Background: Gli3-deficient mice (Gli3Xt-J/Xt-J) show premature suture closure (craniosynostosis).
Results: Gli3Xt-J/Xt-J mice have aberrant cell proliferation and osteogenic differentiation in the sutures. Reducing the dosage of Runx2 (Gli3Xt-J/Xt-J;Runx2+/− mice) rescues the abnormality through canonical Bmp-Smad signaling.
Conclusion: Gli3 represses bone formation Bmp-Smad signaling, which integrates Dlx5/Runx2-II cascade.
Significance: Targeting Runx2 might provide an attractive way of preventing craniosynostosis in patients.
Keywords: Bone, Bone Morphogenetic Protein (BMP), Development, Differentiation, Osteoblasts, Craniosynostosis, Gli3, Runx2
Abstract
Mutations in the gene encoding the zinc finger transcription factor GLI3 (GLI-Kruppel family member 3) have been identified in patients with Grieg cephalopolysyndactyly syndrome in which premature fusion of calvarial suture (craniosynostosis) is an infrequent but important feature. Here, we show that Gli3 acts as a repressor in the developing murine calvaria and that Dlx5, Runx2 type II isoform (Runx2-II), and Bmp2 are expressed ectopically in the calvarial mesenchyme, which results in aberrant osteoblastic differentiation in Gli3-deficient mouse (Gli3Xt-J/Xt-J) and resulted in craniosynostosis. At the same time, enhanced activation of phospho-Smad1/5/8 (pSmad1/5/8), which is a downstream mediator of canonical Bmp signaling, was observed in Gli3Xt-J/Xt-J embryonic calvaria. Therefore, we generated Gli3;Runx2 compound mutant mice to study the effects of decreasing Runx2 dosage in a Gli3Xt-J/Xt-J background. Gli3Xt-J/Xt-J Runx2+/− mice have neither craniosynostosis nor additional ossification centers in interfrontal suture and displayed a normalization of Dlx5, Runx2-II, and pSmad1/5/8 expression as well as sutural mesenchymal cell proliferation. These findings suggest a novel role for Gli3 in regulating calvarial suture development by controlling canonical Bmp-Smad signaling, which integrates a Dlx5/Runx2-II cascade. We propose that targeting Runx2 might provide an attractive way of preventing craniosynostosis in patients.
Introduction
A suture is a fibrous connection between adjacent craniofacial bones, which functions as an articulation, a site of bone deposition, and a shock absorber. Craniosynostosis is the premature fusion of one or more cranial or facial sutures, and ∼30% of those cases are induced by known gene mutations in FGFR1, FGFR2, FGFR3, MSX2, TWIST1, EFNB1, RAB23, and in the zinc finger transcription factor GLI3 (1–3).
For sutures to operate as growth sites, they must remain patent. Bone must be formed at the correct location, in the bones, and at the osteogenic fronts, but osteogenesis also must be repressed in between the bone ends in the midsutural mesenchyme. The processes of osteoblastic cell differentiation and proliferation have to be fine-tuned to occur at the correct speed, at the correct location, and at the appropriate time. The size of the osteoprogenitor cell population is important for intramembranous bone growth, and Bmp proteins acting through the transcription factors Msx1 and Msx2 regulate the size of this population (4, 5). The fine balance between osteoprogenitor cell proliferation and the rate of differentiation are regulated by Fgf signaling, specifically by the temporal-spatial expression of Fgf receptors (6, 7). The Forkhead transcription factor Foxc1 sits at the nexus between Bmp and Fgf signaling during calvarial bone development. Foxc1 is induced by Fgf and regulates the effects of Bmp signaling. It does this independently of the Bmp post-transcriptional repressor, Noggin, which is itself critical for suture patency (8, 9). Foxc1−/− mice exhibit defective calvarial osteogenesis with the calvarial bones remaining rudimentary at the sites of initial osteogenic condensations (10).
We have reported previously that Gli3-deficient mice (Gli3Xt-J/Xt-J) develop craniosynostosis during embryogenesis because of the aberrant enhancement of Runx2 and reduced Twist1 expression in the mid-sutural mesenchymal cells (11). The localization of the phenotype in Gli3Xt-J/Xt-J mice to the interfrontal and lambdoid sutures can be explained by the location specificity of Gli3 expression. In addition, we have shown that the premature ossification in the lambdoid suture can be rescued in vitro by applying FGF2-soaked beads, which normalized Twist1 expression. This rescue was based on the ability of Twist1 to repress Runx2 and thereby stop the excessive differentiation of osteoprogenitors in the suture (11).
Several reports have indicated a close connection between Gli family proteins and bone development. Shimoyama and co-workers (12) have demonstrated the physical interaction between Gli2 and Runx2 and that Indian Hedgehog (Ihh) promotes osteoblast differentiation through enhancement of Runx2 expression by Gli2 but not Gli3. In contrast, Ohba and co-workers have shown that reduction of the repressor form of Gli3 (Gli3R) in Ptch1+/− (Patched1) osteoblasts results in accelerated osteoblast differentiation, and demonstrated the indirect and competitive inhibitory effect of Gli3 against DNA binding by Runx2 in vitro (13). Finally, they concluded that Hh signaling through Ptch plays a critical role in postnatal bone homeostasis (13).
Runx2 is a key regulatory factor in the differentiation of osteoblasts and chondrocytes (14, 15). Runx2 absolutely is required for the initial stage of osteoblast differentiation but must be down-regulated to permit osteoblast maturation (16). Additionally, Runx2 plays a critical role in linking cell fate, cell proliferation, and control of cell growth by regulating genes transcribed by RNA polymerase II and repressing RNA polymerase-mediated ribosomal RNA synthesis (17). Homozygous Runx2-null mice (Runx2−/−) show complete lack of endochondral and membranous bone (14, 15), and their teeth are misshapen and severely hypoplastic (18). Heterozygotes Runx2+/− mice mimic human cleidocranial dysplasia exhibiting hypoplasia of clavicles, delayed ossification of cranial bones, wide anterior and posterior fontanels, and wide cranial sutures (19).
Runx2 has two alternative promoters, the distal P1 promoter and the proximal P2 promoter. The P1 promoter encodes the Runx2-II isoform, also known as Til-1 G1 and Osf2/Cbfa1, whereas the P2 promoter encodes the Runx2-I isoform (Pebp2αA). Runx2-I is expressed in immature osteoprogenitor cells, whereas Runx2-II is expressed in more mature osteoblasts (20). Mice that have a selective loss of Runx2-II unexpectedly form axial, appendicular, and craniofacial bones with the exception of cranial bones derived from endochondral ossification. The expression of Runx2-I is compensatory up-regulated in Runx2-II−/− mice, but they fail to complete osteogenesis (21). Furthermore, the Runx2-I isoform is strongly expressed in mesenchymal cells of sagittal suture and the osteogenic fronts of the parietal bones. In contrast, the Runx2-II isoform expression is limited to the osteogenic fronts and the parietal bones (20). This mutually exclusive expression of the Runx2 isoforms indicates that they have distinct functional significance in the development of the cranial suture. Interestingly, Dlx5 (Distal-less homeobox 5) has been reported to regulate the expression of Runx2-II isoform but not Runx-I in BMP2-induced osteogenic differentiation (22).
In vertebrates, Hh signal transduction is initiated by the binding of Hh ligands to the cell surface receptor Ptch1 (23). In the absence of the Hh ligand, Ptch1 blocks a signal transducer Smoothened. Binding of Hh ligand to Ptch1 releases this repression and inhibits the truncation of Gli to produce the repressor form (GliR) and promotes the production of full-length activator form (GliA). In vertebrates, Gli family consists of Gli1, Gli2, and Gli3. Gli3 and to a lesser extent Gli2 can be proteolytically processed into short GliR in the absence of Hh ligands. Gli2 acts mainly as GliA inducing Gli1 and Ptch expression, which is regarded as readout of Hh signaling as it is also up-regulated by Hh (23).
Our aim in this study was to test the hypothesis that Gli3 signaling is important in keeping Runx2 in check and that Runx2 dosage is important in maintaining the correct balance of osteogenesis in the developing suture. We generated Gli3Xt-J/Xt-J Runx2+/− compound mutant mice to reduce the dosage of Runx2. We discovered that Gli3Xt-J/Xt-J Runx2+/− mice have no craniosynostosis or ectopic ossification centers in either the lambdoid or interfrontal sutures.
