulated by inhibition of its activity and not by reduction in its levels. In conclusion, we have identified a novel mTOR interactor, USP9X, that acts as a negative regulator of mTOR functions. Reduction in USP9X levels results in increased mTORC1 activity upon growth factor and insulin signaling in myoblasts. We also show that USP9X modulates mTORC2 activity in differentiating cells and that this is accompanied by an accelerated differentiation phenotype. The role of USP9X in muscle differentiation is further indicated by its interaction with MAGED1. It is becoming increasingly clear that degenerative diseases associated with aging such as sarcopenia are caused by a reduced capacity for tissue regeneration. Our results show that reducing the activity of USP9X can enhance the activity of mTOR and promote muscle cell differentiation. This suggests that pharmacologic inhibition of USP9X could be of benefit in sarcopenia and other degenerative diseases associated with tissue loss.
Acknowledgments
We thank Michael Hall (University of Basel, Switzerland) for providing mTOR cDNA and Stephan Wood (Griffith University, Australia) for providing USP9X cDNA. We also thank Lorri Reinders for technical help. Danielle Crippen and Cathy Vitelli assisted with confocal imaging and image processing. John Miller and Arvind Ramanathan provided helpful comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant RL1 GM084432 (to R. H.). Mass spectrometry was supported by Center for Research Resources, National Institutes of Health shared instrumentation Grant S10 RR024615 (to B. W. G.) and Geroscience Mass Spectrometry and Imaging Core Grant PL1 AG032118; to B. W. G.).

This article contains supplemental Figs. S1–S9 and Table S1.
R. E. Hughes, unpublished data.
- mTOR
- mammalian target of rapamycin
- mTORC1
- mammalian target of rapamycin complex 1
- mTORC2
- mammalian target of rapamycin complex 2
- HEK293T
- human embryonic kidney 293 T cells
- IP
- immunoprecipitation
- MAGED1
- melanoma antigen family D1
- MyoG
- myogenin
- MHC
- myosin heavy chain
- Raptor
- regulatory-associated protein of mTOR
- Rictor
- rapamycin-insensitive companion of mTOR
- RuvBL1
- Ruv-B like 1
- RuvBL2
- Ruv-B like 2
- S6K1
- S6 kinase 1
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus
- USP9X
- ubiquitin-specific peptidase 9, X-linked.
REFERENCES
- 1. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 2. Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2004) Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 [DOI] [PubMed] [Google Scholar]
- 3. Zoncu R., Efeyan A., Sabatini D. M. (2011) mTOR. From growth signal integration to cancer, diabetes, and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin-insensitive. Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 5. Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 6. Sengupta S., Peterson T. R., Sabatini D. M. (2010) Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell 40, 310–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choo A. Y., Blenis J. (2009) Not all substrates are treated equally. Implications for mTOR, rapamycin resistance, and cancer therapy. Cell Cycle 8, 567–572 [DOI] [PubMed] [Google Scholar]
- 8. Mamane Y., Petroulakis E., LeBacquer O., Sonenberg N. (2006) mTOR, translation initiation, and cancer. Oncogene 25, 6416–6422 [DOI] [PubMed] [Google Scholar]
- 9. Oh W. J., Jacinto E. (2011) mTOR complex 2 signaling and functions. Cell Cycle 10, 2305–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Glass D. J. (2010) PI3-kinase regulation of skeletal muscle hypertrophy and atrophy. Curr. Top. Microbiol. Immunol. 346, 267–278 [DOI] [PubMed] [Google Scholar]
- 11. Ge Y., Wu A. L., Warnes C., Liu J., Zhang C., Kawasome H., Terada N., Boppart M. D., Schoenherr C. J., Chen J. (2009) mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am. J. Physiol. Cell Physiol. 297, C1434–C1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park I. H., Erbay E., Nuzzi P., Chen J. (2005) Skeletal myocyte hypertrophy requires mTOR kinase activity and S6K1. Exp. Cell Res. 309, 211–219 [DOI] [PubMed] [Google Scholar]
- 13. Bentzinger C. F., Romanino K., Cloëtta D., Lin S., Mascarenhas J. B., Oliveri F., Xia J., Casanova E., Costa C. F., Brink M., Zorzato F., Hall M. N., Rüegg M. A. (2008) Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8, 411–424 [DOI] [PubMed] [Google Scholar]