From the analysis of different stages of osteoprogenitor differentiation in the lambdoid suture revealed that the mRNA expression of Dlx5, Runx2-II, and Bmp2 were increased in the mid-sutural mesenchymal cells of Gli3Xt-J/Xt-J but decreased in Gli3Xt-J/Xt-J Runx2+/− compound mutant mice. The expression of Noggin, which is induced by Bmp2 administration (8), was slightly up-regulated at the interfrontal suture of Gli3Xt-J/Xt-J mice indicating the enhancement of Bmp2 expression there. The lambdoid and interfrontal sutures of Gli3Xt-J/Xt-J mice showed high proliferative activity across the broad suture area, whereas low proliferative activity was observed in Gli3Xt-J/Xt-J Runx2+/− mice predominantly at the osteogenic front and was similar to that of WT mice. Finally, phosphorylation of Smad1/5/8 in both calvarial tissue and mid-sutural mesenchymal cells in Gli3Xt-J/Xt-J mice was increased, whereas in Gli3Xt-J/Xt-J Runx2+/− mice, this was normalized. These data suggest that Gli3 has an important role in cranial suture development through the canonical Bmp-Smad pathway involving a Dlx5-Runx2-II cascade. The data also show that targeting Runx2 activity using siRNA might provide an attractive way of preventing craniosynostosis in patients.
EXPERIMENTAL PROCEDURES
Generation of Gli3;Runx2 Compound Mutant Mice
All animal experiments were approved by the University of Helsinki, Helsinki University Hospital and the Southern Finland Council animal welfare and ethics committees. Gli3+/Xt-J mice in C57BL/6 background and Runx2+/− mice were maintained and PCR genotyping was performed using tail or skin DNA as described previously (24, 25). We generated Gli3;Runx2 compound mutant mice by mating Gli3+/Xt-J Runx2+/− mice with Gli3+/Xt-J Runx2+/−mice as described in Table 1. WT littermates were used as a control.
TABLE 1.
Genotypes of offspring generated from crossing Gli3+/Xt-J Runx2+/− male and Gli3+/Xt-J Runx2+/− female mice
| Genotype | No. of embryos | % |
|---|---|---|
| Gli3+/+Runx2+/+ | 4 | 3.8 |
| Gli3+/+Runx2+/− | 22 | 20.8 |
| Gli3+/+Runx2−/− | 2 | 1.9 |
| Gli3+/Xt-J Runx2+/+ | 18 | 17.0 |
| Gli3+/Xt-J Runx2+/− | 17 | 16.0 |
| Gli3+/Xt-J Runx2−/− | 10 | 9.4 |
| Gli3Xt-J/Xt-J Runx2+/+ | 8 | 7.5 |
| Gli3Xt-J/Xt-J Runx2+/− | 22 | 20.8 |
| Gli3Xt-J/Xt-J Runx2−/− | 3 | 2.8 |
| Total | 106 | 100 |
Western Blot Analysis
The expression of Gli3 in embryonic day 15.5 (E15.5)2 calvaria, limb, and brain was determined by Western blot analysis using anti-GLI3 polyclonal antibody (AF3690, R&D Systems, Minneapolis, MN), and the phosphorylation of Smad1/5/8 in E15.5 calvaria was determined using anti-phospho-Smad1 (Ser463/465)/Smad5 (Ser463/465)/Smad8 (Ser426/428) polyclonal antibody (no. 9511, Cell Signaling, Boston, MA). Each sample was lysed in radioimmune precipitation assay buffer (Sigma-Aldrich) supplemented with PhosSTOP phosphatase inhibitor mixture and Complete protease inhibitor mixture (Roche Diagnostics), and 10 μg of each sample was separated by SDS-PAGE (Mini-Protean TGX gels 4–20%, Bio-Rad), transferred to a nitrocellulose membrane (Amersham Biosciences Hybond-ECL, GE Healthcare), and incubated with each primary antibody, and then incubated IRDye800CW secondary antibody (LI-COR, Lincoln, NE). Proteins were visualized using Odyssey detection system (LI-COR). Integrated density values obtained for pSmad levels were normalized against those of α-tubulin (α-tubulin antibody, DM1A, Sigma-Aldrich) and compared against other samples. Statistical values were calculated using Student's t test, with p values below 0.05 considered significant.
In Situ Hybridization
Preparation of 35S-labeled uridine 5′-triphosphate and digoxigenin-UTP-labeled riboprobes, in situ hybridization, and image processing have been described previously (26). The Runx2-I probe used in in situ hybridization was prepared from a 639-bp fragment of murine Runx2 cDNA isolated from C3H10T1/2 cells by RT-PCR with the specific primers: 5′-CGGGATCCTCTCAGCTTTAGCGTCGTCA-3′ for the forward primer and 5′-GCTCTAGACCGCAAGGGACTTGAAGTT-3′ for the reverse primer. The PCR products were digested with BamHI/XbaI and subcloned into pBluescript II KS(-).
Skeletal Staining
For skeletal preparations, the skin of embryos was removed, and the embryos were fixed with cold 96% ethanol overnight. Skeletal elements were stained with 0.3% Alcian blue and 0.03% Alizarin Red-S solution, followed by clarification of the tissues by a glycerol/KOH series.
Cell Proliferation Analysis in Calvarial Sutures by Assessment of BrdU Incorporation
Pregnant female mice (E15.5) were injected intraperitoneally with BrdU solution (10 μl/g body weight, Invitrogen). Two hours after injection, embryos were removed and fixed with 10% neutral buffered formalin. Tissues were processed through alcohol dehydration, embedded in paraffin, and sectioned at 7-μm intervals. The BrdU-incorporated cells were detected using biotinylated monoclonal anti-BrdU antibody and visualized with streptavidin-biotin staining system following the manufacturer's protocol (Invitrogen). Sections were counterstained with hematoxylin. Quantification of BrdU-positive cells in each suture was performed as described previously (11). Results are reported as mean ± S.D. The significance of differences between means was assessed by analysis of variance followed by Mann-Whitney U test with Bonferroni (multiple comparisons). The p value of < 0.05 was considered as significant.
Detection of pSmad1/5/8 by Immunostaining
Whole heads of embryos were dissected, fixed with 4% paraformaldehyde at 4 °C overnight, and sectioned at 7-μm intervals. Immunohistochemical staining was performed using polyclonal anti-phospho-Smad1/Smad5/Smad8 (Ser463/465) antibody (Millipore, Temecula, CA) as the primary antibody and anti-rabbit biotinylated as the secondary antibody. Immunoreactivity was visualized with VECTASTAIN ABC kit (Vector Laboratories, Burlingame, CA) following the manufacturer's instructions. Sections were counterstained with hematoxylin.
RESULTS
Hh Signaling in Osteogenic Front of Calvarial Sutures
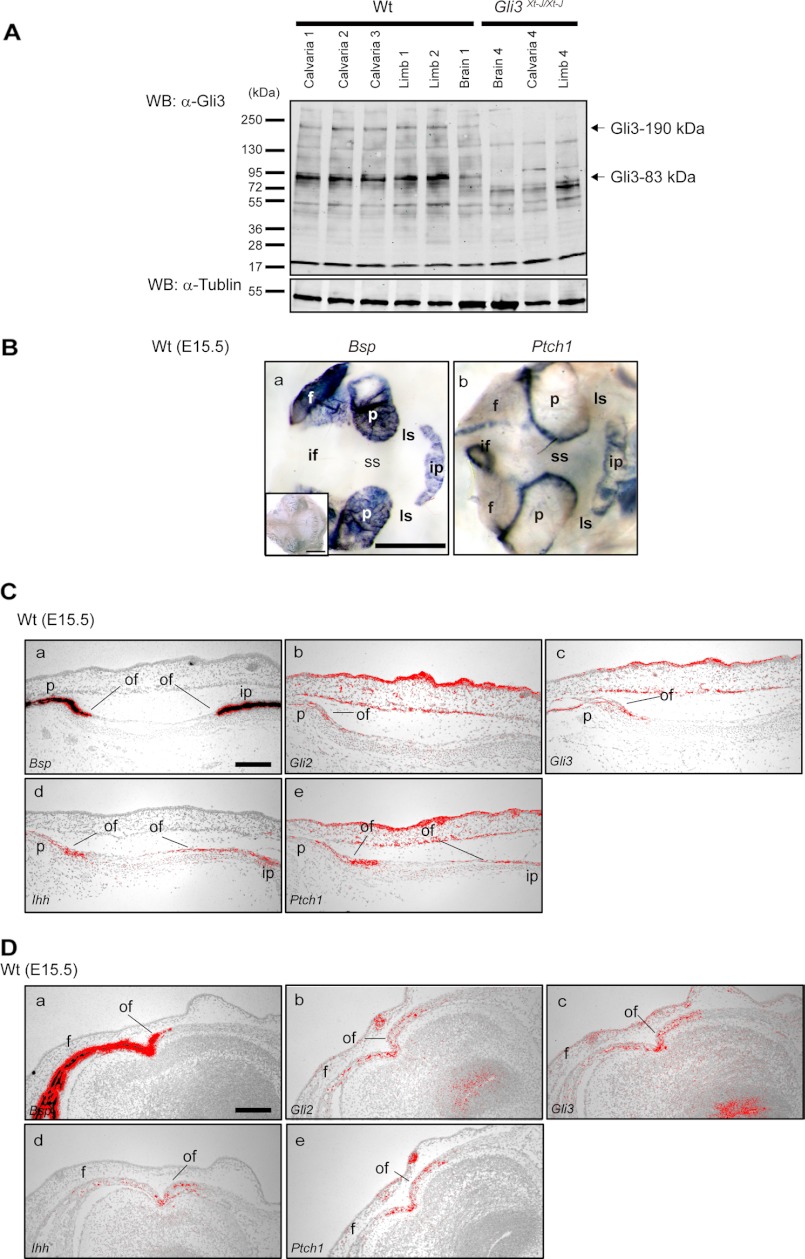
We analyzed the presence of Gli3R in WT calvaria at E15.5 (Fig. 1A) by Western blot analysis using a Gli3 antibody that recognizes both Gli3-full and Gli3R forms. As shown in Fig. 1A, the truncated Gli3R form was detected dominantly in the intact calvarial tissue as well as limb and brain tissue, indicating that processing of the Gli3-full into Gli3R is occurring in calvarial tissue (Fig. 1A). We have shown previously that Gli3 and Ptch1 are expressed in the developing calvaria (11). Ptch1 is a downstream target of Hh and is considered as readout of Hh signaling. Transcript of Ptch1 was observed clearly at the osteogenic fronts of the frontal, parietal, and interparietal bones, by whole mount in situ hybridization in WT E15.5 tissue, indicating active Hh signaling at this location (Fig. 1, B, b panel). In addition to our previous report, the expression of Gli2 and Gli3 mRNA were observed at the osteogenic fronts of the parietal, interparietal, and frontal bones (Fig. 1, C and D), and Ihh mRNA was observed clearly at the osteogenic front of these bones (Fig. 1, C and D).
FIGURE 1.
The expression of repressor form of Gli3 (Gli3–83 kDa) in calvarial tissue and Hh signaling in the calvarial osteogenic front. A, the expression of endogenous Gli3 protein in WT calvaria, limb, and brain was detected by Western blotting using specific antibodies that recognize both the full-length form of Gli3 (Gli3–190 kDa) and the truncated repressor form (Gli3R, Gli3–83 kDa). Predominantly, the Gli3R form was detected in WT calvaria (lanes 1–3), limb (lanes 4 and 5), and brain samples (lane 6). Neither form of Gli3 was detected in Gli3Xt-J/Xt-J brain (lane 7), calvaria (lane 8), or limb (lane 9) samples. α-Tubulin protein was detected equally in all samples. Lanes 1–3, calvaria from three independent WT embryos; lanes 4 and 5, limb from two independent WT embryos; lane 6, brain from WT embryo; lane 7, brain from Gli3Xt-J/Xt-J embryo; lane 8, calvaria from Gli3Xt-J/Xt-J embryo; lane 9, limb from Gli3Xt-J/Xt-J embryo. B, Bsp (a panel) Bsp sense (inset in a panel) and Ptch1 (b panel) expression in E15.5 WT calvaria were detected by whole mount in situ hybridization. Bsp expression was detected in mature osteoblasts (a panel), whereas Ptch1 highly localized to the osteogenic front of the frontal, parietal, and interparietal bones (b panel). These data were confirmed by in situ hybridization on tissue sections of the lambdoid (C) and the interfrontal (D) sutures of WT embryos. Transcripts of Ihh, Gli2, and Gli3 were expressed in osteogenic front in interfrontal and lambdoid sutures (C and D). f, frontal bone; if, interfrontal suture; ip, interparietal bone; ls, lambdoid suture; of, osteogenic front; p, parietal bone; ss, sagittal suture; WB, Western blotting. Scale bars in B, a panel, 1 mm; C, a panel, and D, a panel, 100 μm.
Dlx5 and Runx2-II Isoform Are Aberrantly Expressed in Lambdoid Sutures of Gli3Xt-J/Xt-J Mice
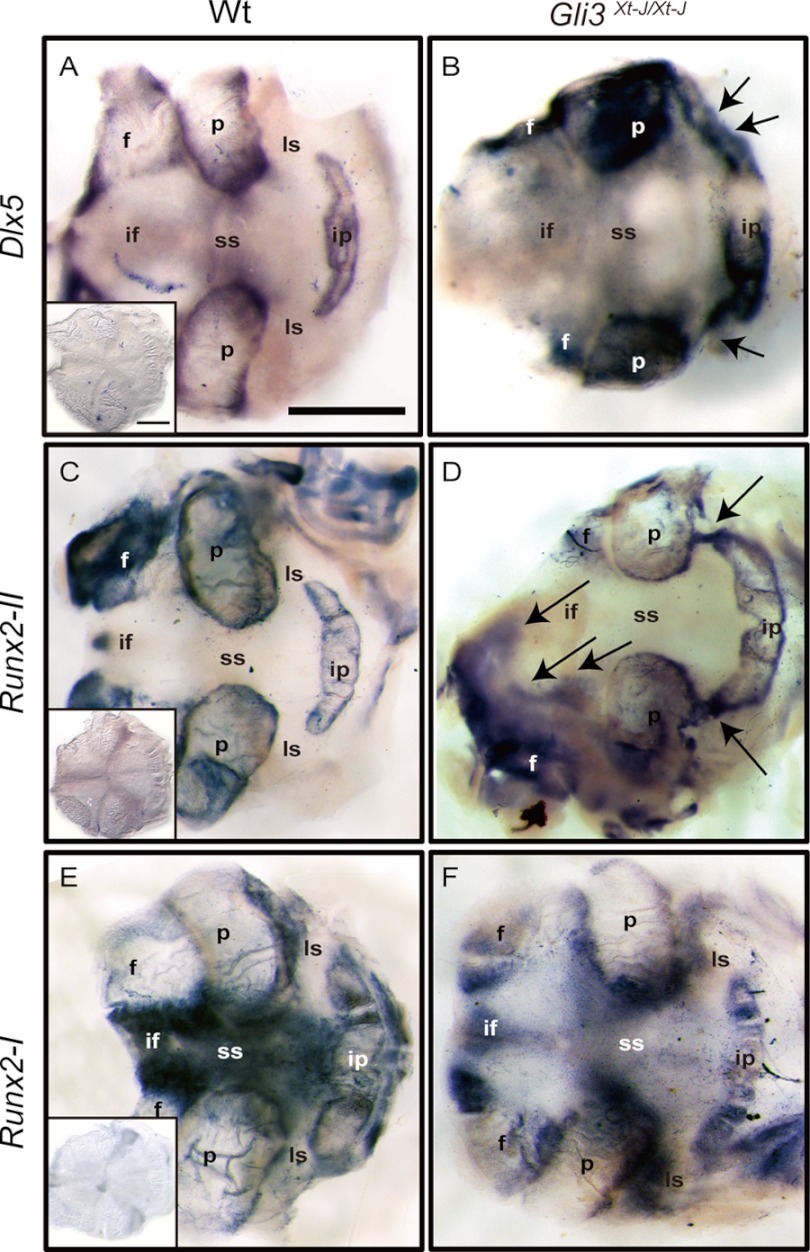
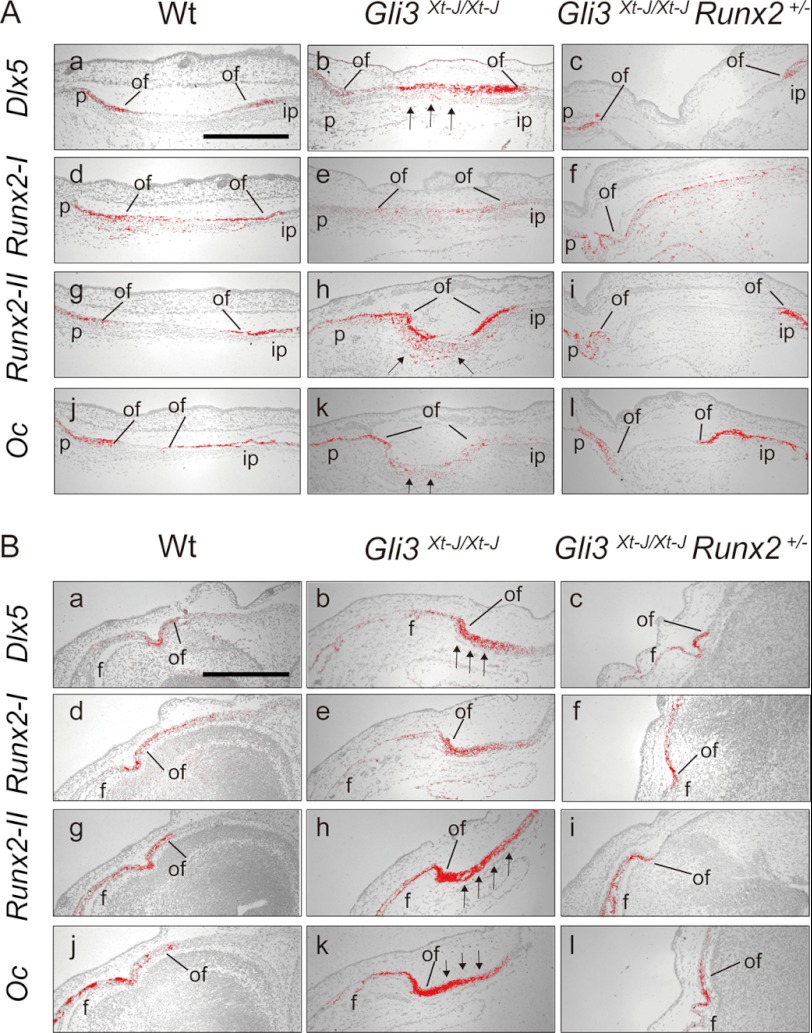
To further elucidate the mechanism of craniosynostosis in Gli3Xt-J/Xt-J mice, we performed whole mount in situ hybridization using riboprobes recognizing Dlx5, Runx2-I, and Runx2-II. We analyzed tissue at E15.5, which is just prior to the onset of craniosynostosis in Gli3Xt-J/Xt-J mice. In WT calvaria, the transcription factor Dlx5 lies downstream of Bmp2 and is important in the regulation of Runx2 by Bmp2 (27), especially the Runx2-II isoform (22). Also, Dlx5 is important in post-proliferative osteoblasts for the initiation of Oc (Osteocalcin) gene transcription (28). Dlx5 transcripts were localized in the osteogenic fronts of WT frontal, parietal, and interparietal bones, but not in the mid-sutural mesenchymal area at E15.5 (Fig. 2A). Runx2-II was expressed in osteogenic fronts and in a more differentiated bone site (Fig. 2C). However, Runx2-I was expressed specifically in the osteogenic fronts and sutural mesenchymal cells some distance from osteogenic fronts (Fig. 2E). Interestingly, Dlx5 and Runx2-II were ectopically expressed in the central mesenchyme of Gli3Xt-J/Xt-J lambdoid and interfrontal sutures (Fig. 2, B and D, arrows), whereas Runx2-I expressed in the osteogenic front and sutural mesenchymal cells of Gli3Xt-J/Xt-J calvaria (Fig. 2F).