- 14. Shu L., Houghton P. J. (2009) The mTORC2 complex regulates terminal differentiation of C2C12 myoblasts. Mol. Cell. Biol. 29, 4691–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Erbay E., Chen J. (2001) The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J. Biol. Chem. 276, 36079–36082 [DOI] [PubMed] [Google Scholar]
- 16. Erbay E., Park I. H., Nuzzi P. D., Schoenherr C. J., Chen J. (2003) IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J. Cell Biol. 163, 931–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pantaleon M., Kanai-Azuma M., Mattick J. S., Kaibuchi K., Kaye P. L., Wood S. A. (2001) FAM deubiquitylating enzyme is essential for preimplantation mouse embryo development. Mech. Dev. 109, 151–160 [DOI] [PubMed] [Google Scholar]
- 18. Murray R. Z., Jolly L. A., Wood S. A. (2004) The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and β-catenin. Mol. Biol. Cell 15, 1591–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taya S., Yamamoto T., Kano K., Kawano Y., Iwamatsu A., Tsuchiya T., Tanaka K., Kanai-Azuma M., Wood S. A., Mattick J. S., Kaibuchi K. (1998) The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J. Cell Biol. 142, 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taya S., Yamamoto T., Kanai-Azuma M., Wood S. A., Kaibuchi K. (1999) The deubiquitinating enzyme Fam interacts with and stabilizes β-catenin. Genes Cells 4, 757–767 [DOI] [PubMed] [Google Scholar]
- 21. Schwickart M., Huang X., Lill J. R., Liu J., Ferrando R., French D. M., Maecker H., O'Rourke K., Bazan F., Eastham-Anderson J., Yue P., Dornan D., Huang D. C., Dixit V. M. (2010) Deubiquitinase USP9X stabilizes MCL1 and promotes tumor cell survival. Nature 463, 103–107 [DOI] [PubMed] [Google Scholar]
- 22. Sato N., Sanjuan I. M., Heke M., Uchida M., Naef F., Brivanlou A. H. (2003) Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev. Biol. 260, 404–413 [DOI] [PubMed] [Google Scholar]
- 23. Van Hoof D., Passier R., Ward-Van Oostwaard D., Pinkse M. W., Heck A. J., Mummery C. L., Krijgsveld J. (2006) A quest for human and mouse embryonic stem cell-specific proteins. Mol. Cell. Proteomics 5, 1261–1273 [DOI] [PubMed] [Google Scholar]
- 24. Jolly L. A., Taylor V., Wood S. A. (2009) USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Mol. Biol. Cell 20, 2015–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bürckstümmer T., Bennett K. L., Preradovic A., Schütze G., Hantschel O., Superti-Furga G., Bauch A. (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013–1019 [DOI] [PubMed] [Google Scholar]
- 26. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 27. Link A. J., Eng J., Schieltz D. M., Carmack E., Mize G. J., Morris D. R., Garvik B. M., Yates J. R., 3rd (1999) Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682 [DOI] [PubMed] [Google Scholar]
- 28. Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 29. Izumi N., Yamashita A., Iwamatsu A., Kurata R., Nakamura H., Saari B., Hirano H., Anderson P., Ohno S. (2010) AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci. Signal. 3, ra27. [DOI] [PubMed] [Google Scholar]
- 30. Kaizuka T., Hara T., Oshiro N., Kikkawa U., Yonezawa K., Takehana K., Iemura S., Natsume T., Mizushima N. (2010) Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 285, 20109–20116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nathan J. A., Sengupta S., Wood S. A., Admon A., Markson G., Sanderson C., Lehner P. J. (2008) The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic 9, 1130–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sasaki A., Masuda Y., Iwai K., Ikeda K., Watanabe K. (2002) A RING finger protein Praja1 regulates Dlx5-dependent transcription through its ubiquitin ligase activity for the Dlx/Msx-interacting MAGE/Necdin family protein, Dlxin-1. J. Biol. Chem. 277, 22541–22546 [DOI] [PubMed] [Google Scholar]
- 33. Fischer-Vize J. A., Rubin G. M., Lehmann R. (1992) The fat facets gene is required for Drosophila eye and embryo development. Development 116, 985–1000 [DOI] [PubMed] [Google Scholar]
- 34. Huang Y., Baker R. T., Fischer-Vize J. A. (1995) Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science 270, 1828–1831 [DOI] [PubMed] [Google Scholar]
- 35. DiAntonio A., Haghighi A. P., Portman S. L., Lee J. D., Amaranto A. M., Goodman C. S. (2001) Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412, 449–452 [DOI] [PubMed] [Google Scholar]