FIGURE 2.
Ectopic mRNA expression of Runx2-II isoform and Dlx5 in the mid-sutural mesenchyme of E15.5. Gli3Xt-J/Xt-J interfrontal and lambdoid sutures. A and B, Dlx5 was expressed in the osteogenic front of each calvarial bone and weakly in the sagittal suture of WT mice. In contrast, in Gli3Xt-J/Xt-J calvaria, Dlx5 was expressed across the lambdoid suture (arrows in B). C and D, Runx2-II was expressed in the osteogenic front and in the mature bone areas of the frontal, parietal, and interparietal bones of WT calvaria. However, in Gli3Xt-J/Xt-J calvaria, Runx2-II was expressed intensely across the lambdoid suture and in the middle of interfrontal suture area (arrows in D). E and F, Runx2-I was specifically expressed in the osteogenic front and sutural-mesenchymal cells especially around the frontal and parietal bones in WT mice. In Gli3Xt-J/Xt-J tissue, Runx2-I was expressed in the sutural-mesenchymal cells similarly to WT tissue but with less intensity in the osteogenic fronts of the parietal bones (insets in A, C, and E). Samples hybridized with sense control probes. f, frontal bone; if, interfrontal suture; ip, interparietal bone; ls; lambdoid suture; p, parietal bone; ss, sagittal suture. Scale bar in A, 1 mm.
Gli3Xt-J/Xt-J Runx2+/− Mice Display Patent Lambdoid Sutures and Have No Ectopic Ossification in Interfrontal Suture
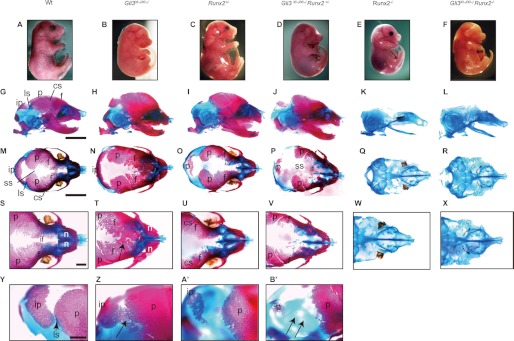
As Runx2 activity is up-regulated in the calvaria of Gli3Xt-J/Xt-J mice, we reduced the dose of Runx2 in compound mutant mice by crossing Gli3+/Xt-J Runx2+/− males with Gli3+/Xt-J Runx2+/− females. At E18.5, both Gli3Xt-J/Xt-J Runx2+/− as well as Gli3Xt-J/Xt-J Runx2−/− embryos had dysplastic eyes similar to those of Gli3Xt-J/Xt-J embryos (Fig. 3, B, D, and F). Alizarin Red-S and Alcian blue staining was performed at E18.5 to observe bones and cartilage (Fig. 3). Gli3Xt-J/Xt-J embryos showed a varying degree of heterotopic bone formation or craniosynostosis of the interfrontal suture (Fig. 3, N and T), and all specimens had craniosynostosis of the lambdoid sutures (Fig. 3Z) as reported previously (11). Runx2+/− embryos displayed wider sagittal sutures and fontanels and hypoplastic parietal and interparietal bones (Fig. 3O) in accordance with previous reports (14). As anticipated, Gli3Xt-J/Xt-J Runx2+/− embryos showed more extensive hypoplasia in frontal, parietal, and interparietal bones (Fig. 3, J and P) and patent lambdoid sutures (Fig. 3B′) (six of six animals). The heterotopic bones observed in the interfrontal suture of Gli3Xt-J/Xt-J embryos were reduced in size and number (Fig. 3V). Additionally, these mice displayed smaller clavicles than either Gli3+/Xt-J or Runx2+/−, a split sternal bone, deformity of scapula, polydactylous hands and feet, deformity of the pubic and ischial bones, and no tibia (supplemental Fig. 1). Skeletal staining of Gli3Xt-J/Xt-J Runx2−/− embryos revealed that they have no mature bone similar to that of Runx2−/− embryos, but they have a widened nasal capsule and bent nasal septum (Fig. 3X), polydactylous digital cartilage, split sternal cartilage, rib deformity, no tibia, and more serious deformity of clavicle and fibula compared with Runx2−/− mice (supplemental Fig. 1).
FIGURE 3.
Reversal of the lambdoid and interfrontal suture phenotypes in Gli3Xt-J/Xt-J Runx2+/− mice compared with Gli3Xt-J/Xt-J mice. A–F, photographs of WT and mutant embryos at E18.5. G–B′, WT and mutant embryos stained with Alizarin Red-S and Alcian blue to stain bone and cartilage, respectively. The lambdoid suture in Gli3Xt-J/Xt-J Runx2+/− mice was patent (arrows in B′), and there was also a wide interfrontal suture and hypoplastic frontal and parietal bones (V), compared with the craniosynostosis and heterotopic bones seen in Gli3Xt-J/Xt-J mice (arrows in T and Z). Similar to Runx2−/− mice, Gli3Xt-J/Xt-J Runx2−/− compound mutant mice showed an absence of mature skeletal elements (L, R, and X) as well as a bent nasal septum and a shortened nasal capsule (X). cs, coronal suture; f, frontal bone; if, interfrontal suture; ip, interparietal bone; ls, lambdoid suture; n, nasal bone; p, parietal bone; ss, sagittal suture. Scale bars (G and M), 5 mm; scale bars (S and Y), 1 mm. The following groups of images are shown in same magnification: A–F, G–L, M–R, S–X, and Y–B′.
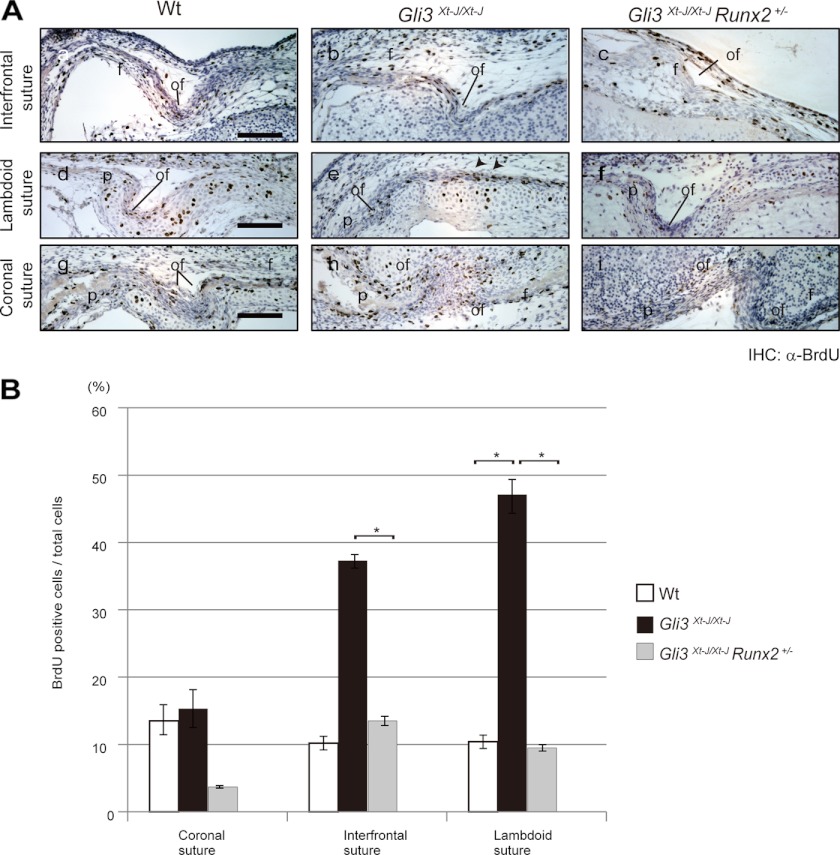
Increased Proliferation in Interfrontal and Lambdoid Sutures of Gli3Xt-J/Xt-J Embryos Was Normalized in Gli3Xt-J/Xt-J Runx2+/− Embryos
To test whether reduced Runx2 dosage would rescue the increased proliferation observed in Gli3Xt-J/Xt-J mice, we pulsed Gli3Xt-J/Xt-J Runx2+/− embryos with BrdU. We analyzed tissue at E15.5, which is just prior to the craniosynostosis. In WT embryos, BrdU-labeled cells were predominantly detected at the osteogenic front and periosteal surface of the frontal, parietal, and interparietal bones (Fig. 4A). In contrast, in Gli3Xt-J/Xt-J embryos, a significant number of BrdU-incorporated cells were observed in mid-sutural mesenchymal cell layer of interfrontal and lambdoid sutures (Fig. 4, A and B). This aberrant cell proliferation was specific to the interfrontal and lambdoid sutures but not to coronal suture of Gli3Xt-J/Xt-J embryos (Fig. 4, A and B). In Gli3Xt-J/Xt-J Runx2+/− embryos, a significant decrease in the number of BrdU-incorporated cells in all sutures assayed was detected compared with Gli3Xt-J/Xt-J embryos (Fig. 4, A and B).
FIGURE 4.
Rescue of the excessive proliferation in Gli3Xt-J/Xt-J embryos by removing one allele of Runx2. A, sections through E15.5 WT and each mutant interfrontal (a–c panels), lambdoid (d–f panels), and coronal sutures (g–i panels). BrdU-positive cells were detected by immunohistochemical staining using anti-BrdU antibody. In WT (a, d, and g panels) samples, BrdU-positive cells were detected predominantly at the osteogenic fronts. In Gli3Xt-J/Xt-J samples, BrdU-positive cells were detected in the mid-sutural mesenchyme of the interfrontal and lambdoid sutures (b panel and arrowheads in e panel) but not in the coronal sutures (h panel). In Gli3Xt-J/Xt-J Runx2+/−, BrdU-positive cells were detected mainly at the osteogenic front of each suture (c, f, and i panels) and not in the mid-sutural mesenchymal cells. B, graphs represent the number of BrdU-positive nuclei as a percentage of the total nuclei counted as described previously (11). In Gli3Xt-J/Xt-J Runx2+/− interfrontal and lambdoid sutures, significant smaller numbers of BrdU-positive cells were detected than in Gli3Xt-J/Xt-J sutures. Results are reported as mean ± S.D. with p < 0.05 considered statistically significant (*, p < 0.05). f, frontal bone; of, osteogenic front; p, parietal bone; IHC, immunohistochemistry. Scale bars in A, a, d, and g panels, 100 μm.
Ectopic and Up-regulated Expression of Osteoblast Differentiation-related Genes Was Normalized in Gli3Xt-J/Xt-J Runx2+/− Mice
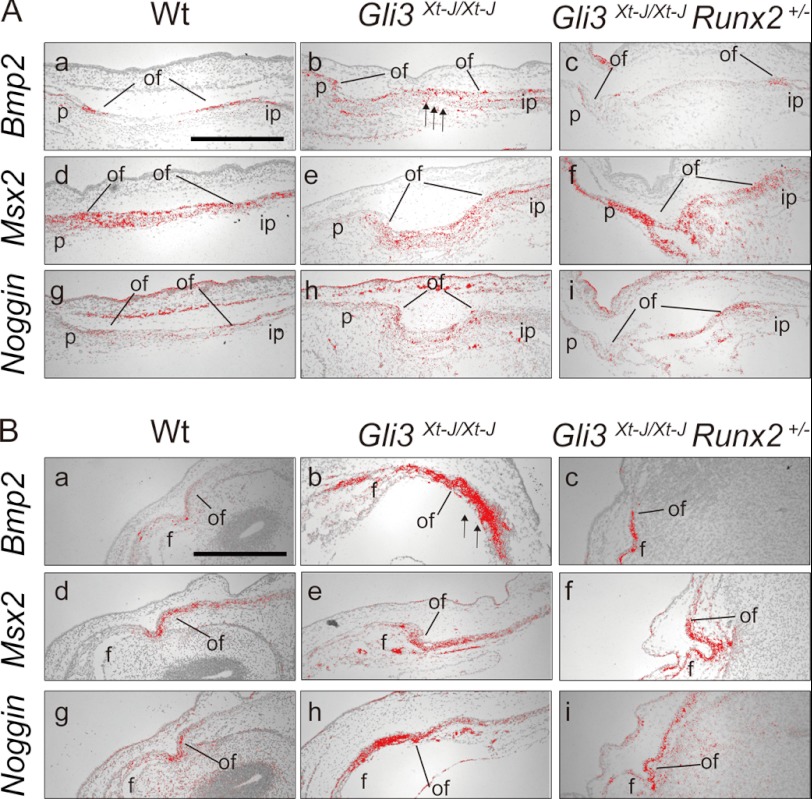
To clarify the mechanism in establishing patent lambdoid sutures and a lack of heterotopic bones in interfrontal suture in Gli3Xt-J/Xt-J Runx2+/− embryos, we performed in situ hybridization using Dlx5, Runx2 I/II, and Osteocalcin riboprobes (Fig. 5). In addition to the ectopic expression of Dlx5 and Runx-II mRNA in Gli3Xt-J/Xt-J in the central of mesenchyme (Fig. 5, A, b and h panels, and B, b and h panels), Oc (Osteocalcin) mRNA was also expressed there (Fig. 5, A, k panel, and B, k panel). Dlx5 has been reported to be down-regulated in Runx2+/− mice (29), so the normalization of Dlx5 in mid-sutural mesenchymal cells in Gli3Xt-J/Xt-J Runx2+/− mice might contribute to reduced expression of Runx2 and its downstream targets. We analyzed the expression of Bmp2 and its downstream target Msx2 and the antagonist Noggin in Gli3Xt-J/Xt-J lambdoid and interfrontal sutures (Fig. 6). Bmp2 transcripts were expressed ectopically in the sutural mesenchyme of Gli3Xt-J/Xt-J mice (Fig. 6, A, b panel, and B, b panel, arrows), and this was normalized in Gli3Xt-J/Xt-J Runx2+/− mice (Fig. 6, A, c panel, and B, c panel). The expression level of Msx2 in sutural mesenchymal cells on WT, Gli3Xt-J/Xt-J, and Gli3Xt-J/Xt-J Runx2+/− mice were similar (Fig. 6, A, d–f panels, and B, d–f panels). However, the expression of Noggin, which is induced by Bmp2 administration (8), was slightly up-regulated at the interfrontal suture of Gli3Xt-J/Xt-J mice, indicating the enhancement of Bmp2 expression (Fig. 6-B-h).
FIGURE 5.