- 36. Castilho R. M., Squarize C. H., Chodosh L. A., Williams B. O., Gutkind J. S. (2009) mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 5, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mouchantaf R., Azakir B. A., McPherson P. S., Millard S. M., Wood S. A., Angers A. (2006) The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem. 281, 38738–38747 [DOI] [PubMed] [Google Scholar]
- 38. Marx C., Held J. M., Gibson B. W., Benz C. C. (2010) ErbB2 trafficking and degradation associated with Lys-48 and Lys-63 polyubiquitination. Cancer Res. 70, 3709–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Al-Hakim A. K., Zagorska A., Chapman L., Deak M., Peggie M., Alessi D. R. (2008) Control of AMPK-related kinases by USP9X and atypical Lys-29/Lys-33-linked polyubiquitin chains. Biochem. J. 411, 249–260 [DOI] [PubMed] [Google Scholar]
- 40. Dupont S., Mamidi A., Cordenonsi M., Montagner M., Zacchigna L., Adorno M., Martello G., Stinchfield M. J., Soligo S., Morsut L., Inui M., Moro S., Modena N., Argenton F., Newfeld S. J., Piccolo S. (2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFβ signaling, controls Smad4 monoubiquitination. Cell 136, 123–135 [DOI] [PubMed] [Google Scholar]
- 41. Harrington L. S., Findlay G. M., Lamb R. F. (2005) Restraining PI3K. mTOR signaling goes back to the membrane. Trends Biochem. Sci. 30, 35–42 [DOI] [PubMed] [Google Scholar]
- 42. Manning B. D. (2004) Balancing Akt with S6K. Implications for both metabolic diseases and tumorigenesis. J. Cell Biol. 167, 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Corradetti M. N., Guan K. L. (2006) Upstream of the mammalian target of rapamycin. Do all roads pass through mTOR? Oncogene 25, 6347–6360 [DOI] [PubMed] [Google Scholar]
- 44. Dibble C. C., Asara J. M., Manning B. D. (2009) Characterization of Rictor phosphorylation sites reveals direct regulation of mTOR complex 2 by S6K1. Mol. Cell. Biol. 29, 5657–5670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Julien L. A., Carriere A., Moreau J., Roux P. P. (2010) mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol. Cell. Biol. 30, 908–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang J., Wu X. J., Zhuo D. X., Liu T., Li W. R., Mao Z. B., Xin Z. C. (2010) Effect of tankyrase 1 on autophagy in the corpus cavernosum smooth muscle cells from ageing rats with erectile dysfunction and its potential mechanism. Asian J. Androl 12, 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Risson V., Mazelin L., Roceri M., Sanchez H., Moncollin V., Corneloup C., Richard-Bulteau H., Vignaud A., Baas D., Defour A., Freyssenet D., Tanti J. F., Le-Marchand-Brustel Y., Ferrier B., Conjard-Duplany A., Romanino K., Bauché S., Hantaï D., Mueller M., Kozma S. C., Thomas G., Rüegg M. A., Ferry A., Pende M., Bigard X., Koulmann N., Schaeffer L., Gangloff Y. G. (2009) Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J. Cell Biol. 187, 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xie X., Zhang D., Zhao B., Lu M. K., You M., Condorelli G., Wang C. Y., Guan K. L. (2011) IκB kinase ϵ and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 108, 6474–6479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sasaki A., Hinck L., Watanabe K. (2005) RumMAGE-D the members. Structure and function of a new adaptor family of MAGE-D proteins. J. Recept. Signal Transduct. Res. 25, 181–198 [DOI] [PubMed] [Google Scholar]
- 50. Reddy E. M., Chettiar S. T., Kaur N., Shepal V., Shiras A. (2010) Dlxin-1, a MAGE family protein, induces accelerated neurite outgrowth and cell survival by enhanced and early activation of MEK and Akt signaling pathways in PC12 cells. Exp. Cell Res. 316, 2220–2236 [DOI] [PubMed] [Google Scholar]
- 51. Wang X., Gao X., Xu Y. (2011) MAGED1. Molecular insights and clinical implications. Ann. Med. 43, 347–355 [DOI] [PubMed] [Google Scholar]
- 52. Kuwajima T., Taniura H., Nishimura I., Yoshikawa K. (2004) Necdin interacts with the Msx2 homeodomain protein via MAGE-D1 to promote myogenic differentiation of C2C12 cells. J. Biol. Chem. 279, 40484–40493 [DOI] [PubMed] [Google Scholar]
- 53. Nguyen P. H., Le T. V., Kang H. W., Chae J., Kim S. K., Kwon K. I., Seo D. B., Lee S. J., Oh W. K. (2010) AMP-activated protein kinase (AMPK) activators from Myristica fragrans (nutmeg) and their anti-obesity effect. Bioorg Med. Chem. Lett. 20, 4128–4131 [DOI] [PubMed] [Google Scholar]