Ectopically-expressed Dlx5 and Runx2-II in Gli3Xt-J/Xt-J calvaria were normalized in the Gli3Xt-J/Xt-J Runx2+/− calvaria. A, para sagittal sections through lambdoid sutures of E15.5 WT (a, d, g, and j panels), Gli3Xt-J/Xt-J (b, e, h, and k panels), Gli3Xt-J/Xt-J Runx2+/− (c, f, i, and l panels) calvaria. In WT and Gli3Xt-J/Xt-J Runx2+/− calvaria, Dlx5 and Runx2-II were expressed in the osteogenic fronts (a, c, g, and i panels). In Gli3Xt-J/Xt-J mice, Dlx5 and Runx2-II were expressed in the osteogenic fronts but also in the mid-sutural mesenchyme (arrows in b and h panels). The expression of Oc was observed in the mature osteoblasts of WT (j panel) and Gli3Xt-J/Xt-J Runx2+/− (l panel), but in the mid-sutural mesenchyme of Gli3Xt-J/Xt-J lambdoid suture (arrows in k panel). B, para coronal sections through interfrontal sutures of E15.5 WT (a, d, g, and j panels), Gli3Xt-J/Xt-J (b, e, h, and k panels), Gli3Xt-J/Xt-J Runx2+/− (c, f, i, and l panels) embryo. Note the ectopic expression of Dlx5 and Runx2-II in the mid-sutural mesenchymal cells in Gli3Xt-J/Xt-J interfrontal suture (arrows in b and h panels). However, in Gli3Xt-J/Xt-J Runx2+/− mice, expression levels were similar to those of WT mice (c and i panels). The expression of Oc in mature osteoblasts in WT and Gli3Xt-J/Xt-J Runx2+/− sutures (j and l panels) was enhanced in the mid-sutural area of Gli3Xt-J/Xt-J interfrontal suture (arrows in k panel). f, frontal bone; ip, interparietal bone; of, osteogenic front; p, parietal bone. Scale bars in A, a panel, 500 μm; scale bar in B, a panel, 500 μm.
FIGURE 6.
Bmp2, Msx2, and Noggin mRNA expression in the developing WT, Gli3Xt-J/Xt-J and Gli3Xt-J/Xt-J Runx2+/− calvaria. A, para sagittal sections through E15.5 WT (a, d, and g panels), Gli3Xt-J/Xt-J (b, e, and h panels), Gli3Xt-J/Xt-J Runx2+/− (c, f, and i panels) lambdoid sutures. Bmp2 was expressed in the osteogenic fronts of the parietal and interparietal bones and in the periosteum of WT (a) and Gli3Xt-J/Xt-J Runx2+/− (c panel) mice. A slight increase in expression was observed in the mid-sutural mesenchymal cell layer in Gli3Xt-J/Xt-J lambdoid suture (arrows in b panel). Msx2 was expressed at the mid-sutural mesenchymal cell layer and osteogenic front of the lambdoid suture in all samples (d–f panels). Noggin was expressed at the osteogenic front and mid-sutural mesenchymal cells almost equally in all samples (g–i panels). B, coronal sections through E15.5 WT (a, d, and g panels), Gli3Xt-J/Xt-J (b, e, and h panels), Gli3Xt-J/Xt-J Runx2+/− (c, f, and i panels) interfrontal suture. Bmp2 was expressed osteogenic fronts of the frontal bones of WT and Gli3Xt-J/Xt-J Runx2+/− (a and c panels) but expressed intensely at the mid-sutural mesenchymal cell layer located away from osteogenic front of frontal bone in Gli3Xt-J/Xt-J embryo (arrows in b panel). Msx2 was expressed at the mid-sutural mesenchymal cell layer and osteogenic front of frontal bones in all samples (d–f panels). Noggin was expressed at the osteogenic front and mid-sutural mesenchymal cells of all samples (g–i panels) but slightly higher in Gli3Xt-J/Xt-J (h panel). f, frontal bone; ip, interparietal bone; of, osteogenic front; p, parietal bone. Scale bars in A (a panel) and B (a panel), 500 μm.
Lack of Gli3R Leads to Activation of Bmp Signaling through Canonical Smad Pathway in Mid-sutural Mesenchyme Resulting in Bone Formation
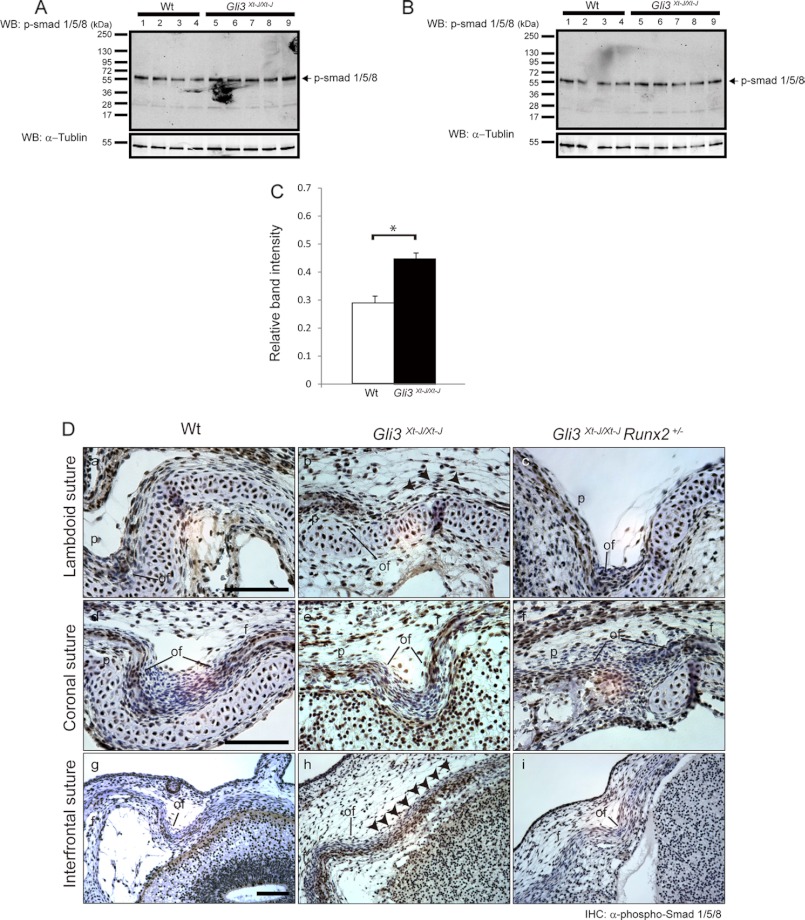
As we found Dlx5 to be up-regulated in Gli3Xt-J/Xt-J sutures and as it has been reported previously that Bmp will induce Dlx5 in avian calvaria (30), we analyzed the distribution of pSmad1/5/8, which is a regulator of Bmp signaling, by Western blot analysis using calvarial tissue from WT (n = 4) and Gli3Xt-J/Xt-J mice (n = 5) (Fig. 7, A and B). Higher phosphorylation of Smad 1/5/8 was detected in calvaria from Gli3Xt-J/Xt-J mice than WT with statistically difference (Fig. 7C). Additionally, we analyzed the phosphorylation of Smad1/5/8 by immunohistochemistry (Fig. 7D). Intensive immune-reactive cells were observed in the osteogenic front and in the periosteum of the frontal, parietal, and interparietal bones but not in the mid-sutural mesenchyme in WT embryos (E15.5) (Fig. 7, D, a, d, and g panels). However, in Gli3Xt-J/Xt-J mice, considerable numbers of pSmad1/5/8-positive cells were spread across the sutural mesenchyme of the interfrontal and lambdoid sutures (Fig. 7, D, b and h panels), but not coronal suture (Fig. 7, D, e panel). These abnormalities were corrected in Gli3Xt-J/Xt-J Runx2+/− specimens (Fig. 7D, c and i panels). As pSmad1/5/8 distribution correlated with expression of Bmp2, Bmp4 (data not shown), the Bmp antagonist Noggin and the Bmp target Msx2 in the sutures of WT, Gli3Xt-J/Xt-J, and Gli3Xt-J/Xt-J Runx2+/− calvaria; these results indicate that Gli3R regulates suture development by keeping Bmp-induced canonical Smad cascade under control (Fig. 7 and data not shown).
FIGURE 7.
Enhanced phosphorylation and nuclear accumulation of pSmad1/5/8 observed in the mid-sutural mesenchymal cells in Gli3Xt-J/Xt-J calvaria was restored in Gli3Xt-J/Xt-J Runx2+/− mice. A and B, equal amount of protein extracts (10 μg) from WT (lanes 1–4) and Gli3Xt-J/Xt-J (lanes 5–9) were analyzed by Western blot using pSmad1/5/8 antibody. C, integrated density values obtained for pSmad1/5/8 levels were normalized against those of α-tubulin and compared against other samples. Higher band intensity of pSmad1/5/8 was observed in Gli3Xt-J/Xt-J in comparison with WT. Statistical values were calculated using Student's t test, with p values < 0.05 considered significant (*, p < 0.05). D, para sagittal sections through E15.5 lambdoid and coronal sutures and coronal sections through E15.5 interfrontal sutures were immunohistochemically stained with anti-phospho-Smad1/5/8 (pSmad1/5/8) antibody. The accumulation of pSmad1/5/8-positive nuclei was observed in the periosteum and osteogenic fronts of parietal and interparietal bones in WT lambdoid suture (D, a panel) and frontal bone in WT interfrontal suture (D, g panel), but mid-sutural mesenchymal cells were negative for pSmad1/5/8. In Gli3Xt-J/Xt-J lambdoid and interfrontal sutures, mid-sutural mesenchymal cells were intensely positive for pSmad1/5/8 throughout the suture area (arrowheads in D, b and h panels). In Gli3Xt-J/Xt-J Runx2+/− lambdoid and interfrontal sutures pSmad1/5/8 was again detected in the periosteum and osteogenic fronts (D, c and i panels). In coronal sutures, pSmad1/5/8 was detected at the osteogenic front and the periosteum of frontal bones of WT (D, d panel), Gli3Xt-J/Xt-J (D, e panel), and Gli3Xt-J/Xt-J Runx2+/− (D, f panel) embryo. Mid-sutural mesenchymal cells in all coronal sutures were negative for pSmad1/5/8 (D, d–f panels). f, frontal bone; ip, interparietal bone; of, osteogenic front; p, parietal bone. Scale bars in D, a, d, and g panels, 500 μm.
DISCUSSION
In this study, we generated Gli3;Runx2 compound mutant mice to correct aberrant osteoblastic cell proliferation and differentiation in Gli3Xt-J/Xt-J suture mesenchymal cells and to rescue the craniosynostosis in these mice (11). Craniosynostosis is an infrequent but important feature of Grieg cephalopolysyndactyly syndrome caused by mutations in GLI3 (2, 3). Most forms of craniosynostosis, including those in Grieg cephalopolysyndactyly syndrome, occur prior to birth and are not detected until birth. Targeting this type of early craniosynostosis is therefore difficult. Treatment often involves the postnatal resection of the prematurely fused sutures and reshaping the calvaria to allow for proper skull growth. However, instead of staying open and allowing normal growth, the affected sutures often re-fuse, committing the child to repeat operations. Targeting RUNX2 may slow down the re-fusion process and reduce the number of operations required. As RUNX2 is a master regulator of osteogenesis, this type of therapy might by applicable to many types of craniosynostosis not just those caused by mutations in GLI3.
We have reported that FGF2-soaked beads applied to the lambdoid suture of Gli3Xt-J/Xt-J embryos will prevent synostosis, and this is by restoring Twist1 expression which, in turn, inhibits Runx2 (11, 26). In this study, we found that Gli3Xt-J/Xt-J mice showed higher expression of Dlx5 in mid-sutural mesenchymal cells in the interfrontal and lambdoid sutures followed by ectopic expression of Runx2-II but not Runx2-I. Although there is no report about direct association between Gli3 and Dlx5, indirect interaction between Shh signaling and Dlx5 through Fgf7 signaling has been shown during palate development and during vestibular morphogenesis through Wnt signaling (31, 32). Dlx3, Dlx5, and Runx2 are expressed by post-proliferative osteoblasts and activate the Oc promoter with the role of Dlx5 being more dominate than Dlx3 in mature osteoblasts at the mineralization stage of differentiation (28). Hassan and co-workers (33) also show that administration of BMP2 to osteogenic fibroblasts induces Dlx3 and Dlx5 expression resulting in the activation of Runx2. Others have reported that Dlx5 specifically transactivates the Runx2-II driven by “bone-related” P1 promoter (22). We suggest that Hedgehog signaling has an effect on bone development through Dlx gene function. Interestingly, the aberrant expression of Dlx5 and Runx2-II in Gli3Xt-J/Xt-J mice calvaria overlapped with the expression of Gli3 (11). These results suggest that Gli3 participates in controlling Dlx5 transcription and that this mechanism contributes to the craniosynostosis in Gli3Xt-J/Xt-J.
Many signaling cascades have been reported to have important roles in calvarial suture development, including Fgf, Bmp, Wnt, Ephrin-Eph, and Hh signaling (4, 10). We focused on Bmp signaling in Gli3Xt-J/Xt-J calvarial sutures and found ectopic expression of Bmp2 in mid-sutural mesenchymal cells. To evaluate functional Bmp signaling, we performed Western blot analysis and immunohistochemistry using pSmad1/5/8. We showed higher phosphorylation of Smad1/5/8 in Gli3Xt-J/Xt-J calvarial tissue. Furthermore, in WT calvaria, pSmad1/5/8 was expressed at the osteogenic front and periosteum, but not at mid-sutural mesenchymal cells, meaning that Bmp signaling is occurring at the place of new bone apposition. The processing of Gli3 and expression of Bmp4 protein has been reported to mediate both cell survival and programmed cell death in the developing limb bud in a position-dependent manner (34). Thus, the absence of Gli3, especially the lack of the repressor form of Gli3 up-regulates Bmp2 and subsequently pSmad1/5/8 in Gli3Xt-J/Xt-J calvarial sutures. Taken together, up-regulation of Bmp signaling contributes to the intramembranous ossification abnormalities in Gli3Xt-J/Xt-J embryos.
It is intriguing that only the interfrontal and lambdoidal sutures exhibit craniosynostosis/heterotopic bones. In patients with craniosynostosis only, some sutures are affected, and only very rarely are all sutures involved. In Gli3Xt-J/Xt-J mice, location specificity of the phenotype is the result of the temporal-spatial expression pattern of Gli3 (11). Here, we show a mechanism which underlies these abnormalities. This involves aberrant osteoprogenitor proliferation in the interfrontal and lambdoidal sutures but not in the coronal suture, which is not affected. We have shown more severe hypoplasia of calvarial bones in Gli3Xt-J/Xt-J Runx2+/− compared with Runx2+/− mice. Additionally, Gli3Xt-J/Xt-J Runx2+/− mice displayed smaller clavicles than either Gli3Xt-J/Xt-J or Runx2+/−. It has been reported that Runx2 controls the cell fate, proliferation, and growth through regulating ribosomal biogenesis (17, 35). In Gli3Xt-J/Xt-J mice, cells fated into mature osteoblasts are abundant in the interfrontal and lambdoidal sutures. Proliferation of these ectopic differentiated osteoblasts might be reduced specifically because of loss of Runx2. Additionally, genetic interaction between Gli3 and Runx2 in morphogenesis of membranous bone might contribute to this matter.
In conclusion, Gli3R plays a key role in repressing bone formation in the suture, thus maintaining suture patency and normal calvarial development. Gli3 inhibits osteogenesis by three different mechanisms. Twist1, which can be induced by FGF2, lies downstream of Gli3 and directly inhibits Runx2 (11, 26, 36). Gli3 also represses Runx2 via a Bmp2/Dlx5 mechanism, specifically targeting Runx2-II (22, 30). Gli3 represses Oc directly thereby inhibiting the action of Runx2 (13). Within the sutural system, the importance of the repression of Runx2 and Oc is that osteogenesis occurs at the correct time and location to ensure proper growth and morphogenesis.
Acknowledgments
We thank Airi Sinkko for technical assistance. We also thank Ritva Rice for critical reading of this manuscript and many helpful comments on this work. We thank Irma Thesleff and Jacqueline Veltmaat for providing the Runx2 and Gli3 mutant mice.
This work was supported by the Academy of Finland, Biocentrum Helsinki, The University of Helsinki, and the Sigrid Jusélius Foundation.

This article contains supplemental Fig. 1.
- E15.5
- embryonic day 15.5
- pSmad
- phospho-Smad.
REFERENCES
- 1. Passos-Bueno M. R., Serti Eacute A. E., Jehee F. S., Fanganiello R., Yeh E. (2008) Genetics of craniosynostosis: Genes, syndromes, mutations, and genotype-phenotype correlations. Front Oral Biol. 12, 107–143 [DOI] [PubMed] [Google Scholar]
- 2. McDonald-McGinn D. M., Feret H., Nah H. D., Bartlett S. P., Whitake L. A., Zackai E. H. (2010) Metopic craniosynostosis due to mutations in GLI3: A novel association. Am. J. Med. Genet. A 152A, 1654–1660 [DOI] [PubMed] [Google Scholar]
- 3. Hurst J. A., Jenkins D., Vasudevan P. C., Kirchhoff M., Skovby F., Rieubland C., Gallati S., Rittinger O., Kroisel P. M., Johnson D., Biesecker L. G., Wilkie A. O. (2011) Metopic and sagittal synostosis in Greig cephalopolysyndactyly syndrome: Five cases with intragenic mutations or complete deletions of GLI3. Eur. J. Hum. Genet. 19, 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim H. J., Rice D. P., Kettunen P. J., Thesleff I. (1998) FGF-, BMP-, and Shh-mediated signaling pathways in the regulation of cranial suture morphogenesis and calvarial bone development. Development 125, 1241–1251 [DOI] [PubMed] [Google Scholar]
- 5. Liu Y. H., Tang Z., Kundu R. K., Wu L., Luo W., Zhu D., Sangiorgi F., Snead M. L., Maxson R. E. (1999) Msx2 gene dosage influences the number of proliferative osteogenic cells in growth centers of the developing murine skull: a possible mechanism for MSX2-mediated craniosynostosis in humans. Dev. Biol. 205, 260–274 [DOI] [PubMed] [Google Scholar]
- 6. Hajihosseini M. K., Duarte R., Pegrum J., Donjacour A., Lana-Elola E., Rice D. P., Sharpe J., Dickson C. (2009) Evidence that Fgf10 contributes to the skeletal and visceral defects of an Apert syndrome mouse model. Dev. Dyn. 238, 376–385 [DOI] [PubMed] [Google Scholar]
- 7. Iseki S., Wilkie A. O., Morriss-Kay G. M. (1999) Fgfr1 and Fgfr2 have distinct differentiation- and proliferation-related roles in the developing mouse skull vault. Development 126, 5611–5620 [DOI] [PubMed] [Google Scholar]
- 8. Rice R., Rice D. P., Thesleff I. (2005) Foxc1 integrates Fgf and Bmp signalling independently of twist or noggin during calvarial bone development. Dev Dyn 233, 847–852 [DOI] [PubMed] [Google Scholar]
- 9. Warren S. M., Brunet L. J., Harland R. M., Economides A. N., Longaker M. T. (2003) The BMP antagonist noggin regulates cranial suture fusion. Nature 422, 625–629 [DOI] [PubMed] [Google Scholar]
- 10. Rice R., Rice D. P., Olsen B. R., Thesleff I. (2003) Progression of calvarial bone development requires Foxc1 regulation of Msx2 and Alx4. Dev. Biol. 262, 75–87 [DOI] [PubMed] [Google Scholar]
- 11. Rice D. P., Connor E. C., Veltmaat J. M., Lana-Elola E., Veistinen L., Tanimoto Y., Bellusci S., Rice R. (2010) Gli3Xt-J/Xt-J mice exhibit lambdoid suture craniosynostosis which results from altered osteoprogenitor proliferation and differentiation. Hum. Mol. Genet. 19, 3457–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shimoyama A., Wada M., Ikeda F., Hata K., Matsubara T., Nifuji A., Noda M., Amano K., Yamaguchi A., Nishimura R., Yoneda T. (2007) Ihh/Gli2 signaling promotes osteoblast differentiation by regulating Runx2 expression and function. Mol. Biol. Cell 18, 2411–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ohba S., Kawaguchi H., Kugimiya F., Ogasawara T., Kawamura N., Saito T., Ikeda T., Fujii K., Miyajima T., Kuramochi A., Miyashita T., Oda H., Nakamura K., Takato T., Chung U. I. (2008) Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev. Cell 14, 689–699 [DOI] [PubMed] [Google Scholar]
- 14. Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. (1997) Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 89, 747–754 [DOI] [PubMed] [Google Scholar]
- 15. Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 16. Hartmann C. (2009) Transcriptional networks controlling skeletal development. Curr. Opin. Genet. Dev. 19, 437–443 [DOI] [PubMed] [Google Scholar]
- 17. Young D. W., Hassan M. Q., Pratap J., Galindo M., Zaidi S. K., Lee S. H., Yang X., Xie R., Javed A., Underwood J. M., Furcinitti P., Imbalzano A. N., Penman S., Nickerson J. A., Montecino M. A., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2007) Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature 445, 442–446 [DOI] [PubMed] [Google Scholar]
- 18. D'Souza R. N., Aberg T., Gaikwad J., Cavender A., Owen M., Karsenty G., Thesleff I. (1999) Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development 126, 2911–2920 [DOI] [PubMed] [Google Scholar]
- 19. Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89, 773–779 [DOI] [PubMed] [Google Scholar]
- 20. Park M. H., Shin H. I., Choi J. Y., Nam S. H., Kim Y. J., Kim H. J., Ryoo H. M. (2001) Differential expression patterns of Runx2 isoforms in cranial suture morphogenesis. J. Bone Miner. Res. 16, 885–892 [DOI] [PubMed] [Google Scholar]
- 21. Xiao Z., Awad H. A., Liu S., Mahlios J., Zhang S., Guilak F., Mayo M. S., Quarles L. D. (2005) Selective Runx2-II deficiency leads to low-turnover osteopenia in adult mice. Dev. Biol. 283, 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee M. H., Kim Y. J., Yoon W. J., Kim J. I., Kim B. G., Hwang Y. S., Wozney J. M., Chi X. Z., Bae S. C., Choi K. Y., Cho J. Y., Choi J. Y., Ryoo H. M. (2005) Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J. Biol. Chem. 280, 35579–35587 [DOI] [PubMed] [Google Scholar]
- 23. Varjosalo M., Taipale J. (2008) Hedgehog: Functions and mechanisms. Genes Dev. 22, 2454–2472 [DOI] [PubMed] [Google Scholar]
- 24. Aberg T., Wang X. P., Kim J. H., Yamashiro T., Bei M., Rice R., Ryoo H. M., Thesleff I. (2004) Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev. Biol. 270, 76–93 [DOI] [PubMed] [Google Scholar]
- 25. Maynard T. M., Jain M. D., Balmer C. W., LaMantia A. S. (2002) High-resolution mapping of the Gli3 mutation extra-toes reveals a 51.5-kb deletion. Mamm Genome 13, 58–61 [DOI] [PubMed] [Google Scholar]
- 26. Rice D. P., Aberg T., Chan Y., Tang Z., Kettunen P. J., Pakarinen L., Maxson R. E., Thesleff I. (2000) Integration of FGF and TWIST in calvarial bone and suture development. Development 127, 1845–1855 [DOI] [PubMed] [Google Scholar]
- 27. Lee M. H., Kim Y. J., Kim H. J., Park H. D., Kang A. R., Kyung H. M., Sung J. H., Wozney J. M., Kim H. J., Ryoo H. M. (2003) BMP-2-induced Runx2 expression is mediated by Dlx5, and TGF-β 1 opposes the BMP-2-induced osteoblast differentiation by suppression of Dlx5 expression. J. Biol. Chem. 278, 34387–34394 [DOI] [PubMed] [Google Scholar]
- 28. Hassan M. Q., Javed A., Morasso M. I., Karlin J., Montecino M., van Wijnen A. J., Stein G. S., Stein J. L., Lian J. B. (2004) Dlx3 transcriptional regulation of osteoblast differentiation: Temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol. Cell Biol. 24, 9248–9261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choi K. Y., Kim H. J., Lee M. H., Kwon T. G., Nah H. D., Furuichi T., Komori T., Nam S. H., Kim Y. J., Kim H. J., Ryoo H. M. (2005) Runx2 regulates FGF2-induced Bmp2 expression during cranial bone development. Dev. Dyn. 233, 115–121 [DOI] [PubMed] [Google Scholar]
- 30. Holleville N., Quilhac A., Bontoux M., Monsoro-Burq A. H. (2003) BMP signals regulate Dlx5 during early avian skull development. Dev. Biol. 257, 177–189 [DOI] [PubMed] [Google Scholar]
- 31. Han J., Mayo J., Xu X., Li J., Bringas P., Jr., Maas R. L., Rubenstein J. L., Chai Y. (2009) Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development 136, 4225–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Riccomagno M. M., Takada S., Epstein D. J. (2005) Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 19, 1612–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hassan M. Q., Tare R. S., Lee S. H., Mandeville M., Morasso M. I., Javed A., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2006) BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J. Biol. Chem. 281, 40515–40526 [DOI] [PubMed] [Google Scholar]
- 34. Bastida M. F., Delgado M. D., Wang B., Fallon J. F., Fernandez-Teran M., Ros M. A. (2004) Levels of Gli3 repressor correlate with Bmp4 expression and apoptosis during limb development. Dev. Dyn. 231, 148–160 [DOI] [PubMed] [Google Scholar]
- 35. Young D. W., Hassan M. Q., Yang X. Q., Galindo M., Javed A., Zaidi S. K., Furcinitti P., Lapointe D., Montecino M., Lian J. B., Stein J. L., van Wijnen A. J., Stein G. S. (2007) Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc. Natl. Acad. Sci. U.S.A. 104, 3189–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bialek P., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D. M., Olson E. N., Justice M. J., Karsenty G. (2004) A twist code determines the onset of osteoblast differentiation. Dev. Cell 6, 423–435 [DOI] [PubMed] [Google Scholar]